Abstract
The Hedgehog (HH) signalling pathway is a cell-cell communication system that controls the patterning of multiple tissues during embryogenesis in metazoans. In adults, HH signals regulate tissue stem cells and regenerative responses. Abnormal signalling can cause birth defects and cancer. The HH signal is received on target cells by Patched (PTCH1), the receptor for HH ligands, and then transmitted across the plasma membrane by Smoothened (SMO). Recent structural and biochemical studies have pointed to a sterol lipid, likely cholesterol itself, as the elusive second messenger that communicates the HH signal between PTCH1 and SMO, thus linking ligand reception to transmembrane signalling.
Introduction
The three HH ligands in amniotes, Sonic, Indian and Desert Hedgehog (SHH, IHH and DHH), function as classical morphogens: they guide the patterning of tissues, such as the limb or spinal cord, by forming spatial and temporal gradients during embryogenesis [1]. Central to their patterning activity is their ability to direct the fate of target cells in a concentration- or exposure-dependent manner. The biogenesis of HH ligands involves the autocatalytic, intein-like cleavage of a ~45 kDa precursor protein into a N-terminal fragment (e.g. ShhN) and the attachment of palmitoyl and cholesteryl moieties to the N- and C-termini, respectively (hereafter pShhNc for palmitoylated and cholesteroylated ShhN) [2–4]. HH ligands initiate signalling in target cells by binding to their receptor Patched1 (PTCH1) [5,6], a multi-pass transmembrane (TM) protein with homology to the resistance-nodulation-division (RND) superfamily of membrane transporters and the cholesterol transporter Niemann-Pick C1 (NPC1) [7](Fig. 1a and b). The interaction with HH ligands inhibits PTCH1, rendering it unable to suppress transmembrane (TM) signalling by Smoothened (SMO), a Class F G protein-coupled receptor (GPCR) most closely related to the Frizzled family of receptors for WNT ligands. Activated SMO then transmits the HH signal across the plasma membrane, eventually leading to the activation of glioma-associated oncogene (GLI) transcription factors and hence HH target gene transcription [8,9].
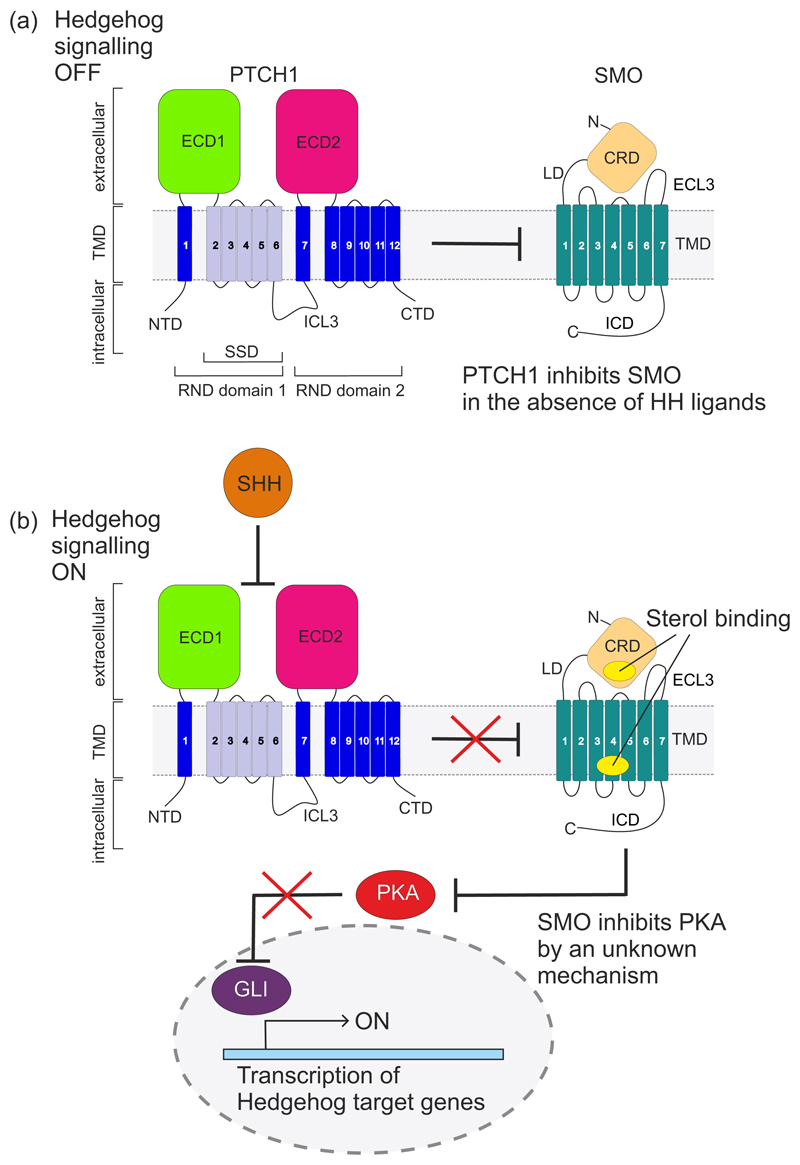
Figure 1. Transmission of the Hedgehog (HH) signal across the plasma membrane.
(a) The receptor for HH ligands, PTCH1 (a 12-pass TM protein) inhibits SMO (a 7-pass TM protein) and thereby prevents it from initiating signalling in the cytoplasm of target cells. The domain architecture of both TM proteins is highlighted. PTCH1 is composed of two tandem RND (resistance-nodulation-division) domains: ECD1, extracellular domain 1; ECD2, extracellular domain 2; NTD, N-terminal domain; CTD, C-terminal domain; ICL3, intracellular loop 3; SSD, sterol-sensing domain (light blue). SMO is composed of an extracellular region which is made up of a cysteine rich domain (CRD) and linker domain (LD) followed by a 7-pass transmembrane bundle characteristic of GPCRs and then a unique intracellular domain (ICD). (b) HH ligands (such as Sonic Hedgehog (SHH)) bind and inhibit PTCH1, allowing SMO activation, likely by sterol ligands. Activated SMO overcomes the negative influence of PKA on the GLI transcription factors, which control gene expression.
