Significance statement
BBX19, a transcription factor with two B-Box motifs, is a suppressor of seed germination that functions as a checkpoint node that optimizes and tailors adaptive responses to distinct developmental and environmental inputs.
Keywords: BBX19, transcriptional regulation, GT1 cis-elements, seed germination, Abscisic acid (ABA), gibberellic acid (GA), ABI5, HY5
Summary
Seed germination is a fundamental process in plant life cycle regulated by functionally opposing internal and external inputs. Here, we explored the role of a negative regulator of photomorphogenesis, a B-box containing protein (BBX19), as a molecular link between the inhibitory action of the phytohormone abscisic acid (ABA) and promoting role of light in germination. We show that seeds of BBX19 overexpressing lines, in contrast to those of BBX19 RNAi, display ABA hypersensitivity, albeit independently of elongated hypocotyl 5 (HY5). Moreover, we establish that BBX19 function is neither via perturbation of the GA signaling, the ABA antagonistic phytohormone, nor through interference with the germination repressors, DELLA proteins. Rather, BBX19 functions as an inducer of ABA INSENSITIVE5 (ABI5) by binding to the light-responsive GT1-motifs in the gene promoter. In summary, we identify BBX19 as a regulatory checkpoint directing diverse developmental processes, and tailoring adaptive responses to distinct endogenous and exogenous signals.
Introduction
Germination is the earliest and the most critical biological step in a plant life cycle, regulated by a myriad of internal and external cues that ultimately enable the seed embryo to resume coordinated metabolic activities required for propagation of next generation of progenies (Chen et al., 2008; Koornneef et al., 2002; Lee et al., 2002; Wang et al., 2016; Xu et al., 2016). The delicate and highly regulated balance between functionally opposing inputs determine timing and speed of seed germination. Major known endogenous hormonal cues are abscisic acid (ABA) and gibberellic acid (GA), an inhibitor and a promotor of germination respectively. In addition, ABA signaling involves several abscisic acid (ABA)-insensitive (ABI) proteins that function in distinct ABA signalling cascades (Finkelstein et al., 1998; Leung et al., 1994; Parcy et al., 1994). Amongst them are ABI3, 4 and 5 that belong to three distinct classes of transcription factors, namely the basic B3, AP2/ERF, and the basic leucine zipper (bZIP) families that display distinct expression patterns, and are implicated in embryonic ABA responses (Finkelstein and Lynch, 2000; Finkelstein et al., 1998; Lim et al., 2013; Lopez-Molina et al., 2001; Parcy and Giraudat, 1997; Parcy et al., 1994). Specifically, ABI4 is expressed only in the embryo while both ABI3 and ABI5 are expressed in both endosperm and embryo, and ABI5 has been reported to act downstream of ABI3(Dai et al., 2013; Lim et al., 2013; Lopez-Molina et al., 2002; Park et al., 2011). It is suggested that ABI3 and ABI5 could act as the ultimate common repressors of germination in response to changes in ABA and GA levels (Lim et al., 2013; Penfield et al., 2006; Piskurewicz et al., 2008).
In addition to endogenous hormones, the external factors such as light alter germination specifically by inducing this physiological process in small-seeded plants including Arabidopsis, lettuce, tomato and tobacco, in part through modulation of GA and ABA levels (Seo et al., 2009).
The B-BOX family of proteins (BBX) is comprised of 32 functionally diverse zinc finger proteins with one or two N-terminal zinc binding BBX domains (Crocco and Botto, 2013; Gangappa and Botto, 2014; Khanna et al., 2009). Eight of the BBX (BBX18 to BBX25) proteins contain two tandem repeat B-box motifs established to be critical for transcriptional regulation as well as homo- and hetero-dimerization with other family members or with nonfamily member proteins (Crocco and Botto, 2013; Gangappa and Botto, 2014; Khanna et al., 2009). Among the group members BBX20, BBX21 and BBX22 are reported to promote photomorphogenesis, whereas BBX18, BBX19, BBX24, and BBX25 are implicated as negative regulators of photomorphogenesis (Datta et al., 2007; Datta et al., 2008; Gangappa and Botto, 2014; Gangappa et al., 2013; Jiang et al., 2012; Wang et al., 2015). There are several reports on the function of BBX family members as integrators of light and other endogenous inputs. An example is the positive regulator of photomorphogenesis, BBX21 (also known as SALT TOLERANCE HOMOLOG 2) that binds to the T/G-box motif in the promoter of elongated hypocotyl 5 (HY5) and induces this b-ZIP type transcription factor in the light (Xu et al., 2016). However, BBX21 also negatively regulates ABI5 expression by interfering with HY5 binding to the ABI5 promoter, and as such is designated as an integrator of light and ABA signaling in Arabidopsis thaliana (Xu et al., 2014).
Recently, we identified BBX19 as a negative regulator of flowering time and a suppressor of photomorphogenesis (Wang et al., 2014; Wang et al., 2015). In this report, we establish BBX19 as a repressor of seed germination by the virtue of inducing the expression of the ABI5. Collectively, our finding expands the function of BBX19 in developmental processes that encompass germination, growth and the flowering time. This report broadens the repertoire of BBX proteins that function as a checkpoint to align endogenous and exogenous inputs, thereby enabling timely and optimal responses of several facets of biological activities.
