There are currently at least 71 million individuals with chronic HCV worldwide and almost two million new infections annually. Although the advent of direct-acting antivirals (DAAs) offers highly effective therapy, considerable remaining challenges argue against reliance on DAAs for HCV elimination, including high drug cost, poorly developed health infrastructure, low screening rates, and significant reinfection rates. Accordingly, development of an effective vaccine is crucial to HCV elimination. An HCV vaccine that elicits T cell immunity in the liver will be highly protective for the following reasons: (i) T cell responses against nonstructural proteins of the virus are associated with clearance of primary infection, and (ii) long-lived liver-resident T cells alone can protect against malaria infection of hepatocytes. Thus, in this study we exploit promising vaccination platforms to highlight strategies that can be used to evoke highly functional and long-lived T cell responses in the liver for protection against HCV.
KEYWORDS: adeno-associated virus vaccine, cytotoxic T cells, DNA vaccine, helper T cells, hepatitis C virus vaccine, liver immunity
ABSTRACT
Hepatitis C virus (HCV) is a significant contributor to the global disease burden, and development of an effective vaccine is required to eliminate HCV infections worldwide. CD4+ and CD8+ T cell immunity correlates with viral clearance in primary HCV infection, and intrahepatic CD8+ tissue-resident memory T (TRM) cells provide lifelong and rapid protection against hepatotropic pathogens. Consequently, we aimed to develop a vaccine to elicit HCV-specific CD4+ and CD8+ T cells, including CD8+ TRM cells, in the liver, given that HCV primarily infects hepatocytes. To achieve this, we vaccinated wild-type BALB/c mice with a highly immunogenic cytolytic DNA vaccine encoding a model HCV (genotype 3a) nonstructural protein (NS5B) and a mutant perforin (pVAX-NS5B-PRF), as well as a recombinant adeno-associated virus (AAV) encoding NS5B (rAAV-NS5B). A novel fluorescent target array (FTA) was used to map immunodominant CD4+ T helper (TH) cell and cytotoxic CD8+ T cell epitopes of NS5B in vivo, which were subsequently used to design a KdNS5B451-459 tetramer and analyze NS5B-specific T cell responses in vaccinated mice in vivo. The data showed that intradermal prime/boost vaccination with pVAX-NS5B-PRF was effective in eliciting TH and cytotoxic CD8+ T cell responses and intrahepatic CD8+ TRM cells, but a single intravenous dose of hepatotropic rAAV-NS5B was significantly more effective. As a T-cell-based vaccine against HCV should ideally result in localized T cell responses in the liver, this study describes primary observations in the context of HCV vaccination that can be used to achieve this goal.
IMPORTANCE There are currently at least 71 million individuals with chronic HCV worldwide and almost two million new infections annually. Although the advent of direct-acting antivirals (DAAs) offers highly effective therapy, considerable remaining challenges argue against reliance on DAAs for HCV elimination, including high drug cost, poorly developed health infrastructure, low screening rates, and significant reinfection rates. Accordingly, development of an effective vaccine is crucial to HCV elimination. An HCV vaccine that elicits T cell immunity in the liver will be highly protective for the following reasons: (i) T cell responses against nonstructural proteins of the virus are associated with clearance of primary infection, and (ii) long-lived liver-resident T cells alone can protect against malaria infection of hepatocytes. Thus, in this study we exploit promising vaccination platforms to highlight strategies that can be used to evoke highly functional and long-lived T cell responses in the liver for protection against HCV.
INTRODUCTION
At least 71 million people are persistently infected with hepatitis C virus (HCV) worldwide, and every year 1.75 million new infections and 399,000 HCV-related deaths are reported (1, 2). Although the new direct-acting antivirals (DAAs) cure >95% of infected individuals, there are still major challenges to overcome in order to achieve the WHO Hepatitis C Global Elimination Goals by 2030. First, only 20% of infected individuals worldwide are diagnosed. Second, DAAs remain prohibitively costly for much of the developing world. Third, the health infrastructure to deliver treatment to marginalized people who inject drugs (PWID) remains underdeveloped. Finally, successful DAA treatment does not prevent reinfection (1, 2). Thus, the development of an effective vaccine is crucial to eliminating HCV worldwide.
HCV is a rapidly mutating RNA virus comprising 8 genotypes and at least 67 subtypes, which vary in regional and global distribution (3), making the development of a prophylactic HCV vaccine extremely challenging. However, approximately 25% of individuals clear the virus during primary infection (4), and consequently, development of a vaccine that can elicit immune responses that correlate with natural clearance represents a rational path for HCV vaccine design (5). Although broadly neutralizing antibodies are targets for HCV vaccine development (6, 7), there is a significant emphasis on developing T-cell-mediated HCV vaccines because robust CD4+ and CD8+ T cell responses that target the conserved nonstructural (NS) proteins of the virus correlate with clearance of the virus from primary infection (8–10). In these individuals, robust and broad T cell responses against HCV NS protein epitopes are thought to eliminate virus-infected cells during acute stages of infection in order to prevent persistent infection (5). This is an acceptable vaccination outcome given that acute infections do not cause serious disease and are often subclinical (5). Furthermore, the only HCV vaccine candidate to have advanced to phase IIb clinical trials thus far exploits vaccine vectors encoding genotype 1 (gt1) NS proteins in a prime/boost regimen to elicit systemic T cell immunity (11).
Since HCV primarily replicates in hepatocytes, a vaccine designed to elicit T cell responses to the NS proteins should also aim to localize these responses to the liver. In this regard, the ability of an HCV vaccine to elicit intrahepatic CD8+ tissue-resident memory T (TRM) cells will be important since several studies in mice and humans suggest that these cells correlate strongly with protection against hepatotropic pathogens, including malaria parasites (12, 13), hepatitis B virus (HBV) (14), and HCV (15). Circulating memory T cells, which include central memory T (TCM) cells and effector memory T (TEM) cells, take some time to migrate to the site of an infection, whereas localized TRM cells, which reside in tissues for the life span of the individual (16), can respond more rapidly. Intrahepatic CD8+ TRM cells produce higher levels of antiviral cytokines such as gamma interferon and are more cytotoxic than CD8+ TEM cells (12). There is a strong genetic bottleneck associated with HCV transmission as only one or a few variants, known as transmitted/founder (T/F) viruses, establish infection following transmission (17). The T/F viruses persist in the new host as the dominant variants mediating infection for ∼100 days before divergent quasispecies emerge, escaping immune selection pressures to establish persistent infection (17). Thus, the presence of intrahepatic CD8+ TRM cells at the time of transmission could facilitate rapid elimination of the virus before the evolving quasispecies facilitate immune escape. Several other studies have shown that CD8+ TRM cells are also crucial for protection against virus infections that occur in different anatomical sites such as the skin, lungs, female reproductive tract, and vagina (18–23).
The first study to describe a vaccination regimen to elicit protective CD8+ TRM cells used a “prime/pull” approach in which systemically primed antiviral CD8+ T cells were “pulled” or recruited to the vagina by intravaginal topical application of the chemokine ligands CXCL9 and CXCL10 (18). The “prime/trap” vaccination regimen, an adaptation of the “prime/pull” approach, was recently shown to generate antigen-specific CD8+ TRM cells in the liver of vaccinated mice that protected against challenge with Plasmodium berghei sporozoites (12). This study involved priming adoptively transferred T cell receptor (TCR) transgenic CD8+ T cells (PbT-I) with a P. berghei peptide immunogen (NVY) fused to an antibody that delivers the immunogen to cross-presenting dendritic cells (DCs). The vast majority of the primed CD8+ PbT-I cells differentiated into TRM cells and consequently were “trapped” in the liver following an intravenous (i.v.) boost with a hepatotropic recombinant adeno-associated virus (rAAV) encoding NVY. Depletion of P. berghei-specific CD8+ TRM cells in the liver confirmed that these cells were crucial for protection against P. berghei sporozoite challenge (12).
We have demonstrated that a cytolytic DNA vaccine encoding a mutant form of perforin (PRF) and an immunogen (ribosomal DNA [rDNA]-PRF) is more effective than canonical DNA vaccines not encoding PRF in eliciting protection against a surrogate human immunodeficiency virus challenge, EcoHIV, of mice (24) and T cell immunity in mice and pigs (25–27). rDNA-PRF, unlike canonical DNA, induces necrosis (mimicking a lytic virus infection) in transfected cells resulting in the release of intrinsic danger-associated molecular patterns (DAMPs), which activate DCs to cross-present antigens to naive CD8+ T cells (28). Consequently, immunization with rDNA-PRF is more effective than canonical DNA in priming naive CD8+ T cells to become effector cells (28). The cytolytic DNA platform was engineered using the pVAX plasmid DNA backbone, which is FDA approved for use in humans and can be used to encode multiple HCV NS proteins and elicit multigenotypic (gt1 and gt3) HCV-specific T cell responses (29).
Although the malaria study (12) provides seminal proof of concept, an analogous vaccination regimen using vaccine vectors encoding native proteins to elicit polyclonal intrahepatic CD8+ TRM cells and CD4+ T cell responses against HCV has not been evaluated or developed. DNA and AAV vectors have been tested extensively and have been shown to be safe and effective for vaccination and gene therapy in humans (30, 31). Thus, in this study, we report the use of cytolytic DNA and a hepatotropic AAV vector encoding a codon-optimized gt3a HCV protein, NS5B, in prime/trap and prime/boost vaccination regimens to elicit T cell responses and intrahepatic CD8+ TRM cells in vivo. Furthermore, we also identified novel immunodominant CD4+ T helper (TH) cell and CD8+ T cell epitopes of NS5B in wild-type BALB/c mice, which were used to thoroughly analyze NS5B-specific TH cell and CD8+ T cell responses in vivo.
RESULTS
Identification of an NS5B immunodominant CD8+ T cell epitope for tetramer synthesis.
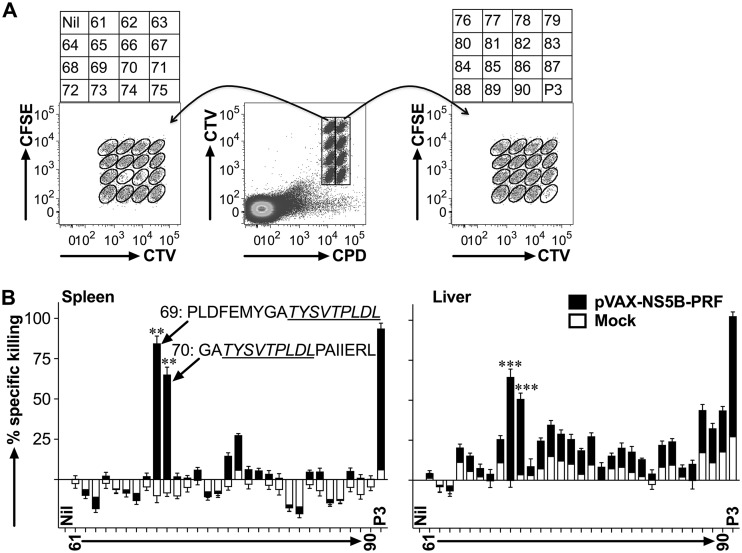
We have previously shown that HCV (gt3a) NS5B is a highly immunogenic protein (29) and can therefore serve as a model antigen to optimize an rDNA/rAAV-based vaccination strategy to elicit HCV-specific T cell immunity. However, the immunodominant CD8+ T cell epitopes of NS5B in BALB/c mice, necessary to synthesize a tetramer in order to detect NS5B-specific CD8+ T cells, are unknown. Consequently, we modified a fluorescent target array (FTA) assay (32–34) to map the most immunodominant TH and CD8+ T cell epitopes of NS5B in vivo in mice vaccinated with cytolytic DNA encoding NS5B (pVAX-NS5B-PRF). The assay introduces fluorescently labeled bar-coded autologous naive splenocytes pulsed with viral peptides (i.e., the FTA) into previously immunized mice to evaluate the magnitude and/or avidity of TH cell and cytotoxic CD8+ T cell responses in vivo (32–34). Traditional peptide-based mapping and T cell stimulation assays such as enzyme-linked immunosorbent spot (ELISpot) and intracellular cytokine staining (ICS) analyses do not distinguish CD4+ and CD8+ T cell responses and/or involve prolonged culture/manipulation of T cells in vitro. We have previously shown that NS5B peptide pool 3 (P3) contains the most immunodominant CD8+ T cell epitopes in BALB/c mice (29). Therefore, we generated an FTA using naive splenocytes pulsed with each or all of the peptides in P3 and injected these cells i.v. into mice previously vaccinated via the intradermal (i.d.) route with pVAX-NS5B-PRF or the pVAX-PRF (mock) control (Fig. 1A). Following recovery of the FTA targets from the spleen and the liver 15 h after the i.v. injection, NS5B-specific T cell responses were assessed by analysis of the population of the FTA cells using flow cytometry (Fig. 1). The greatest NS5B-specific CD8+ cytotoxic T cell responses were detected against targets pulsed with peptides 69 (NS5B442–459) and 70 (NS5B449–466) or P3 (positive control) (Fig. 1B). A common 9-amino-acid (aa) epitope (TYSVTPLDL [NS5B451–459]) was identified in both peptides 69 and 70 using bioinformatics analysis as described in Materials and Methods, which was used to synthesize a KdNS5B451–459 tetramer for the analysis of NS5B-specific CD8+ T cells and intrahepatic CD8+ TRM cells.
