Due to the near monomorphic nature of MHC-E in the human population and inability of many pathogens to inhibit MHC-E-mediated peptide presentation, MHC-E-restricted T cells have become an attractive vaccine target. However, little is known concerning how these cells are induced. Understanding the underlying mechanisms that induce these T cells would provide a powerful new vaccine strategy to an array of neoplasms and viral and bacterial pathogens. Recent studies have indicated a link between TAP inhibition and induction of MHC-E-restricted T cells. The significance of our research is in demonstrating that TAP inhibition alone does not prime MHC-E-restricted T cell generation and suggests that other, currently unknown mechanisms regulate their induction.
KEYWORDS: MHC-E, RhCMV, T cells, transporter associated with antigen processing, major histocompatibility complex
ABSTRACT
Major histocompatibility complex E (MHC-E) is a highly conserved nonclassical MHC-Ib molecule that tightly binds peptides derived from leader sequences of classical MHC-Ia molecules for presentation to natural killer cells. However, MHC-E also binds diverse foreign and neoplastic self-peptide antigens for presentation to CD8+ T cells. Although the determinants of MHC-E-restricted T cell priming remain unknown, these cells are induced in humans infected with pathogens containing genes that inhibit the transporter associated with antigen processing (TAP). Indeed, mice vaccinated with TAP-inhibited autologous dendritic cells develop T cells restricted by the murine MHC-E homologue, Qa-1b. Here, we tested whether rhesus macaques (RM) vaccinated with viral constructs expressing a TAP inhibitor would develop insert-specific MHC-E-restricted CD8+ T cells. We generated viral constructs coexpressing SIVmac239 Gag in addition to one of three TAP inhibitors: herpes simplex virus 2 ICP47, bovine herpes virus 1 UL49.5, or rhesus cytomegalovirus Rh185. Each TAP inhibitor reduced surface expression of MHC-Ia molecules but did not reduce surface MHC-E expression. In agreement with modulation of surface MHC-Ia levels, TAP inhibition diminished presentation of MHC-Ia-restricted CD8+ T cell epitopes without impacting presentation of peptide antigen bound by MHC-E. Vaccination of macaques with vectors dually expressing SIVmac239 Gag with ICP47, UL49.5, or Rh185 generated Gag-specific CD8+ T cells classically restricted by MHC-Ia but not MHC-E. These data demonstrate that, in contrast to results in mice, TAP inhibition alone is insufficient for priming of MHC-E-restricted T cell responses in primates and suggest that additional unknown mechanisms govern the induction of CD8+ T cells recognizing MHC-E-bound antigen.
IMPORTANCE Due to the near monomorphic nature of MHC-E in the human population and inability of many pathogens to inhibit MHC-E-mediated peptide presentation, MHC-E-restricted T cells have become an attractive vaccine target. However, little is known concerning how these cells are induced. Understanding the underlying mechanisms that induce these T cells would provide a powerful new vaccine strategy to an array of neoplasms and viral and bacterial pathogens. Recent studies have indicated a link between TAP inhibition and induction of MHC-E-restricted T cells. The significance of our research is in demonstrating that TAP inhibition alone does not prime MHC-E-restricted T cell generation and suggests that other, currently unknown mechanisms regulate their induction.
INTRODUCTION
The major histocompatibility complex (MHC) gene locus is one of the most diverse regions in the vertebrate genome, with over 13,000 individual MHC-Ia alleles identified in the human population to date (1). In contrast to the profound diversity of MHC-Ia molecules, MHC-E is among the most conserved of all primate MHC genes (2), with only two functional HLA-E molecules expressed in the majority of the human population (3). The constrained diversity of MHC-E is likely a consequence of its primary role as a ligand for the NK cell CD94/NKG2 receptor. Through a mechanism dependent on the transporter associated with antigen presentation (TAP), MHC-E presents the VL9 leader peptides from HLA-A, -B, -C, and -G, thus preventing lysis by NK cells (4–6). However, in a TAP-deficient environment HLA-E is capable of presenting a broad array of peptides (7, 8). Mass spectrometry of peptides eluted from surface-expressed HLA-E on TAP-deficient cells identified over 500 unique self-peptides (8).
Certain viral and bacterial infections, which subvert surveillance via MHC-Ia-restricted CD8+ T cells by modulating MHC-Ia translation, trafficking, or peptide binding, also induce MHC-E to bind virus-derived peptides and subsequently induce MHC-E-restricted T cells. Mycobacterium tuberculosis, Salmonella enterica serovar Typhi, Epstein-Barr virus (EBV), hepatitis C virus, and human cytomegalovirus (HCMV) (9–15) are able to prime HLA-E-restricted T cells in vivo, with the phenotype of HLA-E-restricted CD8+ T cells depending on the specific pathogen. HLA-E-restricted CD8+ T cells induced by M. tuberculosis possess a Th2 phenotype, producing cytolytic enzymes such as granzyme and perforin, but also interleukin-10 (IL-10) and IL-4, thereby facilitating control of M. tuberculosis replication within monocytes (14). HLA-E-restricted CD8+ T cells induced by S. enterica serovar Typhi vaccination are cytolytic and lyse bacterially infected target cells (12). HCMV induces cytolytic, effector memory HLA-E-restricted CD8+ T cells targeting the HCMV UL40-encoded VL9 peptide when this viral mimic peptide is mismatched against the VL9 sequences present in the host HLA-Ia leader sequences (9, 16). Vaccination of rhesus macaques (RM) with rhesus CMV (RhCMV) strain 68-1 expressing simian immunodeficiency virus (SIV) antigens (RhCMV/SIV) induces MHC-E-restricted, SIV-specific CD8+ T cells that exhibit a uniform effector memory phenotype with polyfunctionality for tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), and macrophage inflammatory protein 1β (MIP-1β) (17, 18). These strain 68-1 RhCMV/SIV-induced cellular immune responses stringently control and ultimately clear pathogenic SIV replication in ∼50% of vaccinated RM (18, 19). Thus, there is intense interest in understanding the molecular determinants which give rise to MHC-E-restricted CD8+ T cells in vivo.
Although it remains unclear how pathogens prime MHC-E-restricted CD8+ T cells, recent reports indicate that modulators of TAP play a critical role in their induction. In the mouse model, vaccination with monocyte-derived dendritic cells (moDCs) transduced with the TAP inhibitor UL49.5 from bovine herpesvirus 1 (BHV-1) induces CD8+ T cells restricted by the HLA-E ortholog Qa-1b (20–22). In rhesus macaques, the RhCMV strain 68-1 vector, which induces high-frequency MHC-E-restricted CD8+ T cells, encodes a TAP-inhibiting homologue of the TAP-inhibitory US6 protein of HCMV (23). Finally, both of the human pathogens EBV and HCMV prime HLA-E-restricted CD8+ T cells (9, 11, 16), and both encode potent TAP inhibitors (23–28). These data cumulatively suggest a central role of TAP inhibition in the priming of MHC-E-restricted CD8+ T cells. Thus, we hypothesized that vaccination with a viral construct concomitantly expressing a TAP inhibitor and SIVmac239 Gag would induce Gag-specific, MHC-E-restricted CD8+ T cells. To pursue this hypothesis, we vaccinated rhesus macaques with adenovirus type 5 (Ad5) and adeno-associated virus (AAV) vaccine constructs simultaneously expressing SIVmac239 Gag and a viral TAP inhibitor. Despite the ability of these constructs to inhibit presentation of canonical MHC-Ia-restricted CD8+ T cells in vitro, we observed no induction of MHC-E-restricted, Gag-specific CD8+ T cells in vivo. These results suggest that inhibition of TAP alone is insufficient to elicit nonclassically MHC-E-restricted CD8+ T cells in nonhuman primates.
RESULTS
Vaccine insert construction and functionality testing.
