Recent progress in understanding the human immune response to respiratory syncytial virus has paved the way for new vaccine antigens and therapeutics to prevent and treat disease. Progress toward understanding the immune response to human metapneumovirus (hMPV) has lagged behind, although hMPV is a leading cause of lower respiratory tract infection in children. In this report, we advanced the field by isolating a panel of human mAbs to the hMPV F protein. One potent neutralizing mAb, MPV364, targets antigenic site III on the hMPV F protein and incorporates two protomers into its epitope yet is unique from previously discovered site III mAbs, as it does not cross-react with the RSV F protein. We further examined MPV364 in vivo and found that it limits viral replication in BALB/c mice. Altogether, these data provide new mAb candidates for therapeutic development and provide insights into hMPV vaccine development.
KEYWORDS: human metapneumovirus, monoclonal antibodies
ABSTRACT
Human metapneumovirus (hMPV) is a leading cause of viral lower respiratory tract infection in children. The sole target of neutralizing antibodies targeting hMPV is the fusion (F) protein, a class I viral fusion protein mediating virus-cell membrane fusion. There have been several monoclonal antibodies (mAbs) isolated that neutralize hMPV; however, determining the antigenic sites on the hMPV F protein mediating such neutralizing antibody generation would assist efforts for effective vaccine design. In this report, the isolation and characterization of four new human mAbs, termed MPV196, MPV201, MPV314, and MPV364, are described. Among the four mAbs, MPV364 was found to be the most potent neutralizing mAb in vitro. Binding studies with monomeric and trimeric hMPV F revealed that MPV364 had the weakest binding affinity for monomeric hMPV F compared to the other three mAbs, yet binding experiments with trimeric hMPV F showed limited differences in binding affinity, suggesting that MPV364 targets an antigenic site incorporating two protomers. Epitope binning studies showed that MPV364 targets antigenic site III on the hMPV F protein and competes for binding with previously discovered mAbs MPE8 and 25P13, both of which cross-react with the respiratory syncytial virus (RSV) F protein. However, MPV364 does not cross-react with the RSV F protein, and the competition profile suggests that it binds to the hMPV F protein in a binding pose slightly shifted from mAbs MPE8 and 25P13. MPV364 was further assessed in vivo and was shown to substantially reduce viral replication in the lungs of BALB/c mice. Overall, these data reveal a new binding region near antigenic site III of the hMPV F protein that elicits potent neutralizing hMPV F-specific mAbs and provide a new panel of neutralizing mAbs that are candidates for therapeutic development.
IMPORTANCE Recent progress in understanding the human immune response to respiratory syncytial virus has paved the way for new vaccine antigens and therapeutics to prevent and treat disease. Progress toward understanding the immune response to human metapneumovirus (hMPV) has lagged behind, although hMPV is a leading cause of lower respiratory tract infection in children. In this report, we advanced the field by isolating a panel of human mAbs to the hMPV F protein. One potent neutralizing mAb, MPV364, targets antigenic site III on the hMPV F protein and incorporates two protomers into its epitope yet is unique from previously discovered site III mAbs, as it does not cross-react with the RSV F protein. We further examined MPV364 in vivo and found that it limits viral replication in BALB/c mice. Altogether, these data provide new mAb candidates for therapeutic development and provide insights into hMPV vaccine development.
INTRODUCTION
Human metapneumovirus (hMPV) is a significant respiratory pathogen and is a member of the order Mononegavirales and the Pneumoviridae family of viruses, which also includes respiratory syncytial virus (RSV). The members of this family are single-stranded, nonsegmented, negative-sense RNA viruses and have similar life cycles (1). Infants and the elderly are the major groups for which hMPV infection may require hospitalization (2–6). In addition, hMPV infection is frequent in immunocompromised patients, including lung transplant (7) and hematopoietic stem cell transplant (8–11) recipients, with several deaths associated with viral infection (8–10). hMPV is a significant cause of febrile respiratory illness in HIV-infected patients (12) and has an increased incidence in several HIV-infected age groups (13). hMPV has also been linked to exacerbations of chronic obstructive pulmonary disease (14). hMPV was initially identified in 2001 (15), and the clinical features of hMPV infection display as mid- to upper respiratory tract infection and can be severe enough to cause life-threatening bronchiolitis and pneumonia. A nursing home outbreak of hMPV demonstrates the need for effective vaccines and therapeutics in the elderly (16). There are no licensed vaccines to protect against hMPV, but several candidates have been examined in animal models, including live-attenuated viruses, recombinant viruses, vectored vaccines, and recombinant surface proteins (17). Currently, only one vaccine has been tested in clinical trials (ClinicalTrials.gov identifier NCT01255410); however, results of the trial are not yet available.
The hMPV genome consists of approximately 13,000 nucleotides, with eight genes encoding nine proteins, three of which are surface glycoproteins: the small hydrophobic (SH), attachment (G), and fusion (F) proteins. The hMPV F protein is the sole target of the neutralizing antibody response to hMPV (18). This is in contrast to RSV, where both the RSV G and RSV F proteins elicit neutralizing antibodies (19). The RSV/hMPV F proteins also demonstrate a high degree of conservation between the A and B virus genotypes, a key advantage for developing broadly reactive vaccines and therapeutics (20). The RSV and hMPV F proteins mediate membrane fusion between the virus and host cell membranes; however, the exact mechanism of membrane fusion for RSV and hMPV has yet to be determined. Similar to RSV, hMPV can mediate infection in the absence of the hMPV G protein in vitro, although such viruses are attenuated in vivo (21). The hMPV F protein has an RGD motif hypothesized to interact with α5β1 integrins (22), and heparan sulfate has also been shown to play a potential role in hMPV F protein-mediated attachment (23).
The hMPV F protein is a trimeric, class I viral fusion protein that is initially synthesized as a precursor monomer (F0) that homotrimerizes after proteolytic cleavage into a metastable prefusion conformation. Crystal structures of both conformations of the hMPV F protein have been determined by protein expression in CV-1 cells (24, 25). The hMPV prefusion and postfusion F proteins have been shown to elicit similar antibody responses (24), unlike the RSV F protein (26) and the related parainfluenza virus F proteins (27), where prefusion-elicited antibodies are more abundant and potently neutralizing. Importantly, the hMPV F protein is cleaved by different intracellular enzymes than RSV F (28), and in some strains, low pH is required to activate the fusion mechanism (28). hMPV can fuse with host cells at either the cell membrane or the endosomal membrane (29).
Neutralizing regions in the hMPV F protein have been determined via monoclonal antibody (mAb)-resistant mutant selection using mAbs derived from immunization of Armenian hamsters (18) and BALB/c mice (30). A human mAb, DS7, derived from a phage display library has been discovered (31) and was cocrystallized with a fragment of prefusion hMPV F (32). As RSV F and hMPV F proteins share approximately 30% sequence homology, it is not surprising that mAbs that neutralize both viruses have been isolated (33–36). mAb 54G10 binds near antigenic site IV on the RSV F protein and reduces lung titers of hMPV and RSV in vivo (34). The crystal structure of MPE8 (33) in complex with the RSV F protein has been determined and has a similar binding profile as mAb 25P13 (35). Another human mAb isolated is 17E10, which targets antigenic site IV with a binding pose facilitating cross-reactivity between RSV and hMPV F proteins (36). While several mouse and a few human mAbs that target the hMPV F protein have been isolated, the major antigenic sites on the hMPV F protein and the antibodies elicited in response to hMPV infection are unclear. To advance the field, four new neutralizing human mAbs to the hMPV F protein were isolated in an effort to inform the human antibody response to natural hMPV infection.
RESULTS
Analysis of recombinant hMPV F protein in HEK293F cells.