FIG 1.
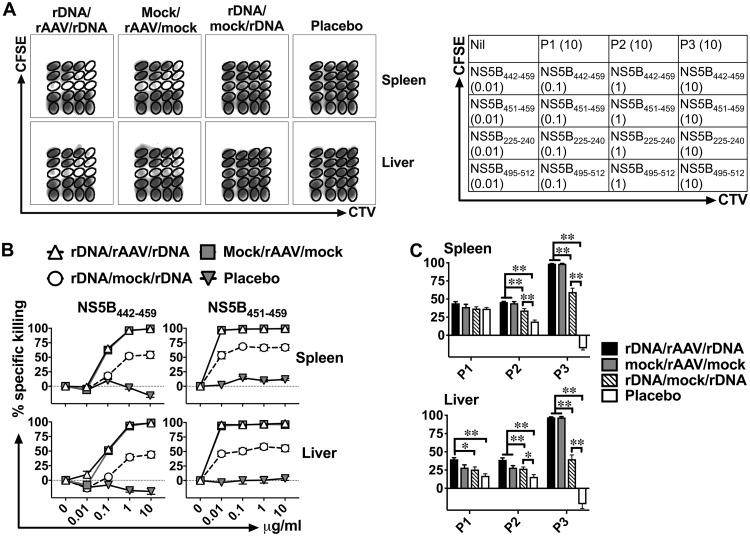
FTA-based mapping analysis reveals immunodominant CD8+ T cell epitopes of gt3a NS5B in vivo. Female BALB/c mice (n = 7 per group) were immunized i.d. on three occasions at fortnightly intervals with 50 μg of rDNA-PRF encoding genotype 3a HCV NS5B (pVAX-NS5B-PRF) or rDNA-PRF lacking this sequence (pVAX-PRF [mock]). Thirteen days after the final immunization, mice were challenged i.v. with naive peptide-pulsed or unpulsed autologous splenocytes labeled with cell tracking dyes (cell trace violet [CTV], carboxyfluorescein succinimidyl ester [CFSE], and cell proliferation dye efluor670 [CPD]). The experimental details regarding how the FTA was constructed are described in the Materials and Methods and Table 1. (A) The representative flow cytometry plots show the fluorescently bar-coded target cell populations that were unpulsed (Nil), pulsed with an individual peptide (peptides 61 to 90) spanning NS5B385–591 or all the peptides spanning NS5B385–591 (P3), and recovered 15 h after the FTA challenge from the spleen of a pVAX-NS5B-PRF-vaccinated mouse. (B) Mean (n = 7) + SEM of the specific killing responses (see Materials and Methods for the formula used for the calculation) detected against each target cell population in the FTA recovered from the spleen (left panel) and the liver (right panel) from immunized mice for the experiment described in panel A. The sequences for the most immunodominant peptides (69 and 70) and the 9-aa sequence (NS5B451–459 [underlined]) used to synthesize a tetramer (KdNS5B451–459) in order to analyze NS5B-specific CD8+ T cells are shown. The asterisks above each bar corresponding to the mapped immunodominant peptides indicate statistically significant data for the comparisons of the killer responses between pVAX-NS5B-PRF and mock. **, P < 0.01; ***, P < 0.001.
Identification of immunodominant TH cell epitopes of NS5B.
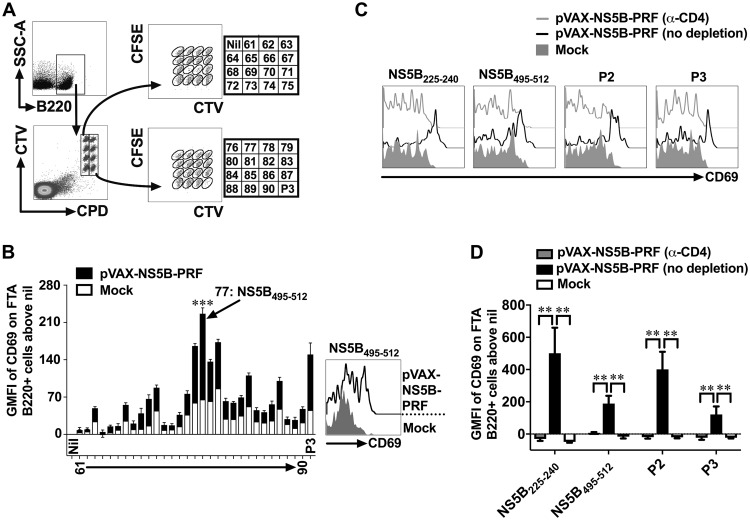
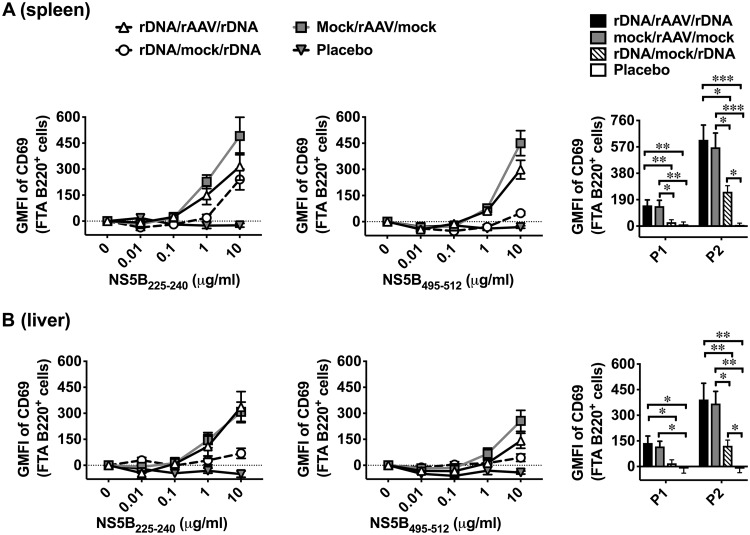
Clinical data suggest that the presence of TH cells that could mobilize CD8+ T cell responses is a robust correlate of clearance from primary HCV infection (9). TH cells recognize cognate peptide:major histocompatibility complex class II (MHC-II) molecules presented on B (B220+) cells and deliver activation signals (i.e., costimulation) to B cells resulting in the upregulation of CD69 on antigen-presenting B220+ cells (29, 33, 34). Therefore, the geometric mean fluorescent intensity (GMFI) of CD69 expression on peptide-pulsed B220+ targets in an FTA is a direct measure of the TH cell responses in vivo (29, 33, 34), and this analysis was used to map the immunodominant TH cell epitope within P3 of NS5B (Fig. 2). We conducted the mapping analysis using B220+ FTA cells recovered from the spleens of vaccinated mice (Fig. 2), given that NS5B-specific TH cell responses were not significantly detected in the liver of pVAX-NS5B-PRF-vaccinated mice compared to the mock control. The data showed that the greatest upregulation of CD69 occurred on B220+ cells pulsed with peptide 77 (NS5B495–512) in pVAX-NS5B-PRF-vaccinated mice compared to other peptides in P3 and the respective peptide-pulsed B220+ cell population in the mock-vaccinated mice (Fig. 2). We have previously reported that TH cell responses were detected against peptide pool 2 (P2) of NS5B (NS5B197–395) (29). Therefore, the immunodominant TH cell epitopes within P2 were also mapped using an FTA analysis similar to that described in the legend to Fig. 2 which showed peptide 36 (NS5B225–240) to be the immunodominant TH cell epitope in P2 (data not shown).
FIG 2.
FTA-based mapping analysis to identify immunodominant TH cell peptides of gt3a NS5B. Female BALB/c mice were vaccinated with pVAX-NS5B-PRF or mock vaccinated and challenged with an FTA comprising target cell populations pulsed with an individual peptide (peptides 61 to 90) or a peptide pool spanning NS5B385–591 (P3) as described in the legend to Fig. 1. (A) Representative dot plots showing how B220+ FTA cells recovered from the spleen of a pVAX-NS5B-PRF-vaccinated mouse were gated for analysis. (B) The bar graph shows the mean (n = 7) + SEM of the specific (beyond nil) expression (GMFI) of CD69 on gated B220+ cells within the FTA (as shown in panel A) recovered from the spleen of vaccinated mice. The representative histogram plot shows the expression of CD69 on gated B220+ FTA cells pulsed with the immunodominant NS5B495–512 peptide. The asterisks above the bar corresponding to the mapped immunodominant peptide indicate statistically significant data for the comparisons of the TH cell responses between pVAX-NS5B-PRF and mock vaccinated. ***, P < 0.001. (C and D) BALB/c mice were vaccinated with pVAX-NS5B-PRF or mock vaccinated as described for panel A. Thirteen days after the final (3rd dose) vaccination, 100 μg of anti-mouse CD4 depletion antibody was injected via the intraperitoneal (i.p.) route into each mouse from a cohort (n = 7 to 9) of pVAX-NS5B-PRF-vaccinated mice 3 days prior to FTA challenge with target cells that were unpulsed or pulsed with NS5B225–240, NS5B495–512, P2, or P3. The representative histogram plots (C) show the expression of CD69 on peptide-pulsed B220+ FTA cells, and the bar graph (D) shows the mean (n = 7 to 9) specific expression (GMFI) of CD69 on B220+ cells depicted in panel C. **, P < 0.01.
In order to confirm whether NS5B225–240 and NS5B495–512 were indeed CD4+ TH cell epitopes, CD4+ T cells were depleted in pVAX-NS5B-PRF-vaccinated mice prior to the FTA challenge (Fig. 2C and D). Data showed that upregulation of CD69 on B220+ FTA cells pulsed with NS5B225–240, NS5B495–512, P2, or P3 was only observed in pVAX-NS5B-PRF-vaccinated mice with an intact CD4+ cell compartment (Fig. 2C and D). The B cell responses in the pVAX-NS5B-PRF-vaccinated mice in which CD4+ cells were depleted was comparable to that of mock-vaccinated mice (Fig. 2C and D). Thus, the mapped NS5B225–240 and NS5B495–512 represent immunodominant CD4+ TH cell epitopes of NS5B in BALB/c mice.
