Abstract
Cholera toxin (Ctx) is an AB-type protein toxin that acts as an ADP-ribosyltransferase to disrupt intracellular signaling in the target cell. It moves by vesicle carriers from the cell surface to the endoplasmic reticulum (ER) of an intoxicated cell. The catalytic CtxA1 subunit then dissociates from the rest of the toxin, unfolds, and activates the ER-associated degradation system for export to the cytosol. Translocation occurs through an unusual ratchet mechanism in which the cytosolic chaperone Hsp90 couples CtxA1 refolding with CtxA1 extraction from the ER. Here, we report that Hsp90 recognizes two peptide sequences from CtxA1: an N-terminal RPPDEI sequence (residues 11–16) and an LDIAPA sequence in the C-terminal region (residues 153–158) of the 192 amino acid protein. Peptides containing either sequence effectively blocked Hsp90 binding to full-length CtxA1. Both sequences were necessary for the ER-to-cytosol export of CtxA1. Mutagenesis studies further demonstrated that the RPP residues in the RPPDEI motif are required for CtxA1 translocation to the cytosol. The LDIAPA sequence is unique to CtxA1, but we identified an RPPDEI-like motif at the N- or C-termini of the A chains from four other ER- translocating toxins that act as ADP-ribosyltransferases: pertussis toxin, Escherichia coli heat- labile toxin, Pseudomonas aeruginosa exotoxin A, and Salmonella enterica serovar Typhimurium ADP-ribosylating toxin. Hsp90 plays a functional role in the intoxication process for most, if not all, of these toxins. Our work has established a defined RPPDEI binding motif for Hsp90 that is required for the ER-to-cytosol export of CtxA1 and possibly other toxin A chains as well.
1. INTRODUCTION
AB protein toxins contain an enzymatic A moiety and a cell-binding B moiety (Zuverink and Barbieri, 2018). They are released into the external environment but attack targets within the cytosol of a host cell. To reach their cytosolic targets, AB toxins bind to the surface of a host cell and are internalized by receptor-mediated endocytosis. Some toxins respond to the acidified lumen of the endosomes with a conformational change in the B subunit that forms a protein-conducting channel in the endosome membrane. The A subunit then utilizes the toxin-generated pore to access the cytosol (Barth et al., 2004). Other AB toxins travel to the endoplasmic reticulum (ER) before holotoxin disassembly and A chain passage into the cytosol through an endogenous translocon pore in the ER membrane (Teter, 2013).
Cholera toxin (Ctx) is a prototypical AB-type, ER-translocating toxin that consists of a catalytic A1 subunit, a helical A2 linker, and a ring-like B homopentamer (Heggelund et al., 2015). CtxAl dissociates from the rest of the toxin in the ER and spontaneously unfolds at physiological temperature (Orlandi, 1997; Tsai et al., 2001; Pande et al., 2007). This activates the ER-associated degradation (ERAD) system for export to the cytosol (Teter and Holmes, 2002; Teter et al., 2003; Massey et al., 2009; Taylor et al., 2011). Toxin translocation occurs through an unusual ratchet mechanism in which the cytosolic chaperones Hsp90 and Hsc70 couple CtxA1 refolding with CtxA1 extraction from the ER (Burress et al., 2014)1. Continued association of Hsp90 with the cytosolic pool of CtxA1 maintains the toxin in a folded, active conformation for the ADP-ribosylation of its G protein target (Banerjee et al., 2014; Burress et al., 2014). The stability of chaperone-associated CtxA1, along with the paucity of CtxA1 lysine residues for ubiquitin conjugation, allows the toxin to evade the ubiquitin-dependent proteasomal degradation that usually accompanies ERAD-mediated export to the cytosol (Hazes and Read, 1997; Rodighiero et al., 2002; Pande et al., 2007).
Hsp90 is also active in the endosome-to-cytosol translocation of AB-type ADP-ribosylating toxins (ADPRTs), but it is not required for the endosomal translocation of most AB toxins with different catalytic activities (Haug et al., 2003; Ratts et al., 2003; Haug et al., 2004; Dmochewitz et al., 2011; Kaiser et al., 2011; Lang et al., 2014; Azarnia Tehran et al., 2017; Schuster et al., 2017; Steinemann et al., 2018). Likewise, Hsp90 is not needed to complete the intoxication process for three ER-translocating toxins that are not ADPRTs: Shiga toxin 1 (Stx1), Shiga toxin 2 (Stx2), and ricin (Spooner et al., 2008; Taylor et al., 2015). It has accordingly been suggested that Hsp90 recognizes a conserved feature in the catalytic A chains of ADPRTs (Barth, 2011; Ernst et al., 2016).