In mice, vaccination with UL49.5-transduced moDCs induced self-reactive CD8+ T cell responses restricted by the murine MHC-E ortholog Qa-1b (22, 29). To test whether delivery of antigen in the setting of deficient TAP function could induce MHC-E-restricted T cells in nonhuman primates, we designed vaccine vectors that coexpress SIVmac239 Gag and one of three herpesvirus-derived TAP inhibitors: ICP47, UL49.5, or Rh185. Herpes simplex virus (HSV)-derived ICP47 blocks the peptide binding site of TAP with high affinity, thus preventing translocation of cytosolic peptides into the endoplasmic reticulum (ER) (30–32). BHV-1-encoded UL49.5 inhibits TAP in two distinct ways: by preventing translocation of TAP-bound peptide into the ER and by targeting the TAP subunits, TAP-1 and TAP-2, for proteasomal degradation (33). Finally, HCMV US6 interferes with TAP function by altering TAP’s conformation, thereby preventing ATP binding and subsequent peptide translocation to the ER (26, 28, 34); the RhCMV US6 ortholog Rh185 also demonstrates potent TAP inhibition although the mechanism has not been elucidated (23). To maximize coexpression of both SIVmac239 Gag and the selected TAP inhibitor, the two genes were separated by a porcine teschovirus 2A site (Fig. 1A). Self-cleaving viral 2A sites induce ribosomal skipping during protein translation, effectively cleaving a nascent polypeptide and allowing for efficient coexpression by a single promoter (35). Of the various newly discovered 2A sites, we selected P2A as it has been frequently found to be the most efficient (36, 37). We placed Gag upstream of our TAP genes as 2A cleavage results in multiple additional amino acids added to the C terminus of the upstream protein, but only one proline is added to the N terminus of the downstream protein, mitigating effects to secondary and tertiary protein structures which could inhibit protein function (Fig. 1A). For tracking purposes, a Flag tag was added to the C terminus of each TAP inhibitor. Previous work showed that C-terminal tagging does not affect TAP inhibition (34, 38).
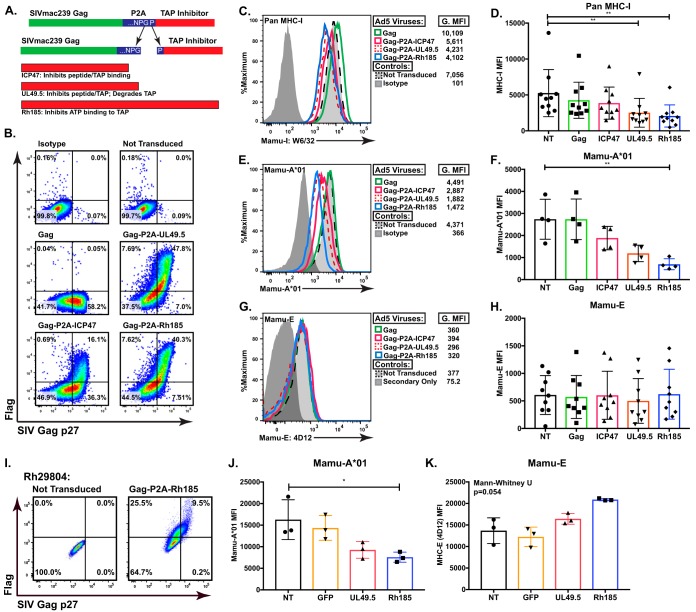
FIG 1.
ICP47, UL49.5, and Rh185 inhibit TAP function in rhesus macaque Cells. (A) Schematic of three vector inserts inclusive of SIVmac239 Gag and the Flag-tagged TAP inhibitors ICP47 (HSV-2), UL49.5 (BHV-1), or Rh185 (RhCMV), separated by 2A peptide derived by porcine teschovirus-1. The resulting cleavage during peptide synthesis yields an additional 18 amino acids attached at the C terminus of Gag but only one proline added to the N terminus of the selected TAP inhibitor. (B) Representative flow cytometry data of SIVmac239 Gag p27 and Flag-tagged TAP inhibitor expression in BLCL transduced at a multiplicity of infection of 2,000 with Ad5/Gag, Gag-P2A-ICP47, Gag-P2A-UL49.5, or Gag-P2A-Rh185. (C and D) Staining of BLCL with pan-MHC-I antibody (W6/32) showed that MHC-I expression was significantly suppressed as a consequence of TAP inhibition. G.MFI, geometric mean fluorescence intensity. Panel C shows representative flow cytometry data from one animal, while panel D shows data compiled from multiple RM. (**, P < 0.001). (E and F) Subset analysis on A*01 surface expression using RM that were positive for the MHC-Ia allele Mamu-A*01. After transduction with Ad5 viral vectors, this subset was stained with an antibody specific to Mamu-A*01. With the smaller subset, only Rh185 transduced BLCL reach statistical significance (**, P < 0.001). Panel E shows representative flow cytometry data from one animal, while panel F shows data compiled from multiple RM. (**, P < 0.001). (G and H) The BLCL from the experiments described in panels C and D were stained with the MHC-E-specific 4D12 antibody, and surface MHC-E was assessed. Panel G shows representative flow cytometry data from one animal, while panel H shows compiled data from multiple RM. (I) Representative flow cytometry data of SIVmac239 Gag p27 and Flag-tagged TAP inhibitor expression in monocyte-derived macrophages (MDM) transduced at a multiplicity of infection of 2,000 with Ad5/Gag-P2A-Rh185, Ad5/Gag-P2A-UL49.5, or Ad5 expressing green fluorescent protein (GFP). Posttransduction MDM from Mamu-A*01-positive RM were stained for surface Mamu-A*01 expression (J) and surface Mamu-E expression (K). Values shown in panels D, F, H, J, and K are background subtracted. NT, not transduced.
After packaging the vaccine inserts into a replication-incompetent Ad5 viral vector, we first tested the cleavage efficiency of our constructs. Transduction of rhesus lymphoblastoid B cell lines (BLCL) yielded high expression of each protein (Fig. 1B). Both the UL49.5 and Rh185 vectors had highly efficient cleavage, with >76% and >72% of all transduced BLCL costaining for the Flag-tagged TAP inhibitor and Gag p27, respectively. In contrast, the ICP47 vector consistently exhibited both lower transduction efficiency and lower dual staining for Gag p27 and Flag (30%) than for the UL49.5 and Rh185 vectors. Despite these variations in expression efficiencies, sufficient coexpression was achieved with all constructs to determine their effect on MHC-mediated antigen presentation to CD8+ T cells.
Next, we examined the impact of TAP inhibition on the expression of classical MHC-Ia molecules on the surface of transduced, Gag p27+ cells using the pan-MHC-I reactive antibody (Ab) W6/32. As expected, we found that expression of SIVmac239 Gag alone had no impact on MHC expression (Fig. 1C and D). In contrast, all three TAP inhibitors reduced surface MHC-I to various degrees. ICP47 exhibited the weakest effect, reducing MHC-I expression by approximately 20% (Fig. 1C). Although there was a consistent trend toward lower MHC-I levels, we found no statistically significant difference in MHC-I regulation between ICP47-expressing and nontransduced BLCL (Fig. 1D). In contrast, expression of UL49.5 or Rh185 efficiently reduced MHC-I surface levels by 40% and 42%, respectively, in a statistically significant manner (Fig. 1C and D).
As W6/32 is a pan-MHC-I antibody staining both classical and nonclassical MHC-I molecules, including HLA-E (39, 40), we examined BLCL from macaques expressing Mamu-A1*001:01 (Mamu-A*01) in order to specifically compare expression of the classical MHC-Ia allele Mamu-A*01 to that of the nonclassical Mamu-E molecule. When analysis was restricted specifically to Mamu-A*01 via use of a Mamu-A*01-specific antibody, reduced surface MHC-Ia expression was more striking. ICP47 remained consistently weak, only lowering Mamu-A*01 levels by 33% (Fig. 1E and F). However, UL49.5 and Rh185 lowered Mamu-A*01 expression by 57% and 66%, respectively (Fig. 1D). To examine Mamu-E surface levels in the setting of TAP inhibition, we utilized the MHC-E binding antibody 4D12, which we previously demonstrated does not bind classical alleles like Mamu-A*01 (17, 41). In contrast to the impact of TAP inhibition on classical MHC-Ia molecules, we observed no significant impact on MHC-E levels due to abrogated TAP function (Fig. 1G and H). These findings confirm that the TAP inhibitors UL49.5 and Rh185 are active in rhesus macaque cells and potently inhibit TAP, which subsequently reduces surface Mamu-Ia expression. In contrast, TAP inhibition did not lower surface levels of the nonclassical Mamu-E molecule in this in vitro system. To confirm these results in a primary cell type, we generated monocyte-derived macrophages (MDM) from Mamu-A*01+ RM and examined the impact of transduction with the Ad5 vectors simultaneously expressing Gag and Rh185 or UL49.5 on surface Mamu-A*01 and Mamu-E expression in these cells (Fig. 1I). Similar to our results in immortalized BLCL, MDM transduced to express TAP inhibitors exhibited lower levels of Mamu-A*01 on the cell surface (Fig. 1J) while no statistically significant change in Mamu-E expression was observed (Fig. 1K); moreover, these results confirmed that Rh185 had a stronger effect in RM cells than UL49.5.