Overall, the FTA-based T cell epitope mapping analyses of gt3a NS5B revealed an immunodominant CD8+ T cell epitope (NS5B451–459) and immunodominant TH cell epitopes (NS5B225–240 and NS5B495–512) for the analysis of NS5B-specific T cell responses in BALB/c mice.
Evaluation of DNA- and AAV-based vaccination regimens to elicit systemic and intrahepatic NS5B-specific CD8+ T cells.
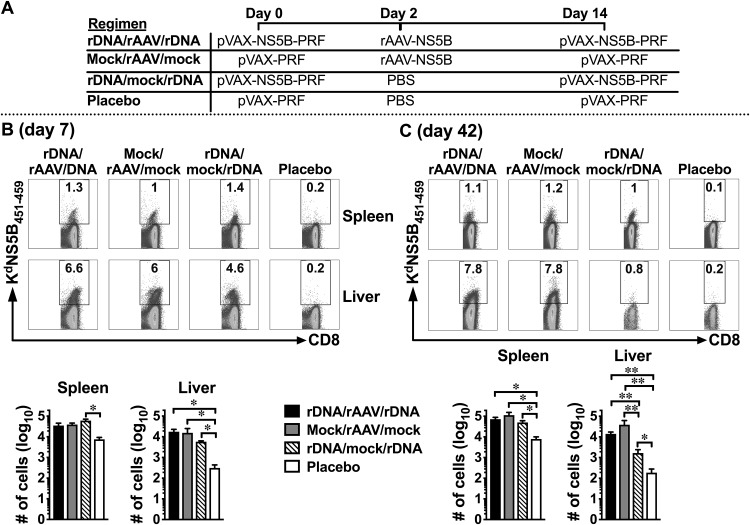
The first “prime and trap” study (12) elicited liver CD8+ TRM cells using a peptide (NVY) antigen from P. berghei and TCR transgenic CD8+ T cells. We evaluated whether this approach could be adapted to a more physiological vaccination setting when rDNA-PRF, which can encode multiple NS antigens (26, 29), was used to prime endogenous polyclonal T cells. Our studies and others have shown that after i.d. immunization of mice with a 50-μg dose of plasmid DNA, the expression of DNA-encoded immunogens peaks within 24 h and DCs initiate priming of naive CD8+ T within 2 days (24, 35, 36). Therefore, following a modified “prime/trap” vaccination protocol we immunized BALB/c mice i.d. with 50 μg of pVAX-NS5B-PRF (prime) 2 days prior to i.v. immunization with 5 × 109 viral genome copies (vgc) of rAAV-NS5B (trap) and 14 days prior to a booster immunization with pVAX-NS5B-PRF (rDNA/rAAV/rDNA) (Fig. 3A). A placebo control regimen and vaccination regimens that only included pVAX-NS5B-PRF (rDNA/mock/rDNA) or rAAV-NS5B (mock/rAAV/mock) were used as relevant comparative controls for the rDNA/rAAV/rDNA regimen (Fig. 3A). We have previously determined that 5 × 109 vgc of rAAV-NS5B results in the transduction of ∼10% of mouse hepatocytes in vivo (data not shown) and have shown that a similar dose of rAAV can elicit robust CD8+ T cell responses in the liver (37). Furthermore, Fernandez-Ruiz et al. (12) showed that an rAAV dose that resulted in ∼10% transduction of hepatocytes was suitable for trapping primed CD8+ T cells.
FIG 3.
rAAV‐NS5B immunization promotes durable, NS5B‐specific intrahepatic CD8+ T cell responses. Female BALB/c mice were immunized with pVAX-NS5B-PRF (“rDNA” on the figure) or rAAV-NS5B (“rAAV” in the figure) and/or mock immunized using the vaccination regimens depicted in panel A. Seven (B) and 42 (C) days after the final vaccination, NS5B-specific CD8+ T cells derived from the spleen and the liver were analyzed using KdNS5B451–459 tetramer staining and flow cytometry. The numbers in the representative dot plots show the percentage of live CD8+ cells in the spleen and the liver that stained positive for the KdNS5B451–459 tetramer. The bar graphs show the mean (n = 3 to 7) absolute number of splenic and intrahepatic NS5B451–459-specific CD8+ T cells. Only comparisons between vaccinated groups that had a statistically significant difference are denoted: *, P < 0.05; and **, P < 0.01.
Using the KdNS5B451–459 tetramer (synthesized as described above), we initially evaluated the number of NS5B-specific CD8+ T cells elicited 7 and 42 days after the final vaccination in each regimen described above (Fig. 3A). At both time points, the number of NS5B-specific CD8+ T cells in the liver was higher after vaccination with regimens that included pVAX-NS5B-PRF and/or rAAV-NS5B compared to the placebo control (Fig. 3). This trend was also observed in the spleen, although this was not statistically significant when the responses in rDNA/rAAV/rDNA- and mock/rAAV/mock-vaccinated mice were compared to those in the placebo control at day 7 (Fig. 3B). Similar numbers of NS5B-specific CD8+ T cells developed in the liver and the spleen of rDNA/rAAV/rDNA, rDNA/mock/rDNA, and mock/rAAV/mock vaccination regimens 7 days after the final vaccination (Fig. 3B). A similar trend was observed in the spleen 42 days after the final vaccination, but rDNA/rAAV/rDNA and mock/rAAV/mock regimens elicited significantly higher numbers of NS5B-specific CD8+ T cells in the liver compared to rDNA/mock/rDNA (Fig. 3C). This suggests that a single exposure to rAAV-NS5B was sufficient to elicit a durable population of intrahepatic NS5B-specific CD8+ T cells, even in the absence of priming and boosting with pVAX-NS5B-PRF.
A vaccination regimen to promote intrahepatic CD8+ TRM cells.
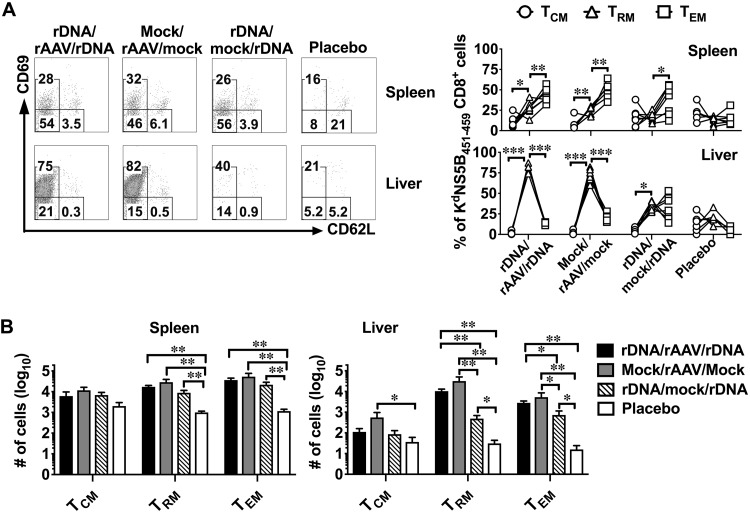
The effectiveness of a vaccine is dependent on its ability to elicit immunological memory and the memory CD8+ T cell population can be broadly categorized into TCM, TEM, and TRM cells (16). Intrahepatic CD8+ TRM cells can be distinguished from circulating memory CD8+ T cells based on the expression of CD69 and CD62L using flow cytometry: TCM (CD69− CD62L+), TRM (CD69+ CD62L−), and TEM (CD69− CD62L−) (12). Thus, using this categorization and flow cytometric analysis of KdNS5B451–459 tetramer+ CD8+ cells, we assessed whether exposure to rAAV-NS5B facilitates the development of high numbers of TRM cells during the memory phase (day 42) following vaccination. The results showed that the majority of the NS5B-specific CD8+ T cells were identified as TRM cells in the liver and TEM cells in the spleen in mice exposed to rAAV-NS5B in the rDNA/rAAV/rDNA and mock/rAAV/mock regimens (Fig. 4A). Furthermore, similar numbers of intrahepatic NS5B-specific CD8+ TRM and TEM cells were elicited between rDNA/rAAV/rDNA and mock/rAAV/mock regimens (Fig. 4B). Although the rDNA/rAAV/rDNA and mock/rAAV/mock regimens elicited the greatest numbers of intrahepatic NS5B-specific CD8+ TRM and TEM cells, the rDNA/mock/rDNA regimen was able to elicit NS5B-specific CD8+ TRM and TEM cells in the liver compared to the placebo control (Fig. 4B). In contrast to the observations in the liver, similar numbers of NS5B-specific CD8+ TCM, TEM, and TRM cells were elicited in the spleen by the rDNA/rAAV/rDNA, mock/rAAV/mock, and rDNA/mock/rDNA regimens (Fig. 4B).
FIG 4.
Evaluation of NS5B-specific memory CD8+ T cells reveals that rAAV-NS5B vaccination is necessary to elicit the highest numbers of intrahepatic TRM cells. Female BALB/c mice were vaccinated as described in the legend to Fig. 3A, and 42 days after the final vaccination, NS5B451–459-specific CD8+ T cells were analyzed for the formation of CD69− CD62L+ (TCM), CD69+ CD62L− (TRM), or CD69− CD62L− (TEM) cells using flow cytometry. (A) Representative dot plots showing the expression of CD69 and CD62L on gated live CD8+ cells that stained positive for the KdNS5B451–459 tetramer in the spleen and the liver. The numbers in each gate denote the percentage of NS5B451–459-specific CD8+ T cells that are TCM, TRM, or TEM based on the expression of CD69 and CD62L as described above. The line graphs depict the percentage of NS5B451–459-specific CD8+ cells that are TCM, TRM, or TEM cells as determined using flow cytometry, with each line representing the analysis of an individual animal used in each vaccination group. (B) Mean (n = 5 to 7) absolute number of NS5B451-459-specific CD8+ TCM, TRM, and TEM cells that formed in the spleen and the liver of vaccinated mice described in panel A. Only comparisons between vaccinated groups that had a statistically significant difference are denoted: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Similar numbers of intrahepatic NS5B-specific CD8+ T cells were elicited 7 days following prime/boost vaccination with pVAX-NS5B-PRF (i.e., in the rDNA/mock/rDNA regimen) compared to the rDNA/rAAV/rDNA and mock/rAAV/mock regimens (Fig. 3A). Therefore, we proposed that rAAV-NS5B vaccination would elicit high numbers of intrahepatic NS5B-specific CD8+ TRM cells more effectively if rAAV-NS5B was delivered 7 days after prime/boost vaccination (at 14-day intervals) with pVAX-NS5B-PRF (i.e., in an rDNA/rDNA/rAAV regimen rather than the rDNA/rAAV/rDNA regimen) compared to a single vaccination with rAAV-NS5B. However, this was not the case, and the numbers of NS5B-specific CD8+ T cells and intrahepatic CD8+ TRM cells elicited by the modified rDNA/rDNA/rAAV regimen and a single vaccination with rAAV-NS5B were similar (data not shown).
Overall, this analysis suggests that the ability of rAAV-NS5B to elicit robust memory intrahepatic NS5B-specific CD8+ T cells (Fig. 3B) correlated with its ability to promote the development of antiviral CD8+ TRM cells in the liver (Fig. 4B).