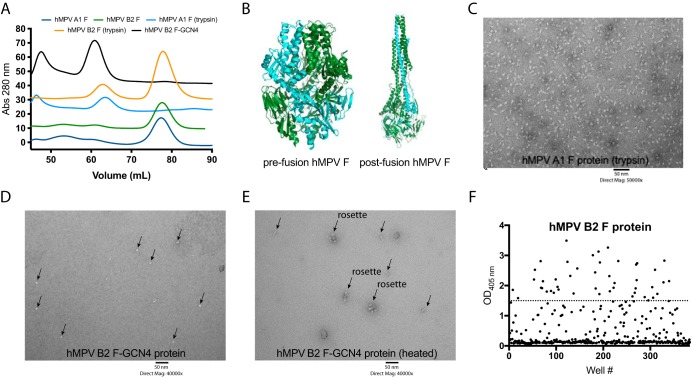
Similar to the RSV F protein, the hMPV F protein is translated as a single-chain protein (F0) that is cleaved into two fragments (F1 and F2), which are joined at disulfide bridges before transport to the cellular membrane. While the RSV F protein is cleaved at two positions by furin to release a small protein fragment, the hMPV F protein contains one cleavage site that is cleaved by a yet-to-be-determined intracellular protease (28). The hMPV F protein can be cleaved by trypsin in vitro, which has been utilized for virus growth and neutralization assays. Three constructs of recombinant hMPV F protein have been previously reported for use in crystallization studies (24, 25, 32). These include (i) hMPV B2 F (strain TN/99-419) linked to a GCN4 trimerization domain (hMPV B2 F-GCN4), where a partial fragment of monomeric hMPV B2 F-GCN4 protein in the prefusion conformation bound to the neutralizing mAb DS7 has been determined (32); (ii) hMPV A1 F (strain NL/1/00) linked to a foldon trimerization domain with a modified cleavage site (hMPV F furin ΔFP), which led to structural determination of hMPV F in the postfusion conformation (25); and (iii) hMPV A1 F (strain NL/1/00) linked to a foldon trimerization domain and containing a modified cleavage site and an A185P mutation in the α4-α5 hinge region (hMPV 115-BV), which facilitated determination of hMPV F in the prefusion conformation (24). hMPV B2 F-GCN4 was expressed in HEK293F cells and was reported to be trimeric and stable enough in the prefusion conformation for Fab-F protein complex formation visible by negative-stain electron microscopy (EM), size exclusion chromatography, and X-ray crystallography. Furthermore, heating of hMPV B2 F-GCN4 led to a transition from the prefusion to the postfusion conformation (32). hMPV F furin ΔFP and hMPV 115-BV were expressed in CV-1 cells using a vaccinia virus expression system. The hMPV F protein cleavage site in hMPV F furin ΔFP and hMPV 115-BV was replaced with the furin cleavage site of the RSV F protein. For these constructs, the addition of a trimerization domain facilitated protein trimerization, and coexpression with furin further enhanced the homogeneity of the protein in the postfusion conformation. In the studies presented here, we utilized several recombinant hMPV F proteins, described in Table 1, based on previously reported constructs. For antibody isolation, four subgroups of the hMPV F protein were expressed in HEK293F cells (hMPV A1, A2, B1, and B2 F), the constructs contained a trimerization domain, and the cleavage site was replaced by the furin cleavage site from the RSV F protein as previously described for the postfusion hMPV F protein (25). All four proteins were expressed well in HEK293F cells, at quantities of at least 1 mg protein/liter of culture volume. Although the proteins contained a trimerization domain and a furin cleavage site that is suitable for cleavage and trimerization of the RSV F protein in HEK293F cells (37), monomeric protein was observed after purification and analysis by size exclusion chromatography (Fig. 1A). This is in contrast to expression in CV-1 cells, where expression of the hMPV F protein with a trimerization domain in the absence of coexpressed furin facilitated protein trimerization (25). As the majority of antigenic sites likely reside on each monomer of the hMPV F protein, this protein was utilized for subsequent antibody isolation and binding studies. To obtain trimeric postfusion hMPV F protein, a previously described methodology was utilized whereby the protein is incubated with trypsin to induce cleavage and subsequent trimerization (25). Indeed, upon trypsin cleavage of hMPV A1 F and hMPV B2 F proteins, a shift in the size exclusion chromatography profile was observed for both proteins, indicating that the proteins had trimerized (Fig. 1A). Further analysis of hMPV A1 F (trypsin) protein by negative-stain electron microscopy revealed a homogeneous population resembling the postfusion conformation of the hMPV F protein (Fig. 1B and C). Recombinant expression of hMPV B2 F-GCN4 produced trimeric hMPV F (Fig. 1A), which resembled the prefusion conformation (Fig. 1B and D) by negative-stain EM analysis, as previously described (32). Upon heating the hMPV B2 F-GCN4 protein, a transition to the postfusion conformation was observed (Fig. 1B and E) based on negative-stain EM analysis as previously described (32). Several rosettes were observed in the heated sample, likely due to aggregation of postfusion hMPV F around the hydrophobic fusion peptide (Fig. 1E).
TABLE 1.
Properties of hMPV F recombinant protein constructs
| Construct | Subgroup | Strain | Size | Trimerization domain |
|---|---|---|---|---|
| hMPV A1 F | A1 | NL/1/00 | Monomer | Foldon |
| hMPV A2 F | A2 | NL/17/00 | Monomer | Foldon |
| hMPV B1 F | B1 | NL/1/99 | Monomer | Foldon |
| hMPV B2 F | B2 | NL/1/94 | Monomer | Foldon |
| hMPV A1 F (trypsin) | A1 | NL/1/00 | Trimer | Foldon |
| hMPV B2 F-GCN4 | B2 | TN/99-419 | Trimer | GCN4 |
FIG 1.
Characterization of recombinantly expressed hMPV F proteins and hMPV F-specific B cell frequency. (A) Size exclusion chromatogram of hMPV F proteins on a Superdex S200 prep-grade column. hMPV B2 F and hMPV A1 F proteins are monomeric based on elution volumes of 78 ml. Upon trypsin cleavage, hMPV A1 F and hMPV B2 F proteins form trimers as determined by a shift in the elution profile to 65 ml. The hMPV B2 F-GCN4 protein is trimeric without the addition of trypsin. The curves are colored by protein construct, as displayed in the key above the graph. Each curve is separated by 10 absorbance (Abs) units to facilitate easier display. (B) X-ray crystal structures of prefusion (PDB accession number 5WB0) and postfusion (PDB accession number 5L1X) hMPV F expressed in CV-1 cells. Structures were rendered in Mac PyMOL. One protomer in each structure is shown in forest green, while the remaining two protomers are displayed in cyan. (C) Negative-stain electron microscopy image of the hMPV A1 F protein after trypsin cleavage. The structure of the protein resembles the crystal structure of the postfusion hMPV F protein in panel B. (D and E) Negative-stain electron microscopy of hMPV B2 F-GCN4 before and after heating. The protein is predominantly in the prefusion conformation (D), while heating (E) causes a transition to the postfusion conformation. Several rosettes were observed upon heating the protein, likely as a result of protein aggregation around the exposed hydrophobic fusion peptide. Arrows indicate proteins on the grid surface. (F) ELISA binding data of bulk B cell supernatants binding to the hMPV B2 F protein from donor 5. Select wells with signals of >1.5 absorbance units were chosen for electrofusion for hybridoma generation. OD405 nm, optical density at 405 nm.
Isolation of human mAbs.
Previous reports have described that the majority of antibodies found in human serum bind both prefusion and postfusion hMPV F proteins (24). This is in contrast to those antibodies binding the RSV F protein, whereby the preponderance of antibodies in sera are prefusion specific (38). In order to expand the knowledge on the human antibody response to the hMPV F protein, four new human mAbs targeting the hMPV F protein were isolated. The majority of individuals are seropositive for hMPV by 5 years of age (39); therefore, healthy donors were recruited for blood draws and subsequent mAb isolation. A high frequency of hMPV F-specific B cells against hMPV B2 F protein was observed from a 19-year-old female (Fig. 1F), and cells from this donor were used in subsequent mAb isolation experiments. B cells from cultures producing antibodies reactive to the hMPV B2 F protein were used to generate stable hybridoma cell lines (40). Hybridoma populations were biologically cloned by single-cell flow cytometric sorting and expanded stepwise before being grown in serum-free medium for antibody purification. Four new mAbs were isolated, MPV196, MPV201, MPV314, and MPV364, and isotyping analysis was completed for each mAb. All four antibodies were of the IgG1 isotype. mAbs MPV201 and MPV314 contain kappa light chains, while mAbs MPV364 and MPV196 utilize lambda light chains.
Binding and neutralization characteristics of hMPV F-specific human mAbs.