TAP inhibition lowers antigen recognition by MHC-Ia-restricted CD8+ T cells but does not impact MHC-E-restricted CD8+ T cell recognition.
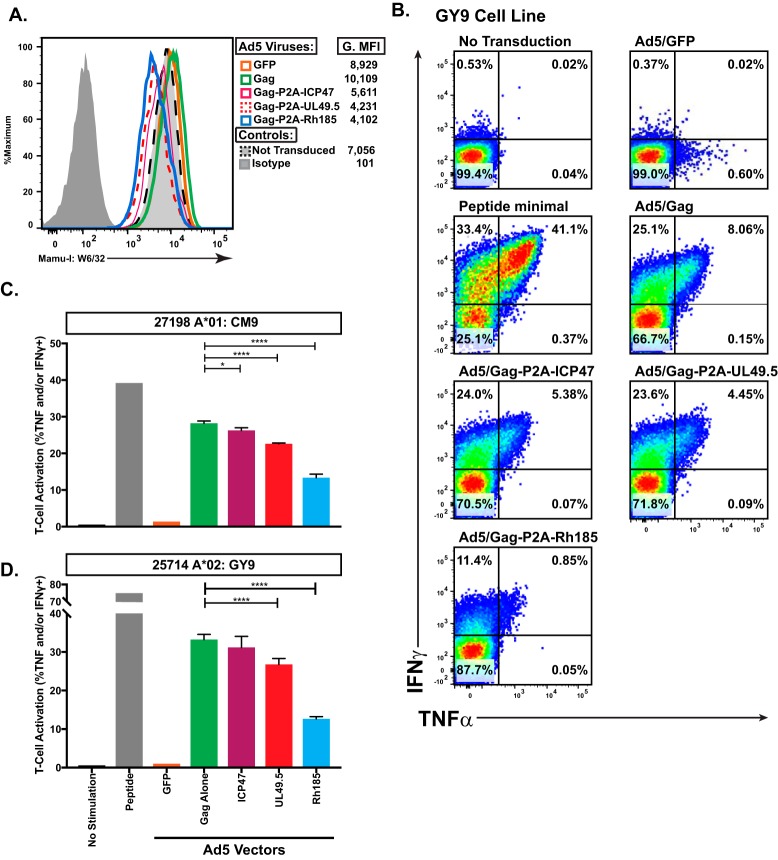
TAP inhibition and subsequent surface MHC-Ia downregulation from the cell surface diminishes CD8+ T cell recognition of MHC-I-bound antigen in both murine and human cells (22, 25, 42). Thus, we next sought to determine if a similar phenomenon would occur in TAP-deficient rhesus macaque cells. To measure the impact of TAP inhibition on CD8+ T cell recognition of MHC-Ia-bound antigen, we coincubated CD8+ T cell lines recognizing either Mamu-A*01-restricted Gag181–189 CM9 (CM9) or Mamu-A1*002:01 (Mamu-A*02)-restricted Gag71–79 GY9 (GY9) epitope with a BLCL line positive for both Mamu-A*01 and Mamu-A*02, which had been transduced with the Ad5 vectors described above. Levels of MHC-Ia downregulation in transduced BLCL were similar to those described above, with Rh185 again showing the most potent effect (Fig. 2A). The GY9 and CM9 lines were most responsive to BLCL pulsed with saturating levels of exogenous peptide, with nearly 75% of all CD8+ T cells within the GY9 line responding to peptide (Fig. 2B and D). Responses to Gag-transduced BLCL were notably lower than those with exogenous peptide but significantly higher than those to BLCL transduced with vectors dually expressing Gag and a TAP inhibitor. Suppression of CD8+ T cell recognition of cognate Gag antigen presented in the context of both Mamu-A*01 and Mamu-A*02 correlated with the magnitude of reduced surface MHC-Ia expression (Fig. 1D and F). ICP47 had the smallest effect on CD8+ T cell recognition, which was statistically significant only in experiments with the CM9 cell line (Fig. 2C). UL49.5 was more effective at blocking CD8+ T cell responsiveness, reducing cytokine secretion by 15% and 20% in CM9 and GY9 experiments, respectively (Fig. 2C). Rh185, which had the greatest impact on surface MHC-Ia expression, was also the most effective at suppressing CD8+ T cell recognition, with responses 50% and 62% lower in CM9 and GY9 lines than the response to BLCL transduced with only Gag (Fig. 2C and D).
FIG 2.
MHC-Ia-restricted T cells have reduced recognition of TAP-inhibited targets. (A) Surface MHC-I levels on the Ad5 transduced heterologous BLCL (Mamu-A*01/A*02+) used as effectors to test the impact of TAP inhibition on MHC-Ia-restricted CD8+ T cell recognition. G.MFI, geometric mean fluorescence intensity. (B) Representative ICS data of Gag GY9-specific CD8+ T cells in response to coincubation with the antigen-presenting cells indicated above each plot. (C) Activation of Gag CM9-specific CD8+ T cells as measured by secretion of TNF-α and/or IFN-γ in response to coincubation with exogenous peptide or BLCL antigen-presenting cells infected with the indicated Ad5 vectors (*, P < 0.01; ****, P < 0.0001). (D) Activation of Gag GY9-specific CD8+ T cells as measured by secretion of TNF-α and/or IFN-γ, in response to coincubation with exogenous peptide or BLCL antigen-presenting cells infected with the indicated Ad5 vectors (*, P < 0.01; ****, P < 0.0001).
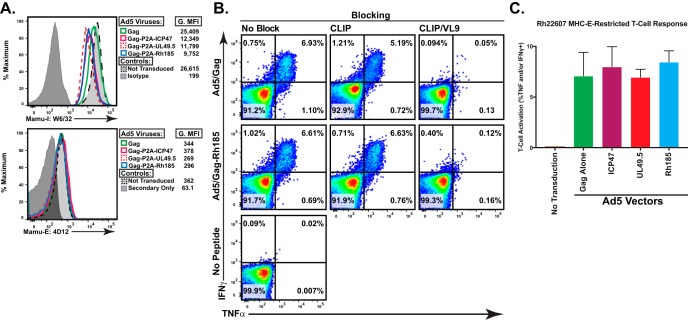
Loading of MHC-E with MHC-Ia-derived leader sequences occurs via a TAP-dependent method (5, 43). However, TAP-independent loading of MHC-E with viral antigen has been reported previously (44, 45). To examine the effect of TAP inhibition on CD8+ T cell recognition of MHC-E-bound antigen, we utilized peripheral blood mononuclear cells (PBMC) from rhesus macaque Rh22607 as effector cells in an intracellular cytokine staining (ICS) assay. We previously demonstrated that this strain 68-1 RhCMV/Gag-vaccinated macaque mounts strong Gag-specific CD8+ T cell responses restricted by either MHC-II or MHC-E (17). Similar to the experiments with classically MHC-Ia-restricted CD8+ T cells described above, we again used Ad5 transduced BLCL as antigen-presenting cells (APCs). As before, TAP inhibitors significantly reduced MHC-Ia expression (Fig. 3A, top panel) with no impact on MHC-E expression (Fig. 3A, bottom panel). In contrast to the diminished recognition of TAP-inhibited cells by MHC-Ia-restricted CD8+ T cells, unconventionally MHC-E-restricted Gag-specific CD8+ T cells responded comparably to BLCL expressing Gag alone or in combination with a TAP inhibitor (Fig. 3C). As immunization with strain 68-1 RhCMV/Gag elicits both MHC-II- and MHC-E-restricted CD8+ T cells, we blocked MHC-II-restricted responses with the class II-associated invariant chain (CLIP) peptide that inhibits MHC-II-bound antigen presentation (17, 46). In the setting of MHC-II blockade, TAP inhibition did not impact MHC-E-restricted CD8+ T cell recognition of Gag antigen (Fig. 3C). Finally, we confirmed that CD8+ T cell recognition in the setting of CLIP blockade was indeed MHC-E restricted by adding saturating exogenous levels of the MHC-Ia-derived leader peptide VL9, which inhibits MHC-E-bound antigen presentation (17). Therefore, in contrast to MHC-Ia-presented peptide antigen, TAP inhibition does not significantly alter presentation of MHC-E-bound peptide to CD8+ T cells.