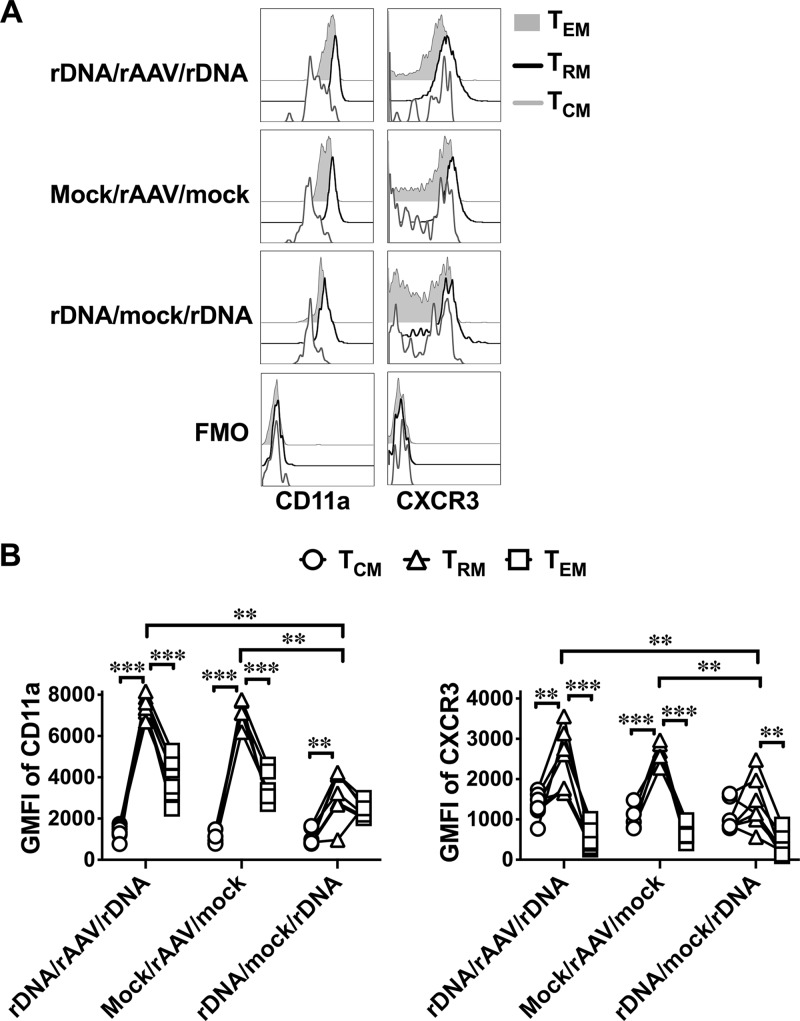
Heightened expression of CD11a and CXCR3 on intrahepatic NS5B-specific CD8+ TRM cells following rAAV-NS5B vaccination.
Although the above studies suggested that CD8+ TRM cells were detected in the liver, elevated expression of CD11a and chemokine (C-X-C motif receptor 3 [CXCR3]) is an authentic characteristic of these cells compared to circulating memory T cells (12, 38). Consequently, we examined the expression of CD11a and CXCR3 on the different populations of NS5B-specific memory CD8+ T cell populations in the liver. Each vaccination regimen (rDNA/rAAV/rDNA, mock/rAAV/mock, and rDNA/mock/rDNA) upregulated CD11a and CXCR3 on intrahepatic NS5B-specific CD8+ TRM cells compared to the respective TEM and TCM populations, although this trend was subtle and not always statistically significant in the rDNA/mock/rDNA group (Fig. 5). Furthermore, CD11a expression on intrahepatic NS5B-specific CD8+ TRM cells that developed from the rDNA/rAAV/rDNA and mock/rAAV/mock regimens was significantly higher than the rDNA/mock/rDNA regimen (Fig. 5). A similar yet more subtle trend was observed with respect to CXCR3 expression on intrahepatic NS5B-specific CD8+ TRM cells that developed in rAAV-NS5B-vaccinated mice compared to mice vaccinated with the rDNA/mock/rDNA regimen. Flow cytometric analysis showed that in the placebo control group, only a small number of events were KdNS5B451-459 tetramer+ CD8+ in the liver (Fig. 3B), while even fewer or no events represented NS5B-specific TEM, TRM, and TCM cells (Fig. 4A). Consequently, these mice were excluded from this analysis. Overall, these results suggest that rAAV-NS5B vaccination is more efficient than pVAX-NS5B-PRF vaccination to ensure heightened expression of CD11a and CXCR3 on intrahepatic NS5B-specific CD8+ TRM cells, which likely facilitates retention of these cells in the liver (see Discussion).
FIG 5.
Among the different memory subsets of intrahepatic NS5B-specific CD8+ T cells, TRM cells express the highest levels of CD11a and CXCR3 in vaccinated mice. Female BALB/c mice vaccinated with pVAX-NS5B-PRF and/or rAAV-NS5B (Fig. 3A) were euthanized 42 days after the final vaccination, and the expression of CD11a and CXCR3 was analyzed on intrahepatic CD69− CD62L+ CD8+ (TCM), CD69+ CD62L− CD8+ (TRM), or CD69− CD62L− CD8+ (TEM) cells that stained positive for KdNS5B451–459 tetramer. (A) Representative histogram plots show the expression of CD11a or CXCR3 on NS5B-specific CD8+ TCM, TRM, or TEM cells. Fluorescent minus one (FMO) plots represent the background staining for CD11a and CXCR3. (B) GMFI of CD11a or CXCR3 in each vaccinated mouse from rDNA/rAAV/rDNA-, mock/rAAV/mock-, and rDNA/mock/rDNA-vaccinated groups. Only comparisons between vaccinated groups that had a statistically significant difference are denoted: **, P < 0.01; and ***, P < 0.001.
rAAV-NS5B vaccination elicits superior NS5B-specific TH cell and CD8+ T cell responses in vivo.
Next, we evaluated the functional activity of NS5B-specific CD8+ T cells (Fig. 6 and Table 3) and TH cells (Fig. 7 and Table 4), not only against the immunodominant epitopes described above (Fig. 1 and 2) but also using the entire NS5B peptide set, 42 days after vaccination with the different regimens. As intrahepatic CD8+ TRM cells survive very poorly following in vitro culture (12), the FTA analysis was used to examine the NS5B-specific T cell responses in vivo. Flow cytometric analysis of the FTA cells recovered from the spleen (Fig. 6B) and the liver (Fig. 6C) suggests that killing responses were significantly higher in rDNA/rAAV/rDNA-, mock/rAAV/mock-, and rDNA/mock/rDNA-vaccinated mice compared to the placebo control against target cells pulsed with P3, immunodominant NS5B442–459, or NS5B451–459. Modest or no responses were detected against target cells pulsed with NS5B peptide pool 1 (NS5B1–207 [P1]) or P2 compared to the placebo control (Fig. 6). Robust killing responses against all target cells pulsed with titrated concentrations of NS5B451–459 were detected after vaccination with the rDNA/rAAV/rDNA, mock/rAAV/mock, and rDNA/mock/rDNA regimens, but the greatest NS5B451–459-specific killing responses were exhibited in rAAV-NS5B-vaccinated mice (Fig. 6). A similar trend was observed against target cells pulsed with the NS5B442–459 peptide, although there was a dose-dependent reduction in killing responses against targets pulsed with 0.01 or 0.1 μg/ml reflective of a lower-avidity response to NS5B442–459- compared to NS5B451–459-pulsed targets (Fig. 6).
FIG 6.
pVAX-NS5B-PRF- and rAAV-NS5B-based vaccination regimens elicit robust NS5B-specific killer CD8+ T cell responses in vivo. Female BALB/c mice vaccinated as described in the legend to Fig. 3A were challenged i.v. 42 days after the final vaccination with peptide-pulsed autologous target cells labeled in an FTA for flow cytometry analysis similar to that described in the legend to Fig. 1. The experimental details regarding how the FTA was constructed are described in Materials and Methods and Table 2. (A) The representative density plots and the table show that the FTA is comprised of target cell populations that were unpulsed (Nil) or pulsed with titrated concentrations of the mapped immunodominant T cell peptides of NS5B or 10 μg/ml of peptide pools spanning NS5B1–207 (P1), NS5B197–395 (P2), or NS5B385–591 (P3). (B and C) Mean (n = 7) specific (above nil targets) killing responses against target cells pulsed with immunodominant CD8+ T cell peptides (NS5B442–459 and NS5B451–459) or peptide pools of NS5B recovered from the spleen (B) and the liver (C) 15 h after the FTA challenge are shown. Only comparisons between vaccinated groups that had a statistically significant difference are denoted: *, P < 0.05; and **, P < 0.01. The P values for the data depicted in the line graphs are shown in Table 3.
TABLE 3.
P values of the comparisons between vaccinated groups depicted in the line graphs of Fig. 6
| Organ and peptide | Peptide concn (μg/ml) |
P value for group comparisona
: |
|||||
|---|---|---|---|---|---|---|---|
| 1 vs 2 | 1 vs 3 | 1 vs 4 | 2 vs 3 | 2 vs 4 | 3 vs 4 | ||
| Spleen | |||||||
| NS5B442–459 | 0.01 | NS | NS | NS | NS | NS | NS |
| 0.1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | NS | |
| 1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 10 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| NS5B451–459 | 0.01 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 |
| 0.1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 10 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| Liver | |||||||
| NS5B442–459 | 0.01 | NS | 0.01 | 0.024 | 0.002 | 0.002 | 0.002 |
| 0.1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 10 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| NS5B451–459 | 0.01 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 |
| 0.1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 1 | NS | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
| 10 | 0.02 | 0.003 | 0.003 | 0.002 | 0.002 | 0.002 | |
Vaccinated groups are numbered as follows: 1, rDNA/rAAV/rDNA; 2, mock/rAAV/mock; 3, rDNA/mock/rDNA; and 4, placebo. NS, not significant (P > 0.05).
FIG 7.
rAAV-NS5B-based vaccination regimens also elicit superior NS5B-specific TH cell responses in vivo. Female BALB/c mice were vaccinated as described in the legend to Fig. 3A and challenged with the peptide-pulsed FTA described in the legend to Fig. 6. Mean (n = 7) specific GMFI of CD69 on B220+ cells in the FTA pulsed with immunodominant TH cell peptides (NS5B225–240 and NS5B495–512) or peptide pools of NS5B recovered from the spleen (A) and the liver (B) 15 h after the FTA challenge are shown. Only comparisons between vaccinated groups that had a statistically significant difference are denoted: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. The P values for the data depicted in the line graphs are shown in Table 4.
TABLE 4.
P values of the comparisons between vaccinated groups depicted in the line graphs of Fig. 7
| Organ and peptide |
Peptide concn (μg/ml) |
P value for group comparisona
: |
|||||
|---|---|---|---|---|---|---|---|
| 1 vs 2 | 1 vs 3 | 1 vs 4 | 2 vs 3 | 2 vs 4 | 3 vs 4 | ||
| Spleen | |||||||
| NS5B225–240 | 0.01 | NS | NS | NS | NS | NS | NS |
| 0.1 | NS | NS | NS | 0.048 | 0.018 | NS | |
| 1 | NS | 0.032 | 0.004 | 0.002 | 0.002 | NS | |
| 10 | NS | NS | 0.015 | NS | 0.002 | 0.02 | |
| NS5B495–512 | 0.01 | NS | NS | NS | NS | NS | NS |
| 0.1 | NS | NS | NS | NS | NS | NS | |
| 1 | NS | 0.000 | 0.000 | 0.000 | 0.000 | NS | |
| 10 | NS | 0.003 | 0.003 | 0.02 | 0.000 | 0.02 | |
| Liver | |||||||
| NS5B225–240 | 0.01 | NS | NS | NS | NS | NS | NS |
| 0.1 | NS | NS | NS | NS | NS | NS | |
| 1 | NS | NS | NS | NS | 0.025 | NS | |
| 10 | NS | 0.009 | 0.000 | 0.015 | 0.000 | 0.000 | |
| NS5B495–512 | 0.01 | NS | NS | NS | NS | NS | NS |
| 0.1 | NS | NS | NS | NS | NS | NS | |
| 1 | NS | NS | NS | NS | 0.035 | NS | |
| 10 | NS | NS | 0.007 | 0.006 | 0.002 | 0.003 | |
Vaccinated groups are numbered as follows: 1, rDNA/rAAV/rDNA; 2, mock/rAAV/mock; 3, rDNA/mock/rDNA; and 4, placebo. NS, not significant (P > 0.05).
