Abstract
Ribosomal DNA (rDNA), the genes encoding the RNA components of ribosomes (rRNA), are highly repetitive in all eukaryotic genomes, containing 100s to 1000s of copies, to meet the demand for ribosome biogenesis. rDNA genes are arranged in large stretches of tandem repeats, forming loci that are highly susceptible to copy loss due to their repetitiveness and active transcription throughout the cell cycle. Despite this inherent instability, rDNA copy number is generally maintained within a particular range in each species, pointing to the presence of mechanism(s) that maintain rDNA copy number in a homeostatic range. In this review, we summarize the current understanding of these maintenance mechanisms and how they sustain rDNA copy number throughout populations.
Introduction: rDNA copy number variations
As the source of ribosomal RNA (rRNA, see Glossary), an essential component of ribosomes, preservation of ribosomal DNA (rDNA, see Glossary) loci consisting of the highly repetitive rRNA genes represents one of the most critical aspects of genome maintenance. The tandemly repeated nature of rDNA copies creates an inherit instability for rDNA loci due to intra-chromatid homologous recombination (see Glossary) between copies (Fig. 1, Key figure) [1]. This instability, combined with the importance of the rDNA loci for cell functionality, demands special attention to maintain these loci. Despite its critical importance, rDNA has escaped genomics-era analysis due to the difficulty in sequencing the loci, because of its repetitiveness and large unit size (8 – 13 kb for each repeat [2–6]), limiting the ability to obtain precise copy number information through genome sequencing. Still, gross assessments of rDNA copy number have led to a widely accepted notion that rDNA loci are dynamic and copy number varies considerably between individuals across a species, and even likely between cells within a single organism [7–11]. The copy number variation probably reflects a combination of the effects of natural copy number loss and the mechanism(s) to recover copy numbers to maintain the functionality of the loci. In this review, we summarize the current understanding of the mechanisms governing rDNA copy number instability and maintenance, mostly focusing on Saccharomyces cerevisiae and Drosophila melanogaster, where these mechanisms have been studied most extensively.
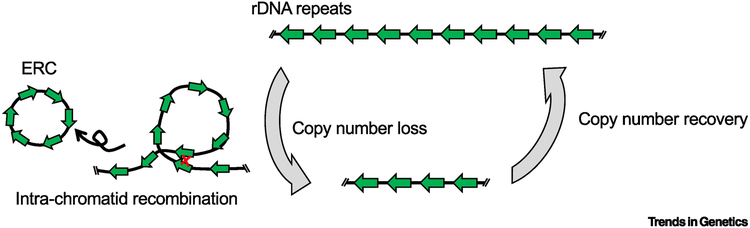
Figure 1. rDNA copy number dynamics through cyclical copy number loss and recovery.
The size of rDNA loci comprised of tandemly duplicated repeats of the rRNA cistron (green arrows) is dynamic due to loss and recovery of individual rDNA repeats. rDNA copies are most notably lost by intra-chromatid recombination between distant rDNA copies, leading to excision of the intervening copies in the form of an extra-chromosomal rDNA circle (ERC). rDNA loci are capable of expanding their copy number to recover lost copies. The multiple proposed mechanisms for rDNA copy number expansion are discussed throughout this review. Overall rDNA copy number is proposed to be maintained through balanced copy number loss and expansion activity.
Sources of rDNA copy number variation
Both genetic and environmental factors have been reported to influence rDNA copy number, providing insights into the underlying mechanisms that create rDNA copy number variation. Only a subset of rDNA copies are needed to be transcribed for normal cellular function, and the unnecessary copies are maintained in the repressed state by heterochromatinization [12]. Silencing of unnecessary rDNA copies likely minimizes transcription-replication conflicts, which can lead to DNA damage and copy number variation. Extensive studies in Arabidopsis established that rDNA silencing is mediated by multitude of epigenetic mechanisms involving DNA methylation and histone modification [13–17], and defects in silencing leads to rDNA copy number changes [18]. Drosophila HP1a mutants that are maintained over many generations have reduced rDNA copy number [19], possibly due to their inability to silence rDNA. It was also shown that a nutrient-rich diet, which likely increases rDNA transcription via target of rapamycin (TOR) [20], results in heritable rDNA copy number loss in Drosophila [21]. Interestingly, it was reported that both mouse tumor model with an activated TOR and human cancer cells with high TOR activity have reduced rDNA copies [22].
Another cause of copy number variation within species is accumulated copy number loss that occurs during aging. rDNA copy number loss during aging has been inferred in studies that measured bulk rDNA amount in tissues from young vs. old animals, e.g. canine brain [23] and human blood cells [24,25]. However, the results from bulk tissues, which likely contain a mixture of cells with different ploidies, allowed limited interpretation. It was reported that mouse hematopoietic stem cells accumulate ‘replicative stress’ in the nucleolus [26], possibly reflecting rDNA copy number loss during aging, although copy number was not measured.