FIG 3.
TAP inhibition does not reduce antigen recognition by MHC-E-restricted T cells. (A) Surface MHC-I and MHC-E levels on the autologous Ad5 transduced BLCL used as effectors to test the impact of TAP inhibition on MHC-E-restricted CD8+ T cell recognition. G.MFI, geometric mean fluorescence intensity. (B) Representative ICS data of CD8+ T cells from strain 68-1 RhCMV/Gag-vaccinated animal Rh22607 in response to coincubation with autologous BLCL transduced with the Ad5 vector as indicated on the y axis. To confirm that responses were indeed MHC-E restricted and not MHC-II restricted, the ICS was performed in the presence of saturating exogenous CLIP or VL9 peptide as indicated. (C) Compiled data of three separate ICS experiments utilizing animal Rh22607 done with saturating exogenous CLIP peptide to specifically examine MHC-E-restricted responses (P = 0.48).
Immunization of nonhuman primates with viral constructs that inhibit TAP does not induce MHC-E-restricted CD8+ T cells.
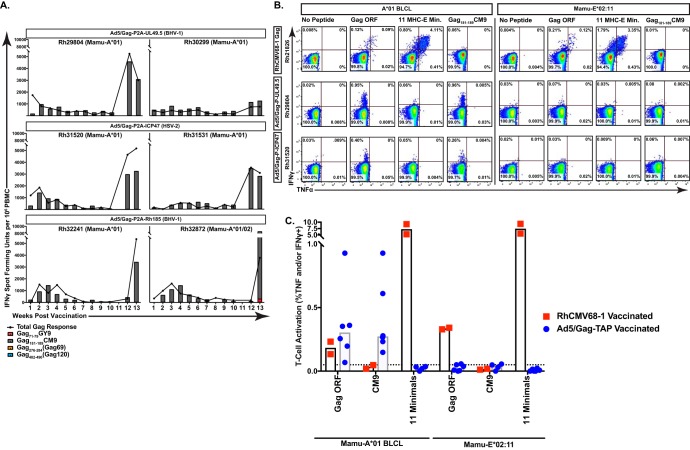
After confirming that our vaccine vectors dually expressing Gag and a TAP inhibitor diminished presentation of MHC-Ia-bound, but not MHC-E-bound, Gag peptide antigen, we next tested whether immunization of nonhuman primates with these vectors would induce Gag-specific, MHC-E-restricted CD8+ T cells. We inoculated six Mamu-A*01+ rhesus macaques, two per group, with viral vectors expressing Gag-ICP47, Gag-UL49.5, or Gag-Rh185 (Fig. 4A). Animals were primed with 1012 viral particles (vp) of each Ad5 vector, followed by a boost of 1013 vp of AAV1 expressing the same construct at 12 weeks postpriming. All macaques developed Gag-specific CD8+ T cells, but the response was almost entirely directed at the Mamu-A*01-restricted, immunodominant Gag CM9 CD8+ T cell epitope (Fig. 4A). One macaque in the study coexpressed Mamu-A*02 and received the Gag-Rh185 vectors. In this animal, we did not detect the immunodominant Mamu-A*02-restricted Gag GY9-specific CD8+ T cell response until after AAV1 boost. However, in contrast to the emergence and longitudinal presence of Gag-specific, MHC-Ia-restricted CD8+ T cell responses in all six vaccinated animals, no MHC-E-restricted CD8+ T cell response was observed in any animal at any time point against the previously described Gag276–284 RL9 (RL9) or Gag482–490 EK9 (EK9) supertopes, epitopes universally targeted by MHC-E-restricted CD8+ T cell responses in strain 68-1 RhCMV/Gag-vaccinated macaques (17). These results suggested that immunization with TAP inhibitor-expressing viral vectors is insufficient to prime MHC-E-restricted CD8+ T cell responses in nonhuman primates.
FIG 4.
Vaccination of rhesus macaques with viral vectors inclusive of a TAP inhibitor does not induce MHC-E-restricted T cells. (A) Macaques were vaccinated with an Ad5 prime followed by AAV1 boost at 12-weeks postpriming using one of the three inserts indicated: Gag-P2A-ICP47, Gag-P2A-UL49.5, or Gag-P2A-Rh185. An IFN-γ ELISpot assay was done weekly using peptide pools spanning the entire Gag protein. These responses were summed to give the total SIV Gag response. Additionally, minimal optimal peptides for SIV-specific responses restricted by Mamu-A*01 (CM9), Mamu-A*02 (GY9), and Mamu-E (RL9 and EK9) were included separately at each time point. (B) Representative ICS results of CD8+ T cells from the strain 68-1 RhCMV/Gag-vaccinated Mamu-A*01+ macaque Rh21826, the Ad5/Gag-P2A-UL49.5-vaccinated Mamu-A*01+ macaque Rh29804, and the Ad5/Gag-P2A-ICP47-vaccinated Mamu-A*01+ macaque Rh31520 are shown following coculture with the indicated antigen-presenting cells pulsed with the indicated peptide(s). E Min, a pool of 11 previously defined minimal optimal SIVmac239 Gag-derived peptides presented by Mamu-E (17). (C) Cumulative results from ICS assays using PBMC from two strain 68-1 RhCMV/Gag-vaccinated macaques and all six macaques that received an Ad5 prime/AAV1 boost regimen concomitantly expressing SIV Gag and a TAP inhibitor following incubation with the indicated antigen-presenting cells pulsed with the indicated peptide(s). The dashed line indicates the limit of detection (median background cytokine secretion) of the ICS assay. 11 Minimals, a pool of 11 previously defined minimal optimal SIVmac239 Gag-derived peptides presented by Mamu-E (17).
It remained possible, however, that vaccination in the setting of TAP inhibition might prime MHC-E-restricted CD8+ T cells responding to Gag epitopes other than the previously described RL9 and EK9 epitopes (17). Therefore, to more stringently test for the presence of MHC-E-restricted CD8+ T cells in animals that received a TAP inhibitor-based vaccine, we next assayed for the presence of any MHC-E-restricted, Gag-specific CD8+ T cell response by performing ICS using a Mamu-E*02:11 transfectant pulsed with either all 124 15-mer peptides spanning the entire SIVmac239 Gag open reading frame (ORF) or all 11 previously identified Mamu-E-restricted CD8+ T cell minimal optimal Gag epitopes (E minimals) (17). We included Gag ORF- or CM9-pulsed Mamu-A*01+ BLCL stimulation to measure bulk MHC-I-restricted responses, and we used PBMC from the vaccinated macaques 1 week post-AAV1 boost as effectors. As expected, CD8+ T cells from the strain 68-1 RhCMV/Gag-vaccinated Mamu-A*01+ animals Rh22607 and Rh21826 did not react against CM9 yet robustly responded to Mamu-E*02:11 pulsed with either Gag ORF or the pool of all previously identified MHC-E-bound Gag minimal epitopes (Fig. 4B, top panels, and Fig. 4C). In contrast, CD8+ T cells from every animal that received a Gag/TAP inhibitor vaccine responded only to BLCL pulsed with Gag ORF and CM9, with no response to Gag peptide presented in the context of Mamu-E*02:11 (Fig. 4B and C). Thus, we did not find evidence that TAP inhibition alone can induce MHC-E-restricted T cells in the macaque model.