Analysis of CD69 expression on B220+ cells in the FTA suggests that mice vaccinated with rDNA/rAAV/rDNA, mock/rAAV/mock, or rDNA/mock/rDNA exhibited TH cell responses significantly above those in the placebo control against target cells pulsed with 10 μg/ml of NS5B225–240, NS5B495–512, or NS5B P2 (NS5B197–395) (Fig. 7). Although the TH cell responses were similar against spleen-derived target cells pulsed with 10 μg/ml of NS5B225–240 peptide in all regimens except the placebo (Fig. 7A), the highest TH cell responses were elicited by the rDNA/rAAV/rDNA and mock/rAAV/mock regimens. Furthermore, only mice in the rDNA/rAAV/rDNA and mock/rAAV/mock regimens elicited TH cell responses against targets pulsed with P1 that were greater than the placebo control. Superior TH cell responses after vaccination with the rDNA/rAAVr/DNA and mock/rAAV/mock regimens were also observed against B220+ FTA cells pulsed with 1 μg/ml of NS5B225–240 or NS5B495–512, particularly those recovered from the spleen (Fig. 7). TH cell responses to target cells pulsed with P3 are not shown as these targets were virtually all killed when introduced into mice vaccinated with a regimen that included rAAV-NS5B, as shown in Fig. 6.
DISCUSSION
The development of a HCV vaccine is a global priority. Modeling studies suggest that using a vaccine that is 50 to 80% effective in a high-risk population can considerably reduce the incidence of persistent HCV infection (39). An effective vaccine is considered essential to meet the WHO’s Sustainable Development Goal of eliminating 90% of incident hepatitis C by 2030 (2). After 2 decades of preclinical studies and phase I HCV vaccine trials, the current lead candidate is under investigation in an NIH-sponsored phase IIb, placebo-controlled trial in high-risk PWID in a prime/boost regimen of gt1 NS antigens delivered via chimpanzee adenovirus and then modified vaccinia virus Ankara vaccines (11). Although this vaccine induces robust systemic T cell immunity to gt1b antigens, it is not known whether similar responses are elicited in the liver (11). The efficacy of any HCV vaccine based on T cell immunity will be dependent on the rapid induction of robust HCV-specific T cell responses in the liver following HCV infection (40), and consequently, a vaccine that elicits intrahepatic HCV-specific CD8+ TRM cells and CD4+ T cell immunity likely has an intrinsic advantage. Recently, robust liver-resident CD8+ T cell responses elicited following vaccination were shown to be crucial for protection against controlled human malaria infection and P. berghei infection in mice (12, 13). Recombinant DNA and rAAV vectors have been regularly used as vectors for vaccination and gene therapy in humans. Thus, for the first time, we evaluated the use of recombinant DNA- and rAAV-based vaccination strategies to elicit intrahepatic HCV-specific T cell immunity and CD8+ TRM cells in vivo.
Previous studies exploiting AAV as a vector to elicit intrahepatic T cell immunity and TRM cells have exploited AAV encoding a single or a few T cell epitopes and TCR transgenic T cells in adoptive transfer settings (12, 37). As native proteins are more susceptible to cross-presentation than a single or a few CD8+ T cell epitopes encoded by vaccine vectors (41), the current study exploited vaccine vectors encoding codon-optimized gt3a NS5B protein and evaluated T cell immunity in a polyclonal repertoire in wild-type mice.
The current study described the use of the FTA assay to map immunodominant T cell epitopes of gt3a NS5B in vivo. Our in vivo FTA-based epitope mapping analysis also led to the primary development of a functional KdNS5B451–459 tetramer and the identification of NS5B225–240 and NS5B495–512 immunodominant peptides for the analysis of NS5B-specific TH cell responses in BALB/c mice. We have previously reported that the FTA is a high-throughput and versatile technique to measure the magnitude and avidity of CD4+ TH cell and CD8+ killer T cell responses in vivo (32–34). FTA overcomes many drawbacks associated with manipulating T cells and their functional parameters in vitro, which are common in conventional T cell stimulation assays such as ELISpot and ICS. Furthermore, CD8+ TRM cells and liver-resident CD8+ TRM cells have been reported to survive poorly following in vitro culture (12, 16), which further highlights the importance of exploiting the FTA for epitope mapping and evaluating T cell responses.
Our data suggest that there is a strong correlation between the number of NS5B-specific CD8+ TRM cells, assessed using the KdNS5B451–459 tetramer, and the magnitude of NS5B451–459-specific cytotoxic CD8+ T cell responses in vaccinated mice as determined by the FTA analysis. These findings are consistent with our previous report showing that i.v. immunization with rAAV can elicit cytotoxic CD8+ T cells capable of killing cognate peptide-pulsed targets in vivo (37). Intravenous delivery is the best-established route to deliver substances to the liver, and the FTA target cells will be readily exposed in the liver following i.v. challenge in order to evaluate intrahepatic T cell responses. However, these target cells can circulate to other sites (e.g., the spleen), making them susceptible to T cell responses systemically. It is important for future studies to test the HCV vaccines, including those developed in the current study, in a challenge model that recapitulates hepatotropic HCV infection in order to gain insight into the protective role of intrahepatic TRM cells. Although a challenge model of this nature has not been developed to evaluate vaccine efficacy in wild-type mice, the recently developed Norway rat hepacivirus infection model (42) has the potential to be adopted for this purpose.
CD4+ TH cell responses as measured by the upregulation of CD69 on FTA B220+ cells were more readily detected in the FTA recovered from the spleen compared to the liver in pVAX-NS5B-PRF- and/or rAAV-NS5B-vaccinated mice. The spleen is the largest secondary lymphoid organ in mammals that is easily accessible for lymphocytes found in the blood and consists of areas that support the development, maturation, and activation of mature B cells, which includes areas (e.g., in the white pulp) where T and B cells interact (43, 44). Under a steady-state condition, the spleen has more T and B cells than the liver, with the vast majority of the lymphocytes in the mouse spleen being B cells, and the liver, by default, functions to tolerize immune responses through the action of liver-resident cells such as Kupffer cells and liver sinusoidal endothelial cells (45). Thus, the higher NS5B-specific activation of FTA B220+ cells in the spleen compared to that of the liver in the current study is likely due to differences in the cellular composition and the microenvironments between the spleen and liver, but this needs to be formally evaluated.
Numerous studies have shown that CXCR3 expression is important for homing of effector CD4+ and CD8+ T cells to inflammatory sites (46). TRM cells are known to express higher levels of CXCR3 (12) and CD11a (38) in the liver compared to TEM and TCM cells, consistent with the observations of the current study. Fernandez-Ruiz et al. (12) reported that CXCR3 was exclusively expressed on CD8+ TRM cells relative to TEM and TCM cell counterparts following vaccination against P. berghei. However, our findings suggest that NS5B-specific CD8+ TEM and TCM cells expressed CXCR3, albeit at lower levels compared to NS5B-specific CD8+ TRM cells in the liver. The reason for this difference is unclear, but it is important to note that Fernandez-Ruiz et al. (12) based their analysis on adoptively transferred TCR transgenic PbT-I cells that were stimulated with vectors encoding or expressing an 8-aa peptide (NVY) immunogen. As noted previously, our study was based on the analysis of NS5B-specifc polyclonal T cells in wild-type mice following vaccination with vectors encoding the full-length gt3a NS5B protein, which consists of multiple CD4+ and CD8+ T cell epitopes. The results of our study are consistent with previous reports which showed that TCM, TEM, and CD11alo and CD11ahi virus-specific CD8+ T cells express CXCR3 following intranasal Sendai virus infection (47, 48). HCV-specific TEM and TCM cells isolated from the blood and the liver of acutely and chronically infected HCV patients express CXCR3 (49). Furthermore, effector memory CD8+ T cells express CXCR3 following intravenous injection of Listeria monocytogenes (50), and lymphocytic choriomeningitis virus (LCMV)-specific P14 TCR transgenic CD8+ T cells universally expressed CXCR3 48 days postinfection with LCMV Armstrong (51).
Several studies have shown that local antigen encounter or deposition usually perpetuates the establishment of high numbers of CD8+ TRM cells in various anatomical sites, including the liver (12, 52–54). Studies in mice and humans suggest that i.v. immunization is the most efficient route to facilitate local antigen encounter in the liver (13, 37, 55). Intravenous delivery of vaccine vectors that can enter hepatocytes and/or the cells surrounding the hepatic tissues has been exploited in prime/trap (13) and prime/target (55) vaccination approaches to recruit or “pull” systemically primed CD8+ T cells into the liver and allow these cells to differentiate into intrahepatic TRM cells. Imaging of live mice injected i.d. or i.v. with P. berghei sporozoites suggested that the sporozoites were reproducibly detected in the liver only after i.v. injection, while the parasite load in the liver was 30-fold higher in the i.v.-injected mice (56). The same study also showed that i.v. compared to i.d. injection of irradiated sporozoites elicited a significantly higher proportion of CD62L− memory CD8+ T cells and increased protection against live sporozoite challenge (56). Intradermal, subcutaneous, and intramuscular immunization of humans with radiation-attenuated Plasmodium falciparum sporozoites (PfSPZ) also appeared to elicit suboptimal immunity and protection against malaria compared to i.v. vaccination (13, 57). Analysis of vaccinated nonhuman primates suggests that the increased efficacy of i.v. PfSPZ vaccination in humans correlated strongly with the ability of this vaccine to elicit long-lived liver-resident CD8+ T cells (13, 57). Furthermore, transduced hepatocytes have the capacity to present AAV-encoded antigens to CD8+ T cells following i.v. vaccination with AAV (37), and analysis of liver biopsy specimens from end-stage hepatitis C patients suggests that HCV infection elicits CD11ahi memory CD8+ T cells (15). Collectively these findings are consistent with our findings, which showed that i.d. vaccination with pVAX-NS5B-PRF was less efficient in eliciting intrahepatic T cell immunity and CD8+ TRM cells compared to i.v. rAAV-NS5B vaccination. Furthermore, antigen presentation following i.d. DNA immunization occurs in the draining cervical lymph nodes (28, 35) and likely does not facilitate local antigen encounter/deposition in the liver to establish high numbers of intrahepatic CD8+ TRM cells. A caveat with this interpretation is that a recent study that analyzed adoptively transferred CD8+ T cells showed that the lodgment of CD8+ TRM cells in the liver is both activation and interleukin-15 (IL-15) dependent, and inflammatory signals (e.g., type I interferon inducers) can enhance the number of intrahepatic CD8+ TRM cells (54). Thus, in addition to facilitating local antigen encounter, rAAV-NS5B vaccination could induce higher levels of IL-15 and relevant inflammatory signals compared to pVAX-NS5B-PRF vaccination in order to increase the numbers of intrahepatic CD8+ TRM cells.