A major unanswered question in HH signalling is the mechanism by which ligand reception by PTCH1 is coupled to transmembrane signalling by SMO. The regulation of SMO by PTCH1 is thought to involve a small molecule intermediate for three reasons: the homology between PTCH1 and RND membrane transporters, the lack of a physical interaction between PTCH1 and SMO and the observation that one molecule of PTCH1 can inhibit multiple molecules of SMO [10–12]. More recent work has led to the proposal that this small molecule is a sterol lipid, likely cholesterol itself [13–16]. This review will discuss our current understanding of this key step in HH signal transduction in the light of the recently obtained structural information on PTCH1 and SMO.
PTCH1 recognises two distinct binding sites on ShhN
The transmembrane domain (TMD) of PTCH1 is composed of 12 TM helices. Two large extracellular domains (ECDs) are anchored to the TMD, one between TM1 and TM2 (ECD1) and the other between TM7 and TM8 (ECD2) (Fig. 1). The TMD consists of two RND domains (TM1-6 and TM7-12), which likely originated from an internal gene duplication during evolution. TMs 2-6 constitute a sterol-sensing domain (SSD), a motif found in other cholesterol transporting (NPC1) and cholesterol-sensing (SREBP cleavage-activating protein; SCAP) proteins [17,18] (Fig. 1a). Several structures of PTCH1 alone and PTCH1 complexed to SHH have been recently determined by cryo-electron microscopy (cryo-EM) at a resolution between 3.5-3.9 Å [19–24] (Fig. 2a). The TMD consists of a compact α-helical bundle with the N- and C-termini of each ECD connected to the TMD by a flexible linker and a neck helix, respectively. Each ECD is composed of two distinct domains: the region closest to the TMD contains a α+β sandwich fold, which is conserved in evolution, and the region distal to the TMD contains several α-helices and lengthy loops (hence the “helical domain”) that bind to SHH [19] (Fig. 2a). This helical domain of the ECD is distinct from analogous domains of PTCH1 homologues and likely evolved in order to carry out HH-specific functions.
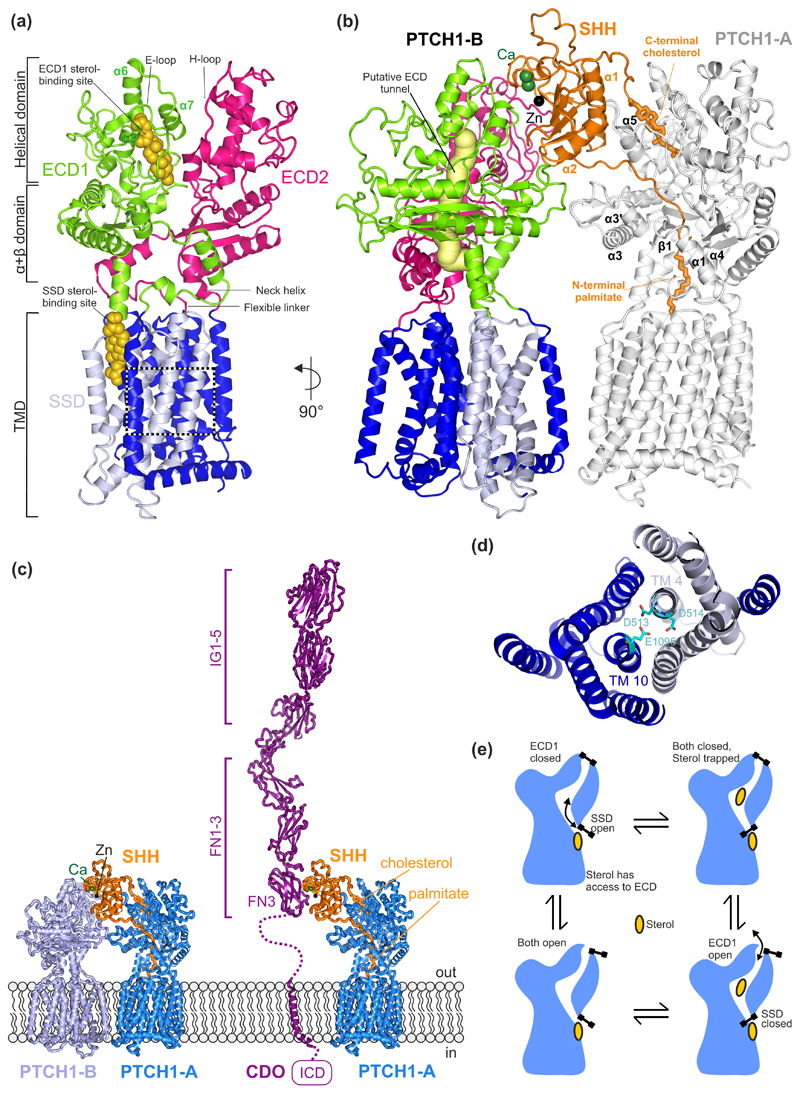
Figure 2. Structures of PTCH1 and their functional implications.