DISCUSSION
To our knowledge, this is the first study investigating the effects of TAP inhibitor-based vaccines on cellular immunity in nonhuman primates. We show that the TAP inhibitors ICP47, UL49.5, and Rh185 are functional and substantively reduce surface levels of MHC-Ia, but not MHC-E, in rhesus macaque BLCL. The ability of TAP inhibition to reduce MHC-Ia surface levels agrees with studies using these inhibitors in human cells (25). Reduced MHC-Ia expression on target cells corresponded with reduced recognition by MHC-Ia-restricted T cells but not MHC-E-restricted T cells. Although TAP inhibition did not abrogate MHC-E-restricted T cell recognition in vitro, vaccination with a TAP inhibitor was not sufficient to induce MHC-E-restricted T cells in rhesus macaques in vivo. These results suggest that mechanisms either separate from, or in addition to, TAP inhibition are responsible for MHC-E-restricted CD8+ T cell priming.
van Hall and colleagues previously reported that vaccination of mice with TAP-deficient dendritic cells primed CD8+ T cells restricted by Qa-1b, the mouse homologue of MHC-E (21, 47, 48). These CD8+ T cells specifically recognized and lysed TAP-deficient cancer cells in a Qa-1b-restricted fashion, and the epitopes targeted by these Qa-1b-restricted T cells were collectively termed T cell epitopes associated with impaired peptide processing (TEIPP) (29). While TEIPP-specific CD8+ T cells recognizing neoantigens from TAP-deficient tumors exist within the human population, these unique CD8+ T cells are restricted by classical HLA-Ia molecules and not HLA-E (42, 49). The results of our TAP inhibition vaccine study agree with these observations and suggest either that specific cell subsets must be directly targeted or that the requirements for priming MHC-E-restricted CD8+ T cells are sharply divergent between mice and primates. Independent of the mechanism, the study presented here indicates that loss of TAP function alone is insufficient to elicit MHC-E-restricted T cells in primates.
Additional evidence that TAP deficiency alone is insufficient to induce MHC-E-restricted CD8+ T cells comes from clinical case studies of an extremely rare autosomal recessive, primary immunodeficiency caused by mutations in the TAP1 or TAP2 subunit. TAP deficiency syndrome, or bare lymphocyte syndrome (BLS), is defined by lack of expression of either TAP1 or TAP2 subunits, which completely abrogates formation of a functional TAP heterodimer (50). Although these patients exhibit low frequencies of αβ T cells, they retain functional cellular immunity and do not suffer from chronic viral infections. Indeed, antiviral EBV-specific CD8+ T cells exist and can be isolated from BLS patients (51, 52). Despite being primed in a completely TAP-deficient environment, an EBV-specific T cell clone generated from a BLS patient was restricted by HLA-B, a classical MHC-Ia allele, and not by HLA-E. Of note, BLS patients are not devoid of surface HLA-E expression, with equivalent expression of HLA-E on monocytes derived from a BLS patient compared to the levels of healthy controls that have intact TAP function (53). While our results of unchanged Mamu-E levels on the surface of MDM transduced to express the Rh185 TAP inhibitor agree with these results from monocytes isolated from TAP-deficient BLS patients, it is possible that these observations are cell type dependent. Although no exhaustive studies focused on identifying HLA-E-restricted CD8+ T cells in BLS patients have been performed to date, the published studies referenced above are in alignment with our vaccination results that TAP deficiency alone is insufficient to elicit MHC-E-restricted T cells in humans. In further support of our conclusion that mechanisms other than TAP inhibition are involved in the priming of MHC-E-restricted T cells, we previously demonstrated that Δ182–189 RhCMV, a strain 68-1 vector missing the critical immune evasion genes Rh182 to Rh189 including the TAP inhibitor Rh185, still gives rise to unconventionally MHC-restricted CD8+ T cells (46). Of note, the Δ182–189 RhCMV/Gag vector elicited classically MHC-Ia-restricted Gag-specific CD8+ T cells in addition to, not instead of, unconventionally MHC-restricted T cells. Therefore, the generation of unconventionally MHC-E-restricted and classically MHC-Ia-restricted CD8+ T cells does not appear to be mutually exclusive.
While TAP inhibition alone is insufficient to induce MHC-E-restricted CD8+ T cells, it remains unknown if TAP inhibition is involved in priming MHC-E-restricted T cells in primates. Although loading of the MHC-Ia leader sequences into MHC-E is TAP dependent (54), the results presented here demonstrate that alternate, TAP-independent routes of antigen loading exist. Further studies focused on strain 68-1 RhCMV, which induces a high frequency of MHC-E-restricted CD8+ T cells in rhesus macaques (17), will likely be key to unravelling the precise mechanism involved. In addition to the TAP-inhibiting US6 family of proteins, RhCMV, like HCMV, includes multiple immunoregulatory gene families impacting all aspects of antigen processing and presentation (55). Indeed, the HCMV UL40 glycoprotein encodes its own VL9 leader sequence, which upregulates HLA-E expression on HCMV-infected cells (45). Future work examining the role of the myriad of CMV-encoded immunoevasion, cell tropism, and chemokine genes will be critical in understanding how MHC-E-restricted CD8+ T cell responses are elicited in vivo.
MHC-E-restricted CD8+ T cells remain a highly attractive target for cancer and infectious disease immunotherapy. T cells specific to antigens expressed on solid and nonsolid tumor cells have shown great promise in the clinic. Injection of the bacillus Calmette-Guérin (BCG) vaccine into bladder cancer tumors to induce tumor-specific T cells has been standard care for decades (56). Moreover, tumor-specific antigen vaccines have successfully led to tumor regression in multiple clinical trials (57). However, the necessity of sustained MHC-Ia expression to present antigen to cancer- or pathogen-specific αβ T cells is a significant roadblock to such therapies. Loss of MHC-Ia expression on tumors cells with subsequent immune escape is extremely common (58). For example, up to 90% of examined cervical cancer tumors exhibit partial or complete loss of MHC-Ia expression (59). Conversely, increased expression of the nonclassical MHC-E molecule is extremely common in multiple cancers, including renal cell and colorectal carcinomas (60, 61) and ovarian and cervical cancers (62). This heightened expression serves to suppress NK cell function against tumor cells through interaction with CD94/NKG2A. Likewise, many viruses, including human immunodeficiency virus (HIV) and herpesviruses dampen CD8+ T cell and NK surveillance by lowering MHC-Ia and increasing MHC-E expression, respectively (24, 45, 63, 64). While upregulation of MHC-E expression on the surface of neoplastic and virally infected cells evades NK-mediated cytotoxicity, it leaves them vulnerable to lysis by MHC-E-restricted CD8+ T cells. Therefore, MHC-E-restricted CD8+ T cells represent a powerful tool for immunotherapy, and understanding the determinants of their priming is critical for harnessing them for clinical applications.
MATERIALS AND METHODS
Animals.
Nine nonhuman primates were utilized in this study. All animals were cared for at the Oregon National Primate Research Center (ONPRC) with the approval of the ONPRC Animal Care and Use Committee using the standards of the NIH-approved Guide for the Care and Use of Laboratory Animals (65). All procedures were conducted under anesthesia (intramuscular administered ketamine, 10 to 20 mg/kg).
Viral constructs.
SIVmac239 Gag AAV65320.1, Rh185 YP_068274, ICP47 (HSV-2 YP_009137225.1), and UL49.5 NP_045309 sequences were obtained from GenBank and codon optimized for human using Integrated DNA Technology’s online tool. Genes with the following modifications were synthesized by GeneScript: EcoRI-SIVMac239Gag-BamHI_P2A_KpnI and each TAP inhibitor were synthesized with KpnI-(TAP inhibitor)-AgeI-Flag-NotI. SIVMac239 Gag-P2A was first cloned into AAV using EcoRI and KpnI. Each TAP inhibitor-Flag construct was then cloned into the SIVMac239 Gag-P2A AAV vector using KpnI and NotI sites, creating SIVMac239 Gag-P2A-(TAP inhibitor)-Flag constructs. Subsequently, those constructs were cloned into ViraQuest Ad5 CMVK vectors using EcoRI and NotI sites. Ad5 and matching AAV1 vectors were produced by Viraquest and the University of North Carolina vector core, respectively, as previously described (66, 67). For the three vaccine groups, Ad5 prime was intramuscularly injected over the lateral aspect of the upper arm and lateral in 500-μl aliquots of Ad5 vector containing 5 × 1011 vp each using a 3-ml syringe fitted with a 23-gauge, 1-inch needle. Using the same procedure an AAV1 dose of 1013 vp with matching vaccine insert was delivered as a boost 12 weeks subsequent to priming.