The prime/trap malaria study (12) is the only study to have evaluated TRM cell formation following i.v. immunization with AAV (i.e., rAAV-NVY), which was performed only 35 days after rAAV-NVY injection. The time point at which we evaluated NS5B-specific CD8+ TRM cells was 42 days after the final pVAX-PRF (mock) or pVAX-NS5B-PRF vaccination, but it was 56 days after the rAAV-NS5B vaccination in the rDNA/rAAV/rDNA and mock/rAAV/mock regimens. Thus, this is the longest duration following rAAV immunization in which a study has evaluated TRM cell formation. The results clearly show that a robust population of TRM cells can be elicited at least 56 days following i.v. rAAV immunization. Furthermore, the numbers of intrahepatic NS5B-specific CD8+ T cells that formed 7 and 42 days after the final vaccination in regimens involving rAAV-NS5B vaccinations were similar (Fig. 3), with an overwhelming majority of the intrahepatic NS5B-specific CD8+ T cells elicited at day 42 displaying a TRM phenotype in this instance (Fig. 4). This was not the case with prime/boost vaccination with pVAX-NS5B-PRF (i.e., in the rDNA/mock/rDNA regimen), which elicited higher numbers of intrahepatic NS5B-spcific CD8+ T cells at day 7 compared to day 42 (Fig. 3). Collectively, these studies suggest that a stable and durable population of NS5B-specific CD8+ TRM cells has formed following rAAV-NS5B vaccination. A caveat is that a recent study showed that TRM cells that form in the liver in recipient mice adoptively transferred with in vitro-activated OT-I CD8+ T cells or PbT-I cells that are later primed with radiation-attenuated sporozoite (RAS) vaccination have a half-life of 36 or 28 days, respectively (54). Thus, future studies could also determine the half-life of the intrahepatic CD8+ TRM cells that form following rAAV vaccination, which will be important for understanding the limitations and strengths of using rAAV to elicit a stable population of intrahepatic CD8+ TRM cells.
CXCR3 expression is required for T cells to home to inflamed sites and is likely to be important for the retention of CD8+ TRM cells in the liver (12), although CXCR6 rather than CXCR3 appears to be crucial for the establishment of liver-resident CD8+ T cells (58). A recent study also showed that there is a threshold level of expression of CD11a necessary to retain CD8+ TRM cells in the liver, as heightened expression of CD11a was crucial for CD8+ TRM cells to remain in the liver (38). We observed that CD11a expression was significantly lower on NS5B-specific CD8+ TRM cells in mice vaccinated with pVAX-NS5B-PRF (rDNA/mock/rDNA) compared to mice vaccinated with rAAV-NS5B, although a similar yet more subtle trend was observed relative to CXCR3 expression. This could also explain the inability of pVAX-NS5B-PRF vaccination (rDNA/mock/rDNA) to elicit high numbers of intrahepatic CD8+ TRM cells or augment the number of these cells in the rDNA/rAAV/rDNA regimen compared to rAAV-NS5B vaccination alone (i.e., mock/rAAV/mock). Thus, future studies should investigate the factors associated with the upregulation of CD11a on intrahepatic CD8+ TRM cells as such investigations have the potential to revaluate the manner in which a T-cell-mediated vaccine is designed against hepatotropic pathogens.
Overall, we evaluated rDNA- and rAAV-based vaccination strategies that can be exploited to elicit systemic and intrahepatic HCV-specific T cell immunity, which have ramifications for the design of effective vaccines against hepatotropic pathogens such as malaria, HBV, and HCV. This study also highlights the importance of exploiting hepatotropic vaccine vectors to elicit robust and durable intrahepatic cytotoxic CD8+ T cell immunity and TH cell responses against HCV in vivo.
MATERIALS AND METHODS
Mice.
All studies used 6- to 8-week-old age-matched female BALB/c mice, which were purchased from The University of Adelaide Laboratory Animal Services and housed in individually HEPA-filtered cages at The Queen Elizabeth Hospital, Woodville, Australia. Furthermore, all the animal studies were approved and conducted under the guidelines of The University of Adelaide and SA Pathology animal ethics committees.
Vaccines and immunizations.
We have published several studies (24, 26, 27, 29) describing how immunogens such as HCV NS proteins were encoded in the bicistronic rDNA-PRF vector, which was used to construct pVAX-NS5B-PRF. The DNA used for vaccinations was isolated from transformed Escherichia coli DH5α cells and purified using a Qiagen EndoFree Plasmid Giga kit with removal of bacterial endotoxins as per the manufacturer’s instructions. For each i.d. vaccination with DNA, mice were anesthetized using isoflurane and injected with 50 μg of pVAX-NS5B-PRF or an equimolar amount of pVAX-PRF in 40 μl of phosphate-buffered saline (PBS) as we described previously (24, 26, 27, 29).
rAAV-NS5B was constructed as we described previously (37). In summary, codon-optimized gt3a NS5B and enhanced green fluorescent protein (eGFP) were cloned into the multiple-cloning site of pAM2AA (59) under the control of a liver-specific enhancer/promoter that is flanked by cis-acting inverted terminal repeats. Protease cleavage between NS5B and eGFP was achieved by introducing the foot and mouth disease virus 2A (FMDV2A) protease; eGFP was included to facilitate analysis of the efficiency of rAAV transduction of hepatocytes in vivo as we described previously (37). The rAAV-NS5B was packaged and pseudoserotyped to capsid 8 following transfection of HEK293T cells with pAM2AA encoding NS5B, pXX6 helper plasmid (containing adenoviral E2a, E4Orf6, and VA RNA genes), and p5E1VD2/8 plasmid (containing the Cap gene from AAV-8 and the Rep gene from AAV-2) (60). The molar ratio of the plasmids for the transfection was 1:3:1 pAM2AA-NS5B to pXX6 to p5E1VD2/8. rAAV-NS5B was harvested and purified using a modified polyethylene glycol 8000 (PEG 8000)-NaCl precipitation method as described previously (61). The yield of the purified virus was determined using the AAVpro titration kit for real-time PCR version 2 (TaKaRa) following the protocol for quantitative real-time PCR. For each vaccination with rAAV-NS5B, 5 × 109 vgc of the virus in 200 μl of PBS was delivered i.v. into the lateral tail vein of the mice.
Isolation of cells from the liver and the spleen for analysis.
The livers of euthanized mice were perfused with 10 to 15 ml of PBS via the portal vein, and the perfused livers were meshed through 70-μm-pore cell strainers (BD Biosciences). The cells were then resuspended in a 36% isotonic Percoll (Sigma-Aldrich) gradient and centrifuged (500 × g at 4°C with no brakes) for 20 min. The pelleted cells, comprising lymphocytes, were then harvested, and red blood cells (RBCs) were lysed before analysis. Isolated spleens were meshed through 70-μm-pore cell strainers, and RBC-depleted splenocytes were analyzed.
Analysis of KdNS5B451–459 tetramer-positive cells using flow cytometry.
A total of 1 × 106 to 4 × 106 RBC-depleted splenocytes and lymphocytes derived from the liver were seeded in each well of 96-well U-bottom well cell culture plates (Sigma-Aldrich) and stained with 40 μl/well of 1:500-diluted fixable viability stain 620 (FV620; BD Biosciences) for 15 min at room temperature (RT). The cells were then washed twice with PBS and stained with 1:50 diluted allophycocyanin (APC)-conjugated KdNS5B451–459 tetramer (Biomolecular Resource Facility, The John Curtin School of Medical Research, Canberra, Australia) in PBS for 45 min at RT in the dark. The cells were then washed twice with flow buffer (PBS plus 2% FBS plus 25 mM HEPES plus 5 mM EDTA) prior to cell surface staining of the samples as described previously (62). A cocktail of fluorochrome-conjugated antibodies to mouse proteins CD69 (clone H1.2F3; BD Biosciences), CD8 (clone 53-6.7; eBioscience), CD62L (clone MEL-14), CXCR3 (clone CXC3-173; BD Biosciences), and CD11a (clone M17/4; BD Biosciences) was used for the cell surface stain. The fluorochromes conjugated to the antibodies in the cocktail include phycoerythrin (PE)-Cy7 (CD69), APC-efluor 780 (CD8), PE (CD62L), brilliant violet 421 (CXCR3), and brilliant violet 510 (CD11a). The cells were washed twice with 0.5% paraformaldehyde and resuspended in 0.5% paraformaldehyde prior to analysis in a Becton, Dickinson FACS Canto II flow cytometer. The flow cytometry plots were constructed using the FlowJo software (version 8.8.7), and FV620+ (dead) events and doublets were excluded prior to analysis of NS5B-specific CD8+ T cells.
FTA analysis.
For the FTA in Fig. 1 and 2, naive autologous BALB/c splenocytes were serially labeled with the indicated concentrations (Table 1) of cell proliferation dye (CPD), cell trace violet (CTV), and carboxyfluorescein succinimidyl ester (CFSE) at RT for 5 min as we described previously (29). The dye-labeled cells were unpulsed (nil) or pulsed with 10 μg/ml of the indicated gt3a NS5B peptide or peptide pool (Table 1) spanning NS5B385–591 (peptides 61 to 90) for 4 h at 37°C with 5% CO2 as we described previously (29). The 90-peptide array spanning the gt3a (K3a/650) NS5B was a gift from The National Institutes of Health and Biodefence and Emerging Infections (BEI) Research Resources, and the sequence for each peptide can be viewed using the BEI Research Resources website (www.beiresources.org) under catalogue no. NR-4070. A total of 48 × 106 dye-labeled and peptide-pulsed target cells (1.5 × 106 per target cell cluster) were resuspended in 200 μl of PBS and injected i.v. into the lateral tail vein of mice 13 days after the final vaccination with pVAX-NS5B-PRF or placebo as described in the legends to Fig. 1 and 2.
TABLE 1.
Fluorescent bar-coding scheme for the peptide-pulsed target cells in the FTA used for analysis in Fig. 1 and 2
| Target labeling combination |
Bar code with: |
|||
|---|---|---|---|---|
| 0.6 μM CTV | 6.2 μM CTV | 23 μM CTV | 85 μM CTV | |
| 10 μM CPD +: | ||||
| 17 μM CSFE | Nil | 61 | 62 | 63 |
| 4.8 μM CSFE | 64 | 65 | 66 | 67 |
| 1.4 μM CSFE | 68 | 69 | 70 | 71 |
| 0.4 μM CSFE | 72 | 73 | 74 | 75 |
| 39 μM CPD +: | ||||
| 17 μM CSFE | 76 | 77 | 78 | 79 |
| 4.8 μM CSFE | 80 | 81 | 82 | 83 |
| 1.4 μM CSFE | 84 | 85 | 86 | 87 |
| 0.4 μM CSFE | 88 | 89 | 90 | P3 |
For the FTA in Fig. 6 and 7, naive BALB/c splenocytes were serially labeled with CPD, CTV, and CFSE as described above and pulsed with the indicated concentrations of the peptides or peptide pools of gt3a NS5B as shown in Table 2. Subsequently, 20 × 106 dye-labeled and peptide-pulsed target cells (1 × 106 per target cell cluster) were resuspended in 200 μl of PBS and injected i.v. into the lateral tail vein of mice 42 days after vaccination.
TABLE 2.
Fluorescent bar-coding scheme for the peptide-pulsed target cells in the FTA used for analysis in Fig. 6 and 7
| Target labeling combination |
Bar code witha
: |
|||
|---|---|---|---|---|
| 0.2 μM CTV | 0.6 μM CTV | 6.2 μM CTV | 23 μM CTV | |
| 39 μM CPD +: | ||||
| 17 μM CSFE | Nil | P1 (10) | P2 (10) | P3 (10) |
| 4.8 μM CSFE | NS5B442–459 (0.01) | NS5B442–459 (0.1) | NS5B442–459 (1) | NS5B442–459 (10) |
| 1.4 μM CSFE | NS5B451–459 (0.01) | NS5B451–459 (0.1) | NS5B451–459 (1) | NS5B451–459 (10) |
| 0.4 μM CSFE | NS5B225–440 (0.01) | NS5B225–240 (0.1) | NS5B225–240 (1) | NS5B225–240 (10) |
| 0 μM CSFE | NS5B495–512 (0.01) | NS5B495–512 (0.1) | NS5B495–512 (1) | NS5B495–512 (10) |
In parentheses is indicated the concentration (μg/ml) of the peptide or peptide pool used for pulsing the target cells.