(a) The overall structural arrangement of monomeric PTCH1 (PDB 6DMB [19]). Each ECD is connected to the TMD through a flexible linker at the N-terminus and a neck helix at the C-terminus. Each ECD contains an α+β domain, located immediately above the TM, followed by a helical domain consisting of α-helices and lengthy loops. The E loop on ECD1 and the H loop on ECD2 contain residues important for SHH binding. Two sterol-like molecules have been identified in the structures, one located in ECD1 and another one in the SSD at the level of the outer leaflet of the membrane. (b) Cryo-EM structure of the 2:1 PTCH1:SHH complex (PDB 6E1H [21]) containing two PTCH1 molecules labelled PTCH1-A and PTCH1-B. A putative tunnel (yellow) through PTCH1 is proposed to be the conduit for sterol flow from the ECD1 to the SSD. The N-terminal palmitoyl appendage of SHH occludes this tunnel and the C-terminal cholesterol appendage occludes the sterol binding site in ECD1, presumably blocking transport. These lipidic appendages, together with α1 and α2 of SHH, form the major SHH-PTCH1-A binding interface. SHH interacts with PTCH1-B through an interface organized by the Ca2+-binding site in SHH. (c) Structural model of the CDO:SHH:PTCH1 co-receptor complex. CDO occupies a similar position as PTCH-B. The model of CDO:SHH was generated using the HHPRED [63] webserver as well as the crystal strcture structure of the CDO-FN3:SHH complex (PDB ID. 3D1M [25]). Ig: immunoglobulin-like domain, FN: fibronectin type III-like domain, ICD: intracellular domain. (d) Close-up of a charged triad, formed by PTCH1 residues D513, D514 (TM4) and E1095 (TM10) that may facilitate cation flow down a concentration gradient to power sterol transport. Mutations in this region have been shown to abolish the function of PTCH1. (e) Ordered conformational cycling of PTCH1, driven by cation flow, may enforce the directionality of sterol movement through PTCH1. An “alternating gate” mechanism is proposed where conformational changes (stylized as gates) either lead to the opening of the gate in the membrane, near the SSD site, or the mouth of the ECD1. Sterol flow could be in either direction, from the SSD site to the ECD1 (as shown) or vice versa.
SHH interacts with the PTCH1 ECD using two distinct binding interfaces [19,20]. An un-lipidated form of ShhN (ShhN24-197) uses its conserved calcium and zinc binding sites to interact with the helical domain of PTCH1 (hereafter the “protein-protein interface”) (Fig. 2b). Contacts are primarily formed by polar and charged residues in the E loop and α3 of ECD1, the H loop of ECD2 and a patch of Arg and Lys residues located near the centre of the Ca2+-binding site in SHH [19]. Interestingly, the surface of SHH that interacts with the ECD of PTCH1 overlaps significantly with the surface that binds to heparin-containing glycosaminoglycans as well as other SHH receptors, including cell adhesion molecule-related/down-regulated by oncogenes (CDO) and HH-interacting protein (HHIP) [25–30]. In contrast, a structure of PTCH1 in complex with dually-lipidated pShhNc shows that the first 15 amino acid residues of the palmitoylated SHH N-terminus insert in between ECD1 and ECD2 of PTCH1 (hereafter the “lipid-based interface”) (Fig. 2b) [20]. To accommodate the palmitoyl moiety, several regions within ECD1 and ECD2 undergo small structural shifts: ECD1 α3 and α3’ move slightly towards the TMD, whereas the movement of ECD1 α1 and β1 allow the palmitate to insert into the space between ECD1 α1 and ECD2 α4. This interface is stabilised by an extensive network of van der Waals interactions mediated by the palmitoyl moiety [20]. Unlike the protein-protein interface, this arrangement between PTCH1 and SHH would allow simultaneous binding of SHH receptors, a prediction confirmed experimentally by pull-down assays with an anti-SHH antibody [20]. The potential formation of a HH co-receptor signalling complex such as CDO:SHH:PTCH1 (Fig. 2c) may increase the local HH concentration at the surface of the plasma membrane and facilitate inactivation of PTCH1 function by the lipid-based interface of SHH. On the other hand, HHIP, a secreted HH signalling agonist with low nanomolar affinity for SHH might be able to sequester extracellular HH ligands away from the membrane surface, making it unavailable for PTCH1 binding. In any case, it seems that the interaction of SHH with the known co-receptors HH receptors (other than PTCH1) is independent of the lipidation state of SHH. The structural observations of the PTCH1-SHH interaction have also been validated by mutational studies. Disruption of either the protein-protein interface or the lipid-based interface of PTCH1-SHH reduces binding or responsiveness to SHH [19,20]. In addition, the palmitoyl moiety of SHH has been shown to be critical for PTCH1 inactivation in cellular signalling assays [2,31,32].
Which of these two interfaces between SHH and PTCH1 is most relevant for signalling? This question was partially resolved by a third structure of an asymmetric 2:1 PTCH1:SHH assembly [21] (Fig. 2b). Two PTCH1 molecules (PTCH1-A, PTCH1-B) simultaneously engage one pShhNc at both its metal-ion and lipid-based interfaces. The majority of interactions between PTCH1-A and the palmitoylated N-terminus of SHH are formed by ECD1, with minor support from ECD2. In comparison to the corresponding 1:1 PTCH1:pShhNc complex there is a slight shift in the lipid-based interface, which may be induced by the formation of a protein-protein interface with PTCH1-B [20,21]. The binding interface between SHH and PTCH1-B is consistent with the previously reported structure of PTCH1 and ShhN24-197 [19,21]. The 2:1 structure further revealed that the TMDs of the PTCH1 molecules are rotated 150° relative to each other. Mutations in either of the binding interfaces reduced HH signalling activity, suggesting that the formation of a 2:1 complex is essential for maximum signalling output in vertebrates [21]. These studies raise the interesting question of whether invertebrate HH signalling utilises such a 2:1 complex. While functional studies have reached different conclusions about the roles of the co-receptor Interference HH (IHOG) and its paralogue Brother of IHOG (BOI) in the binding and sequestering of HH ligands, a heterotrimeric complex between HH, PTCH and IHOG/BOI has been proposed to initiate signalling in target cells [33,34].
There are several limitations of the presented structures that are worth highlighting. In all structures, most information about the intracellular regions of PTCH1 (the NTD, CTD and ICL3, Fig. 1a) is missing. Except for the helices directly preceding TM1 and TM7, the structural arrangement of the NTD and ICL3 could not be determined, possibly due to intrinsic flexibility of the domains. The PTCH1 CTD was truncated in all of the constructs in order to reach expression levels suitable for structural studies. While it has been reported that the SHH N-terminal palmitoyl moiety inserts into a cavity between PTCH1 ECD1 and ECD2, the C-terminal cholesterol-modification on pShhNc could initially not be resolved [20,21]. However, the close proximity of the C-terminus of pShhNc to a hydrophobic pocket formed by α3, α6,α7 and the E loop of PTCH1 ECD1 suggests that the cholesterol moiety may interact with this domain, a possibility that is supported by cryo-EM density that extends from the C-terminus of pShhNc into the cavity bounded by these helices [24] (Fig. 2b).