Results
BBX19 induction of ABA hypersensitivity is independent of the GA-signaling cascade
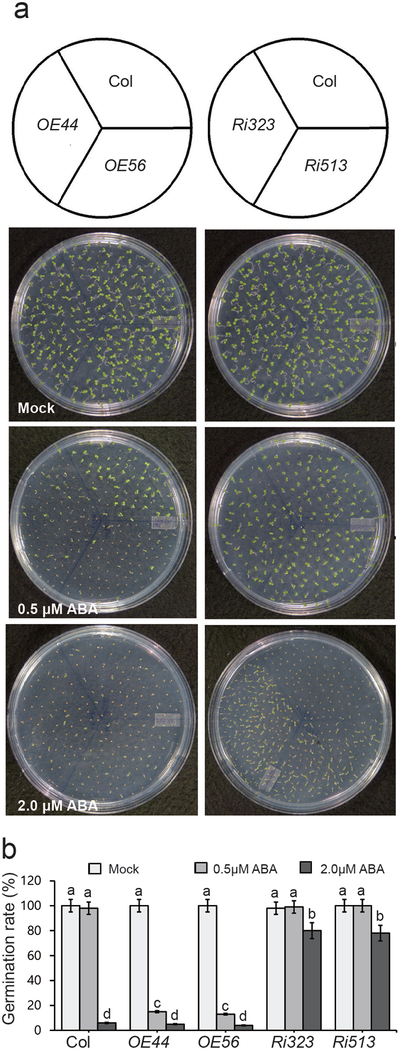
The opposing functions of light and ABA signaling in promoting and suppressing seed germination respectively (Nakamura et al., 2001; Xu et al., 2014), led us to question the potential role of the negative regulator of photomorphogenesis, BBX19, in this fundamental biological process. Thus, we examined germination rates of BBX19 over-expressing (OE44, OE56), and RNAi (Ri323, Ri513) seeds, on various ABA concentration. Initially we examined whether the basal levels of germination in seeds of plants with altered BBX19 levels are modified. The similar germination rates among the various genotypes implies that under standard conditions modulation of BBX19 levels does not alter the timing of seed germination (Fig. S1). Next, we examined whether the germination rate among the aforementioned genotypes might be altered in the presence of ABA. This data established genotype-dependent differential sensitivity of seed germination in response to different concentrations of ABA (Fig. S2). Specifically, in the presence of inhibitory concertation of ABA (2 μM), Ri323, Ri513 seeds display up to 80% germination rate, whereas, these rates both in the control and in the overexpressing OE44, OE56 seeds is reduced to only ~5–10% (Fig. 1a-b). Moreover, OE44, OE56 seeds display enhanced sensitivity to the presence of otherwise non-inhibitory concentration of ABA (0.5μM), and the fact that longer incubation time does not overcome this hypersensitivity (Fig. S3), indicating that the response is not simply due to delayed germination. Collectively, the result establishes BBX19-mediated altered sensitivity of seeds to ABA (Fig. 1a-b).
Fig. 1. BBX19 induces ABA hypersensitivity.
(a) Representative germination phenotypes of 5-d-old seedlings from various genotypes grown on plates with 1/2 MS media, in the absence (mock) and the presence of ABA (0.5 and 2.0 μM).
(b) Germination rates of mock and ABA treated seeds of various genotypes. Germination rate was based on three biological replicates, and three technical repeats each with at least 100 seeds (n>100). Data are means ± SD, letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
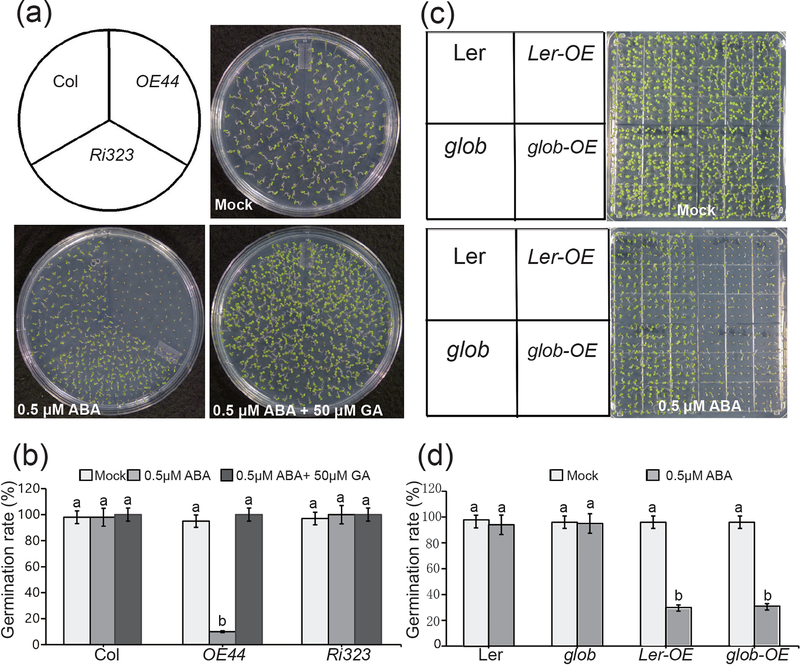
The antagonistic function of the two phytohormones GA and ABA, prompted us to question whether this altered sensitivity of seed germination to ABA might be mediated through an interference of BBX19 with the GA signaling cascade. To address this question, we exploited seeds from OE44 and Ri323 plants as representative transgenic lines with opposing levels of BBX19 expression. Germination analyses of their respective seeds treated with ABA (0.5 μM) in the presence and absence of GA3 (50 μM) clearly exhibited recovery of OE44 hypersensitivity to the ABA in the presence of GA (Fig. 2a-b). This data support the functionality of GA in promoting germination in OE44 seeds, thus illustrating that the BBX19 mode of action is not through perturbation of the GA-signaling cascade.
Fig. 2. BBX19 induction of ABA hypersensitivity is DELLA independent.
(a) The representatives of seed germination from various genotypes, grown for 5 days after stratification on 1/2 MS germination medium plates in the absence (mock), and the presence of ABA
(b) Germination rates of mock, ABA (0.5 μM) alone or together with GA3 (50 μM) treated seeds from various genotypes.
(c) Representative phenotypes of seed germination from various genotypes including LER, the global-DELLA mutant (glob; gai-t6/rga-t2/rgl1–1/rgl2–1/rgl3–4), Ler-OE and glob-OE grown on 1/2 MS germination medium plates in the absence (mock) or the presence of ABA (0.5 μM) 5-days after stratification.
(d) Germination rates of the above genotypes determined from three biological replicates, and three technical replicates, each with at least 100 seeds (n>100). Data are means ± SD, letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
Next, we questioned whether BBX19-mediated induction of ABA hypersensitivity might be through stabilization of the germination repressors, DELLA proteins. These proteins are known to be targeted to 26S proteasome-mediated degradation in response to increasing GA levels which in turn promote seed germination (Fuentes et al., 2012; Lee et al., 2002; Piskurewicz et al., 2008). To examine this possibility, we introduced the 35S:BBX19 construct into the global-DELLA (gai-t6/rga-t2/rgl1–1/rgl2–1/rgl3–4) mutant lacking all the five DELLA proteins (Nakamura et al., 2001), and the corresponding Landsberg erecta (Ler) wild type background. Notable reduction in germination rates of BBX19-overexpressing Ler (Ler-OE) and global mutant (glob-OE) seeds to otherwise non-inhibitory ABA concentration (0.5 μM) in their respective backgrounds clearly support an ecotype-independent and a DELLA-independent mechanism of BBX19 action (Fig. 2c-d).