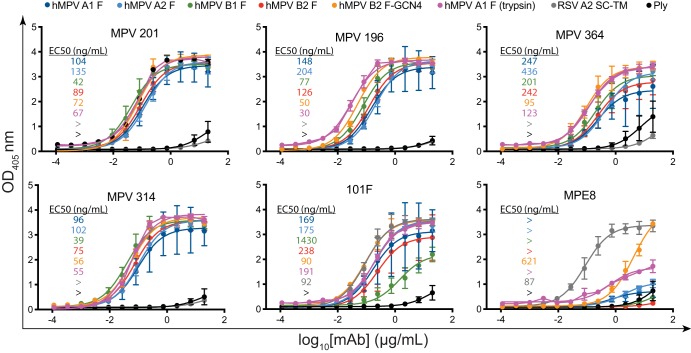
Previously isolated mouse mAbs to the hMPV F protein were shown to neutralize the virus and protect against viral replication in vivo; however, the majority of the mAbs did not neutralize across subgroups (30). In contrast, the limited number of human mAbs isolated that were shown to neutralize both RSV and hMPV show broad activity across hMPV subgroups (33–36). To determine the breadth of binding of the isolated human mAbs, binding was examined by an enzyme-linked immunosorbent assay (ELISA) for monomeric hMPV F protein from each of the four subgroups of hMPV F as well as the trimeric hMPV A1 F protein isolated following trypsin cleavage and trimeric hMPV B2 F-GCN4 (Fig. 2). Two control mAbs previously discovered to cross-react between RSV F and hMPV F proteins were included, the humanized mouse mAb 101F (41) and the human mAb MPE8 (33). The epitope for 101F is known to include a conserved GIIK motif on both the RSV F and hMPV F proteins (25) and is shared by the site IV cross-reactive mAb 17E10 (36). Notably, MPE8 is prefusion F protein specific and binds to an antigenic site across two protomers of RSV F (and likely hMPV F) (24, 33). As expected, MPE8 showed no binding to the monomeric hMPV F protein constructs or to trimeric hMPV A1 F (trypsin), which is in the postfusion conformation (Fig. 1B and C). MPE8 showed binding to RSV F SC-TM, as expected, yet showed limited binding to hMPV B2 F-GCN4, suggesting that the protein does not completely recapitulate the antigenic sites on prefusion hMPV F, although it resembles prefusion hMPV F by negative-stain EM. mAb 101F showed binding to all constructs of hMPV F and to RSV F single-chain triple mutants (SC-TMs), as the epitope for 101F at antigenic site IV completely resides on a single protomer of hMPV F and RSV F (36).
FIG 2.
ELISA binding curves of the hMPV F protein-specific mAbs. (A) ELISA binding curves of the isolated mAbs with recombinant hMPV F protein constructs. Prefusion RSV F protein (SC-TM) was utilized to determine if any mAb cross-reacts with RSV F. No cross-reactivity was observed except for mAbs 101F and MPE8. The pneumolysin toxin from Streptococcus pneumoniae was used as a negative binding control. Each point represents the average of data from three replicates, and error bars represent 95% confidence intervals. Data are representative of results from two independent experiments. EC50 values are inlaid in each graph and are color-coded according to the key at the top of the curves. Recombinant hMPV F protein constructs are hMPV A1 F (dark blue), hMPV A2 F (light blue), hMPV B1 F (green), hMPV B2 F (red), hMPV B2 F-GCN4 (orange), hMPV A1 F (trypsin) (purple), RSV A2 SC-TM (gray), and Ply (black).
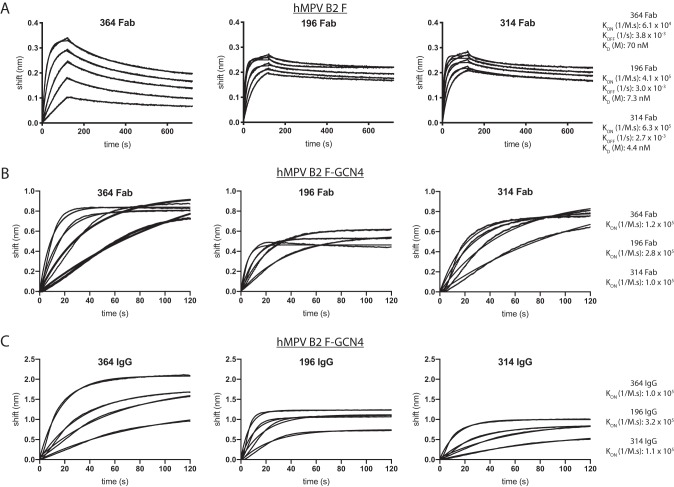
All four newly discovered mAbs bound to monomeric hMPV F proteins from each of the four hMPV F protein subgroups, and none of the four mAbs showed cross-reactivity with the prefusion RSV F protein A2 SC-TM (42). mAbs MPV196, MPV314, and MPV201 showed equivalent binding to monomeric hMPV F proteins, while MPV364 binding was lower based on the 50% effective concentration (EC50) determined by an ELISA, suggesting that the affinity for MPV364 is lower for monomeric hMPV F protein (Fig. 2A). The lower binding of MPV364 was abrogated when binding to the trimeric hMPV F constructs hMPV A1 F (trypsin) and hMPV B2 F-GCN4, suggesting that the full epitope for MPV364 incorporates two protomers of the hMPV F protein. To confirm these results, we performed affinity studies via biolayer interferometry incorporating monomeric hMPV B2 F and trimeric hMPV B2 F-GCN4 (Fig. 3). First, MPV364, MPV196, and MPV314 were cleaved to Fab fragments by papain digestion, and affinity studies were performed against hMPV B2 F (Fig. 3A). MPV364 has a 10-fold-higher KD (equilibrium dissociation constant) than MPV196 and MPV314 for hMPV B2 F, which is primarily due to a slower on rate of the MPV364 Fab for the hMPV F protein. Next, binding of the three Fabs to trimeric hMPV B2 F-GCN4 was assessed (Fig. 3B). Limited dissociation was observed when incorporating trimeric hMPV B2 F-GCN4, so a KD could not be determined. However, the differences between the Kon (second-order rate constant of the binding reaction) rates of MPV364 and those of MPV196 and MPV314 were much less pronounced, with MPV364 and MPV314 having nearly identical Kon rates, further supporting our hypothesis that the full epitope for MPV364 incorporates an antigenic site incorporating two protomers. The Kon rates of full-length IgG molecules of MPV364, MPV196, and MPV314 binding to trimeric hMPV B2 F-GCN4 were also assessed, and similar results were obtained as for binding of Fabs to hMPV B2 F-GCN4 (Fig. 3C).
FIG 3.
Binding affinity of hMPV F protein-specific mAbs. (A) Association and dissociation curves for MPV364, MPV196, and MPV314 Fab fragments using biolayer interferometry. The hMPV B2 F protein was loaded onto anti-penta-His biosensors. Curves include association for 120 s followed by dissociation for 600 s. Kon, Koff, and KD values are displayed below each curve and were calculated as the averages of values calculated from each binding curve. mAb concentrations for each experiment are 666 nM, 333 nM, 167 nM, 83 nM, and 42 nM. A reference curve containing buffer only was subtracted from each curve. (B) Association curves for MPV364, MPV196, and MPV314 Fab fragments binding to hMPV B2 F-GCN4. Association occurred over 120 s, and the Kon values are displayed on the right. (C) Identical experiment as in panel B except that IgGs of each mAb were tested for binding to hMPV B2 F-GCN4 instead of Fab fragments. In panels B and C, the mAb concentrations are 666 nM, 333 nM, 167 nM, and 83 nM. Kon, second-order rate constant of the binding reaction; Koff, first-order rate constant for dissociation of the protein-ligand complex; KD, equilibrium dissociation constant.
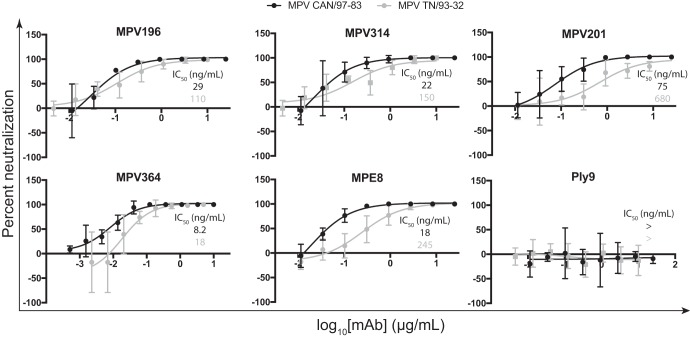
To determine if the isolated mAbs neutralize hMPV infection, plaque neutralization assays were performed using immunostaining for plaque visualization (Fig. 4). mAbs were tested for neutralization against hMPV CAN/97-83 and hMPV TN/93-32, which are representative of the A and B genotypes, respectively. All four mAbs neutralized both genotypes of hMPV and overall neutralized MPV CAN/87-83 slightly better. MPV364 was the most potent neutralizing mAb, with 50% inhibitory concentration (IC50) values of 8 and 18 ng/ml for MPV CAN/97-83 and MPV TN/93-32, respectively. The IC50 is at least 1 log lower than those of the other three mAbs for the MPV TN/93-32 strain, suggesting that MPV364 better inhibits the fusogenic activity of hMPV F or blocks hMPV F attachment more efficiently. These data led to experiments to probe the antigenic regions targeted by each mAb to determine if the epitope was important for neutralization potency. This has been previously shown for the RSV F protein, whereby prefusion F protein-specific mAbs D25 (26) and hRSV90 (43) exhibit greater neutralizing activity than mAbs that bind both prefusion and postfusion F protein conformations.
FIG 4.
Neutralization profiles of the hMPV F protein-specific mAbs. Shown are neutralization curves for the human mAbs and controls. IC50 values are inlaid on each graph and are color-coded according to the key at the top of each curve. Data represent the averages from three replicates, and error bars are 95% confidence intervals. Data are representative of results from two independent experiments.