APC preparation.
B lymphoblastoid cell lines (BLCL) were generated by infecting macaque PBMC with herpesvirus papio as previously described (46). A mammalian expression vector for Mamu-E*02:11 was generated by ligating the full-length Mamu-E*02:011 coding sequence into pCEP4 HindIII/NotI restriction sites. The plasmid was cloned in DH5α Escherichia coli (Life Technologies), the sequence was confirmed, and it was electroporated into MHC-I-negative K562 cells using an Amaxa Nucleofector kit C (Lonza), program G-016. Transfected cells were maintained on drug selection (hygromycin B; Corning) and routinely confirmed for surface expression of MHC-I by staining with pan-MHC-I Ab clone W6/32 alongside negative-control K562 cells. BLCL were transduced with Ad5 utilizing ViroMag transduction reagent (Ozbiosciences, CA) according to the manufacturer’s recommendation. Briefly, cells were plated in 24-well plates at 2 × 106 per ml in complete medium (RPMI 1640 medium with 10% fetal bovine serum [FBS]). ViroMag beads were incubated with 4 × 109 viral particles (vp) of Ad5 for 15 min at room temperature. Beads/Ad5 were then added to BLCL, and the plate was briefly spun at 1,600 × g. The plate was then placed on top of a manufacturer’s magnet and incubated at 37°C with 5% CO2 for 15 min. Transduction was assessed at 2 days postinfection. Cells were stained with SIV Gag p27 antibody conjugated in-house to fluorescein isothiocyanate (FITC) using a FluoReporter FITC protein labeling kit (Invitrogen, CA) and anti-DYKDDDDK (Miltenyi Biotec). Live/Dead Fixable Near Infra-Red Dead Cell Stain (Life Technologies) was used to assess cell viability. MHC-E staining was done as previously described with antibody 4D12 (HLA-E), grown and purified in-house as described previously (68).
Monocyte-derived macrophages.
Monocyte derived macrophages (MDM) were derived as previously described (69). Briefly, CD14+ monocytes were isolated via magnetically activated cell sorting (MACS) using CD14 microbeads (Miltenyi Biotec) and incubated in 50% fresh R10 medium (RPMI medium plus 10% fetal calf serum [FCS]) and 50% KPB-M15-conditioned R10 medium supplemented with 10 ng/ml macrophage colony-stimulating factor (M-CSF; Sigma) for 6 days, with feeding every other day. MDM were consistently >95% CD14+.
T cell tetramer staining and assays.
Approximately 1 × 106 to 2 × 106 cells were placed in 50 to 100 μl of RPMI 1640 medium (with 10% FBS) for tetramer staining purposes. Tetramer was added at a final concentration of 100 nM, and cells were incubated in the dark at room temperature for 1 h. Surface-staining antibodies were then added, and cells were incubated for an additional 30 min in the dark at room temperature. Cells were then washed once with 1× phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde (PFA). CD8+ T cell lines were generated as previously described (70). Briefly, PBMC from a Mamu-A*01 or Mamu-A*02 SIV-infected RM were stimulated weekly with irradiated autologous BLCL pulsed with the peptide of interest and cultured in R15 medium (RPMI 1640 with 10% fetal bovine serum) supplemented with 500 U/ml IL-2 (National Institutes of Health AIDS Reagent Program). Mamu-A*01-restricted Gag181–189 CM9 and Mamu-A*02-restricted Gag71–79 GY9 T cell lines were 91% and 87% specific for their cognate antigens as determined by MHC tetramer staining, respectively.
Intracellular cytokine staining.
PBMC or CD8+ T cell lines were incubated with peptide-pulsed APCs or transduced BLCL and the costimulatory molecules CD28 and CD49d (BD Biosciences) for 1 h, followed by addition of brefeldin A (Sigma-Aldrich) for an additional 8 h. Costimulation with unpulsed APCs served as background controls. The MHC association (MHC-E or MHC-II) of response(s) against transduced BLCL was determined by preincubating BLCL for 1 h at 37°C (prior to combining effector and target cells and incubation per the standard ICS assay) with the following blockers: (i) the MHC-II-blocking CLIP peptide (MHC-II-associated invariant chain, amino acids [aa] 89 to 100; 20 μM) and (ii) the MHC-E-blocking VL9 peptide (VMAPRTLLL; 20 μM), alone or in combination (blocking reagents were not washed but remained throughout the assay). Stimulated cells were stained, collected, and analyzed as previously described (17). Briefly, cells were washed with 1× PBS, surface stained for 30 min, washed with PBS, fixed with 2% paraformaldehyde, permeabilized with medium B buffer (ThermoFisher), and stained intracellularly for 1 h. Antibodies used in this study included the following: anti-CD3 (clone SP34-2, conjugated to Pacific Blue; BD Biosciences), anti-CD8 (clone SK1, conjugated to TruRed; BD Biosciences), anti-CD4 (clone L200, conjugated to phycoerythrin [PE]-Cy7; BD Biosciences), anti-IFN-γ (clone B27 conjugated to FITC; BD Biosciences), and anti-TNF (monoclonal antibody 11 [MAb11] conjugated to allophycocyanin [APC]; BD Biosciences). Live/Dead Fixable Near Infra-Red Dead Cell Stain (Life Technologies) was used to assess cell viability. Sample collection was performed on an LSR-II instrument (BD Biosciences), and analysis was conducted with FlowJo software (Tree Star).
IFN-γ ELISpot assay.
Gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assays were done as prescribed by the manufacturer (Mabtech, Sweden). Briefly 96-well ELISpot plates were washed with 1× PBS and blocked with complete medium (RMPI 1640 medium with 10% FBS). Longitudinal PBMC (105) samples were added in duplicate to plate wells. Cells were stimulated by SIVmac239 Gag peptide pools obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH. After 17 h of incubation, cells were removed, and plates were incubated with biotinylated IFN-γ detection antibody (7-B6-1) for 2 h at room temperature. The plates were washed, streptavidin-alkaline phosphate (ALP) was added, and the plates incubated for an additional hour. Following a subsequent wash, plates were developed with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT) substrate. The reaction was stopped with water after clear spots developed. Plates were read with an AID ELISpot plate reader (AID, Germany).
Statistical analysis.
Due to small sample sizes, we did not assess the normality of our data and utilized nonparametric tests throughout our analyses. For group comparisons, we used a Kruskal-Wallis test followed by a post hoc Dunn’s test to evaluate differences between each column.
ACKNOWLEDGMENTS
We thank Daniel E. Geraghty for the MHC-E MAb 4D12 hybridoma, the Nonhuman Primate Reagent Resource for providing Mamu-A*01-specific antibody, and the NIH AIDS Reagent Program for providing recombinant IL-2.
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI117802, R01AI117802-S, and R01AI140888 (awarded to J.B.S.) and P51OD011092 (awarded to the Oregon National Primate Research Center).