BBX19 induces expression of selected ABIs and functions upstream of ABI3 and ABI5
To further examine the underlying mechanism of BBX19 action in regulation of seed germination, we explored the potential function of BBX19 as a regulator of ABA biosynthetic pathway by analyzing ABA levels in dry and imbibed seeds of Col and OE44 and Ri323 (Fig. S4). Similarly high ABA levels in dry seeds followed by the expected but equally reduced levels in 3-d imbibed seeds of all genotypes examined, excludes BBX19 function in the ABA biosynthetic pathway.
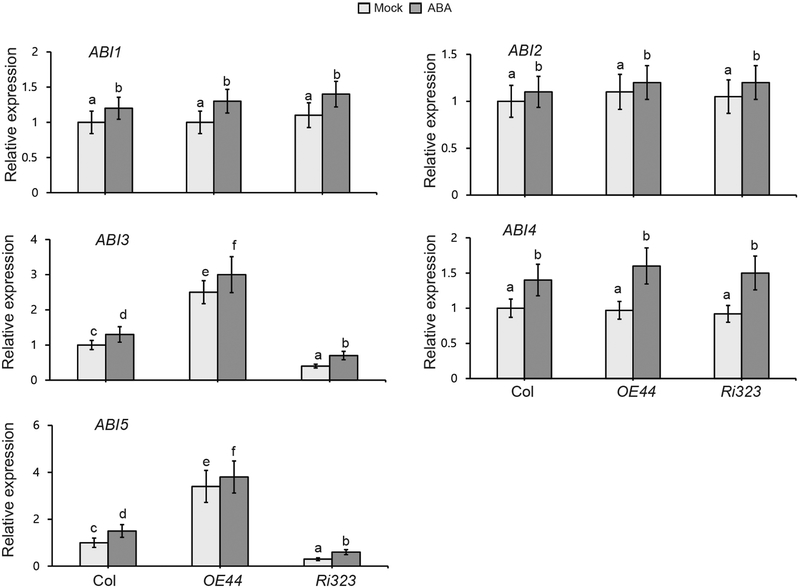
The above findings led us to examine the BBX19 site of action in the ABA signaling cascade, with an initial focus on the germination repressors, ABI proteins. We first examined the transcript levels of all five ABIs (ABI1-to-ABI5) in mock or ABA (0.5 μM) treated 3-d-old seedlings of various genotypes (Col, OE44 and Ri323) (Fig. 3). The data show increased transcript levels of these genes in response to ABA treatment regardless of the genotype, albeit at different degrees. Moreover, the analyses display heightened and exclusive induction of basal and ABA-treated transcript levels of ABI3 and ABI5 in OE44 as compared to the Col background (Fig. 3). Conversely, the transcript levels of the ABI3 and ABI5 are notably reduced in Ri323 line as compared to the control and to the OE44 lines. This selective BBX19-medited alteration of transcript levels of the two common repressors of germination (ABI3 and ABI5) points at the function of BBX19 in the ABA signaling cascade.
Fig. 3. BBX19 is an inducer of ABI3 and ABI5.
Total RNA was extracted from 3-d-old seedlings of wild-type Col-0, OE44 and Ri323 grown in the absence (mock) and the presence of ABA (0.5 μM), and subjected to real-time quantitative PCR analysis. The transcript levels were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. Data are mean fold differences ± SD of three biological replicates each with three technical repeats, letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
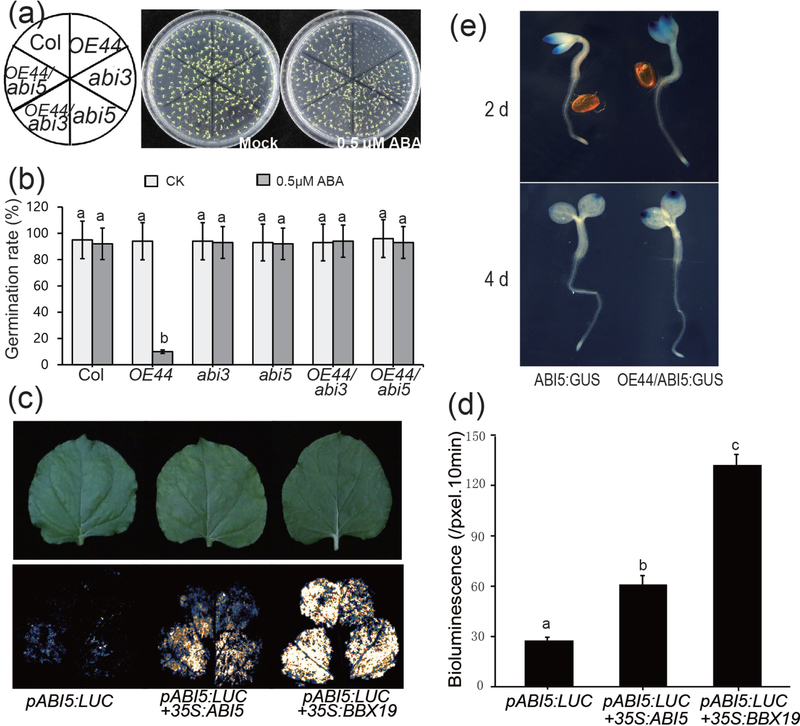
To genetically assess the involvement of BBX19 in the ABA signalling pathway, we generated OE44 homozygous lines crossed to abi null mutants, namely abi3 (SALK_023411C) and abi5 (SALK_013163C). The outcome of these analyses using these genotypes display similar germination rates for mocked and ABA treated single (abi3 and abi5) and homozygous abi3/ and abi5/OE44 seeds (Fig. 4a-b), suggesting that BBX19 site of action is most likely upstream of ABI3 and ABI5 in the ABA signaling pathway.
Fig. 4. BBX19 is epistatic to ABI3 and ABI5 and binds the promoter of ABI5.
(a) The representatives of germination rates of Col, OE44, abi3, abi5, OE44/abi3, OE44/abi5 seeds sown on plates containing 1/2MS germination medium in the absence (mock) or the presence of ABA (0.5 μM) 5 days after stratification.
(b) Germination rates of mock and ABA treated genotypes (Col, OE44, abi3, abi5, OE44/abi3, OE44/abi5). Germination rate was determined from three biological replicates, and three technical replicates with at least 100 seeds in each repeat (n>100). Data are means ± SD, letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
(c) Representative images of transient expression assays in N. benthamiana displayed by bright- (upper panel) and dark-field (lower panel), and (d) intensities of the LUC bioluminescence of leaves expressing pABI5:LUC alone and pABI5:LUC+35S:BBX19, and expression of pABI5:LUC+35S:ABI5 as an established positive control. Intensities of the LUC bioluminescence were measured using Andor Solis image analysis software. Data are means ± SD (n = 20), letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
(e) The representatives of ABI5:GUS and OE44/ABI5:GUS seedlings grown on 1/2 MS germination medium plates for 2- and 4-d after stratification show enhanced GUS intensity as a proxy for ABI5 protein levels in the OE44 background.