Epitope determination of the newly isolated mAbs.
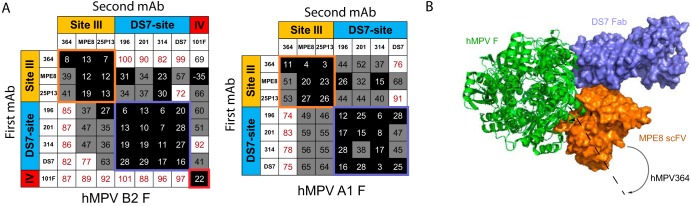
To determine the antigenic site targeted by each isolated human mAb, epitope binning was performed, where antibodies are tested in a pairwise combinatorial manner for those that compete for the same binding region, which are then grouped together into bins using the OctetRED384 system. Previously discovered human mAbs were utilized to identify the general antigenic regions on the hMPV F protein. The hMPV/RSV cross-reactive humanized mouse mAb 101F (41, 44) was used to identify antigenic site IV, and the cross-reactive human mAbs MPE8 (33) and 25P13 (35) were used to identify antigenic site III. The previously discovered mAb DS7 (32) was also incorporated, which was derived from a human phage display library. Each mAb was competed for binding against itself and other mAbs in the group. Anti-penta-His biosensors were loaded with the hMPV F protein, after which mAbs were loaded as either the first or second mAb in different experiments. Although MPE8 did not show substantial binding to the hMPV F protein by an ELISA, significant binding was observed via biolayer interferometry at a concentration of 100 μg/ml, suggesting that the MPE8 epitope is at least partially conserved in the postfusion conformation of the hMPV F protein. The mAbs were observed to fall into three groups (Fig. 5A). mAbs MPV201, MPV314, and MPV196 all competed primarily with mAb DS7 and partially with MPE8 and 25P13 at antigenic site III. Antigenic site III lies in close proximity to the DS7 antigenic site, and mAbs 25P13 and DS7 show partial competition. mAb MPV364 instead competes primarily with mAbs MPE8 and 25P13 at antigenic site III. While mAbs MPE8 and 25P13 partially compete with mAbs MPV196, MPV201, and MPV314, MPV364 shows no competition with these mAbs except one-directionally on the hMPV A1 F protein. The crystal structure of MPE8 in complex with the RSV F protein was recently determined (35). Based on a structural overlay of the prefusion RSV and hMPV F proteins, MPV364 likely binds in a slightly altered angle compared to MPE8, shifting away from the DS7 site (Fig. 5B). Both MPE8 and 25P13 are cross-reactive between hMPV and RSV F proteins; however, no such cross-reactivity was observed for MPV364. It is therefore surmised that the altered binding pose of MPV364 reduces the cross-reactivity with RSV F, instead binding at a nearby antigenic site, which is termed here antigenic site IIIa.
FIG 5.
Epitope binning analysis for mAbs binding hMPV F protein. (A) Epitope binning for mAbs binding hMPV B2 F and hMPV A1 F proteins. Data indicate the percent binding of the competing antibody in the presence of the primary antibody, compared with the competing antibody alone. Cells filled in black indicate full competition, in which ≤33% of the uncompeted signal was observed; cells in gray indicate intermediate competition, in which the signal was between 33% and 66%; and cells in white indicate noncompetition, where the signal was ≥66%. Antigenic sites are highlighted at the top and side based on competition binding with the control mAbs MPE8 and 25P13 (site III), DS7, and 101F (site IV). (B) Crystal structure of the RSV F protein in complex with MPE8 (PDB accession number 5U68) overlaid with the crystal structure of the hMPV F protein in complex with DS7 Fab (PDB accession number 4DAG). Based on the competition profile of MPV364, the mAb likely binds at an angle shifted away from DS7, since no competition was observed between DS7 and MPV364. This is in contrast to site III mAbs MPE8 and 25P13, both of which compete at least one-directionally with DS7.
Sequence determinants of the hMPV F-specific mAbs.
To identify the sequence determinants of the newly isolated mAbs, we performed hybridoma RNA isolation followed by reverse transcription-PCR (RT-PCR) of the variable regions. IMGT/V-Quest predictions for each mAb compared to previously discovered mAbs are shown in Table 2. All four of the mAbs utilize newly discovered hMPV F-specific V genes, and MPV201 and MPV314 are closely related due to their shared VH and VL gene usage. MPV196, MPV201, and MPV314 bind near DS7 yet utilize a unique V gene compared to DS7, which suggests that the DS7 epitope can stimulate B cell receptors from at least three V genes. MPV364 utilizes VH1-3, JH5, and DH2-8 genes for the heavy chain and VL3-21 and JL1 genes for the light chain. Both heavy and light chain V regions are heavily mutated, with percent identity to the respective VH genes being 91.67% and to VL being 90.32%, which is generally consistent with the hypothesis that a higher somatic mutation rate leads to higher neutralizing activity, as MPV364 exhibits the most potent neutralizing activity. These data suggest that MPV364 is the result of multiple hMPV infections in this particular donor. Previously discovered antigenic site III-targeting human mAbs isolated against RSV, which also cross-react with hMPV, utilize VH3-21, VH1-46, VH3-11, and VH1-69 (33, 35, 45). The sequence of MPV364 provides a new V gene in an antibody sequence that can recognize antigenic site III of the hMPV F protein and does not provide cross-reactivity with the RSV F protein.
TABLE 2.
Sequence determinants of the isolated human mAbs and previously discovered mAbsa
| mAb | VH (% mutation) | DH | JH | VL (% mutation) | JL | HC CDR3 sequence | LC CDR3 sequence |
|---|---|---|---|---|---|---|---|
| MPV364 | 1-3 (91.7) | J5 | D2-8 | V3-21 (90.3) | J1 | ARVDQYCIGGVCYGGKNWFDP | QVWDRDSDHPYV |
| MPV196 | 4-34 (96.5) | J4 | D2-21 | V3-1 (95.0) | J2 | ARGVRGGYNLWHFDV | QAWDGRTAVV |
| MPV201 | 3-30 (96.5) | J4 | D3-22 | V3-15 (95.3) | J1 | AKDQGRRYYYSSGYLDY | QQYNRWPPWT |
| MPV314 | 3-30 (96.2) | J1 | D3-10 | V3-15 (96.4) | J2 | AKDESRRYYYSSGIHSH | HQYNYWPPGT |
| MPE8 | 3-21 (94.8) | J4 | D5-24 | V1-40 (96.5) | J1 | ARARATGYSSITPYFDI | QSYDRNLSGV |
| 25P13 | 3-21 (90.6) | J5 | D1-26 | V1-40 (94.1) | J2 | ARDENTGISHYWFDP | QSYDRSLNWV |
| DS7 | 3-66 | J3 | D1-26 | V3-01 | J2/3 | VLSRASGMPDAFDI | QAWDSSTAV |
Analysis was performed using IMGT/V-Quest. The nucleotide sequence of DS7 is not publicly available, so no percent mutation could be calculated. HC, heavy chain; LC, light chain.
mAb MPV364 reduces viral replication hMPV in vivo.
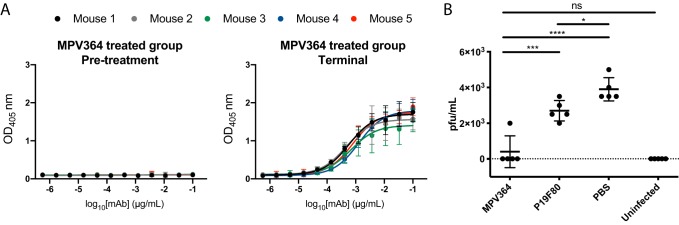
As MPV364 exhibits potent neutralizing activity in vitro, mAb efficacy was determined in vivo when administered before hMPV infection. Prophylactic administration of palivizumab is recommended for infants at high risk of RSV infection (46), and a similar therapeutic strategy could be utilized for hMPV. MPV364 is a fully human antibody and would likely not require extensive optimization to reduce immunogenicity in human patients. For these studies, BALB/c mice were utilized, as these animals were previously shown to be susceptible to hMPV replication (47–49). One day before hMPV infection, mice were treated with 10 mg/kg of body weight of MPV364, and three additional groups were included as controls: an uninfected group, a phosphate-buffered saline (PBS)-treated group, and a group treated with an IgG1 isotype control non-hMPV-specific mAb, P19F80. After treatment, mice were infected the next day and sacrificed on day 5. Protection from hMPV infection is likely mediated by the Fab region of MPV364, which would inhibit the attachment and/or fusogenic activity of the hMPV F protein. Therefore, we maintained the human Fc region of MPV364 in the mouse model, which would not stimulate mouse Fc receptor activity, instead of isotype switching MPV364 to the corresponding mouse Fc. Intramuscular delivery of MPV364 was tracked by an ELISA against MPV B2 F in each mouse, and as expected, a robust signal correlating to an average 50% endpoint serum titer of 1/1,318 was determined in terminal samples, indicating that intramuscular delivery of MPV364 was sufficient to allow transfer to the circulatory system (Fig. 6A). To determine if MPV364 exhibited efficacy in reducing viral replication in the lungs of infected mice, lungs were harvested and homogenized in PBS 5 days after viral infection. Viral titers were determined using an immunostaining plaque assay. A substantial reduction in lung viral titers was observed for the MPV364-treated group compared to the PBS-treated or negative-control antibody-treated groups (Fig. 6B). A minimal nonspecific decrease in lung viral titers was also observed for the control antibody P19F80. No substantial difference was observed between the MPV364-treated and the uninfected mice. Overall, these data indicate that administration of MPV364 before viral infection limits virus replication in the lungs of BALB/c mice.