Structural evidence that PTCH1 is a sterol transporter
Putative sterol-binding sites were identified in all reported structures of PTCH1, located in ECD1, between ECD1 and ECD2, and at two locations in the TMD (in the SSD and in the symmetry-related segment composed of TM helices 8-12) [19–24] (Fig. 2). A hydrophobic conduit has been proposed to run through the ECD, which connects the ECD1 site to two openings: one above the SSD and one above the symmetry-related TM8-12 segment [19,21–23](Fig. 2b, left molecule). These openings may function as gates for sterol entry into the tunnel (or exit out of the tunnel). Insertion of the pShhNc palmitoyl moiety into PTCH1-A blocks this proposed hydrophobic tunnel (Fig. 2b) and mutations predicted to obstruct this hydrophobic tunnel reduce PTCH1 activity [21,22]. The importance of the SSD sterol-binding site in PTCH1 is supported by data from engineered mutations, as well as from mutations found in HH-driven cancers. The SSD sterol in TM6 is in close proximity to residue D599, which correspond to residues in Drosophila PTC (D584) and yeast SCAP (D443) important for SMO repression and cholesterol sensitivity, respectively [35,36]. Other known mutations, such as P504L, a mutation found in Gorlin’s syndrome, also map closely to sterol-binding sites seen in the structures [19].
The structures of PTCH1 also confirm its similarity to the RND superfamily of transporters, which use the energy provided by an ion gradient to pump small molecules. Firstly, the 2-fold pseudo-symmetry between TM1-6 and TM7-12 is typical for RND transporters (Fig. 2b) [7,19–23]. Second, a triad of acidic residues is present in PTCH1 TM4 and TM10 (Fig. 2d), formed by human PTCH1 residues D513, D514 and E1095. In RND-family proteins, such as the multidrug efflux pump AcrB and the hopanoid biosynthesis transporter HpnN, the protonation/deprotonation of these acidic residues is thought to be coupled to conformational changes, thereby coupling proton flow to substrate transport [37]. In animal cells, flow of sodium ions rather than protons may drive the PTCH1 conformational cycle [38]. As in RND transporters, mutations in PTCH1 that abolish these charges also attenuate the ability of PTCH1 to inhibit SMO [12,19,22]. Such a transport mechanism fuelled by cation flow down a gradient would enable PTCH1 to transport sterols against a concentration gradient.
Taken together, the structural data are consistent with an “alternating gate” mechanism for sterol transport by PTCH1 (Fig. 2e). The two gates are located at the mouth of ECD1 and the SSD (or pseudo-symmetrically-related TM8-12) and connected by a conduit through the ECD. PTCH1 cycles between distinct conformational states in which each of these gates are open or closed. In one conformation, the SSD gate is open and the ECD1 gate is closed, allowing a cholesterol (and/or another sterol) to move from the outer leaflet of the plasma membrane into the interior of the ECD. The SSD gate then closes, leading to a second conformation where cholesterol is trapped in the interior of the ECD. In a third conformational state, the ECD1 gate opens, allowing the sterol to escape from the apex of PTCH1 to a protein or membrane acceptor. Ion transport imparts order to the transitions between these conformational states, ensuring directional transport of the sterol. Of course, the direction of sterol transport could just as likely be the opposite of what is described above - entering at the ECD1 and exiting at the SSD. HH ligands interrupt this conformational cycle by binding to one state, inserting their lipidic appendages to block the conduit for transport. This idea is supported by the observation that mutations that inactivate PTCH1, presumably by blocking conformational cycling, can also reduce the affinity for SHH [19,32].
Sterol lipids can bind and regulate SMO
A remarkable hypothesis published nearly a decade ago suggested that the HH pathway was adapted to cell-cell communication from an ancient pathway designed to regulate cellular levels of hopanoids, pentacyclic sterol-like molecules found in bacteria [39,40]. In this ancient pathway, a SMO-like molecule would be a sensor for hopanoids and regulate the transcription of a PTCH1-like transporter tasked with pumping hopanoids out of cells. A rise in hopanoid levels would activate the sensor, inducing the transcription of the ancestral transporter to expel hopanoids from the cell and thus cause activity of sensor protein to drop. In the modern HH pathway the transcription of PTCH1 is indeed a direct target of SMO activity and PTCH1 is likely to be a sterol transporter, based on both structural studies (described above) and more limited, indirect functional data [22,41]. The above model predicts that SMO activity should be positively regulated by a sterol that is a substrate for PTCH1 transport; PTCH1 would inhibit SMO by depleting the levels of such a sterol, likely in a local membrane compartment or microdomain. Consistent with the Hausmann et al. model [39], SMO is indeed a sterol-responsive protein as we discuss below. Notably, the pharmacological or genetic depletion of cholesterol in target cells significantly dampens SMO signalling (reviewed in [42]).
SMO is composed of four domains: the extracellular cysteine-rich (CRD) and linker domains (LD), the stereotypical GPCR heptahelical TMD and the C-terminal intracellular domain (ICD) (Fig. 1a). The ICD is necessary for SMO ciliary localisation and activation of canonical downstream HH signalling but has been removed from all existing structural models due to its intrinsically disordered nature [43]. SMO contains multiple ligand binding sites, two of which have been confirmed through X-ray crystallography (Fig. 3a). Multiple structures of either the CRD, TMD or multi-domain CRD-TMD have been determined [15,16,44–50]. They highlight a canonical TMD binding site seen in many GPCRs, in addition to a shallow hydrophobic groove in the CRD that can bind to both oxysterols and cholesterol [15,50]. Numerous natural and synthetic small molecules are known to interact at these sites, including the anticancer drug vismodegib at the TMD and the plant steroidal alkaloid cyclopamine at the TMD and the CRD [51,52]. Two additional sterol binding sites at the TMD have recently been proposed using computational docking studies [53,54] (Fig. 3a). The first of these sites is located between the extracellular ends of TM2 and TM3, on the exterior of the helical bundle. Molecular dynamics simulations show this to be a defined sterol interaction site and reveal the possibility of direct exchange of cholesterol between SMO and the outer membrane leaflet [54]. The second proposed site is located at the cytoplasmic face of the TMD, named the cytoplasmic binding pocket (CBP), and involves the binding of oxysterols (oxidized metabolites of cholesterol) to residues from TM1, TM3, TM6 and TM7 [53].