Fifteen hours after the FTA challenge, RBC-depleted cells isolated from the livers and spleens of vaccinated mice were stained with PE-Cy7-conjugated anti-mouse CD69 (clone H1.2F3; BD Biosciences) and Alexa Fluor 700-conjugated anti-mouse B220 (RA3-6B2; Invitrogen) for flow cytometric analysis as described above. The percentage of specific killing of target cells was calculated based on the percentage of FTA cells recovered using the formula [(nil target value % – peptide-pulsed target value %)/nil target value %] × 100. The plotted values for GMFI of CD69 on B220+ FTA targets were calculated using the formula B220+ peptide-pulsed target value (GMFI of CD69) − B220+ nil target value (GMFI of CD69).
In vivo depletion of CD4+ T cells and FTA analysis.
Six- to 9-week-old female BALB/c mice were vaccinated with three equimolar doses (50 μg/dose) of pVAX-PRF (mock) or pVAX-NS5B-PRF at fortnightly intervals as described previously (Fig. 1 and 2). Thirteen days after the 3rd dose, 7 to 9 mice from the pVAX-NS5B-PRF-vaccinated group were injected i.p. with 100 μg/mouse of anti-mouse CD4 depletion antibody (clone GK1.5; InVivoMab). Three days after the depletion, FTA cells that were unpulsed or pulsed with 10 μg/ml of NS5B225–240, NS5B495–512, P2, or P3 were injected i.v. into vaccinated mice 15 h prior to recovery of the injected targets from the spleens of mice for the analysis of CD4+ TH cell responses as described in the legend to Fig. 2.
Epitope prediction and selection.
Fourteen- to 19-aa-long peptide sequences (peptides 61 to 90) from the gt3a NS5B385–591 region described above were used as input data for epitope prediction using IEDB MHC class I epitope prediction algorithm (available at www.iedb.org). H2-kd was selected as the parameter for MHC type, and epitope prediction length was restricted at 9 amino acids. The “IEDB Recommended” approach was the algorithm selected to perform the prediction. The result of this prediction was downloaded in .csv format. These data were then put through an in-house script to match with epitopes that have been experimentally validated. The epitope (KdNS5B451–459) that matched the experimentally validated epitope found within peptides 69 and 70 and had high prediction score (where scores closer to 0.0 indicate a better predicted epitope) was selected for tetramer synthesis.
Statistical analysis.
Graphs were constructed using the GraphPad Prism 8 software. IBM SPSS Statistics software (version 24) was used to perform the normality tests of the data distribution and statistical significance of the data, with P values of <0.05 considered to be statistically significant. Data were initially analyzed using a Shapiro-Wilk test, and normality was assumed for data sets with P values of >0.05. A parametric Levene’s test was performed on normally distributed data, and homogeneity of variance was assumed for data sets with P values of >0.05. To determine the statistical significance for comparisons related to normally distributed and homoscedastic data sets, one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test was used. For all comparisons related to heteroscedastic data sets, the Welch ANOVA with Games-Howell post hoc test was used to determine the statistical significance. For nonnormally distributed data sets (P < 0.05 by the Shapiro-Wilk test), a nonparametric Levene’s test was performed to evaluate the homogeneity of variance of the data. If homogeneity of variance was assumed (P > 0.05), a Kruskal-Wallis H test was used to determine the statistical significance of the data. All paired comparisons between TRM versus TCM cells and TRM versus TEM cells in each vaccination group shown in Fig. 4A and Fig. 5B were performed using a paired t test. P values for comparisons of the groups shown in Fig. 6 and 7 are given in Tables 3 and 4, respectively.
ACKNOWLEDGMENTS
The Hospital Research Foundation (THRF), The Australian Centre for HIV and Hepatitis Virology (ACH2), and the Australia-India Biotechnology Fund provided grant funding support. THRF also provided early career research fellowships to Danushka K. Wijesundara and Ashish C. Shrestha. Andrew R. Lloyd is supported by a fellowship from the National Health and Medical Research Council (NHMRC [no. 1043067]).
REFERENCES
- 1.WHO. 2017. Global Hepatitis Report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/.
- 2.WHO. 2016. Global health sector strategies for viral hepatitis 2016–2021. https://www.who.int/hepatitis/strategy2016-2021/ghss-hep/en/.
- 3.Borgia SM, Hedskog C, Parhy B, Hyland RH, Stamm LM, Brainard DM, Subramanian MG, McHutchison JG, Mo H, Svarovskaia E, Shafran SD. 2018. Identification of a novel hepatitis C virus genotype from Punjab, India: expanding classification of hepatitis C virus into 8 genotypes. J Infect Dis 218:1722–1729. doi: 10.1093/infdis/jiy401. [DOI] [PubMed] [Google Scholar]
- 4.Sacks-Davis R, Grebely J, Dore GJ, Osburn W, Cox AL, Rice TM, Spelman T, Bruneau J, Prins M, Kim AY, McGovern BH, Shoukry NH, Schinkel J, Allen TM, Morris M, Hajarizadeh B, Maher L, Lloyd AR, Page K, Hellard M, InC3 Study Group. 2015. Hepatitis C virus reinfection and spontaneous clearance of reinfection—the InC3 Study. J Infect Dis 212:1407–1419. doi: 10.1093/infdis/jiv220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumert TF, Fauvelle C, Chen DY, Lauer GM. 2014. A prophylactic hepatitis C virus vaccine: a distant peak still worth climbing. J Hepatol 61:S34–S44. doi: 10.1016/j.jhep.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Bailey JR, Flyak AI, Cohen VJ, Li H, Wasilewski LN, Snider AE, Wang S, Learn GH, Kose N, Loerinc L, Lampley R, Cox AL, Pfaff JM, Doranz BJ, Shaw GM, Ray SC, Crowe JE Jr. 2017. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. doi: 10.1172/jci.insight.92872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst TR, Pierce BG, Keck ZY, Foung SKH. 2017. Designing a B cell-based vaccine against a highly variable hepatitis C virus. Front Microbiol 8:2692. doi: 10.3389/fmicb.2017.02692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wedemeyer H, He XS, Nascimbeni M, Davis AR, Greenberg HB, Hoofnagle JH, Liang TJ, Alter H, Rehermann B. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol 169:3447–3458. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 9.Smyk-Pearson S, Tester IA, Klarquist J, Palmer BE, Pawlotsky JM, Golden-Mason L, Rosen HR. 2008. Spontaneous recovery in acute human hepatitis C virus infection: functional T-cell thresholds and relative importance of CD4 help. J Virol 82:1827–1837. doi: 10.1128/JVI.01581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Delft A, Humphreys IS, Brown A, Pfafferott K, Lucas M, Klenerman P, Lauer GM, Cox AL, Gaudieri S, Barnes E. 2016. The broad assessment of HCV genotypes 1 and 3 antigenic targets reveals limited cross-reactivity with implications for vaccine design. Gut 65:112–123. doi: 10.1136/gutjnl-2014-308724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swadling L, Capone S, Antrobus RD, Brown A, Richardson R, Newell EW, Halliday J, Kelly C, Bowen D, Fergusson J, Kurioka A, Ammendola V, Del Sorbo M, Grazioli F, Esposito ML, Siani L, Traboni C, Hill A, Colloca S, Davis M, Nicosia A, Cortese R, Folgori A, Klenerman P, Barnes E. 2014. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci Transl Med 6:261ra153. doi: 10.1126/scitranslmed.3009185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V, Cozijnsen A, Collins N, Li J, Davey GM, Kato Y, Devi S, Skandari R, Pauley M, Manton JH, Godfrey DI, Braun A, Tay SS, Tan PS, Bowen DG, Koch-Nolte F, Rissiek B, Carbone FR, Crabb BS, Lahoud M, Cockburn IA, Mueller SN, Bertolino P, McFadden GI, Caminschi I, Heath WR. 2016. Liver-resident memory CD8(+) T cells form a front-line defense against malaria liver-stage infection. Immunity 45:889–902. doi: 10.1016/j.immuni.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Ishizuka AS, Lyke KE, DeZure A, Berry AA, Richie TL, Mendoza FH, Enama ME, Gordon IJ, Chang L-J, Sarwar UN, Zephir KL, Holman LA, James ER, Billingsley PF, Gunasekera A, Chakravarty S, Manoj A, Li M, Ruben AJ, Li T, Eappen AG, Stafford RE, Kc N, Murshedkar T, DeCederfelt H, Plummer SH, Hendel CS, Novik L, Costner PJM, Saunders JG, Laurens MB, Plowe CV, Flynn B, Whalen WR, Todd JP, Noor J, Rao S, Sierra-Davidson K, Lynn GM, Epstein JE, Kemp MA, Fahle GA, Mikolajczak SA, Fishbaugher M, Sack BK, Kappe SHI, Davidson SA, Garver LS, Björkström NK, Nason MC, Graham BS, Roederer M, Sim BKL, Hoffman SL, Ledgerwood JE, Seder RA. 2016. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med 22:614–623. doi: 10.1038/nm.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallett LJ, Davies J, Colbeck EJ, Robertson F, Hansi N, Easom NJW, Burton AR, Stegmann KA, Schurich A, Swadling L, Gill US, Male V, Luong T, Gander A, Davidson BR, Kennedy PTF, Maini MK. 2017. IL-2(high) tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J Exp Med 214:1567–1580. doi: 10.1084/jem.20162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heydtmann M, Hardie D, Shields PL, Faint J, Buckley CD, Campbell JJ, Salmon M, Adams DH. 2006. Detailed analysis of intrahepatic CD8 T cells in the normal and hepatitis C-infected liver reveals differences in specific populations of memory cells with distinct homing phenotypes. J Immunol 177:729–738. doi: 10.4049/jimmunol.177.1.729. [DOI] [PubMed] [Google Scholar]
- 16.Rosato PC, Beura LK, Masopust D. 2017. Tissue resident memory T cells and viral immunity. Curr Opin Virol 22:44–50. doi: 10.1016/j.coviro.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, Lloyd AR. 2011. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog 7:e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin H, Iwasaki A. 2012. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature 491:463–467. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuburu N, Graham BS, Buck CB, Kines RC, Pang YY, Day PM, Lowy DR, Schiller JT. 2012. Intravaginal immunization with HPV vectors induces tissue-resident CD8+ T cell responses. J Clin Invest 122:4606–4620. doi: 10.1172/JCI63287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller SN, Mackay LK. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 16:79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- 21.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, Fife BT, Masopust D. 2018. Intravital mucosal imaging of CD8(+) resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol 19:173–182. doi: 10.1038/s41590-017-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, Carbone FR, Tscharke DC, Heath WR, Mueller SN, Mackay LK. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 19:183–191. doi: 10.1038/s41590-017-0027-5. [DOI] [PubMed] [Google Scholar]
- 23.Pizzolla A, Nguyen TH, Sant S, Jaffar J, Loudovaris T, Mannering SI, Thomas PG, Westall GP, Kedzierska K, Wakim LM. 2018. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J Clin Invest 128:721–733. doi: 10.1172/JCI96957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gargett T, Grubor-Bauk B, Garrod TJ, Yu W, Miller D, Major L, Wesselingh S, Suhrbier A, Gowans EJ. 2014. Induction of antigen-positive cell death by the expression of perforin, but not DTa, from a DNA vaccine enhances the immune response. Immunol Cell Biol 92:359–367. doi: 10.1038/icb.2013.93. [DOI] [PubMed] [Google Scholar]