BBX19 induces ABI5 by direct binding to GT1-motifs in the gene promoter
It is established that ABI5 acts downstream of ABI3 to repress germination (Lopez-Molina et al., 2002; Nakamura et al., 2001). As such, we focused on the potential direct role of BBX19 in regulation of ABI5. Moreover, it is reported that another BBX family member, BBX21, promotes germination by direct interaction with ABI5 protein and thus interfering with binding of the ABI5 to its own promoter (Xu et al., 2016). Accordingly, we initially examined potential binding of ABI5 protein to BBX19 as a possible coactivator inducing expression of ABI5, by a combination of transient transfection assays in tobacco leaves and Y2H analysis (Fig. S5 and S6). This data confirmed the earlier finding regarding BBX21 interacting with ABI5, and further established lack of interaction between BBX19 and ABI5.
Next, we examined whether BBX19 is a direct regulator of ABI5 expression levels. To address this possibility, we employed ABI5 promoter-driven LUC activity as an activation proxy in the presence and absence of 35S:BBX19 construct using transient transfection assays in tobacco leaves (Fig. 4c-d). The result show 5-fold increase in the basal levels of LUC activity in leaves co-infiltrated with pABI5:LUC+35S:BBX19, as compared with the 2-fold increase in the presence of the established positive regulator pABI5:LUC+35S:ABI5 (Xu et al., 2014). This suggests that BBX19 binds to the ABI5 promoter, and induces the expression of the gene. This notion is further supported by studies using stably transformed Arabidopsis line expressing pABI5:ABI5:GUS alone or in conjunction with OE44 (OE44/ABI5:GUS) displaying enhanced GUS intensity as a proxy for ABI5 protein levels in the OE44 background (Fig. 4e). Collectively, these results identify BBX19 as a transcriptional inducer of ABI5.
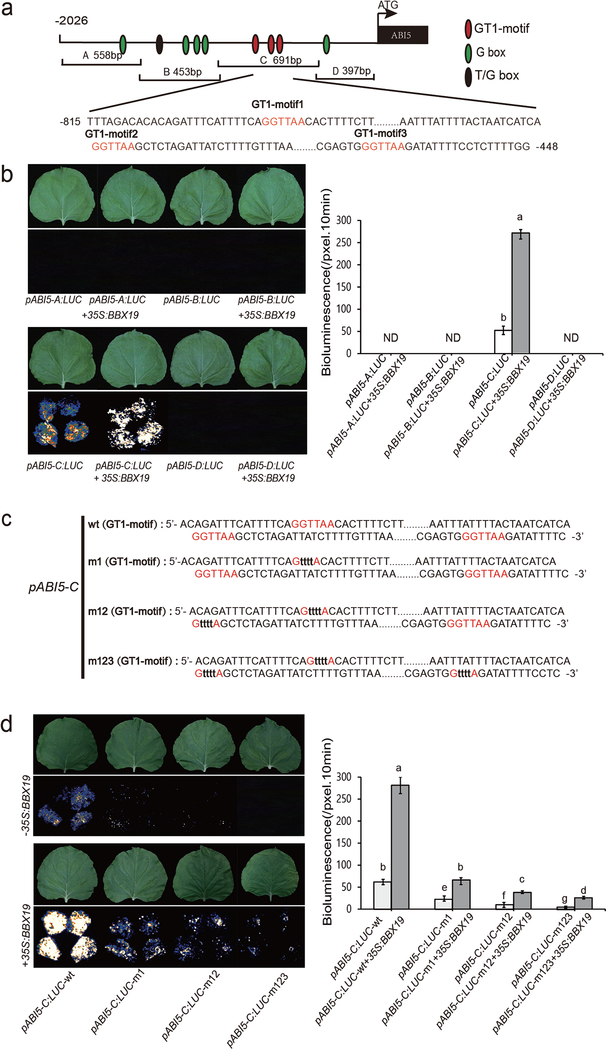
To identify the BBX19 targeted cis-elements in the promoter of ABI5, we cloned four overlapping fragments of the ABI5 promoter, designated A, B, C, and D (Fig. 5a), and generated LUC reporter fusion constructs for each fragment. The rationale for selecting these fragments was the presence of known cis-elements in each segment, namely: i. the presence one G-box (CACGTG) essential for HY5 binding (Chen et al., 2008) in fragment A; ii. The presence of three G-boxes necessary for ABI5 binding in fragment B (Xu et al., 2014) as well as a T/G box known to be present in the promotors of light regulated genes such as HY5 (Xu et al., 2016); iii. the presence of three typical GT1-motifs (GGTTAA) designated as light responsive element (Chattopadhyay et al., 1998; Gilmartin et al., 1992) in fragment C; and iv. the presence of one G-Box in fragment D (Fig. 5a). These promoter fragments were employed in a yeast-one-hybrid (Y1H) as well as in transient expression assays in tobacco leaves. The collective data clearly show exclusive binding of BBX19 to the C fragment which contains three GT1 motifs (Fig.5b and Fig. S7a). To provide further evidence of binding and to determine the biological relevance and contribution of each of the GT1 boxes to this binding, we performed a combination of EMSA and transient expression assays using the intact and mutated GT1 boxes (GGTTAA mutated to GttttA) individually and in various combinations (designated m1, m12 and m123) (Fig. S8 and Fig. 5c). The EMSA assay clearly show direct binding of BBX19 to each of the three GT1 boxes (Fig. S8). Moreover, transient expression assays show that with increasing number of mutated GT1-motifs there is a further reduction in LUC activity measured in transient expression as well as in β-galactosidase activity in Y1H assays (Fig. 5d and Fig. S7b). Collectively, these analyses reveal the direct binding of the BBX19 to each of the three GT1 boxes, and establish the critical role of all three GT1 motifs in the induction of ABI5.
Fig. 5. BBX19 directly binds to GT1-motifs in the promoter of ABI5.
(a) Schematic diagram of the ABI5 promoter depicting overlapping fragments containing known functional cis-elements. The GT1 motif sequences are illustrated below. The adenine residue of the start codon (ATG) was assigned position +1, and the numbers flanking the sequences of fragments were based on this number. A, B, C, and D indicate the corresponding promoter fragments used in binding assays.