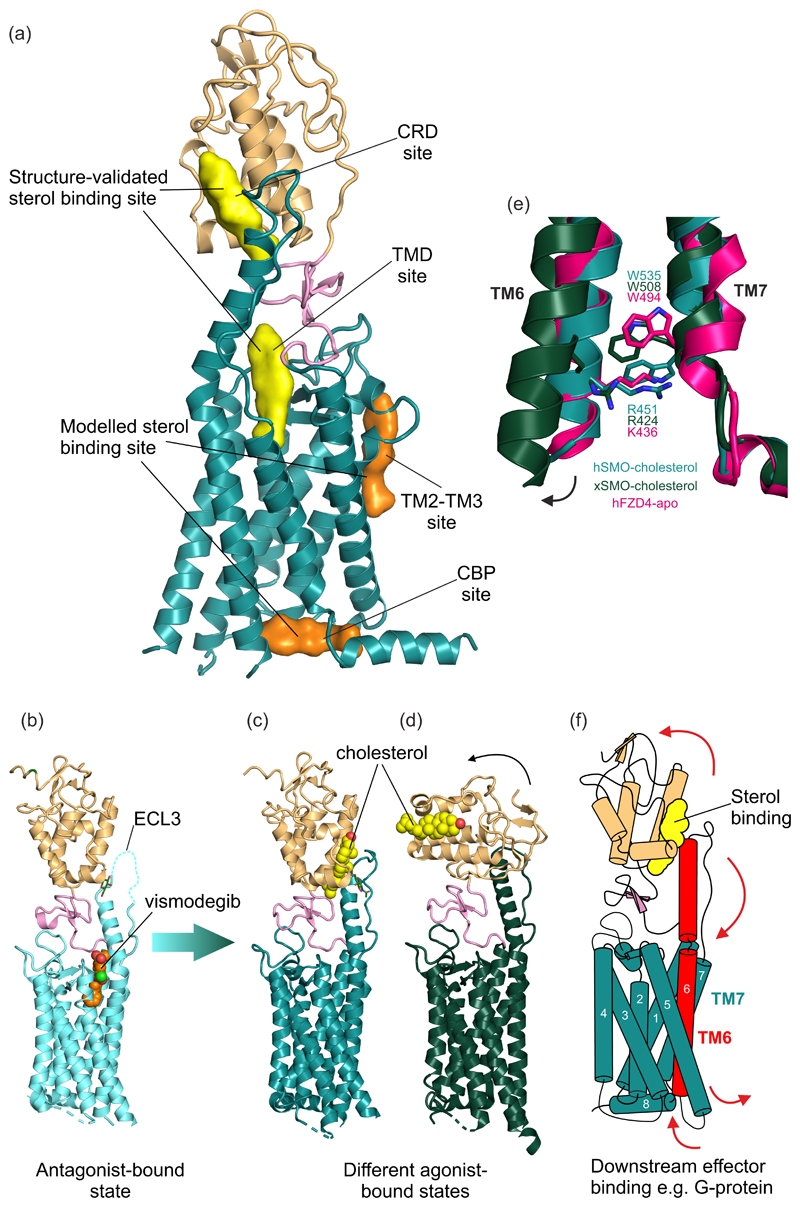
Figure 3. Structure of the HH signal transducer Smoothened.
(a) Multiple sterol binding sites have been proposed in SMO. In biochemical assays and crystal structures, cholesterol and oxysterols have been shown to bind to the CRD and the plant sterol-like molecule cyclopamine has been shown to bind in the TMD (shown in yellow). Two additional TMD sites (shown in orange) have been implicated using computational methods. (b) The structure of inhibited human SMO (hSMO) in complex with the antagonist vismodegib (PDB 5L7I [15]). Extracellular loop 3 (ECL3) is partially disordered (dashed line). Two potential active-state structures of SMO in complex with the agonist cholesterol are shown in (c) and (d). (c) The hSMO-cholesterol complex structure (PDB 5L7D [15]) shows SMO in an agonist-bound state, with the ordered ECL3 forming contacts with the CRD. (d) Xenopus SMO (xSMO)-cholesterol structure (PDB 6D35 [50]) reveals a more dramatic rotation of the CRD compared to the hSMO structure in (c), likely facilitated by removal of native glycans to facilitate crystallization. In (d) the TM6 moves outwards, as seen for other “canonical” GPCRs. (e) The cation-π lock is formed between a conserved tryptophan and basic residue at the cytoplasmic end of TM6 and 7 in class F GPCRs. hSMO-cholesterol (teal) shows an intact lock, while xSMO-cholesterol (dark green) shows the ruptured lock, perhaps indicative of activation. The lock is also seen in apo-Frizzled (PDB 6BD4 [61], hot pink). (f) Proposed model of transmembrane signalling by SMO. Sterol binding to the CRD causes its rotation relative to the TMD, causing a conformational change that is transmitted from the helical extension in ECL3 to TM6. This induces outward movement of TM6, rupturing the cation-π lock and exposing a new cytoplasmic surface for interaction with downstream effectors such as G-proteins.
Side-chain oxysterols were shown to activate HH signalling even in the absence of HH ligands [55–57], an effect that was subsequently shown to be mediated through a direct interaction with SMO [14]. Oxysterols seem to function primarily by binding to a shallow hydrophobic groove in the CRD [44,58,59], though recent work has also implicated the CBP [53]. In addition to oxysterols, cholesterol itself was shown to be both necessary and sufficient to activate SMO signalling in cells [13,15,16], first raising the possibility that cholesterol, rather than oxysterols, was the physiological SMO ligand. Cholesterol activates SMO by binding to the CRD in the same groove as that occupied by oxysterols [15,16]. CRD mutations that prevent cholesterol-mediated SMO activation (but have no effect on oxysterol-mediated activation) attenuate SHH-mediated signalling, supporting the model that cholesterol is the sterol lipid most relevant to endogenous signalling [13,15,16,60]. Indeed, cholesterol and HH ligands act synergistically to activate SMO [15,16].