REFERENCES
- 1.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh S. 2015. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knapp LA, Cadavid LF, Watkins DI. 1998. The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J Immunol 160:189–196. [PubMed] [Google Scholar]
- 3.Grimsley C, Ober C. 1997. Population genetic studies of HLA-E: evidence for selection. Hum Immunol 52:33–40. doi: 10.1016/S0198-8859(96)00241-8. [DOI] [PubMed] [Google Scholar]
- 4.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. 1998. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med 187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 6.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol 160:4951–4960. [PubMed] [Google Scholar]
- 7.Kraemer T, Celik AA, Huyton T, Kunze-Schumacher H, Blasczyk R, Bade-Döding C. 2015. HLA-E: presentation of a broader peptide repertoire impacts the cellular immune response-implications on HSCT outcome. Stem Cells Int 2015:346714. doi: 10.1155/2015/346714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lampen MH, Hassan C, Sluijter M, Geluk A, Dijkman K, Tjon JM, de Ru AH, van der Burg SH, van Veelen PA, van Hall T. 2013. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol Immunol 53:126–131. doi: 10.1016/j.molimm.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, Moretta L, Mingari MC. 2003. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci U S A 100:10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulte D, Vogel M, Langhans B, Krämer B, Körner C, Nischalke HD, Steinberg V, Michalk M, Berg T, Rockstroh JK, Sauerbruch T, Spengler U, Nattermann J. 2009. The HLA-ER/HLA-ER genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8+ T cells. J Infect Dis 200:1397–1401. doi: 10.1086/605889. [DOI] [PubMed] [Google Scholar]
- 11.Jørgensen PB, Livbjerg AH, Hansen HJ, Petersen T, Höllsberg P. 2012. Epstein-Barr virus peptide presented by HLA-E is predominantly recognized by CD8bright cells in multiple sclerosis patients. PLoS One 7:e46120. doi: 10.1371/journal.pone.0046120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salerno-Gonçalves R, Fernandez-Viña M, Lewinsohn DM, Sztein MB. 2004. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol 173:5852–5862. doi: 10.4049/jimmunol.173.9.5852. [DOI] [PubMed] [Google Scholar]
- 13.McMurtrey C, Harriff MJ, Swarbrick GM, Duncan A, Cansler M, Null M, Bardet W, Jackson KW, Lewinsohn DA, Hildebrand W, Lewinsohn DM. 2017. T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLoS One 12:e0188288. doi: 10.1371/journal.pone.0188288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff THM, Joosten SA. 2015. Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLoS Pathog 11:e1004671. doi: 10.1371/journal.ppat.1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joosten SA, van Meijgaarden KE, van Weeren PC, Kazi F, Geluk A, Savage NDL, Drijfhout JW, Flower DR, Hanekom WA, Klein MR, Ottenhoff T. 2010. Mycobacterium tuberculosis peptides presented by HLA-E molecules are targets for human CD8 T-cells with cytotoxic as well as regulatory activity. PLoS Pathog 6:e1000782. doi: 10.1371/journal.ppat.1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouand N, Bressollette-Bodin C, Gérard N, Giral M, Guérif P, Rodallec A, Oger R, Parrot T, Allard M, Cesbron-Gautier A, Gervois N, Charreau B. 2018. HCMV triggers frequent and persistent UL40-specific unconventional HLA-E-restricted CD8 T-cell responses with potential autologous and allogeneic peptide recognition. PLoS Pathog 14:e1007041. doi: 10.1371/journal.ppat.1007041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, López CA, Früh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ. 2016. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351:714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen SG, Piatak M, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Früh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. 2013. Immune clearance of highly pathogenic SIV infection. Nature 502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotterill LA, Stauss HJ, Millrain MM, Pappin DJ, Rahman D, Canas B, Chandler P, Stackpoole A, Simpson E, Robinson PJ, Dyson PJ. 1997. Qa-1 interaction and T cell recognition of the Qa-1 determinant modifier peptide. Eur J Immunol 27:2123–2132. doi: 10.1002/eji.1830270902. [DOI] [PubMed] [Google Scholar]
- 21.Chambers B, Grufman P, Fredriksson V, Andersson K, Roseboom M, Laban S, Camps M, Wolpert EZ, Wiertz E, Offringa R, Ljunggren H-G, van Hall T. 2007. Induction of protective CTL immunity against peptide transporter TAP-deficient tumors through dendritic cell vaccination. Cancer Res 67:8450–8455. doi: 10.1158/0008-5472.CAN-07-1092. [DOI] [PubMed] [Google Scholar]
- 22.van Hall T, Laban S, Koppers-Lalic D, Koch J, Precup C, Asmawidjaja P, Offringa R, Wiertz E. 2007. The varicellovirus-encoded TAP inhibitor UL49.5 regulates the presentation of CTL epitopes by Qa-1b1. J Immunol 178:657–662. doi: 10.4049/jimmunol.178.2.657. [DOI] [PubMed] [Google Scholar]
- 23.Pande NT, Powers C, Ahn K, Früh K. 2005. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J Virol 79:5786–5798. doi: 10.1128/JVI.79.9.5786-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verweij MC, Horst D, Griffin BD, Luteijn RD, Davison AJ, Ressing ME, Wiertz E. 2015. Viral inhibition of the transporter associated with antigen processing (TAP): a striking example of functional convergent evolution. PLoS Pathog 11:e1004743. doi: 10.1371/journal.ppat.1004743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oosten LEM, Koppers-Lalic D, Blokland E, Mulder A, Ressing ME, Mutis T, van Halteren AGS, Wiertz E, Goulmy E. 2007. TAP-inhibiting proteins US6, ICP47 and UL49.5 differentially affect minor and major histocompatibility antigen-specific recognition by cytotoxic T lymphocytes. Int Immunol 19:1115–1122. doi: 10.1093/intimm/dxm082. [DOI] [PubMed] [Google Scholar]
- 26.Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJ, Ploegh HL, Peterson PA, Yang Y, Früh K. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613–621. doi: 10.1016/S1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 27.Hengel H, Koopmann JO, Flohr T, Muranyi W, Goulmy E, Hämmerling GJ, Koszinowski UH, Momburg F. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623–632. doi: 10.1016/S1074-7613(00)80350-7. [DOI] [PubMed] [Google Scholar]
- 28.Lehner PJ, Karttunen JT, Wilkinson GW, Cresswell P. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc Natl Acad Sci U S A 94:6904–6909. doi: 10.1073/pnas.94.13.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hall T, Wolpert EZ, van Veelen P, Laban S, van der Veer M, Roseboom M, Bres S, Grufman P, de Ru A, Meiring H, de Jong A, Franken K, Teixeira A, Valentijn R, Drijfhout JW, Koning F, Camps M, Ossendorp F, Kärre K, Ljunggren H-G, Melief CJM, Offringa R. 2006. Selective cytotoxic T-lymphocyte targeting of tumor immune escape variants. Nat Med 12:417–424. doi: 10.1038/nm1381. [DOI] [PubMed] [Google Scholar]
- 30.Ahn K, Meyer TH, Uebel S, Sempé P, Djaballah H, Yang Y, Peterson PA, Früh K, Tampé R. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J 15:3247–3255. doi: 10.1002/j.1460-2075.1996.tb00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomazin R, Hill AB, Jugovic P, York I, van Endert P, Ploegh HL, Andrews DW, Johnson DC. 1996. Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J 15:3256–3266. doi: 10.1002/j.1460-2075.1996.tb00690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Früh K, Ahn K, Djaballah H, Sempé P, van Endert PM, Tampé R, Peterson PA, Yang Y. 1995. A viral inhibitor of peptide transporters for antigen presentation. Nature 375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 33.Koppers-Lalic D, Reits EAJ, Ressing ME, Lipinska AD, Abele R, Koch J, Rezende MM, Admiraal P, van Leeuwen D, Bienkowska-Szewczyk K, Mettenleiter TC, Rijsewijk FAM, Tampe R, Neefjes J, Wiertz EJHJ. 2005. Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc Natl Acad Sci U S A 102:5144–5149. doi: 10.1073/pnas.0501463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hewitt EW, Gupta SS, Lehner PJ. 2001. The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J 20:387–396. doi: 10.1093/emboj/20.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. 2001. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip.” J Gen Virol 82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 36.Kim JH, Lee S-R, Li L-H, Park H-J, Park J-H, Lee KY, Kim M-K, Shin BA, Choi S-Y. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibrahimi A, Vande Velde G, Reumers V, Toelen J, Thiry I, Vandeputte C, Vets S, Deroose C, Bormans G, Baekelandt V, Debyser Z, Gijsbers R. 2009. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther 20:845–860. doi: 10.1089/hum.2008.188. [DOI] [PubMed] [Google Scholar]