Budding yeast is the best-studied model system to address rDNA copy number instability during aging. Mother cells age progressively with each division and eventually die after ~20 cell divisions due to rDNA instability caused by intra-chromatid recombination that reduces chromosomal rDNA copy number and generates extrachromosomal rDNA circles (ERCs, see Glossary) (Fig. 1) [27,28]. These ERCs are selectively retained within mother cells during division and proposed to contribute to mother cell aging [28,29], whereas the daughter (bud) cells can reset their age and divide another ~20 times [27]. rDNA copy loss counterbalanced by restoration results in a relatively constant size of the locus throughout the population over infinite generations (Fig. 1).
While copy number dynamics are well characterized in yeast, little is known about the source of individual-to-individual rDNA copy number variation in multicellular organisms. The restriction of rDNA loci to the sex chromosomes and availability of genetic tools has made Drosophila a useful model to examine rDNA loci in multicellular organisms [30]. ERC formation also causes rDNA loss in Drosophila [31,32]. Observation that individual rDNA copies (uniquely marked by transposon insertions) are frequently lost in a span of several generations [33,34] confirms the notion that rDNA copies are lost during cell divisions/generations in Drosophila as well. It was recently shown that rDNA copy number is decreased in the Drosophila male germline during aging: although the semi-quantitative nature of copy number estimation does not provide precise copy numbers, the data indicate that rDNA copy number might decrease by half during 40 days of Drosophila aging [35]. This copy number decrease in the germline leads to inheritance of reduced rDNA copy number by the offspring [35]. Strikingly, rDNA copy number is recovered in the germline of the subsequent generation [35], indicating Drosophila germ cells also have mechanisms to restore rDNA copy number. These results collectively suggest that Drosophila rDNA is also normally maintained by cyclical copy number loss and recovery. Given the common feature of rDNA across species (e.g. tandemly repeated, unstable rDNA loci) [36], knowledge obtained from yeast and Drosophila likely applies to other eukaryotes, including humans.
The mechanisms that recover rDNA copy number
The age-related reduction in rDNA copy number raises an interesting question as to how organisms can maintain rDNA through generations. As described above, the reduced rDNA copy number is inherited to the offspring, and continued rDNA loss from generation to generation would soon create loci with too few rDNA copies to sustain cellular/organismal viability. Therefore, mechanisms to restore rDNA copies and counteract the loss during aging must be in place.
Budding yeast has been an instrumental model system to dissect the precise mechanism for rDNA copy number expansion, and detailed description of this mechanism is found in [36]. Briefly, rDNA copy number expansion in yeast requires binding of the replication fork barrier protein Fob1 to rDNA intergenic spacer sequence (IGS, see Glossary) 1 downstream of the rRNA cistron (see Glossary) (Fig. 2A) [37]. This binding blocks the progression of replication forks, leading to the formation of a double-strand break (DSB, see Glossary) in the leading strand [37–39] (Fig. 2A). In this model, DSB formation in the leading strand leads to strand invasion and repair using its sister chromatid (lagging strand) as a template [40–43]. Owing to the repetitiveness of rDNA loci, strand invasion can occur in a way that completion of repair will increase rDNA copies in a process called unequal sister chromatid recombination (USCR, see Glossary) (Fig. 2A) [42,43]. Interestingly, the ability to expand rDNA copies is dependent on the expression of non-coding RNA transcribed from the E-pro promoter within IGS1. E-pro transcription displaces cohesin from the nearby rDNA, allowing the newly replicated sister chromatids to misalign during recombination (Fig. 2B) [44]. On the contrary, when rDNA copy expansion is not required, Sir2, a NAD+-dependent histone deacetylase [45,46], represses E-pro transcription and maintains cohesin binding [42,44], thereby preventing USCR and rDNA copy expansion (Fig. 2B).
Figure 2. Model of rDNA copy number expansion in budding yeast.
(A) The mechanism of rDNA copy number expansion in budding yeast through unequal sister chromatid recombination (USCR). Replication is initiated at the replication origin in IGS2 region (i). The replication fork that moves rightwards is arrested at replication fork barrier (RFB) in a Fob1-dependent manner (ii). The fork arrest creates double-strand break (DSB) in the leading strand (iii). The DSB repair by gene conversion between sister chromatids leads to copy number increase on the leading strand while maintaining the copy number on the lagging strand (iv–v). (B) The mechanism that regulates the decision of unequal vs. equal sister chromatid recombination. (i) In the absence of the need of copy number expansion, Sir2 represses the expression of IGS1-derived non-coding RNA (E-pro). (ii) When copy number is decreased, Sir2 protein is reduced, leading to E-pro expression, which displaces cohesin from the nearby rDNA, allowing the newly replicated sister chromatids to misalign and leading to USCR.