- 25.Gargett T, Grubor-Bauk B, Miller D, Garrod T, Yu S, Wesselingh S, Suhrbier A, Gowans EJ. 2014. Increase in DNA vaccine efficacy by virosome delivery and co-expression of a cytolytic protein. Clin Transl Immunol 3:e18. doi: 10.1038/cti.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gummow J, Li Y, Yu W, Garrod T, Wijesundara D, Brennan AJ, Mullick R, Voskoboinik I, Grubor-Bauk B, Gowans EJ. 2015. A multiantigenic DNA vaccine that induces broad hepatitis C virus-specific T-cell responses in mice. J Virol 89:7991–8002. doi: 10.1128/JVI.00803-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubor-Bauk B, Yu W, Wijesundara D, Gummow J, Garrod T, Brennan AJ, Voskoboinik I, Gowans EJ. 2016. Intradermal delivery of DNA encoding HCV NS3 and perforin elicits robust cell-mediated immunity in mice and pigs. Gene Ther 23:26–37. doi: 10.1038/gt.2015.86. [DOI] [PubMed] [Google Scholar]
- 28.Wijesundara DK, Yu W, Quah BJC, Eldi P, Hayball JD, Diener KR, Voskoboinik I, Gowans EJ, Grubor-Bauk B. 2017. Cytolytic DNA vaccine encoding lytic perforin augments the maturation of—and antigen presentation by—dendritic cells in a time-dependent manner. Sci Rep 7:8530. doi: 10.1038/s41598-017-08063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wijesundara DK, Gummow J, Li Y, Yu W, Quah BJ, Ranasinghe C, Torresi J, Gowans EJ, Grubor-Bauk B. 2018. Induction of genotype cross-reactive, hepatitis C virus-specific, cell-mediated immunity in DNA-vaccinated mice. J Virol 92:e02133-17. doi: 10.1128/JVI.02133-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trimble CL, Morrow MP, Kraynyak KA, Shen X, Dallas M, Yan J, Edwards L, Parker RL, Denny L, Giffear M, Brown AS, Marcozzi-Pierce K, Shah D, Slager AM, Sylvester AJ, Khan A, Broderick KE, Juba RJ, Herring TA, Boyer J, Lee J, Sardesai NY, Weiner DB, Bagarazzi ML. 2015. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet 386:2078–2088. doi: 10.1016/S0140-6736(15)00239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naso MF, Tomkowicz B, Perry WL III, Strohl WR. 2017. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quah BJ, Wijesundara DK, Ranasinghe C, Parish CR. 2012. Fluorescent target array killing assay: a multiplex cytotoxic T-cell assay to measure detailed T-cell antigen specificity and avidity in vivo. Cytometry A 81:679–690. doi: 10.1002/cyto.a.22084. [DOI] [PubMed] [Google Scholar]
- 33.Quah BJ, Wijesundara DK, Ranasinghe C, Parish CR. 2013. Fluorescent target array T helper assay: a multiplex flow cytometry assay to measure antigen-specific CD4+ T cell-mediated B cell help in vivo. J Immunol Methods 387:181–190. doi: 10.1016/j.jim.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Wijesundara DK, Ranasinghe C, Jackson RJ, Lidbury BA, Parish CR, Quah BJ. 2014. Use of an in vivo FTA assay to assess the magnitude, functional avidity and epitope variant cross-reactivity of T cell responses following HIV-1 recombinant poxvirus vaccination. PLoS One 9:e105366. doi: 10.1371/journal.pone.0105366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hovav AH, Panas MW, Rahman S, Sircar P, Gillard G, Cayabyab MJ, Letvin NL. 2007. Duration of antigen expression in vivo following DNA immunization modifies the magnitude, contraction, and secondary responses of CD8+ T lymphocytes. J Immunol 179:6725–6733. doi: 10.4049/jimmunol.179.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elnekave M, Furmanov K, Nudel I, Arizon M, Clausen BE, Hovav AH. 2010. Directly transfected langerin+ dermal dendritic cells potentiate CD8+ T cell responses following intradermal plasmid DNA immunization. J Immunol 185:3463–3471. doi: 10.4049/jimmunol.1001825. [DOI] [PubMed] [Google Scholar]
- 37.Tay SS, Wong YC, McDonald DM, Wood NA, Roediger B, Sierro F, McGuffog C, Alexander IE, Bishop GA, Gamble JR, Weninger W, McCaughan GW, Bertolino P, Bowen DG. 2014. Antigen expression level threshold tunes the fate of CD8 T cells during primary hepatic immune responses. Proc Natl Acad Sci U S A 111:E2540–E2549. doi: 10.1073/pnas.1406674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNamara HA, Cai Y, Wagle MV, Sontani Y, Roots CM, Miosge LA, O'Connor JH, Sutton HJ, Ganusov VV, Heath WR, Bertolino P, Goodnow CG, Parish IA, Enders A, Cockburn IA. 2017. Up-regulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci Immunol 2:eaaj1996. doi: 10.1126/sciimmunol.aaj1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn JA, Wylie D, Dill J, Sanchez MS, Lloyd-Smith JO, Page-Shafer K, Getz WM. 2009. Potential impact of vaccination on the hepatitis C virus epidemic in injection drug users. Epidemics 1:47–57. doi: 10.1016/j.epidem.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin EC, Park SH, Demino M, Nascimbeni M, Mihalik K, Major M, Veerapu NS, Heller T, Feinstone SM, Rice CM, Rehermann B. 2011. Delayed induction, not impaired recruitment, of specific CD8(+) T cells causes the late onset of acute hepatitis C. Gastroenterology 141:686–695.e1. doi: 10.1053/j.gastro.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, Gibbs J, Bennink JR, Yewdell JW. 2004. CD8+ T cell cross-priming via transfer of proteasome substrates. Science 304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 42.Billerbeck E, Wolfisberg R, Fahnoe U, Xiao JW, Quirk C, Luna JM, Cullen JM, Hartlage AS, Chiriboga L, Ghoshal K, Lipkin WI, Bukh J, Scheel TKH, Kapoor A, Rice CM. 2017. Mouse models of acute and chronic hepacivirus infection. Science 357:204–208. doi: 10.1126/science.aal1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loder F, Mutschler B, Ray RJ, Paige CJ, Sideras P, Torres R, Lamers MC, Carsetti R. 1999. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp Med 190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nolte MA, Arens R, Kraus M, van Oers MH, Kraal G, van Lier RA, Mebius RE. 2004. B cells are crucial for both development and maintenance of the splenic marginal zone. J Immunol 172:3620–3627. doi: 10.4049/jimmunol.172.6.3620. [DOI] [PubMed] [Google Scholar]
- 45.Kubes P, Jenne C. 2018. Immune responses in the liver. Annu Rev Immunol 36:247–277. doi: 10.1146/annurev-immunol-051116-052415. [DOI] [PubMed] [Google Scholar]
- 46.Groom JR, Luster AD. 2011. CXCR3 in T cell function. Exp Cell Res 317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. 2007. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med 204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohlmeier JE, Miller SC, Smith J, Lu B, Gerard C, Cookenham T, Roberts AD, Woodland DL. 2008. The chemokine receptor CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity 29:101–113. doi: 10.1016/j.immuni.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirozyan MR, Nguyen N, Cameron B, Luciani F, Bull RA, Zekry A, Lloyd AR. 2019. Chemokine-regulated recruitment of antigen-specific T cell sub-populations to the liver in acute and chronic hepatitis C infection. J Infect Dis 219:1430–1438. doi: 10.1093/infdis/jiy679. [DOI] [PubMed] [Google Scholar]
- 50.Guarda G, Hons M, Soriano SF, Huang AY, Polley R, Martin-Fontecha A, Stein JV, Germain RN, Lanzavecchia A, Sallusto F. 2007. l-Selectin-negative CCR7− effector and memory CD8+ T cells enter reactive lymph nodes and kill dendritic cells. Nat Immunol 8:743–752. doi: 10.1038/ni1469. [DOI] [PubMed] [Google Scholar]
- 51.Hu JK, Kagari T, Clingan JM, Matloubian M. 2011. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc Natl Acad Sci U S A 108:E118–E127. doi: 10.1073/pnas.1101881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuburu N, Wang K, Goodman KN, Pang YY, Thompson CD, Lowy DR, Cohen JI, Schiller JT. 2015. Topical herpes simplex virus 2 (HSV-2) vaccination with human papillomavirus vectors expressing gB/gD ectodomains induces genital-tissue-resident memory CD8+ T cells and reduces genital disease and viral shedding after HSV-2 challenge. J Virol 89:83–96. doi: 10.1128/JVI.02380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies B, Prier JE, Jones CM, Gebhardt T, Carbone FR, Mackay LK. 2017. Cutting edge. Tissue-resident memory T cells generated by multiple immunizations or localized deposition provide enhanced immunity. J Immunol 198:2233–2237. doi: 10.4049/jimmunol.1601367. [DOI] [PubMed] [Google Scholar]
- 54.Holz LE, Prier JE, Freestone D, Steiner TM, English K, Johnson DN, Mollard V, Cozijnsen A, Davey GM, Godfrey DI, Yui K, Mackay LK, Lahoud MH, Caminschi I, McFadden GI, Bertolino P, Fernandez-Ruiz D, Heath WR. 2018. CD8(+) T cell activation leads to constitutive formation of liver tissue-resident memory T cells that seed a large and flexible niche in the liver. Cell Rep 25:68–79.e4. doi: 10.1016/j.celrep.2018.08.094. [DOI] [PubMed] [Google Scholar]
- 55.Gola A, Silman D, Walters AA, Sridhar S, Uderhardt S, Salman AM, Halbroth BR, Bellamy D, Bowyer G, Powlson J, Baker M, Venkatraman N, Poulton I, Berrie E, Roberts R, Lawrie AM, Angus B, Khan SM, Janse CJ, Ewer KJ, Germain RN, Spencer AJ, Hill A. 2018. Prime and target immunization protects against liver-stage malaria in mice. Sci Transl Med 10:eaap9128. doi: 10.1126/scitranslmed.aap9128. [DOI] [PubMed] [Google Scholar]
- 56.Nganou-Makamdop K, Ploemen I, Behet M, Van Gemert GJ, Hermsen C, Roestenberg M, Sauerwein RW. 2012. Reduced Plasmodium berghei sporozoite liver load associates with low protective efficacy after intradermal immunization. Parasite Immunol 34:562–569. doi: 10.1111/pim.12000.x. [DOI] [PubMed] [Google Scholar]
- 57.Epstein JE, Tewari K, Lyke KE, Sim BK, Billingsley PF, Laurens MB, Gunasekera A, Chakravarty S, James ER, Sedegah M, Richman A, Velmurugan S, Reyes S, Li M, Tucker K, Ahumada A, Ruben AJ, Li T, Stafford R, Eappen AG, Tamminga C, Bennett JW, Ockenhouse CF, Murphy JR, Komisar J, Thomas N, Loyevsky M, Birkett A, Plowe CV, Loucq C, Edelman R, Richie TL, Seder RA, Hoffman SL. 2011. Live attenuated malaria vaccine designed to protect through hepatic CD8(+) T cell immunity. Science 334:475–480. doi: 10.1126/science.1211548. [DOI] [PubMed] [Google Scholar]
- 58.Tse SW, Radtke AJ, Espinosa DA, Cockburn IA, Zavala F. 2014. The chemokine receptor CXCR6 is required for the maintenance of liver memory CD8(+) T cells specific for infectious pathogens. J Infect Dis 210:1508–1516. doi: 10.1093/infdis/jiu281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cunningham SC, Dane AP, Spinoulas A, Alexander IE. 2008. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol Ther 16:1081–1088. doi: 10.1038/mt.2008.72. [DOI] [PubMed] [Google Scholar]
- 60.Xiao X, Li J, Samulski RJ. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 72:2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo P, El-Gohary Y, Prasadan K, Shiota C, Xiao X, Wiersch J, Paredes J, Tulachan S, Gittes GK. 2012. Rapid and simplified purification of recombinant adeno-associated virus. J Virol Methods 183:139–146. doi: 10.1016/j.jviromet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holmes K, Lantz LM, Fowlkes BJ, Schmid I, Giorgi JV. 2001. Preparation of cells and reagents for flow cytometry. Curr Protoc Immunol Chapter 5:Unit 5.3. doi: 10.1002/0471142735.im0503s44. [DOI] [PubMed] [Google Scholar]