FIG 6.
Prophylactic delivery of MPV364 in BALB/c mice to prevent hMPV replication. (A) Mouse serum collected before MPV364 treatment and after MPV364 treatment was tested for the presence of human anti-hMPV B2 F protein-specific mAbs by an ELISA. Data represent the averages from three replicates, and error bars are 95% confidence intervals. Data are representative of results from two independent experiments. Each curve represents one mouse and is colored according to the key. (B) Viral titers in the lung homogenates of BALB/c mice in each treatment group (n = 5 mice per group). Mice were treated with 10 mg/kg of MPV364 24 h before hMPV infection. ****, P < 0.0001; ***, P = 0.0001; *, P = 0.0348; ns, not significant. The dashed line indicates the limit of detection.
DISCUSSION
While hundreds of human mAbs to the RSV F protein have been isolated, and the antigenic sites have been well characterized, the major neutralizing and nonneutralizing epitopes on the hMPV F protein targeted by the human immune system have not been determined. The isolation of human mAbs to the RSV F protein has led to the design of new vaccine constructs currently being examined in preclinical and clinical trials. Research on the epitopes targeted by human mAbs on the hMPV F protein has lagged behind. In this report, four new human mAbs were isolated, which bind to the hMPV F protein and neutralize both genotypes of hMPV in vitro. The mAbs were mapped to two distinct antigenic sites, antigenic site III, targeted by MPV364, while the remaining mAbs competed with the previously discovered phage display-derived human mAb DS7. Previously discovered mAbs that neutralize hMPV via binding at antigenic site III have been identified by mAb isolation against the RSV F protein and were subsequently found to neutralize hMPV as well. MPV364 is an hMPV F protein-specific human mAb that binds near antigenic site III and exhibits potent neutralizing activity. Furthermore, MPV364 limits virus replication in a mouse model of infection when administered prophylactically. The antigenic site for MPV364 likely incorporates amino acid residues from across two protomers, similar to the site III-targeting mAb MPE8. However, MPV364 binds quite well overall to monomeric hMPV F, which suggests that the majority of the antigenic-site residues are on one protomer. Also supporting this idea is the fact that we isolated MPV364 using monomeric hMPV F. The ability of MPV364 to neutralize hMPV more effectively than the other three mAbs in vitro, despite weaker binding to monomeric hMPV F, further supports the idea that the MPV364 antigenic site incorporates residues across two protomers, as the majority of hMPV F on the surface of the virus is likely in the trimeric form. Binding to antigenic site III may also be more effective at mediating virus neutralization through prevention of hMPV F fusogenic activity; however, the isolation of additional site III mAbs and further experimentation will be required to address this idea.
Future experiments to crystallize MPV364 in complex with the hMPV F protein, coupled with escape mutant experiments, will further elucidate the mechanism of MPV364 binding and neutralization. Structural characterization of MPV364 would provide the conformational epitope required for elicitation of MPV364-like mAbs, which would inform the development of structure-based vaccines for hMPV. For example, designing an epitope-focused vaccine incorporating only the MPV364 epitope could elicit MPV364-like mAbs while bypassing elicitation of other mAbs with less efficacy. Additionally, experiments in higher species, such as nonhuman primates, will be critical to advancing MPV364 as a therapeutic to prevent hMPV infection.
The isolation of human mAbs that neutralize hMPV has great potential for use in humans. The humanized mouse mAb palivizumab is administered to infants at high risk of RSV infection (46). MPV364 and the additional mAbs described here are fully human and should be examined in additional preclinical models of infection. Both RSV and hMPV remain problematic viruses for vaccine design, and chemical inactivation of the viruses has been linked to the induction of Th2-skewed immune responses and, for RSV, sensitization to vaccine-induced enhanced disease upon RSV challenge (50, 51). These pathogens are a concern for the very young, the elderly, and the immunocompromised, so while an effective vaccine would certainly be a breakthrough for human health, human mAbs could serve as a means to control infection and disease in these populations. Currently, the generation of mAbs for clinical use remains costly; however, as new methodologies for gene delivery are refined, human mAbs for the treatment and prevention of infectious diseases may gain more widespread use.
MATERIALS AND METHODS
Blood draws and informed consent.
This study was approved by the University of Georgia Institutional Review Board as STUDY00005127. Healthy human donors were recruited to the University of Georgia Clinical and Translational Research Unit. After obtaining informed consent, 90 ml of blood was drawn by venipuncture into 9 heparin-coated tubes, and 10 ml of blood was collected into a serum separator tube. Peripheral blood mononuclear cells (PBMCs) were isolated from human donor blood samples using Ficoll-Histopaque density gradient centrifugation, and PBMCs were frozen in the liquid nitrogen vapor phase until further use.
Production and purification of recombinant hMPV F proteins.
Plasmids encoding cDNAs for hMPV A1, A2, B1, and B2 F proteins and the hMPV B2 F-GCN4 protein were synthesized (GenScript) and cloned into the pcDNA3.1+ vector. The plasmids were expanded by transformation in Escherichia coli DH5α cells using 100 μg/ml of ampicillin (Thermo Fisher Scientific) for selection. Plasmids were purified using either the ZymoPure II maxiprep kit (Zymo Research) or the E.Z.N.A. plasmid maxi kit (Omega BioTek), both according the manufacturers’ protocols. For each liter of protein expressed, 1 mg of plasmid DNA was mixed with 4 mg of 25,000-molecular-weight polyethylenimine (PEI; PolySciences Inc.) in Opti-MEM I cell culture medium (Thermo Fisher Scientific). After 30 min, the DNA-PEI mixture was added to Freestyle HEK293F cells (Thermo Fisher Scientific) at 1 × 106 cells/ml in Freestyle 293 expression medium (Thermo Fisher Scientific). After 4 to 6 days, the cultures were centrifuged to pellet the cells, and the supernatants were filtered through a 0.45-μm sterile filter. Recombinant proteins were purified directly from the filtered culture supernatants using HisTrap Excel columns (GE Healthcare Life Sciences). Each column was stored in 20% ethanol and washed with 5 column volumes (CV) of 20 mM Tris (pH 7.5)–500 mM NaCl before loading samples onto the column. After sample application, columns were washed with 10 CV of a solution containing 20 mM Tris (pH 7.5), 500 mM NaCl, and 20 mM imidazole. Proteins were eluted from the column with 5 CV of a solution containing 20 mM Tris (pH 7.5), 500 mM NaCl, and 250 mM imidazole. Proteins were concentrated and buffer exchanged using Amicon Ultra-15 centrifugal filter units with a 30-kDa cutoff (Millipore Sigma) and buffer exchanged into phosphate-buffered saline (PBS).
Production of trimeric postfusion hMPV F.
In order to generate homogeneous trimeric postfusion hMPV F, trypsin-tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) (Thermo Scientific) was dissolved in double-distilled water (ddH2O) at 2 mg/ml. hMPV A1 F and B2 F proteins were incubated with 5 TAME (p-toluene-sulfonyl-l-arginine methyl ester) units/mg of trypsin-TPCK (l-1-tosylamido-2-phenylethyl chloromethyl ketone) for 2 h at 37°C. Trimeric postfusion hMPV A1 and B2 F proteins were purified from the digestion reaction mixture by size exclusion chromatography on a Superdex S200, 16/600 column (GE Healthcare Life Sciences) in 50 mM Tris (pH 7.5)–100 mM NaCl. The trimer was identified by a shift in the elution profile from monomeric hMPV A1 and B2 F proteins. The fractions containing the trimers were concentrated using 30-kDa Spin-X UF concentrators (Corning) before subjecting them to negative-stain electron microscopy and binding ELISAs.
Human hybridoma generation.