In addition to USCR, reintegration of ERCs has been proposed as a potential mechanism to increase chromosomal rDNA copy number [47]. ERCs can be produced by intra-chromatid recombination evens (described above), then amplified using an internal replication origin. Alternatively, it has been proposed that excision of rDNA copies from replication bubbles can generate ERCs without reducing chromosomal copy number, through DNA replication that fills the excised copies[47–49]. Irrespective of the mechanism to produce ERCs, their reintegration can expand rDNA copy number on the chromosomal rDNA loci. It has been reported that cells with fewer rDNA copies produce more ERCs in a replication dependent manner [47,50,51], implying that those amplified ERCs may be integrated to increase the chromosomal copy number. However, ERC insertion events are reported to be very infrequent [28,47], making it unclear how much this mechanism can contribute to rDNA copy number recovery.
In multicellular organisms, where rDNA copy number loss has not been extensively characterized, the mechanisms of rDNA recovery have not gained much attention. Given the conservation in the structure of rDNA loci from yeast to humans [36,52], it is plausible that similar mechanisms operate in multicellular organisms. The phenomenon of ‘rDNA magnification’ (see Glossary) that was discovered in the Drosophila male germline over 50 years ago [53] provided a paradigm for understanding rDNA expansion in multicellular organisms. Drosophila strains with large rDNA deletions have a characteristic developmental defect called the bobbed phenotype, exhibiting disrupted abdominal cuticle and thin bristles [30]. The offspring of bobbed males were frequently found to develop normally due to expansion - or ‘magnification’ - of the rDNA locus [53,54], demonstrating the ability of flies to expand rDNA copies in the male germline. Although the physiological relevance of this phenomenon remained unknown, some of the genes known to be required for rDNA magnification (mus-101, the TOPBP1 homolog, and mei-41, the ATM homolog) [55,56] are also found to be required for normal rDNA maintenance from generation to generation [35,55], suggesting that rDNA magnification may be a manifestation of the normal physiological rDNA maintenance mechanism.
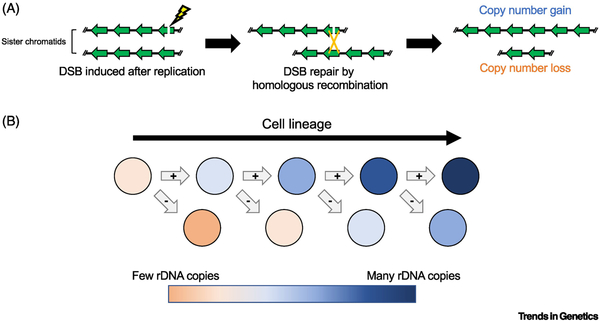
Similar to rDNA expansion in yeast, it has been proposed that rDNA magnification is mediated by unequal recombination between sister chromatids [57,58] and/or integration of amplified ERCs [59]. ERC amplification/reintegration was initially an attractive model because it easily explained the surprisingly large number of rDNA copies that are gained during rDNA magnification [60]. However, the distinct sequence variations between X- and Y-chromosome rDNA loci [61,62] and mapping of uniquely sized rDNA transposon insertions revealed that newly-gained rDNA copies are tandem duplications of stretches of the existing copies within the locus [63], disfavoring the model of integration of amplified ERCs. Instead, the data suggested that new rDNA copies are synthesized from local copies, likely through the process of recombination. Furthermore, circular ring X-chromosomes fail to expand rDNA copies and instead form dicentric chromosomes in magnifying conditions [57,64], indicating that magnification requires DNA exchange between sister chromatids. The requirement for sister chromatid exchange prompted the model that rDNA magnification is achieved by a process called unequal sister chromatid exchange (Fig. 3A) (USCE, see Glossary) [57,58], where recombination between misaligned sister chromatids causes one sister chromatid to acquire rDNA copies from the other sister chromatid. USCE is somewhat similar to USCR in yeast described above, but distinct in that USCE results in reciprocal gain vs. loss of copy numbers between sister chromatids, whereas USCR is similar to gene conversion (see Glossary) and one sister chromatid gains copy number without the other sister chromatid losing any copies.
Figure 3. rDNA copy number expansion by unequal sister chromatid exchange in mitotic Drosophila germ cells.
(A) Unequal sister chromatid exchange (USCE) can create reciprocal rDNA copy number gain and loss between sister chromatids. When a double-stranded DNA break (DSB) occurs in an rDNA region post replication, the DSB can be repaired by homologous recombination with the sister chromatid. Homologous recombination between misaligned rDNA copies resolved by chromatid exchange increases rDNA copies on one sister while reducing the copies on the other sister. (B) USCE over successive cell divisions can rapidly expand rDNA copies (range from orange to blue). Repeated inheritance of the sister chromatid that gains copies during USCE will lead to rapid expansion of rDNA copies, beyond what could be gained by a single USCE event.