(b) Representative images of transient expression assays in N. benthamiana displayed by bright- (upper panel) and dark- (lower panel) field (left) and intensities of the LUC bioluminescence (right) of leaves expressing pABI5-A:LUC, pABI5-B:LUC, pABI5-C:LUC and pABI5-D:LUC alone and together with 35S:BBX19, respectively. Intensities of the LUC bioluminescence were measured using Andor Solis image analysis software. Data are means ± SD (n = 20), letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
(c) Nucleotide sequences of the wild type (wt) and mutants (mu, nucleotide substitutions shown in lowercased black letters) of GT1 motifs (ABI5, fragment C) used in transient transfection assays in N. benthamiana leaves.
(d) Representative images of transient expression assays in N. benthamiana displayed by bright- (upper panel) and dark-(lower panel) field (left) and intensities of the LUC bioluminescence (right) of leaves expressing pABI5-C:LUC, pABI5-C:LUC-m1, pABI5-C:LUC-m12 and pABI5-C:LUC-m123 alone and together with 35S:BBX19, respectively. Intensities of the LUC bioluminescence were measured using Andor Solis image analysis software. Data are means ± SD (n = 20), letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
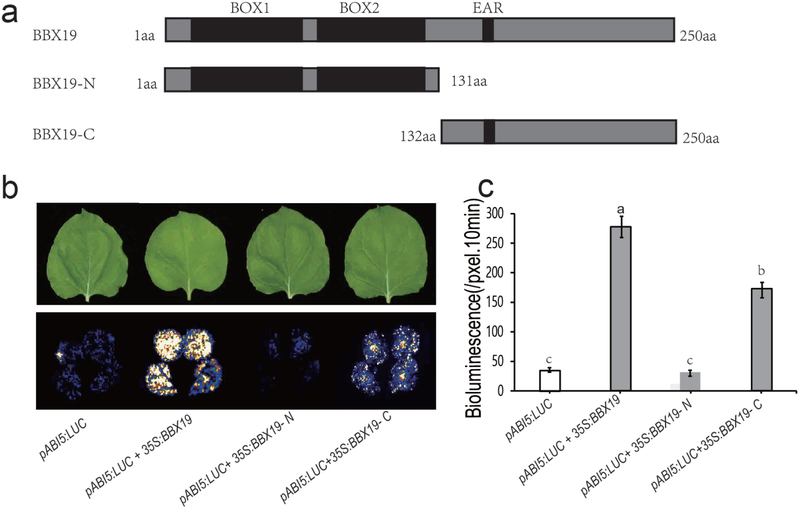
Next, we further utilized transient transfection assays in tobacco leaves and Y1H to identify the DNA-binding domain of BBX19. These assays were performed with the full-length, as well as amino- (1–131 amino acids) and carboxyl-terminus (132–250 amino-acids) of BBX19 (Fig. 6a-c, Fig. S9). These analyses show LUC activity with both full-length and with the C-terminus portion of the BBX19. However, the strongest noted LUC activity, especially in transient expression assay, was in the presence of the full-length BBX19. This is likely due to the necessity for additional DNA-binding regions potentially upstream of the current C-terminus. However, the absence of signal with the N-terminus, eliminates potential function of both B-boxes as DNA binding domains of BBX19.
Fig. 6. The C-terminal of BBX19 binds to GT1 motifs.
(a) Schematic diagram of the BBX19 full-length and segmented protein depicting BOX1 and BOX2 at the N-terminus (1–131 aa) and the EAR motif in the C-terminal (132–250 aa).
(b) Representative images of transient expression assays in N. benthamiana displayed by dark- and bright-field, and (c) LUC intensities of leaves expressing pABI5:LUC, pABI5:LUC+BBX19, pABI5:LUC+BBX19-N, pABI5:LUC+BBX19-C, respectively. Intensities of the LUC bioluminescence were measured using Andor Solis image analysis software. Data are means ± SD (n = 20), letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
Collectively, the finding identifies BBX19 as a transcriptional regulator that induces ABI expression through direct binding to the three GT1-motifs of the gene promoter.
BBX19-mediated suppression of seed germination is HY5-independent
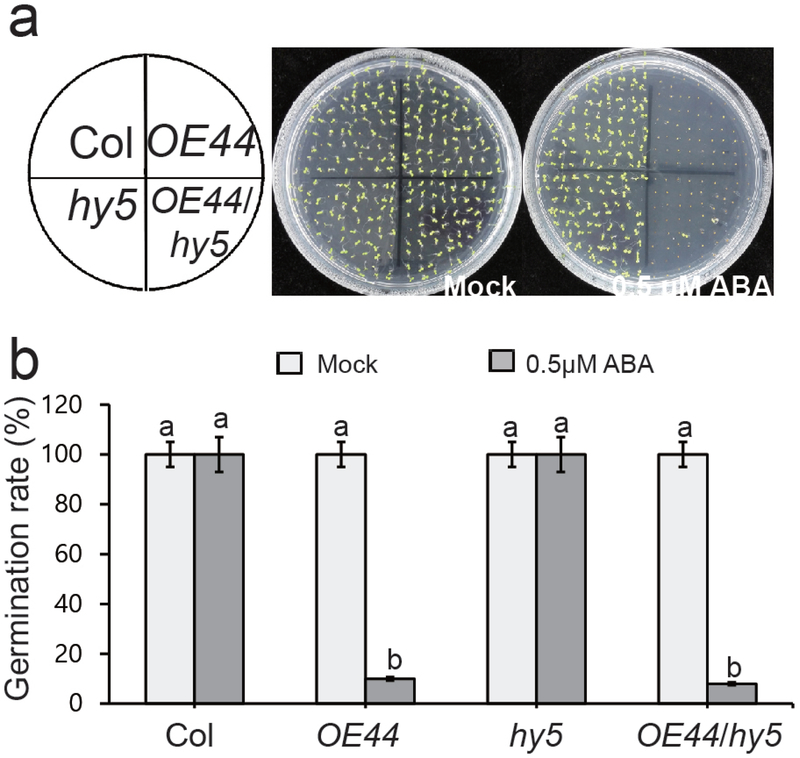
The HY5-mediated ABA responses in seed germination via direct binding to G-Box motif required for the expression of ABI5 and ABI5-targeted genes is well-established (Chen et al., 2008). Accordingly, we sought to genetically investigate a potential interaction between HY5 and BBX19 in ABA regulation of seed germination. To this end, we crossed and generated hy5 with OE44 progenies and examined their respective germination rates. For these analyses, we specifically utilized mock and ABA treated genotypes of Col, OE44, hy5 alone or together with OE44 (OE44/hy5) (Fig. 7). The result illustrates indistinguishable germination rates of mock and ABA-treated hy5 and Col seeds. However, the identical germination rates of mock treated hy5 and OE44 and OE44/hy5 differ notably from those of the ABA-treated seeds. Specifically, germination rates of ABA-treated seeds at ~ 10% in all OE44 backgrounds is in sharp contrast to the 100 % germination of hy5 seeds (Fig. 7). This finding verifies HY5-independent function of BBX19 in induction of ABA hypersensitivity and by extension suppression of seed germination.