For antibody generation experiments, 8 million previously frozen and irradiated NIH 3T3 cells modified to express human CD40L, human interleukin-21 (IL-21), and human BAFF (gift from Deepta Bhattacharya, Washington University) were mixed with 10 million PBMCs in 80 ml StemCell medium A (StemCell Technologies) containing 6.3 μg/ml of CpG (phosphorothioate-modified oligodeoxynucleotide ZOEZOEZZZZZOEEZOEZZZT; Invitrogen) and 1 μg/ml of cyclosporine (Sigma), and cells were plated in four 96-well plates at 200 μl per well in StemCell medium A. After 6 days, culture supernatants were screened by an ELISA for binding to recombinant hMPV B2 F protein, and cells from positive wells were electrofused with a nonsecreting myeloma cell line as previously described (40). Cells from each cuvette were resuspended in 20 ml StemCell medium A containing 1× HAT (hypoxanthine-aminopterin-thymidine; Sigma-Aldrich), 0.2× HT (hypoxanthine-thymidine; Corning), and 0.3 μg/ml ouabain (Thermo Fisher Scientific) and plated at 50 μl per well in a 384-well plate. After 7 days, cells were fed with 25 μl of StemCell medium A. Hybridomas were screened after 2 weeks for antibody production by an ELISA, and cells from wells with reactive supernatants were expanded to 48-well plates for 1 week in 0.5 ml of StemCell medium E (StemCell Technologies), before being screened again by an ELISA and then subjected to single-cell fluorescence-activated sorting. After cell sorting into 384-well plates containing StemCell medium A, hybridomas were screened by an ELISA before further expansion.
Human mAb and Fab production and purification.
Plasmids encoding cDNAs for the protein sequences of mAbs 101F, MPE8, 25P13, and DS7 were synthesized (GenScript), and heavy and light chain sequences were cloned into vectors encoding human IgG1 and lambda or kappa light chain constant regions, respectively. Large-scale DNA was isolated, and mAbs were obtained by transfection into Freestyle HEK293F cells as described above. For hybridoma-derived mAbs, hybridoma cell lines were expanded in StemCell medium A until 80% confluent in 75-cm2 flasks. For antibody production, cells from one 75-cm2 cell culture flask were collected with a cell scraper and expanded to four 225-cm2 cell culture flasks in serum-free medium (Hybridoma-SFM; Thermo Fisher Scientific). Recombinant cultures were stopped after 4 to 6 days, hybridoma cultures were stopped after 30 days, and culture supernatants were sterile filtered using 0.45-μm-pore-size filter devices. mAbs were purified directly from culture supernatants using HiTrap protein G columns (GE Healthcare Life Sciences) according to the manufacturer’s protocol. To obtain Fab fragments, papain digestion was performed using the Pierce Fab preparation kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Fab fragments were purified by removing IgG and Fc contaminants using a HiTrap MabSelectSure column according to the manufacturer’s protocol.
Negative-stain electron microscopy analysis.
All samples were purified by size exclusion chromatography on a Superdex S200, 16/600 column (GE Healthcare Life Sciences) in 50 mM Tris (pH 7.5)–100 mM NaCl before applying samples to grids. The hMPV B2 F-GCN4 protein was heated at 55°C for 20 min before application to grids to induce hMPV F transition from the prefusion to the postfusion conformation. Carbon-coated copper grids were overlaid with proteins at 100 μg/ml for 3 min. The grid was washed in water twice and then stained with 0.75% uranyl formate for 1 min. Negative-stain electron micrographs were acquired using a JEOL JEM1011 transmission electron microscope equipped with a high-contrast 2K-by-2K AMT midmount digital camera at a 50,000× magnification.
Enzyme-linked immunosorbent assay for binding to hMPV F proteins.
For recombinant protein capture ELISAs, 384-well plates (catalog number 781162; Greiner Bio-One) were treated with 2 μg/ml of antigen in PBS for 1 h at 37°C or overnight at 4°C. Following this, plates were washed once with water before blocking for 1 h with 2% milk supplemented with 2% goat serum in PBS with 0.05% Tween 20 (PBS-T). Primary mAbs or culture supernatants were applied to wells for 1 h following three washes with water. Plates were washed with water three times before applying 25 μl secondary antibody (goat anti-human IgG Fc; Meridian Life Science) at a dilution of 1:4,000 in blocking solution. After incubation for 1 h, the plates were washed five times with PBS-T, and 25 μl of a PNPP (p-nitrophenyl phosphate) solution (1 mg/ml PNPP in 1 M Tris base) was added to each well. The plates were incubated at room temperature for 1 h before reading the optical density at 405 nm on a BioTek plate reader. Binding assay data were analyzed in GraphPad Prism using a nonlinear regression curve fit and the log(agonist)-versus-response function to calculate the binding EC50 values.
Isotype determination for human mAbs.
For determination of mAb isotypes, 96-well Immulon HB 4× ELISA plates (Thermo Fisher Scientific) were coated with 2 μg/ml of each mAb in PBS. The plates were incubated at 37°C for 1 h or overnight at 4°C and then washed with 1× water. Plates were blocked with 2% nonfat milk block with 2% goat serum in PBS-T and then left to incubate for 1 h at room temperature. After incubation, the plates were washed three times with PBS-T. Isotype-specific antibodies obtained from Southern Biotech (goat anti-human kappa-alkaline phosphatase [AP] [catalog number 100244-340], goat anti-human lambda-AP [catalog number 100244-376], mouse anti-human IgG1 [Fc]-AP [catalog number 100245714], mouse anti-human IgG2 [Fc]-AP [catalog number 100245-734], mouse anti-human IgG3 [hinge]-AP [catalog number 100245-824], and mouse anti-human IgG4 [Fc]-AP [catalog number 100245-812]) were diluted 1:1,000 in blocking solution, and 50 μl of each solution was added to the respective wells. Plates were incubated for 1 h at room temperature and then washed five times with PBS-T. The PNPP substrate was prepared in a 1-mg/ml solution in 1 M Tris base, and 100 μl of this solution was added to each well. Plates were incubated for 1 h and read at 405 nm on a BioTek plate reader.
RT-PCR for hybridoma mAb variable gamma chain and variable light chain.
RNA was isolated from expanded hybridoma antibodies by using the Zymo Research quick RNA miniprep kit according to the manufacturer’s protocol. A Qiagen OneStep RT-PCR kit was used for cDNA synthesis and PCR amplification. For RT-PCRs, 50-μl reaction mixtures were designed with the following final concentrations: 1× Qiagen OneStep RT-PCR buffer, 400 μM deoxynucleoside triphosphate (dNTP) mix, 0.6 μM primer mix, 2 μl of Qiagen OneStep RT-PCR enzyme mix, 600 ng total of the template RNA, and RNase-free water. Three separate sets of primer mixes were used: gamma and kappa forward and reverse primers as previously described (52) and lambda chain primers as previously described (53). After mixing, the RT-PCR mixtures were placed in a thermocycler with the following program: 30 min at 50°C, 15 min at 95°C, and then a 3-step cycle with 30 repeats of denaturation for 30 s at 94°C, annealing for 30 s at 50°C, and extension for 1 min at 72°C, followed by 10 min of final extension at 72°C. Samples were analyzed by agarose gel electrophoresis and submitted directly for sequencing to Genewiz or cloned into the pCR2.1 vector using the Original TA cloning kit (Thermo Fisher Scientific) according to the manufacturer’s protocol before submitting the samples to Genewiz for sequencing. Sequences were analyzed using IMGT/V-Quest (54).
Growth of hMPV.
hMPV B2 strain TN/93-32 was obtained from BEI Resources (catalog number NR-22240), and hMPV A2 strain CAN/97-83 was a kind gift from Ralph Tripp. Viruses were grown in LLC-MK2-7.1 cells (ATCC CCL-7.1). Cells were grown to 80% confluence in 185-cm2 flasks in Opti-MEM I supplemented with 2% fetal bovine serum (FBS). For virus infection, cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS) and then coated with 6 ml of diluted virus. The flasks were rocked for 1 h at room temperature to allow adsorption of virus. Following this, 10 ml of Opti-MEM I supplemented with 5 μg/ml trypsin-EDTA and 100 μg/ml CaCl2 was added to the flask. Cells were incubated for 4 to 5 days before harvesting virus. For virus harvest, the medium was removed from the flask, and 5 ml of cold 25% (wt/vol) sterile-filtered sucrose was added to the flask. The flask was transferred to −80°C until the solution was frozen. The flask was then moved to thaw at room temperature, followed by another freeze-thaw cycle. Cell lysates were scraped down, and all cells and the sucrose solution were transferred to a sterile tube and centrifuged at 1,100 rpm for 5 min. The clarified supernatant containing hMPV was aliquoted and flash frozen for later use.
hMPV plaque neutralization experiments.