The USCE model is supported by direct observation of sister chromatid exchange in meiotic germ cells under magnifying conditions [64]. Moreover, the emergence of a small fraction of offspring that had further reduced rDNA copy number during magnification supported that magnification involves reciprocal exchange of sister chromatids [57,65]. Despite the attraction of the USCE model, the observation that some offspring magnified rDNA >4-fold disfavored the USCE model, based on the rational that a single USCE event would generate a 2-fold expansion at most (by receiving all rDNA copies from the sister chromatid) [60]. However, a more than 2-fold increase in copy number could be explained by multiple USCE events occurring over successive cell divisions (Fig 3B) [65]. Indeed, mapping of magnified chromosomes indicated multiple duplication steps occur during magnification [63]. Therefore, it is likely that multiple USCE events occur in mitotically amplifying germ cells (germline stem cells (GSCs) and spermatogonia (SGs)), leading to >2-fold increase in rDNA copy number. GSCs are the most attractive candidate, where successive USCE events may lead to effective increase in rDNA copy number, particularly if GSCs can consistently inherit magnified copies. (Fig 3B). Moreover, GSCs can produce many differentiating cells/gametes with improved copy number through their self-renewing ability. In contrast, SGs, which committed to differentiation, will undergo only 4 rounds of mitotic divisions prior to meiotic commitment, thereby limiting the opportunity for USCEs. This model would explain why large clusters of offspring with expanded rDNA copies are produced at once by individual fathers [57] and how multiple magnified offspring can inherit identical rDNA loci [66].
The mechanisms that monitor rDNA locus size
Whereas the above mechanisms may explain how reduced copy number can be recovered to sustain rDNA integrity through generations, they raise questions as to how cells know within what range to maintain the rDNA copy number. There is a wide distribution in the number of rDNA copies across wild populations, yet individuals with insufficient or excessive copy numbers are rarely observed [67–69]. The majority of individuals within a population/species cluster around a median number of rDNA copies [68,70,71], suggesting that there are mechanisms that function as a ‘rheostat’ for rDNA copy number.
Recent work reported an elegant rDNA ‘counting mechanism’ in yeast to maintain a set number of rDNA copies. The Pol I UAF complex binds to rDNA promoters to promote Pol I transcription [72–74], but when rDNA copies are low, promoter binding is saturated and free UAF complexes instead bind to the Sir2 promoter and terminator [75]. UAF binding to the Sir2 locus represses Sir2 transcription [75], preventing E-pro repression which in turn leads to copy number expansion as described above (Fig. 2B) [44]. UAF preferentially binds to the rDNA promoter over the Sir2 locus, and accordingly the Sir2 locus is depleted of UAF as rDNA number increases, forming an autonomous feedback loop to maintain rDNA copy number [75]. In this way, the amount of UAF complexes in the cell sets the rDNA locus size, and manipulating the amount of UAF will adjust the number of rDNA copies [75].
Since the molecular factors responsible for rDNA expansion in Drosophila are still unclear, it is unknown if Sir2 orthologues and UAF factors have a similar function serving as an rDNA rheostat in the Drosophila germline. However, the size of the Drosophila rDNA loci has been proposed to be defined by the balance of opposing forces that contract and expand rDNA copies [69,71,76]. In this model, the upper limit of rDNA size is constrained by the inherent instability of these loci, where larger rDNA loci have a higher chance for rDNA loss to occur, while the lower limit is set by selective pressure and the frequency of USCE events. rDNA magnification only occurs under specific context [54,57,58,77] and the frequency of magnification events is dependent on the severity of rDNA insufficiency [65], indicating the presence of a mechanism to sense rDNA copy number and actively induce USCE when copy number is critically low. The observation that Y-chromosomes containing a large rDNA deletion are required for magnification [78,79] and are sufficient to induce magnification of normal-sized loci [80] suggest that the sensor and / or USCE inducer is a Y-chromosome factor. It remains unclear what these factors may be or how they may detect insufficient rDNA copy number and promote USCE.