Regulation of SMO signalling by multiple ligand-binding sites
The contribution of the various ligand-binding sites on SMO for endogenous HH signalling remains to be fully resolved, with both the CRD and TMD sites being implicated (summarized in [51]). However, it is likely that SMO activation involves movement of the CRD relative to the TMD, with TM6 facilitating communication between these two domains (Fig. 3b and c). The deletion of the CRD (SMOΔCRD) or mutations that disrupt the CRD-LD interface activate SMO, suggesting this region is important for stabilization of its inactive state [15,16]. Indeed, the CRD is rotated relative to the TMD in agonist bound structures relative to antagonist bound structures [15,50] (Fig. 3b-d). This conformational change may provide a structural method of communication between the CRD and TMD sites, allowing ligand binding to SMO to be translated into transmembrane signalling (Fig. 3f).
How does SMO activation lead to downstream signalling in the cytoplasm? Recent work has highlighted that SMO activation is associated with rupture of a π-cation bond between the cytoplasmic ends of TM6 and TM7 [50]. The residues forming this π-cation lock (R451 and W535 in human SMO) are evolutionarily conserved in SMO and more generally across class F GPCRs, including the Frizzled family of WNT receptors [61,62] (Fig. 3e). These residues are frequently mutated in various cancers, suggesting that they may be involved in activation of Class F GPCRs [62]. Thus, the activation of SMO may involve breakage of this lock, allowing the cytoplasmic end of TM6 to move outwards, revealing a new surface for interaction with a downstream effector in the cytoplasm (Fig. 3f).
Conclusions and perspectives
The structural and biochemical data supporting the function of PTCH1 as a sterol transporter and SMO as a sterol-activated protein suggests the concept that PTCH1 regulates SMO by restricting its access to an activating sterol. There is also considerable data suggesting that this sterol is cholesterol itself, though it remains possible that a less abundant sterol, such as an oxysterol, is the relevant species. However, further work will be required to show that PTCH1 has sterol transport activity, particularly in an in vitro purified transporter assay. Several possible mechanisms can be envisioned for how PTCH1 may control sterol access to SMO (summarized in Fig. 4). PTCH1 could regulate either the overall membrane levels or the trans-bilayer distribution of cholesterol, thus preventing cholesterol access to either the CRD and/or TMD of SMO. In the model where PTCH1 depletes a membrane compartment of sterols, it would have to transfer the sterol to a membrane or protein acceptor. Alternatively, PTCH1 could strip a sterol from SMO, much like NPC1 accepts cholesterol from NPC2 in the lumen of the lysosome.
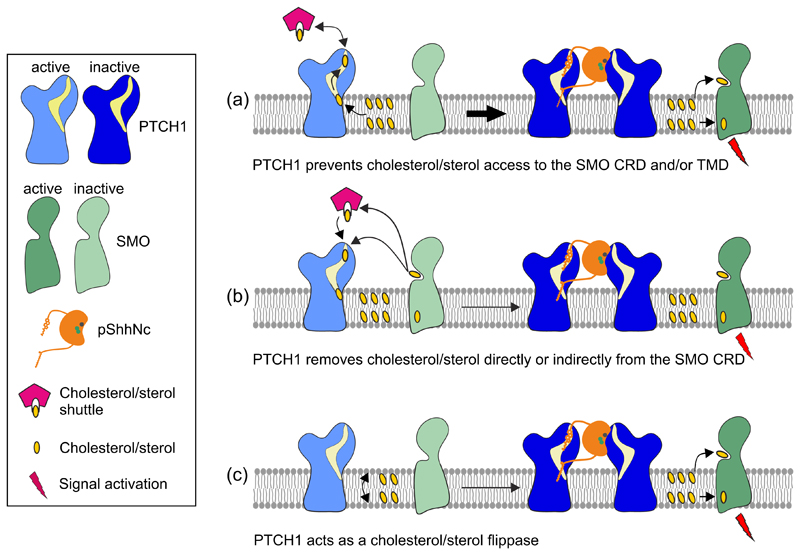
Figure 4. Models for how PTCH1 regulates SMO by controlling its access to sterols.
(a) PTCH1 prevents the access of sterols to the SMO CRD and/or TMD, thereby inhibiting HH signalling. Upon SHH-binding, PTCH1 is rendered inactive, allowing sterol-bound SMO to activate downstream signalling. (b) PTCH1 removes sterols from the SMO CRD either directly or with the help of a yet unidentified sterol-binding protein, thereby inhibiting HH signalling. (c) PTCH1 acts as a sterol flippase, moving sterols from the outer to the inner membrane leaflet, or vice versa. This prevents sterol supply to the SMO CRD or TMD, respectively. These proposed mechanisms are likely confined to the membrane of the primary cilium or a ciliary microdomain.
Finally, an important issue that remains to be resolved is how membrane trafficking and subcellular localization of PTCH1 and SMO are related to their biochemical interaction. The membrane of the primary cilium is thought to play a central role [55]. SMO likely has to localise in primary cilia to engage downstream signalling components and hence PTCH1 could regulate SMO by altering the sterol composition in primary cilia or a ciliary microdomain. Thus, integration of cellular data with the structural information presented herein will be required to gain a complete understanding of how these two unique membrane proteins regulate initiation of HH signalling in target cells.
Acknowledgements
C.K. was supported by Cancer Research UK studentship (C20724/A16135), R.E.W. by a Wellcome Trust studentship (203726/Z/16/Z) and M.K. by a pre-doctoral fellowship from the National Science Foundation. RR was supported by grants from the National Institutes of Health (GM118082 and GM106078). CS was supported by grants from Cancer Research UK (C20724/A14414 and C20724/A26752) and a European Research Council grant (647278). The Wellcome Centre for Human Genetics, Oxford, is funded by Wellcome Trust Core Award 203852/Z/16/2.