Fig. 7. BBX19-mediated suppression of seed germination is HY5-independent.
(a) The representative images of seed from various genotypes germinated on 1/2 MS medium plates in the absence (mock) and the presence of ABA (0.5 μM) 5-d after stratification.
(b) Germination rates of mock and ABA treated seeds from Col, OE44, hy5, OE44/hy5 determined from three biological replicates, with three technical replicates each with at least 100 seeds (n>100). Data are means ± SD, letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
Discussion
BBX family of proteins belong to the class of transcription factors that through protein-protein interaction and by transcriptional activity can regulate several facets of biological processes ranging from photomorphogenesis, photoperiodic regulation of flowering, shade avoidance and responses to environmental stresses (Gangappa and Botto, 2014). In addition to the involvement of BBX proteins in light regulated processes, several lines of evidence illustrate their function in hormonal signaling pathways (Huang et al., 2012; Luo et al., 2010; Sun et al., 2010; Wang et al., 2011; Weller et al., 2009). These reports collectively support the notion that BBX proteins function as integrators of internal and external inputs to optimize plant responses to the prevailing environmental conditions and developmental status. This and established role of light as a critical regulator of seed germination by modulating endogenous levels of gibberellin (GA) and abscisic acid (ABA) led us to examine a potential role of the BBX19, previously identified as a negative regulator of flowering time and a suppressor of photomorphogenesis (Wang et al., 2014; Wang et al., 2015), in seed germination. Our finding clearly illustrate that BBX19 is a suppressor of germination via increasing hypersensitivity of seeds to ABA. This enhanced sensitivity to ABA could be the result of suppression of the signalling of the antagonistic hormone GA (Piskurewicz et al., 2008), or due to enhancement of ABA levels/signalling or both. Here we establish that BBX19 function is neither through perturbation of the GA signaling cascade, nor is through interference with the germination repressors, DELLA proteins, the known negative regulators of the GA pathway (Zentella et al., 2007). Moreover, BBX19 function is not via intrusion with ABA biosynthetic pathway and alteration of ABA levels. We show that, BBX19 rather functions as an inducer of ABI5, an essential protein for execution of ABA dependent arrest of germination (Lopez-Molina et al., 2001; Lopez-Molina et al., 2002). We specifically show that BBX19 binding to the GT1 motifs in the ABI5 promoter result in induced expression and hence increased levels of ABI5 protein, and by extension suppression of seed germination. This mode of BBX19 action is in contrast to dual mechanisms by which BBX21 suppresses ABI5 expression (Xu et al., 2014). In one mechanism, BBX21 direct interaction with ABI5 protein prohibits binding of the ABI5 to its own promoter, and in the other, BBX21 interferes with HY5 binding to the ABI5 promoter. Our data clearly establishes that BBX19 induction of ABI5 is through direct interaction with the GT1 motifs and is independent of HY5. As such, the result verifies distinct and antagonistic roles of different BBX family members in the same physiological process, devised for optimal responses tailored to the cues perceived.
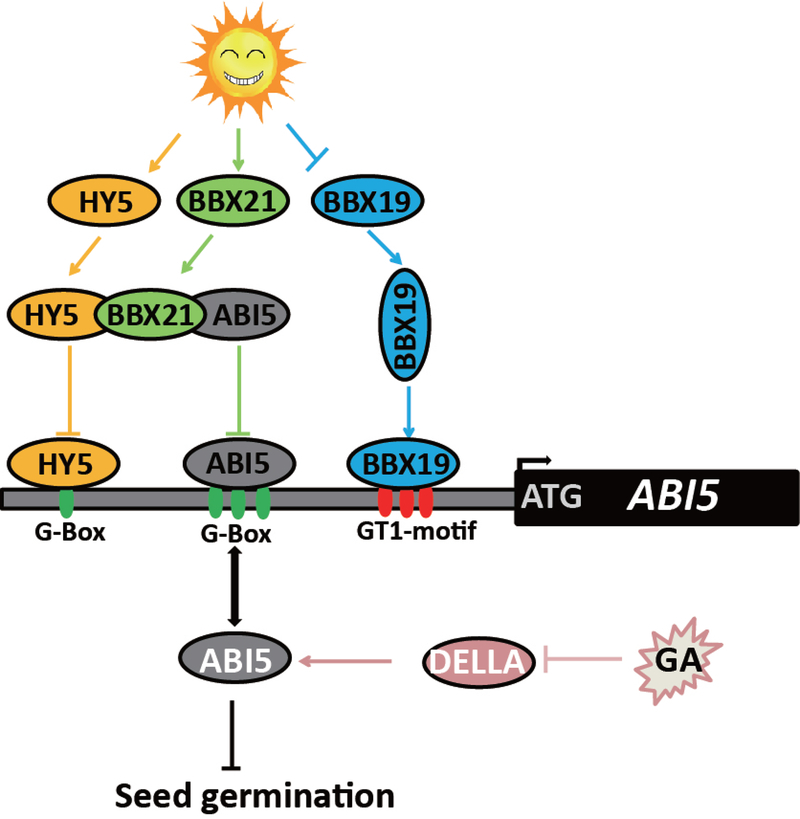
The summary of our finding is captured in the proposed model depicting BBX19 as a transcriptional regulator that directly binds to the GT1-motifs in the ABI5 promoter and induces the expression of this gene, which in turn results in enhanced ABA hypersensitivity and ultimately suppression of seed germination (Fig. 8). This model further depicts the highly regulated and quintessential balance between the intricate networks regulating one of the most critical biological steps in a plant life cycle. The intricate circuitry is comprised of the two opposing networks regulated by a promoter of photomorphogenesis, HY5 and a suppressor of photomorphogenesis BBX19 that control seed germination by repressing and inducing ABI5 respectively. These regulatory circuitries in coordination with the networks that establish the delicate balance between GA levels and degradation of DELLA, control seed germination.