Global rDNA transcriptional state is an additional feature that might function as a sensor for rDNA copy number. As rDNA copy number decreases, a larger fraction of the remaining rDNA copies will be expressed to keep up with the demand of ribosome biogenesis. Transcription of a greater portion of rDNA increases the probability of transcription-replication collisions and DSB formation [49], which could initiate USCE-mediated magnification. Additionally, the regulation of the choice of active rDNA loci (the phenomenon called ‘nucleolar dominance’) may function as a sensor. Nucleolar dominance is a phenomenon, where a subset of rDNA loci is actively transcribed, whereas the other loci are silenced. Although nucleolar dominance was originally observed in interspecies hybrids [81,82], it has been shown/inferred to occur within species (i.e. in the non-hybrid context) ranging from Arabidopsis [14,82], Drosophila [35,83] and mammals [84,85]. In male Drosophila cells, using transcription-dependent deposition of histone H3.3-GFP onto the rDNA loci as well as single nucleotide polymorphism (SNP) in situ RNA hybridization that differentially labels X vs. Y rDNA transcripts, it has been shown that the Y rDNA locus is predominantly active, while X rDNA is silent [35,83]. As Y rDNA copy number decreases in GSCs during aging, transcription of X rDNA gradually increases [35], which may signal a need for rDNA expansion to the cell. Cells may further utilize distinct rDNA gene structures between X and Y rDNA [61,62] to sense the expression of X rDNA to trigger magnification. It is tempting to speculate that nucleolar dominance serves as a mechanism to regulate rDNA magnification. In this model, certain rDNA loci may be marked as ‘active’ and others as ‘back-ups’ by the mechanism of nuclear dominance, and the activation of the ‘back-up’ loci may serve as an indication of the need to expand rDNA copies.
Despite the apparent robust mechanism to expand rDNA copies in the Drosophila male germline, rDNA copies are progressively lost from the germline during aging [35]. Given that rDNA appears to be maintained by a balance of rDNA copy loss and expansion, the net loss observed during aging is likely due to a shift in this balance. An increase in rDNA transcription, as has been observed in some aged cultured cells and human muscle tissue samples [86,87], may enhance copy number loss in older GSCs through frequent replication-transcription collisions (more than they help increase the copy number). Alternatively, the accumulated rDNA loss during aging may be due to a reduction in the amount of expansion, or a reduced sensitivity to recognize rDNA copy loss and activate expansion. Homologous recombination repair activity is diminished in the GSCs of old males compared to young males [88], raising the possibility that USCE activity is reduced in old animals and thus copy number expansion is reduced. These models of rDNA dynamics during aging are not mutually exclusive. Further understanding the molecular factors that regulate and execute rDNA copy expansion will help to uncover how maintenance is disrupted during aging.
Concluding remarks
Like telomeres, rDNA is an essential but unstable genomic element that requires active maintenance. However, the mechanisms that maintain rDNA copy number have been underexplored except for budding yeast, despite the marked importance of maintenance through generations of multicellular organisms. Similar to telomeres, rDNA maintenance is likely a prerequisite of supporting ‘immortality’ of cell lineage, and accordingly all immortal cells, including germline and cancer cells, may have specialized mechanisms to actively maintain rDNA copy number. Reflecting its importance, rDNA and its transcription and the nucleolus as an organelle for rRNA biogenesis are subjected to a multitude of regulation. Here we described the current proposed models for how rDNA copy number is regulated and maintained from generation to generation. Understanding how these mechanisms of regulation may be interconnected to each other for the ultimate purpose of supporting cellular viability awaits future investigation.
Supplementary Material
Recent highlights.
Ribosomal DNA (rDNA) copy number is highly variable across individuals of a species and dynamically changes within a few generations
Both genetic and environmental factors can alter rDNA copy number
rDNA loci are inherently unstable and copy loss naturally occurs during aging
rDNA copy number can be expanded during cell division via variations of homologous recombination mechanisms to offset rDNA loss
The factors that initiate rDNA copy number expansion are not conserved across eukaryotes
Age-related rDNA copy number loss in the germline is inherited to the next generation, but can be restored in their germline of Drosophila.
Sensors may exist to monitor the rDNA copy number such that the mechanisms to increase copy number can be activated as necessary.
Outstanding questions.
The molecular factors and mechanisms to expand rDNA copies are well characterized in S. cerevisiae, yet homologues of the key components are not yet identified in multicellular organisms. Unequal sister chromatid exchange (USCE) is proposed to expand rDNA copies in Drosophila, but the molecular factors that initiate and execute this process for rDNA expansion remain largely unknown.
An expected consequence of rDNA copy expansion by USCE is a reciprocal rDNA copy loss inherited by the sister cell, however there is a much smaller portion of offspring observed to inherit reduced rDNA copies than those with expanded copies. The underlying mechanism for this phenomenon is entirely unknown.
rDNA copy numbers are maintained within a certain range in many organisms. The mechanisms that sense the ‘appropriate’ copy number such that the copy number restoration mechanism(s) can be turned on/off remains largely unknown, particularly in multicellular organisms.
rDNA copy loss occurs in yeast and Drosophila germ cells during aging despite robust mechanisms to expand rDNA copies. How the factors that mediate rDNA copy loss and expansion are altered over time so that loss accumulates during aging, and why these factors cannot infinitely maintain rDNA within a single cell / individual, remains unknown.