References and Recommended Reading
** of outstanding interest: [13], [15], [16], [19]–[24], [25], [49], [50], [61], [62]
- 1.Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–372. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- 2.Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski K, et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273:14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 3.Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV, et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 4.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 5.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 6.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 7.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 8.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 9.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem Biol. 2017;24:252–280. doi: 10.1016/j.chembiol.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 11.Ingham PW, Nystedt S, Nakano Y, Brown W, Stark D, van den Heuvel M, Taylor AM. Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr Biol. 2000;10:1315–1318. doi: 10.1016/s0960-9822(00)00755-7. [DOI] [PubMed] [Google Scholar]
- 12.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 13.Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, Covey DF, Siebold C, Rohatgi R. Cholesterol activates the G-protein coupled receptor Smoothened to promote morphogenetic signaling. Elife. 2016;5 doi: 10.7554/eLife.20304. [** Shows cholesterol is both necessary and sufficient to activate cellular Hedgehog signalling via SMO.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nachtergaele S, Mydock LK, Krishnan K, Rammohan J, Schlesinger PH, Covey DF, Rohatgi R. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat Chem Biol. 2012;8:211–220. doi: 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrne EF, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016;535:517–522. doi: 10.1038/nature18934. [** The first crystal structures of multi-domain SMO. Highlights the rotation between the CRD and TMD upon agonist/antagonist binding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, Salic A. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell. 2016;166:1176–1187 e1114. doi: 10.1016/j.cell.2016.08.003. [** Shows that cholesterol is both necessary and sufficient to activate cellular Hedgehog signalling via SMO.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies JP, Ioannou YA. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer SR. NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J Biol Chem. 2019;294:1706–1709. doi: 10.1074/jbc.TM118.004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, Yan N. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science. 2018;361 doi: 10.1126/science.aas8935. [** Cryo-EM structure highlights PTCH1-SHH interaction, mediated by the metal-binding site on SHH. It furthermore shows that mutations near the sterol binding sites cause conformational changes in PTCH1 which interferes with SHH binding ability.] [DOI] [PubMed] [Google Scholar]
- 20.Qi X, Schmiege P, Coutavas E, Wang J, Li X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature. 2018;560:128–132. doi: 10.1038/s41586-018-0308-7. [** Cryo-EM structure highlights PTCH1-SHH interaction, mediated by the palmitoylated N-terminus of SHH. Shows the structural basis for co-operative binding between SHH, PTCH1 and the HH co-receptors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi X, Schmiege P, Coutavas E, Li X. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science. 2018;362 doi: 10.1126/science.aas8843. [** Cryo-EM structure of 2:1 PTCH1:SHH complex confirms that both binding mechanisms occur simultaneously. It further suggests a putative tunnel through PTCH1, which is disrupted by the SHH palmitate insertion and mutations that mimic the palmitate inhibit PTCH1 function. Biological assays show that the 2:1 complex formation is essential for maximum HH signalling output.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, Myers BR, Cho W, Cheng Y, Beachy PA. Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell. 2018;175:1352–1364 e1314. doi: 10.1016/j.cell.2018.10.026. [** Cryo-EM structure of PTCH1 highlights putative tunnel. Mutational studies show importance of the integrity of the tunnel and charged triad for HH signalling. Biochemical evidence suggests that PTCH1 reduces inner membrane cholesterol to inhibit SMO.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi C, Di Minin G, Vercellino I, Wutz A, Korkhov VM. Structural basis of sterol recognition by human hedgehog receptor PTCH1. bioRxiv. 2018 doi: 10.1126/sciadv.aaw6490. 508325. [** Cryo-EM structure of PTCH1-SHH shows putative tunnel as a proposed mechanism of sterol transport through PTCH1.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian H, Cao P, Hu M, Gao S, Yan N, Gong X. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. bioRxiv. 2018 doi: 10.1038/s41467-019-10234-9. 461491. [** Cryo-EM structure of PTCH1-SHH shows that the C-terminal cholesterol-moiety of SHH inserts into a hydrophobic cavity on PTCH1.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455:979–983. doi: 10.1038/nature07358. [** Crystal structure of the SHH:CDO complex. The authors discovered the crucial function fo the SHH calcium binding site for receptor interactions and provided the structural basis for SHH co-receptor interactions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem. 285:24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop B, Aricescu AR, Harlos K, O'Callaghan CA, Jones EY, Siebold C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat Struct Mol Biol. 2009;16:698–703. doi: 10.1038/nsmb.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosanac I, Maun HR, Scales SJ, Wen X, Lingel A, Bazan JF, de Sauvage FJ, Hymowitz SG, Lazarus RA. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat Struct Mol Biol. 2009;16:691–697. doi: 10.1038/nsmb.1632. [DOI] [PubMed] [Google Scholar]
- 29.Whalen DM, Malinauskas T, Gilbert RJ, Siebold C. Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci U S A. 2013;110:16420–16425. doi: 10.1073/pnas.1310097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24:2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor FR, Wen D, Garber EA, Carmillo AN, Baker DP, Arduini RM, Williams KP, Weinreb PH, Rayhorn P, Hronowski X, et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry. 2001;40:4359–4371. doi: 10.1021/bi002487u. [DOI] [PubMed] [Google Scholar]