Fig. 8. Schematic model of the circuitry network regulating seed germination.
The model depicts ABI5 as a convergence node through which various independent or interdependent circuity networks regulate seed germination. In one network, germination is promoted through HY5 binding to BBX21 which, in turn binds to and sequester ABI5 required for the induction of its own gene. In the other, BBX19 binding to the GT1 motif induces ABI5 and thus suppresses germination. Lastly, enhanced GA levels leads to degradation of DELLA, that otherwise enhances ABA synthesis and block germination. The tightly controlled integration of these circuitries determines optimal timing of seed germination.
Taken together, our collective approaches establish BBX19 as a suppressor of a spectrum of developmental processes through a complex regulatory function that leads to delayed flowering time, and suppression of photomorphogenesis and seed germination. Regulation of these distinct biological processes throughout the plant life cycle are either by BBX19 targeted interactions with hetero-proteins resulting in delayed flowering time and suppression of photomorphogenesis (Wang et al., 2014; Wang et al., 2015), or through its direct binding to functional cis-elements, as is the case of suppression of seed germination. Thus, BBX19 is a checkpoint node that regulates a spectrum of biological processes through optimizing and tailoring adaptive responses to distinct developmental and environmental inputs.
Experimental procedures
Plant materials and growth Conditions
BBX19 over-expression and RNAi transgenic Arabidopsis thaliana lines in ecotype Col-0 (OE44, OE56, Ri323, Ri513), and over-expression in Ler background (Ler-OE, glob-OE) were generated as previously described(Wang et al., 2014). The DELLA global mutant seeds in Ler background were gifts from Lars Ostergaard (UK). pABI5:ABI5:GUS transgenic seeds were provided by Dr. JZ Wang from University of California, Riverside. abi3 (SALK_023411C), abi5 (SALK_013163C) and hy5 (SALK_056405C) seeds were purchased from ABRC (Arabidopsis Biological Resource Center).
Surface-sterilized Arabidopsis seeds were planted on half-strength Murashige and Skoog medium (2.16 g/L Murashige and Skoog salts, pH 5.75, and 8 g/L agar) without and with various concentration of ABA. To synchronize germination, the plates were kept for 3 days at 4°C prior to their transfer to growth chambers maintained at 22°C under LD (16-h-light/8-h-dark cycle) conditions. GA application was by adding GA3 (50 μM) to the 1/2 MS medium.
Analyses of seed germination and quantification of endogenous ABA levels
Seed germination assays were performed with minimum sample size of 100 seeds/genotype sown on 1/2MS medium supplemented with various ABA concentrations (0, 0.5 and 2.0 μM). Germination was defined as emergence of radicle’s tip and scored daily up to the 6th day post synchronization. Each experiment was performed on at least three independent replicates.
Endogenous ABA levels in dry seeds and 3-d old seedlings were analyzed by gas chromatography coupled to electrospray tandem mass spectrometry as described previously (Savchenko et al., 2014).
Expression Analyses
Expression analyses using RT-qPCR were conducted as described previously (Walley et al., 2008). Briefly, total RNA from 3-d old seedlings was isolated by TRIzol extraction (Life Technologies) and further purified using the Qiagen RNeasy kit with on-column DNase treatment (Qiagen) to eliminate DNA contamination. One microgram of RNA was reverse transcribed using SuperScript III (Invitrogen). RT-qPCR was performed using At4g34270 and At4g26410 as reference genes for the internal controls as described previously for transcript normalization. The primers used are listed in Supplemental Table 1.
Nicotiana benthamiana transient transformation, promoter activity and protein/protein interaction assays
Agrobacterium (GV3101) infiltration assays using Nicotiana benthamiana leaves were carried out as previously described (Wang et al., 2014) and promoter:LUC activity assay were performed by measuring luciferase activity using a CCD camera (Andor Technology) as previously described(Wang et al., 2014). Promoter:Luc activity assays were conducted with the full-length pABI5 promoter (2026 bp) or a selected fragments (A, B, C and D) obtained by amplifying genomic DNA from Col-0 ecotype using the listed primers (Supplemental Table 1), followed by cloning the DNA fragments into pENTR-D-TOPO vector (Invitrogen). Point mutations in GT1-motif were introduced using Site-Directed Mutagenesis Kit (NEB) according to the manufacturer’s instructions. Expression plasmids (pABI5:LUC, pABI5-A:LUC, pABI5-B:LUC, pABI5-C:LUC and pABI5-D:LUC) were prepared by LR recombination between the entry vector and the binary vector pBGWL7. The various combination of full-length and promoter fragments (pABI5:LUC) constructs together with 35S:BBX19 were vacuum infiltrated into Nicotiana benthamiana leaves, and luciferase activities were determined 72 h thereafter by a CCD camera (Andor Technology).
For identification of the DNA-binding domain of BBX19, the amino- (1–131 amino acids) and carboxyl-terminus (132–250 amino-acids) segments of the protein were cloned using the listed primers (Supplemental Table 1). The generation of 35S:BBX19, 35S:BBX19-N and 35S:BBX19-C construct used in co-infiltration assays were as previously described (Wang et al., 2014). Split luciferase system was used in protein/protein interaction analysis. 35S:BBX19:LUC-C, 35S:BBX21:LUC-C, and 35S:ABI5:LUC-N were generated by multiple LR reactions as we described previously (Wang et al., 2014). 35S:BBX19:LUC-C or 35S:BBX21:LUC-C with 35S:ABI5:LUC-N were co-infiltrated into Nicotiana benthamiana leaves to test the possible interaction. The luciferase activities as a measure of reconstitution of an active luciferase were determined 72 h thereafter.
Yeast-hybrid experiments
Yeast- (Saccharomyces cerevisiae) one-hybrid assay used to identify the promoter binding site, was performed with yeast strain Y187 cells (Clontech) according to the manufacturer’s instructions. BBX19, BBX19-N and BBX19-C were cloned into the pGADT7 vector, and the full length as well as different overlapping fragments (A, B, C and D) of ABI promoter were inserted into the pHIS2 vector. Different combinations of recombinant and empty vectors were co-transformed into yeast Y187 cells, and the interactions were examined on SD medium lacking Trp, Leu, and His (SD/−Trp/−Leu/−His) with 15 and 25 mM of 3-amino-1, 2, 4-triazole.