The specific mechanisms to expand rDNA copies are not conserved between yeast and Drosophila despite the importance of rDNA maintenance for all eukaryotes. It remains unknown whether either of these mechanisms is more widely utilized in other eukaryotes, or whether the pathways for rDNA copy expansion are divergent across species.
Acknowledgements
We apologize to colleagues whose work could not be cited due to space limitations. We thank Yamashita laboratory members for discussion. The research in the Yamashita laboratory is supported by Howard Hughes Medical Institute and National Institute of General Medical Sciences (R01GM118308).
Glossary
- rRNA
Ribosomal RNA, the RNA components of the ribosome
- rDNA
Ribosomal DNA, loci comprised of hundreds of tandem repeats of a large (8 – 13kb) sequence motif containing the genes encoding the RNA components of the ribosome
- Intra-chromatid homologous recombination
DNA recombination between homologous sequences within the same chromatid
- ERC
Extra-chromosomal rDNA circle, a loop of one or more rDNA repeats that has been excised and circularized from the chromosomal rDNA due to intra-chromatid recombination between two rDNA copies in the same rDNA locus
- IGS
Intergenic spacer sequence, the sequence in between repeated copies of the rDNA cistron that is not transcribed as part of the cistron
- rRNA cistron
The single transcriptional unit that contains the 18S, 5.8S, and 28S rRNAs. These rRNAs are separated and flanked by transcribed spacer sequences that are spliced out after transcription. Each rDNA copy contains a cistron, its promoter, and intergenic spacer sequences
- DSB
Double-strand DNA breaks, DNA damage that creates breaks in both strands of a DNA molecule
- USCR
Unequal sister chromatid recombination (at the rDNA locus), the recombination between two sister chromatids of rDNA, where the leading strand with a DSB invades the lagging strand to increase the rDNA copy number, while maintaining the rDNA copy number of the lagging strand
- rDNA magnification
The phenomenon of rapid expansion of many rDNA copies within a single generation
- USCE
Unequal sister chromatid exchange (at the rDNA locus), the exchange of sister chromatids at misaligned rDNA copies, which results in one chromatid gaining rDNA copies while the sister has a reciprocal loss of rDNA copies
- Gene conversion
The process of one sequence replacing another near-homologous sequence by recombination between the two sequences during homologous recombination-mediated repair of double-strand DNA breaks
- Nucleolar dominance
The phenomenon of entire rDNA loci being transcriptionally active or silenced and rRNAs are transcribed exclusively from specific rDNA loci
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Park PU et al. (1999) Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol 19, 3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klootwijk J et al. (1979) The primary transcript of the ribosomal repeating unit in yeast. Nucleic Acids Res. 6, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawid IB et al. (1978) Ribosomal DNA in Drosophila melanogaster. I. Isolation and characterization of cloned fragments. J Mol Biol 126, 749–768 [DOI] [PubMed] [Google Scholar]
- 4.Wellauer PK and Dawid IB (1974) Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol 89, 379–395 [DOI] [PubMed] [Google Scholar]
- 5.Schibler U et al. (1975) Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J Mol Biol 94, 503–517 [DOI] [PubMed] [Google Scholar]
- 6.Wellauer PK and Dawid IB (1973) Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci USA 70, 2827–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabanal FA et al. (2017) Unstable Inheritance of 45S rRNA Genes in Arabidopsis thaliana. G3 (Bethesda) 7, 1201–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lofgren LA et al. (2019) Genome- based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol Ecol 133, 203–10 [DOI] [PubMed] [Google Scholar]
- 9.Henderson AS et al. (1974) The chromosomal location of ribosomal DNA in the mouse. Chromosoma 49, 155–160 [DOI] [PubMed] [Google Scholar]
- 10.Miller L and Knowland J (1972) The number and activity of ribosomal RNA genes in Xenopus laevis embryos carrying partial deletions in both nucleolar organizers. Biochem. Genet 6, 65–73 [DOI] [PubMed] [Google Scholar]
- 11.Schmidtke J et al. (1976) Gene action in fish of tetraploid origin. IV. Ribosomal DNA amount in clupeoid and salmonoid fish. Biochem. Genet 14, 293–297 [DOI] [PubMed] [Google Scholar]
- 12.Srivastava R et al. (2016) The Epigenetic Pathways to Ribosomal DNA Silencing. Microbiol. Mol. Biol. Rev 80, 545–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ZJ and Pikaard CS (1997) Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes & Development 11, 2124–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence RJ et al. (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Molecular Cell 13, 599–609 [DOI] [PubMed] [Google Scholar]