LLC-MK2-7.1 cells were maintained in Opti-MEM I (Thermo Fisher Scientific) supplemented with 2% fetal bovine serum and grown in 24-well plates at 37°C in a CO2 incubator. Two days prior to neutralization assays, 40,000 cells/well were seeded into 24-well plates. On the day of the experiment, serially diluted mAbs isolated from hybridoma supernatants were incubated 1:1 with a suspension of infectious hMPV B2 strain TN/93-32 or hMPV A2 strain CAN/97-83 for 1 h. Following this, cells were inoculated with 50 μl of the antibody-virus mixture for 1 h with rocking at room temperature. Cells were then overlaid with 1 ml of 0.75% methylcellulose dissolved in Opti-MEM I supplemented with 5 μg/ml trypsin-EDTA and 100 μg/ml CaCl2. Cells were incubated for 4 to 5 days, after which the cells were fixed with 10% neutral buffered formalin. The cell monolayers were blocked with 2% nonfat milk supplemented with 2% goat serum for 1 h. The plates were washed with water, 200 μl of mouse anti-hMPV N primary antibody (catalog number C01851M; Meridian Biosciences) diluted 1:1,000 in blocking solution was added to each well, and the plates were incubated for 1 h. The plates were then washed three times with water, after which 200 μl of goat anti-mouse IgG-horseradish peroxidase (HRP) secondary antibody (catalog number 5220-0286; SeraCare) diluted 1:1,000 in blocking solution was added to each well for 1 h. Plates were then washed five times with water, and 200 μl of TrueBlue peroxidase substrate (SeraCare) was added to each well. Plates were incubated until plaques were clearly visible. Plaques were counted by hand under a stereomicroscope and compared to a virus-only control, and the data were analyzed in GraphPad Prism using a nonlinear regression curve fit and the log(inhibitor)-versus-response function to calculate the IC50 values.
Experimental setup for biolayer interferometry.
After obtaining an initial baseline in running buffer (PBS, 0.5% bovine serum albumin [BSA], 0.05% Tween 20, 0.04% thimerosal), 10 μg/ml of His-tagged hMPV F protein was immobilized on anti-penta-His biosensor tips (FortéBio) for 120 s. The baseline signal was measured again for 60 s before biosensor tips were immersed into wells containing 100 μg/ml primary antibody for 300 s. Following this, biosensors were immersed into wells containing 100 μg/ml of a second mAb for 300 s. Percent binding of the second mAb in the presence of the first mAb was determined by comparing the maximal signal of the second mAb after the first mAb was added to the maximum signal of the second mAb alone. mAbs were considered noncompeting if maximum binding of the second mAb was ≥66% of its uncompeted binding. A level of between 33% and 66% of its uncompeted binding was considered intermediate competition, and ≤33% was considered competition. For affinity studies, hMPV B2 F or hMPV B2 F-GCN4 proteins were loaded as described above, and decreasing concentrations of Fabs or IgGs were analyzed for binding by association for 120 s and dissociation for 600 s. Octet data analysis software was used to analyze the data. Values for reference wells containing no antibody were subtracted from the data, and affinity values were calculating using the local and partial fit curves function. Binding curves were independently graphed in GraphPad Prism for data visualization.
Mouse experiments.
All animal experiments were approved by the University of Georgia Institutional Animal Care and Use Committee. To examine the efficacy of hMPV364, a BALB/c mice model of hMPV infection was utilized. One day prior to virus infection, mice were bled via tail nick for baseline serum collection. Mice were then prophylactically treated with 10 mg/kg of MPV364, 10 mg/kg of the isotype control antibody P19F80, or PBS, and one group was untreated. The next day, the MPV364, P19F80, and PBS groups were intranasally inoculated with 100 μl of 105 PFU of hMPV CAN/97-83. Five days following infection, mice were euthanized via 2,2,2-tribromoethanol (Avertin), followed by cervical dislocation, and serum and lungs were collected. Lungs were harvested, homogenized using a gentleMACS dissociator, flash frozen in liquid nitrogen, and stored at −80°C until use. Lung viral titers were determined by plaque immunostaining as previously described (49). ELISAs for mouse serum were conducted using hMPV B2 F, using the protocol described above. Data were analyzed in GraphPad Prism using the ordinary one-way analysis of variance (ANOVA) function to calculate P values.
ACKNOWLEDGMENTS
These studies were supported by the University of Georgia Office of the Vice President for Research, Department of Infectious Diseases, and Center for Vaccines and Immunology, and by National Center for Advancing Translational Sciences award number UL1TR002378. J.J.M. was supported by National Institutes of Health award number 1K01OD026569-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Georgia Electron Microscopy at the University of Georgia for assistance with negative-stain electron microscopy. We also thank the University of Georgia Clinical and Translational Research Unit for assistance with donor identification and blood draws.
REFERENCES
- 1.Schildgen V, van den Hoogen B, Fouchier R, Tripp RA, Alvarez R, Manoha C, Williams J, Schildgen O. 2011. Human metapneumovirus: lessons learned over the first decade. Clin Microbiol Rev 24:734–754. doi: 10.1128/CMR.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panda S, Mohakud NK, Pena L, Kumar S. 2014. Human metapneumovirus: review of an important respiratory pathogen. Int J Infect Dis 25:45–52. doi: 10.1016/j.ijid.2014.03.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falsey AR, Erdman D, Anderson LJ, Walsh EE. 2003. Human metapneumovirus infections in young and elderly adults. J Infect Dis 187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- 4.van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 188:1571–1577. doi: 10.1086/379200. [DOI] [PubMed] [Google Scholar]
- 5.Madhi SA, Ludewick H, Abed Y, Klugman KP, Boivin G. 2003. Human metapneumovirus-associated lower respiratory tract infections among hospitalized human immunodeficiency virus type 1 (HIV-1)-infected and HIV-1-uninfected African infants. Clin Infect Dis 37:1705–1710. doi: 10.1086/379771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haas LEM, Thijsen SFT, van Elden L, Heemstra KA. 2013. Human metapneumovirus in adults. Viruses 5:87–110. doi: 10.3390/v5010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larcher C, Geltner C, Fischer H, Nachbaur D, Müller LC, Huemer HP. 2005. Human metapneumovirus infection in lung transplant recipients: clinical presentation and epidemiology. J Heart Lung Transplant 24:1891–1901. doi: 10.1016/j.healun.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Cane PA, van den Hoogen BG, Chakrabarti S, Fegan CD, Osterhaus AD. 2003. Human metapneumovirus in a haematopoietic stem cell transplant recipient with fatal lower respiratory tract disease. Bone Marrow Transplant 31:309–310. doi: 10.1038/sj.bmt.1703849. [DOI] [PubMed] [Google Scholar]
- 9.Englund JA, Boeckh M, Kuypers J, Nichols WG, Hackman RC, Morrow RA, Fredricks DN, Corey L. 2006. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann Intern Med 144:344–349. doi: 10.7326/0003-4819-144-5-200603070-00010. [DOI] [PubMed] [Google Scholar]
- 10.Dokos C, Masjosthusmann K, Rellensmann G, Werner C, Schuler-Lüttmann S, Müller KM, Schiborr M, Ehlert K, Groll AH. 2013. Fatal human metapneumovirus infection following allogeneic hematopoietic stem cell transplantation. Transpl Infect Dis 15:E97–E101. doi: 10.1111/tid.12074. [DOI] [PubMed] [Google Scholar]
- 11.Shah DP, Shah PK, Azzi JM, El Chaer F, Chemaly RF. 2016. Human metapneumovirus infections in hematopoietic cell transplant recipients and hematologic malignancy patients: a systematic review. Cancer Lett 379:100–106. doi: 10.1016/j.canlet.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein MB, Yang H, DelBalso L, Carbonneau J, Frost E, Boivin G. 2010. Viral pathogens including human metapneumovirus are the primary cause of febrile respiratory illness in HIV‐infected adults receiving antiretroviral therapy. J Infect Dis 201:297–301. doi: 10.1086/649587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groome MJ, Moyes J, Cohen C, Walaza S, Tempia S, Pretorius M, Hellferscee O, Chhagan M, Haffejee S, Dawood H, Kahn K, Variava E, Cohen AL, von Gottberg A, Wolter N, Venter M, Madhi SA. 2015. Human metapneumovirus-associated severe acute respiratory illness hospitalisation in HIV-infected and HIV-uninfected South African children and adults. J Clin Virol 69:125–132. doi: 10.1016/j.jcv.2015.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan-o K, Ramirez R, Macdonald MI, Rolph M, Rudd PA, Spann KM, Mahalingam S, Bardin PG, Thomas BJ. 2017. Human metapneumovirus infection in chronic obstructive pulmonary disease: impact of glucocorticosteroids and interferon. J Infect Dis 215:1536–1545. doi: 10.1093/infdis/jix167. [DOI] [PubMed] [Google Scholar]