Protein-protein interaction between BBX19/BBX21 and ABI5 was examined by yeast-two-hybrid assay conducted according to the manufacturer’s instructions (Clontech, Mountain View, CA, USA). The coding sequences were inserted into the pGBKT7 vector (bait, BD), and the pGADT7 vector (prey, AD). All recombinant vectors were co-transformed into yeast strain Y2H Gold cells using the lithium acetate method. The cells were cultured on synthetic defined (SD) medium lacking leucine and tryptophan (SD/−Leu/−Trp). Putative transformants were transferred to SD medium lacking adenine, histidine, Leu, and Trp (SD/−Ade/−His/−Leu/−Trp; Clontech) with or without X-α-gal.
GUS Histochemical Assay
GUS staining were performed on 2-d and 4-d old seedlings grown on 1/2MS medium at long day (16-h-light/8-h-dark cycle) condition. The seedlings were stained in buffer containing 100 mM phosphate buffer, pH 7, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl-b-D-glucuronic acid at 37°C for 24 h before de-staining in ethanol.
Electrophoretic mobility shift assay
An electrophoretic mobility shift assays (EMSA) were performed using biotinylated probes and the LightShift Chemiluminescence EMSA kit (Thermo Scientific, Waltham, MA, USA). The AtBBX19-His fusion protein and probes (biotin-labeled, unlabeled, and mutated probes) were mixed and incubated at 24 °C for 20 min. The unlabeled probes were used as control in competition assay. The free and bound probes were separated on an acrylamide gel electrophoresis.
Statistical Analyses
To determine statistical significance, we employed Tukey’s honesty significant difference (HSD) test. The difference was considered significant at P < 0.05.
Supplementary Material
Fig. S1 Modulated BBX19 levels does not alter basal timing of seed germination
Fig. S2 Modulation of BBX19 levels alters seeds sensitivity to ABA.
Fig. S3 BBX19 overexpresser seeds remain hypersensitive to ABA
Fig. S4 BBX19 is not a modulator of the ABA production
Fig. S5 ABI5 interact with BBX21 in transient expression system in N. benthamiana
Fig. S6 ABI5 interacts with BBX21 in yeast-two-hybrid assay.
Fig. S7 BBX19 binds to GT1 motifs in ABI5 promoter in yeast-one-hybrid assay
Fig. S8 BBX19 binds to the GT1-motifs in the promoter of ABI5
Fig. S9 The C-terminal of BBX19 binds to the ABI5 promoter
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (KYYJ201706), the Key Research and Development Project of Jiangsu Province (BE2016377), and the NSFC-XINJIANG joint foundation (U1803102) granted to Changquan Wang, and the China Postdoctoral Science Foundation (2016M600425) and the NSFC (31801890) to Jinyi Liu, and National Science Foundation (NSF) IOS-1036491, NSF IOS-1352478, and National Institutes of Health (NIH) R01GM107311 to KD.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- Chattopadhyay S, Puente P, Deng XW and Wei N (1998) Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J 15, 69–77. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW and Xiong L (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci U S A 105, 4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco CD and Botto JF (2013) BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531, 44–52. [DOI] [PubMed] [Google Scholar]
- Dai M, Xue Q, McCray T, Margavage K, Chen F, Lee JH, Nezames CD, Guo L, Terzaghi W, Wan J, Deng XW and Wang H (2013) The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25, 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H and Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19, 3242–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V and Holm M (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR and Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S and Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S, Ljung K, Sorefan K, Alvey E, Harberd NP and Ostergaard L (2012) Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 24, 3982–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN and Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19, 460–470. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M and Botto JF (2013) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. The Plant cell 25, 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Memelink J, Hiratsuka K, Kay SA and Chua NH (1992) Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell 4, 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Weng X, Wang L and Xie W (2012) The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS One 7, e48242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li QF, Bjorn LO, He JX and Li SS (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22, 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T and Wu SH (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21, 3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L and Hilhorst H (2002) Seed dormancy and germination. Current opinion in plant biology 5, 33–36. [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP and Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes & development 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F and Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264, 1448–1452. [DOI] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N and Choi G (2013) ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S and Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT and Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32, 317–328. [DOI] [PubMed] [Google Scholar]
- Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, Cheng M, Hao Y, Oh E, Tian M, Liu L, Zhang M, Xie Q, Chong K and Wang ZY (2010) Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Developmental cell 19, 872–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ and Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Parcy F and Giraudat J (1997) Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J 11, 693–702. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M and Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee N, Kim W, Lim S and Choi G (2011) ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S and Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y and Lopez-Molina L (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T, Kolla VA, Wang CQ, Nasafi Z, Hicks DR, Phadungchob B, Chehab WE, Brandizzi F, Froehlich J and Dehesh K (2014) Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol 164, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G and Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69, 463–472. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang WQ, He K, Zhu JY, He JX, Bai MY, Zhu SW, Oh E, Patil S, Kim TW, Ji HK, Wong WH, Rhee SY and Wang ZY (2010) Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Developmental cell 19, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D and Dehesh K (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS pathogens 4, e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK and Dehesh K (2014) BBX19 interacts with CONSTANS to REPRESS FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26, 3589–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Sarmast MK, Jiang J and Dehesh K (2015) The Transcriptional Regulator BBX19 Promotes Hypocotyl Growth by Facilitating COP1-Mediated EARLY FLOWERING3 Degradation in Arabidopsis. Plant Cell 27, 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zeng J, Deng K, Tu X, Zhao X, Tang D and Liu X (2011) DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 233, 13–23. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen F, Li X, Cao H, Ding M, Zhang C, Zuo J, Xu C, Xu J, Deng X, Xiang Y, Soppe WJ and Liu Y (2016) Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nature communications 7, 13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Hecht V, Vander Schoor JK, Davidson SE and Ross JJ (2009) Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21, 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M and Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci U S A 113, 7655–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW and Holm M (2014) Convergence of Light and ABA signaling on the ABI5 promoter. PLoS genetics 10, e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y and Sun TP (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Modulated BBX19 levels does not alter basal timing of seed germination
Fig. S2 Modulation of BBX19 levels alters seeds sensitivity to ABA.
Fig. S3 BBX19 overexpresser seeds remain hypersensitive to ABA
Fig. S4 BBX19 is not a modulator of the ABA production
Fig. S5 ABI5 interact with BBX21 in transient expression system in N. benthamiana
Fig. S6 ABI5 interacts with BBX21 in yeast-two-hybrid assay.
Fig. S7 BBX19 binds to GT1 motifs in ABI5 promoter in yeast-one-hybrid assay
Fig. S8 BBX19 binds to the GT1-motifs in the promoter of ABI5
Fig. S9 The C-terminal of BBX19 binds to the ABI5 promoter