- 15.Earley KW et al. (2010) Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes & Development 24, 1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earley K et al. (2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes & Development 20, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preuss SB et al. (2008) Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Molecular Cell 32, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pontvianne F et al. (2012) Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes & Development 26, 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aldrich JC and Maggert KA (2014) Simple quantitative PCR approach to reveal naturally occurring and mutation-induced repetitive sequence variation on the Drosophila Y chromosome. PLoS ONE 9, e109906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grewal SS et al. (2007) Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J. Cell Biol. 179, 1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aldrich JC and Maggert KA (2015) Transgenerational inheritance of diet-induced genome rearrangements in Drosophila. PLoS Genet. 11, e1005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu B et al. (2017) Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 13, e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson R and Strehler BL (1972) Loss of genes coding for ribosomal RNA in ageing brain cells. Nature 240, 412–414 [DOI] [PubMed] [Google Scholar]
- 24.Malinovskaya EM et al. (2018) Copy Number of Human Ribosomal Genes With Aging: Unchanged Mean, but Narrowed Range and Decreased Variance in Elderly Group. Front. Genet 9, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stults DM et al. (2008) Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 18, 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flach J et al. (2014) Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi T (2011) How does genome instability affect lifespan?: roles of rDNA and telomeres. Genes Cells 16, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinclair DA and Guarente L (1997) Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 29.Shcheprova Z et al. (2008) A mechanism for asymmetric segregation of age during yeast budding. Nature 454, 728–734 [DOI] [PubMed] [Google Scholar]
- 30.Ritossa FM et al. (1966) A molecular explanation of the bobbed mutants of Drosophila as partial deficiencies of“ ribosomal” DNA. Genetics 54, 819–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen S et al. (2003) Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13, 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graziani F et al. (1977) Circular ribosomal DNA during ribosomal magnification in Drosophila melanogaster. J Mol Biol 112, 49–63 [DOI] [PubMed] [Google Scholar]
- 33.Pérez-González CE and Eickbush TH (2002) Rates of R1 and R2 Retrotransposition and Elimination From the rDNA Locus of Drosophila melanogaster. Genetics 162, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-González CE et al. (2003) R1 and R2 Retrotransposition and Deletion in the rDNA Loci on the X and Y Chromosomes of Drosophila melanogaster. Genetics 165, 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu KL et al. (2018) Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. Elife 7, e109906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi T (2014) Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci 90, 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T et al. (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes & Development 12, 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brewer BJ et al. (1992) The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71, 267–276 [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T (2003) The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol 23, 9178–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weitao T et al. (2003) Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem 278, 22513–22522 [DOI] [PubMed] [Google Scholar]
- 41.Burkhalter MD and Sogo JM (2004) rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Molecular Cell 15, 409–421 [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi T et al. (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117, 441–453 [DOI] [PubMed] [Google Scholar]
- 43.Gangloff S et al. (1996) Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15, 1715–1725 [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T and Ganley ARD (2005) Recombination Regulation by Transcription-Induced Cohesin Dissociation in rDNA Repeats. Science 309, 1581–1584 [DOI] [PubMed] [Google Scholar]
- 45.Imai S et al. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 46.Smith JS et al. (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA 97, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansisidor A et al. (2018) Genomic Copy-Number Loss Is Rescued by Self-Limiting Production of DNA Circles. Molecular Cell 72, 583–593.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi Y et al. (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes & Development 17, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganley A et al. (2009) The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Molecular Cell 35, 638–693 [DOI] [PubMed] [Google Scholar]
- 50.Ivessa AS et al. (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100, 479–489 [DOI] [PubMed] [Google Scholar]
- 51.Defossez PA et al. (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Molecular Cell 3, 447–455 [DOI] [PubMed] [Google Scholar]
- 52.Mirkin EV and Mirkin SM (2007) Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev 71, 13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritossa FM (1968) Unstable redundancy of genes for ribosomal RNA. Proceedings of the National Academy of … 60, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boncinelli E et al. (1972) rDNA magnification at the bobbed locus of the Y chromosome in Drosophila melanogaster. Cell Differ. 1, 133–142 [DOI] [PubMed] [Google Scholar]