- 15.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peña SA, Davis SS, Lu X, Sakthivel SKK, Peret TCT, Rose EB, Smelser C, Schneider E, Stone ND, Watson J. 2019. Severe respiratory illness associated with human metapneumovirus in nursing home, New Mexico, USA. Emerg Infect Dis 25:383–384. doi: 10.3201/eid2502.181298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shafagati N, Williams J. 2018. Human metapneumovirus—what we know now. F1000Res 7:135. doi: 10.12688/f1000research.12625.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulbrandt ND, Ji H, Patel NK, Barnes AS, Wilson S, Kiener PA, Suzich JA, McCarthy MP. 2008. Identification of antibody neutralization epitopes on the fusion protein of human metapneumovirus. J Gen Virol 89:3113–3118. doi: 10.1099/vir.0.2008/005199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tripp RA, Power UF, Openshaw PJM, Kauvar LM. 2018. Respiratory syncytial virus: targeting the G protein provides a new approach for an old problem. J Virol 92:e01302-17. doi: 10.1128/JVI.01302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo-Neto E, De Swart RL, Osterhaus ADME, Fouchier RAM. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis 10:658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biacchesi S, Skiadopoulos MH, Yang L, Lamirande EW, Tran KC, Murphy BR, Collins PL, Buchholz UJ. 2004. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol 78:12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox RG, Livesay SB, Johnson M, Ohi MD, Williams JV. 2012. The human metapneumovirus fusion protein mediates entry via an interaction with RGD-binding integrins. J Virol 86:12148–12160. doi: 10.1128/JVI.01133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang A, Masante C, Buchholz UJ, Dutch RE. 2012. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol 86:3230–3243. doi: 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battles MB, Más V, Olmedillas E, Cano O, Vázquez M, Rodríguez L, Melero JA, McLellan JS. 2017. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun 8:1528. doi: 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Más V, Rodriguez L, Olmedillas E, Cano O, Palomo C, Terrón MC, Luque D, Melero JA, McLellan JS. 2016. Engineering, structure and immunogenicity of the human metapneumovirus F protein in the postfusion conformation. PLoS Pathog 12:e1005859. doi: 10.1371/journal.ppat.1005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart-Jones GBE, Chuang G-Y, Xu K, Zhou T, Acharya P, Tsybovsky Y, Ou L, Zhang B, Fernandez-Rodriguez B, Gilardi V, Silacci-Fregni C, Beltramello M, Baxa U, Druz A, Kong W-P, Thomas PV, Yang Y, Foulds KE, Todd J-P, Wei H, Salazar AM, Scorpio DG, Carragher B, Potter CS, Corti D, Mascola JR, Lanzavecchia A, Kwong PD. 2018. Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1–4. Proc Natl Acad Sci U S A 115:12265–12270. doi: 10.1073/pnas.1811980115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J Virol 80:10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox RG, Mainou BA, Johnson M, Hastings AK, Schuster JE, Dermody TS, Williams JV. 2015. Human metapneumovirus is capable of entering cells by fusion with endosomal membranes. PLoS Pathog 11:e1005303. doi: 10.1371/journal.ppat.1005303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulbrandt ND, Ji H, Patel NK, Riggs JM, Brewah YA, Ready S, Donacki NE, Folliot K, Barnes AS, Senthil K, Wilson S, Chen M, Clarke L, MacPhail M, Li J, Woods RM, Coelingh K, Reed JL, McCarthy MP, Pfarr DS, Osterhaus ADME, Fouchier RAM, Kiener PA, Suzich JA. 2006. Isolation and characterization of monoclonal antibodies which neutralize human metapneumovirus in vitro and in vivo. J Virol 80:7799–7806. doi: 10.1128/JVI.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JV, Chen Z, Cseke G, Wright DW, Keefer CJ, Tollefson SJ, Hessell A, Podsiad A, Shepherd BE, Sanna PP, Burton DR, Crowe JE, Williamson RA. 2007. A recombinant human monoclonal antibody to human metapneumovirus fusion protein that neutralizes virus in vitro and is effective therapeutically in vivo. J Virol 81:8315–8324. doi: 10.1128/JVI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen X, Krause JC, Leser GP, Cox RG, Lamb RA, Williams JV, Crowe JE, Jardetzky TS. 2012. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat Struct Mol Biol 19:461–463. doi: 10.1038/nsmb.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, Piralla A, Percivalle E, Sallusto F, Baldanti F, Lanzavecchia A. 2013. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 34.Schuster JE, Cox RG, Hastings AK, Boyd KL, Wadia J, Chen Z, Burton DR, Williamson RA, Williams JV. 2015. A broadly neutralizing human monoclonal antibody exhibits in vivo efficacy against both human metapneumovirus and respiratory syncytial virus. J Infect Dis 211:216–225. doi: 10.1093/infdis/jiu307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE, Jardetzky TS. 2017. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol 2:16272. doi: 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mousa JJ, Binshtein E, Human S, Fong RH, Alvarado G, Doranz BJ, Moore ML, Ohi MD, Crowe JE. 2018. Human antibody recognition of antigenic site IV on pneumovirus fusion proteins. PLoS Pathog 14:e1006837. doi: 10.1371/journal.ppat.1006837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang G, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GBE, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards KM, Zhu Y, Griffin MR, Weinberg GA, Hall CB, Szilagyi PG, Staat MA, Iwane M, Prill MM, Williams JV. 2013. Burden of human metapneumovirus infection in young children. N Engl J Med 368:633–643. doi: 10.1056/NEJMoa1204630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mousa JJ, Sauer MF, Sevy AM, Finn JA, Bates JT, Alvarado G, King HG, Loerinc LB, Fong RH, Doranz BJ, Correia B, Kalyuzhniy O, Wen X, Jardetzky TS, Schief WR, Ohi MD, Meiler J, Crowe JE. 2016. Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein. Proc Natl Acad Sci U S A 113:E6849–E6858. doi: 10.1073/pnas.1609449113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S-J, Schmidt A, Beil EJ, Day ND, Branigan PJ, Liu C, Gutshall LL, Palomo C, Furze J, Taylor G, Melero JA, Tsui P, Del Vecchio AM, Kruszynski M. 2007. Characterization of the epitope for anti-human respiratory syncytial virus F protein monoclonal antibody 101F using synthetic peptides and genetic approaches. J Gen Virol 88:2719–2723. doi: 10.1099/vir.0.82753-0. [DOI] [PubMed] [Google Scholar]
- 42.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJM, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JPM. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mousa JJ, Kose N, Matta P, Gilchuk P, Crowe JE. 2017. A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol 2:16271. doi: 10.1038/nmicrobiol.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLellan JS, Chen M, Chang J-S, Yang Y, Kim A, Graham BS, Kwong PD. 2010. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol 84:12236–12244. doi: 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilman MSA, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, McLellan JS, Walker LM. 2016. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol 1:eaaj1879. doi: 10.1126/sciimmunol.aaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.IMpact-RSV Study Group. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:531–537. doi: 10.1542/peds.102.3.531. [DOI] [PubMed] [Google Scholar]
- 47.Alvarez R, Harrod KS, Shieh WJ, Zaki S, Tripp RA. 2004. Human metapneumovirus persists in BALB/c mice despite the presence of neutralizing antibodies. J Virol 78:14003–14011. doi: 10.1128/JVI.78.24.14003-14011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Haas DL, Poore S, Isakovic S, Gahan M, Mahalingam S, Fu ZF, Tripp RA. 2009. Human metapneumovirus establishes persistent infection in the lungs of mice and is reactivated by glucocorticoid treatment. J Virol 83:6837–6848. doi: 10.1128/JVI.00379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez R, Tripp RA. 2005. The immune response to human metapneumovirus is associated with aberrant immunity and impaired virus clearance in BALB/c mice. J Virol 79:5971–5978. doi: 10.1128/JVI.79.10.5971-5978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Swart RL, van den Hoogen BG, Kuiken T, Herfst S, van Amerongen G, Yüksel S, Sprong L, Osterhaus ADME. 2007. Immunization of macaques with formalin-inactivated human metapneumovirus induces hypersensitivity to hMPV infection. Vaccine 25:8518–8528. doi: 10.1016/j.vaccine.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 51.Murphy BR, Walsh EE. 1988. Formalin-inactivated respiratory syncytial virus vaccine induces antibodies to the fusion glycoprotein that are deficient in fusion-inhibiting activity. J Clin Microbiol 26:1595–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thornburg NJ, Zhang H, Bangaru S, Sapparapu G, Kose N, Lampley RM, Bombardi RG, Yu Y, Graham S, Branchizio A, Yoder SM, Rock MT, Creech CB, Edwards KM, Lee D, Li S, Wilson IA, García-Sastre A, Albrecht RA, Crowe JE. 2016. H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J Clin Invest 126:1482–1494. doi: 10.1172/JCI85317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, Wilson PC. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]