- 32.Tukachinsky H, Petrov K, Watanabe M, Salic A. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc Natl Acad Sci U S A. 2016;113:E5866–E5875. doi: 10.1073/pnas.1606719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X, Mann RK, Sever N, Beachy PA. Genetic and biochemical definition of the Hedgehog receptor. Genes Dev. 24:57–71. doi: 10.1101/gad.1870310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camp D, Haitian He B, Li S, Althaus IW, Holtz AM, Allen BL, Charron F, van Meyel DJ. Ihog and Boi elicit Hh signaling via Ptc but do not aid Ptc in sequestering the Hh ligand. Development. 2014;141:3879–3888. doi: 10.1242/dev.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strutt H, Thomas C, Nakano Y, Stark D, Neave B, Taylor AM, Ingham PW. Mutations in the sterol-sensing domain of Patched suggest a role for vesicular trafficking in Smoothened regulation. Curr Biol. 2001;11:608–613. doi: 10.1016/s0960-9822(01)00179-8. [DOI] [PubMed] [Google Scholar]
- 36.Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 37.Guan L, Nakae T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J Bacteriol. 2001;183:1734–1739. doi: 10.1128/JB.183.5.1734-1739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers BR, Neahring L, Zhang Y, Roberts KJ, Beachy PA. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci U S A. 2017;114:E11141–E11150. doi: 10.1073/pnas.1717891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hausmann G, von Mering C, Basler K. The hedgehog signaling pathway: where did it come from? PLoS Biol. 2009;7:e1000146. doi: 10.1371/journal.pbio.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bazan JF, de Sauvage FJ. Structural ties between cholesterol transport and morphogen signaling. Cell. 2009;138:1055–1056. doi: 10.1016/j.cell.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Bidet M, Joubert O, Lacombe B, Ciantar M, Nehme R, Mollat P, Bretillon L, Faure H, Bittman R, Ruat M, et al. The hedgehog receptor patched is involved in cholesterol transport. PLoS One. 2011;6:e23834. doi: 10.1371/journal.pone.0023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blassberg R, Jacob J. Lipid metabolism fattens up hedgehog signaling. BMC Biol. 2017;15:95. doi: 10.1186/s12915-017-0442-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Nachtergaele S, Whalen DM, Mydock LK, Zhao Z, Malinauskas T, Krishnan K, Ingham PW, Covey DF, Siebold C, Rohatgi R. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife. 2013;2:e01340. doi: 10.7554/eLife.01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rana R, Carroll CE, Lee HJ, Bao J, Marada S, Grace CR, Guibao CD, Ogden SK, Zheng JJ. Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat Commun. 2013;4 doi: 10.1038/ncomms3965. 2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Wu H, Evron T, Vardy E, Han GW, Huang XP, Hufeisen SJ, Mangano TJ, Urban DJ, Katritch V, et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat Commun. 2014;5 doi: 10.1038/ncomms5355. 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weierstall U, James D, Wang C, White TA, Wang D, Liu W, Spence JC, Bruce Doak R, Nelson G, Fromme P, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun. 2014;5 doi: 10.1038/ncomms4309. 3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Zhao F, Wu Y, Yang J, Han GW, Zhao S, Ishchenko A, Ye L, Lin X, Ding K, et al. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat Commun. 2017;8 doi: 10.1038/ncomms15383. 15383. [** Crystal structure of a multi-domain human SMO protein, bound and stabilized by the designed ligand TC114. The authors identified key structural elements in SMO – the TMD helix 6 and the ECL3 loop – that are important for communication between the extracellular and transmembrane domains.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang P, Zheng S, Wierbowski BM, Kim Y, Nedelcu D, Aravena L, Liu J, Kruse AC, Salic A. Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell. 2018;175:295–297. doi: 10.1016/j.cell.2018.09.003. [** Crystal structures of unglycosylated Xenopus SMO in complex with cholesterol or cyclopamine. Highlights breakage of the conserved cation-lock, thought to beimportant for activation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrne EF, Luchetti G, Rohatgi R, Siebold C. Multiple ligand binding sites regulate the Hedgehog signal transducer Smoothened in vertebrates. Curr Opin Cell Biol. 2018;51:81–88. doi: 10.1016/j.ceb.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharpe HJ, Wang W, Hannoush RN, de Sauvage FJ. Regulation of the oncoprotein Smoothened by small molecules. Nat Chem Biol. 2015;11:246–255. doi: 10.1038/nchembio.1776. [DOI] [PubMed] [Google Scholar]
- 53.Raleigh DR, Sever N, Choksi PK, Sigg MA, Hines KM, Thompson BM, Elnatan D, Jaishankar P, Bisignano P, Garcia-Gonzalo FR, et al. Cilia-Associated Oxysterols Activate Smoothened. Mol Cell. 2018;72:316–327 e315. doi: 10.1016/j.molcel.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hedger G, Koldsø H, Chavent M, Siebold C, Rohatgi R, Sansom MSP. Cholesterol Interaction Sites on the Transmembrane Domain of the Hedgehog Signal Transducer and Class F G Protein-Coupled Receptor Smoothened. Structure. 2019;27:549–559.e542. doi: 10.1016/j.str.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 56.Corcoran RB, Scott MP. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci U S A. 2006;103:8408–8413. doi: 10.1073/pnas.0602852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, Parhami F. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J Biol Chem. 2007;282:8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- 58.Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, Bazan JF, Beachy PA. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev Cell. 2013;26:346–357. doi: 10.1016/j.devcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat Chem Biol. 2013;9:557–564. doi: 10.1038/nchembio.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao X, Tang JJ, Peng C, Wang Y, Fu L, Qiu ZP, Xiong Y, Yang LF, Cui HW, He XL, et al. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Mol Cell. 2017;66:154–162 e110. doi: 10.1016/j.molcel.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 61.Yang S, Wu Y, Xu TH, de Waal PW, He Y, Pu M, Chen Y, DeBruine ZJ, Zhang B, Zaidi SA, et al. Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature. 2018;560:666–670. doi: 10.1038/s41586-018-0447-x. [** First crystal structure of the transmembrane domain of a class F Frizzed receptor.] [DOI] [PubMed] [Google Scholar]
- 62.Wright SC, Kozielewicz P, Kowalski-Jahn M, Petersen J, Bowin CF, Slodkowicz G, Marti-Solano M, Rodríguez D, Hot B, Okashah N, et al. A conserved molecular switch in Class F receptors regulates receptor activation and pathway selection. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08630-2. 667. [** Identification of a conserved cation-π lock across Class F GPCRs which is thought to regulate activation and downstream signalling.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]