- 38.Jarosinski KW, Hunt HD, Osterrieder N. 2010. Down-regulation of MHC class I by the Marek's disease virus (MDV) UL49.5 gene product mildly affects virulence in a haplotype-specific fashion. Virology 405:457–463. doi: 10.1016/j.virol.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 39.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 40.Braud V, Jones EY, McMichael A. 1997. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol 27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 41.Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, Geraghty DE. 2003. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J Immunol 171:1376–1384. doi: 10.4049/jimmunol.171.3.1376. [DOI] [PubMed] [Google Scholar]
- 42.Lampen MH, Verweij MC, Querido B, van der Burg SH, Wiertz E, van Hall T. 2010. CD8+ T cell responses against TAP-inhibited cells are readily detected in the human population. Ji 185:6508–6517. doi: 10.4049/jimmunol.1001774. [DOI] [PubMed] [Google Scholar]
- 43.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, López-Botet M, Geraghty DE. 1998. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U S A 95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorente E, Infantes S, Abia D, Barnea E, Beer I, García R, Lasala F, Jiménez M, Mir C, Morreale A, Admon A, López D. 2012. A viral, transporter associated with antigen processing (TAP)-independent, high affinity ligand with alternative interactions endogenously presented by the nonclassical human leukocyte antigen E class I molecule. J Biol Chem 287:34895–34903. doi: 10.1074/jbc.M112.362293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 46.Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, Malouli D, Xu G, Richards R, Whizin N, Reed JS, Hammond KB, Fischer M, Turner JM, Legasse AW, Axthelm MK, Edlefsen PT, Nelson JA, Lifson JD, Früh K, Picker LJ. 2013. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340:1237874–1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doorduijn EM, Sluijter M, Querido BJ, Seidel UJE, Oliveira CC, van der Burg SH, van Hall T. 2018. T cells engaging the conserved MHC class Ib molecule Qa-1b with TAP-independent peptides are semi-invariant lymphocytes. Front Immunol 9:60. doi: 10.3389/fimmu.2018.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliveira CC, van Veelen PA, Querido B, de Ru A, Sluijter M, Laban S, Drijfhout JW, van der Burg SH, Offringa R, van Hall T. 2010. The nonpolymorphic MHC Qa-1b mediates CD8+ T cell surveillance of antigen-processing defects. J Exp Med 207:207–221. doi: 10.1084/jem.20091429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marijt KA, Blijleven L, Verdegaal EME, Kester MG, Kowalewski DJ, Rammensee H-G, Stevanović S, Heemskerk MHM, van der Burg SH, van Hall T. 2018. Identification of non-mutated neoantigens presented by TAP-deficient tumors. J Exp Med 215:2325–2337. doi: 10.1084/jem.20180577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gadola SD, Moins-Teisserenc HT, Trowsdale J, Gross WL, Cerundolo V. 2000. TAP deficiency syndrome. Clin Exp Immunol 121:173–178. doi: 10.1046/j.1365-2249.2000.01264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.la Salle de H, Hanau D, Fricker D, Urlacher A, Kelly A, Salamero J, Powis SH, Donato L, Bausinger H, Laforet M. 1994. Homozygous human TAP peptide transporter mutation in HLA class I deficiency. Science 265:237–241. doi: 10.1126/science.7517574. [DOI] [PubMed] [Google Scholar]
- 52.la Salle de H, Houssaint E, Peyrat MA, Arnold D, Salamero J, Pinczon D, Stevanovic S, Bausinger H, Fricker D, Gomard E, Biddison W, Lehner P, UytdeHaag F, Sasportes M, Donato L, Rammensee HG, Cazenave JP, Hanau D, Tongio MM, Bonneville M. 1997. Human peptide transporter deficiency: importance of HLA-B in the presentation of TAP-independent EBV antigens. J Immunol 158:4555–4563. [PubMed] [Google Scholar]
- 53.Furukawa H, Yabe T, Akaza T, Tadokoro K, Tohma S, Inoue T, Tokunaga K, Yamamoto K, Geraghty DE, Juji T. 1999. Cell surface expression of HLA-E molecules on PBMC from a TAP1-deficient patient. Tissue Antigens 53:292–295. doi: 10.1034/j.1399-0039.1999.530310.x. [DOI] [PubMed] [Google Scholar]
- 54.Braud VM, Allan DS, Wilson D, McMichael AJ. 1998. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol 8:1–10. doi: 10.1016/S0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 55.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol 77:6620–6636. doi: 10.1128/jvi.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Askeland EJ, Newton MR, O'Donnell MA, Luo Y. 2012. Bladder cancer immunotherapy: BCG and beyond. Adv Urol 2012:181987–181913. doi: 10.1155/2012/181987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Disis ML, Bernhard H, Jaffee EM. 2009. Use of tumour-responsive T cells as cancer treatment. Lancet 373:673–683. doi: 10.1016/S0140-6736(09)60404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, van Hall T. 2016. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol 39:44–51. doi: 10.1016/j.coi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koopman LA, Corver WE, van der Slik AR, Giphart MJ, Fleuren GJ. 2000. Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J Exp Med 191:961–976. doi: 10.1084/jem.191.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seliger B, Jasinski-Bergner S, Quandt D, Stoehr C, Bukur J, Wach S, Legal W, Taubert H, Wullich B, Hartmann A. 2016. HLA-E expression and its clinical relevance in human renal cell carcinoma. Oncotarget 7:67360–67372. doi: 10.18632/oncotarget.11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benevolo M, Mottolese M, Tremante E, Rollo F, Diodoro MG, Ercolani C, Sperduti I, Monaco Lo E, Cosimelli M, Giacomini P. 2011. High expression of HLA-E in colorectal carcinoma is associated with a favorable prognosis. J Transl Med 9:184. doi: 10.1186/1479-5876-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gooden M, Lampen M, Jordanova ES, Leffers N, Trimbos JB, van der Burg SH, Nijman H, van Hall T. 2011. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8+ T lymphocytes. Proc Natl Acad Sci U S A 108:10656–10661. doi: 10.1073/pnas.1100354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dirk BS, Pawlak EN, Johnson AL, Van Nynatten LR, Jacob RA, Heit B, Dikeakos JD. 2016. HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci Rep 6:37021. doi: 10.1038/srep37021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, Rockstroh J, Weiss EH, Sauerbruch T, Spengler U. 2005. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther 10:95–107. [DOI] [PubMed] [Google Scholar]
- 65.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 66.Burwitz BJ, Wettengel JM, Mück-Häusl MA, Ringelhan M, Ko C, Festag MM, Hammond KB, Northrup M, Bimber BN, Jacob T, Reed JS, Norris R, Park B, Moller-Tank S, Esser K, Greene JM, Wu HL, Abdulhaqq S, Webb G, Sutton WF, Klug A, Swanson T, Legasse AW, Vu TQ, Asokan A, Haigwood NL, Protzer U, Sacha JB. 2017. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat Commun 8:2146. doi: 10.1038/s41467-017-01953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sacha JB, Kim I-J, Chen L, Ullah JH, Goodwin DA, Simmons HA, Schenkman DI, Pelchrzim von F, Gifford RJ, Nimityongskul FA, Newman LP, Wildeboer S, Lappin PB, Hammond D, Castrovinci P, Piaskowski SM, Reed JS, Beheler KA, Tharmanathan T, Zhang N, Muscat-King S, Rieger M, Fernandes C, Rumpel K, Gardner JP, Gebhard DH, Janies J, Shoieb A, Pierce BG, Trajkovic D, Rakasz E, Rong S, McCluskie M, Christy C, Merson JR, Jones RB, Nixon DF, Ostrowski MA, Loudon PT, Pruimboom-Brees IM, Sheppard NC. 2012. Vaccination with cancer- and HIV infection-associated endogenous retrotransposable elements is safe and immunogenic. J Immunol 189:1467–1479. doi: 10.4049/jimmunol.1200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu HL, Wiseman RW, Hughes CM, Webb GM, Abdulhaqq SA, Bimber BN, Hammond KB, Reed JS, Gao L, Burwitz BJ, Greene JM, Ferrer F, Legasse AW, Axthelm MK, Park BS, Brackenridge S, Maness NJ, McMichael AJ, Picker LJ, O’Connor DH, Hansen SG, Sacha JB. 2018. The role of MHC-E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J Immunol 200:49–60. doi: 10.4049/jimmunol.1700841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gersuk G, Hiraoka A, Marr KA. 2005. Human monocytes differentiate into macrophages under the influence of human KPB-M15 conditioned medium. J Immunol Methods 299:99–106. doi: 10.1016/j.jim.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 70.Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, Allison DB, Adnan S, Hoji A, Wilson NA, Friedrich TC, Lifson JD, Yang OO, Watkins DI. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J Immunol 178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]