- 55.Hawley RS and Tartof KD (1983) THE EFFECT OF mei-41 ON rDNA REDUNDANCY IN DROSOPHILA MELANOGASTER. Genetics 104, 63–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hawley RS et al. (1985) Repair-defect mutations inhibit rDNA magnification in Drosophila and discriminate between meiotic and premeiotic magnification. Proc Natl Acad Sci USA 82, 8095–8099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tartof KD (1974) Unequal Mitotic Sister Chromatid Exchange as the Mechanism of Ribosomal RNA Gene Magnification. Proc Natl Acad Sci USA 71, 1272–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tartof KD (1974) Unequal Mitotic Sister Chromatid Exchange and Disproportionate Replication as Mechanisms Regulating Ribosomal RNA Gene Redundancy. Cold Spring Harb Symp Quant Biol 38, 491–500 [DOI] [PubMed] [Google Scholar]
- 59.RITOSSA F (1972) Procedure for Magnification of Lethal Deletions of Genes for Ribosomal RNA. Nature New Biology 240, 109–111 [DOI] [PubMed] [Google Scholar]
- 60.de Cicco DV and Glover DM (1983) Amplification of rDNA and type I sequences in Drosophila males deficient in rDNA. Cell 32, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 61.Tartof KD and Dawid IG (1976) Similarities and differences in the structure of X and Y chromosome rRNA genes of Drosophila. Nature 263, 27–30 [DOI] [PubMed] [Google Scholar]
- 62.Yagura T et al. (1979) Drosophila melanogaster has different ribosomal RNA sequences on X and Y chromosomes. J Mol Biol 133, 533–547 [DOI] [PubMed] [Google Scholar]
- 63.Bianciardi A et al. (2012) Ribosomal DNA organization before and after magnification in Drosophila melanogaster. Genetics 191, 703–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Endow SA et al. (1984) Ring chromosomes and rDNA magnification in Drosophila. Genetics 108, 969–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawley RS and Tartof KD (1985) A TWO-STAGE MODEL FOR THE CONTROL OF rDNA MAGNIFICATION. Genetics 109, 691–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terracol R (1987) Differential magnification of rDNA gene types in bobbed mutants of Drosophila melanogaster. Molec. Gen. Genet 208, 168–176 [DOI] [PubMed] [Google Scholar]
- 67.Schweizer E et al. (1969) The redundancy of ribosomal and transfer RNA genes in Saccharomyces cerevisiae. J Mol Biol 40, 261–277 [DOI] [PubMed] [Google Scholar]
- 68.Lyckegaard EM and Clark AG (1989) Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA 86, 1944–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyckegaard E and Clark AG (1991) Evolution of ribosomal RNA gene copy number on the sex chromosomes of Drosophila melanogaster. Mol. Biol. Evol 8, 458–474 [DOI] [PubMed] [Google Scholar]
- 70.Long EO and Dawid IB (1980) Repeated genes in eukaryotes. Annu. Rev. Biochem 49, 727–764 [DOI] [PubMed] [Google Scholar]
- 71.Zhou J et al. (2013) A population genetic model for the maintenance of R2 retrotransposons in rRNA gene loci. PLoS Genet. DOI: 10.1371/journal.pgen.1003179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nomura M (2001) Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol 66, 555–565 [DOI] [PubMed] [Google Scholar]
- 73.Planta RJ (1997) Regulation of ribosome synthesis in yeast. Yeast 13, 1505–1518 [DOI] [PubMed] [Google Scholar]
- 74.Goetze H et al. (2010) Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol. Cell. Biol 30, 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iida T and Kobayashi T (2018) RNA Polymerase I Activators Count and Adjust Ribosomal RNA Gene Copy Number. Molecular Cell DOI: 10.1016/j.molcel.2018.11.029 [DOI] [PubMed] [Google Scholar]
- 76.Zhang X et al. (2008) Role of recombination in the long-term retention of transposable elements in rRNA gene loci. Genetics 180, 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malva C et al. (1980) Comparison between rDNA magnification and bb lethal mutation frequencies in Drosophila melanogaster. Molec. Gen. Genet 180, 511–515 [DOI] [PubMed] [Google Scholar]
- 78.Komma DJ and Endow SA (1987) Incomplete Y chromosomes promote magnification in male and female Drosophila. Proc Natl Acad Sci USA 84, 2382–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Komma DJ and Endow SA (1986) Magnification of the ribosomal genes in female Drosophila melanogaster. Genetics 114, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Komma DJ et al. (1993) Constitutive magnification by the Y[bb-] chromosome of Drosophila melanogaster. Genetics research 62, 205–212 [DOI] [PubMed] [Google Scholar]
- 81.McStay B (2006) Nucleolar dominance: a model for rRNA gene silencing. Genes & Development 20, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 82.Preuss S and Pikaard CS (2007) rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim. Biophys. Acta 1769, 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greil F and Ahmad K (2012) Nucleolar dominance of the Y chromosome in Drosophila melanogaster. Genetics 191, 1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conconi A et al. (1989) Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57, 753–761 [DOI] [PubMed] [Google Scholar]
- 85.Schlesinger S et al. (2009) Allelic inactivation of rDNA loci. Genes & Development 23, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mehta IS et al. (2007) Alterations to nuclear architecture and genome behavior in senescent cells. Ann. N. Y. Acad. Sci 1100, 250–263 [DOI] [PubMed] [Google Scholar]
- 87.Stec MJ et al. (2015) The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J Appl Physiol (1985) 119, 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delabaere L et al. (2017) Aging impairs double-strand break repair by homologous recombination in Drosophila germ cells. Aging Cell 16, 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.