Hsc70 recognizes a YYIYVI sequence in CtxA1 that spans residues 83–88 of the 192 amino acid protein1. Specific CtxA1 binding motifs for Hsp90 have not yet been identified. Here, we report that Hsp90 recognizes two distinct peptide sequences from CtxA1: an N-terminal RPPDEI sequence (residues 11–16) and an LDIAPA sequence in the C-terminal region (residues 153–158) of the protein. This is the first time Hsp90 has been shown to interact with defined amino acid sequences. The LDIAPA sequence is unique to CtxA1, but we identified an RPPDEI-like motif at the N- or C-termini of the A chains from several ER-translocating ADPRTs. In contrast, the RPPDEI motif was absent from ER-translocating toxins that are not ADPRTs and all endosome-translocating toxins. This indicates Hsp90 recognizes a conserved sequence in ER-translocating ADPRTs but interacts with a different feature of endosome- translocating ADPRTs. The location of the Hsp90 and Hsc70 binding sites in CtxA1 suggests a model of CtxA1 translocation that involves (i) extraction of CtxA1 from the ER in an N-terminal to C-terminal direction; (ii) initial contact of Hsp90 with the N-terminus of CtxA1 as the toxin emerges from the translocon pore; (iii) subsequent interaction between Hsc70 and an internal region of CtxA1; and (iv) further interaction between Hsp90 and a C-terminal region of CtxA1. Identification of the Hsp90 binding sites in CtxA1 has thus provided additional insight into Hsp90 chaperone biology as well as the mechanism of toxin translocation from the ER to the cytosol.
2. RESULTS
2.1. Hsp90 recognizes two distinct sites in CtxA1
To identify the CtxA1 binding site(s) for Hsp90, we generated a series of peptides that contained overlapping amino acid sequences from the entire length of the CtxA1 subunit. Peptides mimic the conformation of CtxA1 as it emerges from the cytosolic face of the ER translocon pore in an unfolded/linearized state. As summarized in Table 1, Hsp90 bound two pairs of consecutive peptides spanning CtxA1 amino acid residues 1–26 and 143–168. Hsp90 did not bind to any of these peptides in the absence of ATP (not shown), which was consistent with the ATP-dependent nature of Hsp90 chaperone function (Li et al., 2012) and confirmed the specificity of our observed interactions. The lack of chaperone interaction with the majority of screened peptides further suggested the binding of Hsp90 to select, consecutive peptides was a specific event.
TABLE 1.
Hsp90 recognizes two discrete regions in the CtxA1 polypeptide. Hsp90/ATP was perfused over SPR sensor slides coated with each of the listed CtxA1-derived peptides. Plus signs indicate Hsp90 binding to the peptide. Hsp90 did not bind to any peptide in the absence of ATP.
| CtxA1 residues | Peptide sequence | Hsp90 binding |
|---|---|---|
| 1–16 | NDDKLYRADSRPPDEI | + |
| 11–26 | RPPDEIKQ SGGLMPRG | + |
| 21–36 | GLMPRGQ SEYFDRGTQ | − |
| 31–46 | FDRGTQMNINLYDHAR | − |
| 41–56 | LYDHARGTQTGFVRHD | − |
| 51–66 | GFVRHDDGYVSTSISL | − |
| 63–78 | SISLRSAHLVGQTILS | - |
| 73–88 | GQTILSGHSTYYIYVI | − |
| 83–98 | YYIYVIATAPNMFNVN | − |
| 93–108 | NMFNVNDVLGAY SPHP | − |
| 103–118 | AYSPHPDEQEVSALGG | − |
| 113–128 | VSALGGIPYSQIYGWY | − |
| 123–138 | QIYGWYRVHFGVLDEQ | − |
| 133–148 | GVLDEQLHRNRGYRDR | − |
| 143–158 | RGYRDRYYSNLDIAPA | + |
| 153–168 | LDIAPAADGYGLAGFP | + |
| 163–177 | GLAGFPPEHRAWREE | − |
| 173–188 | AWREEPWIHHAPPGCG | − |
| 179–192 | WIHHAPPGCGNAPR | − |
Each pair of consecutive peptides identified as hits from the primary screen shared an overlapping sequence of six amino acids: RPPDEI or LDIAPA. In a secondary screen (Table 2), we documented Hsp90 binding to an RPPDEI hexapeptide but not to the sequences flanking RPPDEI (residues 1–10 and 17–26). Hsp90 only bound to RPPDEI in the presence of ATP (not shown), which again demonstrated the specificity of the interaction. The second overlapping sequence for Hsp90 recognition (LDIAPA) could not be generated as a soluble hexapeptide, most likely due to its hydrophobic character. However, GYRDRYYSN and ADGYGLAGFP peptides that lacked the shared LDIAPA motif (residues 143–152 and 159–168) could not bind to Hsp90 (Table 2). We therefore concluded that Hsp90 specifically recognizes the RPPDEI and LDIAPA sequences from CtxA1.
TABLE 2.
Specific CtxA1 sequences recognized by Hsp90. The indicated peptide sequences from CtxA1 were appended to SPR sensor slides. Hsp90/ATP was then perfused over each slide, with a positive interaction indicated by a plus sign. Hsp90 binding to the RPPDEI hexapeptide did not occur in the absence of ATP.
| CtxA1 residues | Peptide sequence | Hsp90 binding |
|---|---|---|
| 1–10 | NDDKLYRADS | − |
| 11–16 | RPPDEI | + |
| 17–26 | KQSGGLMPRG | − |
| 143–152 | RGYRDRYYSN | − |
| 159–168 | ADGYGLAGFP | − |
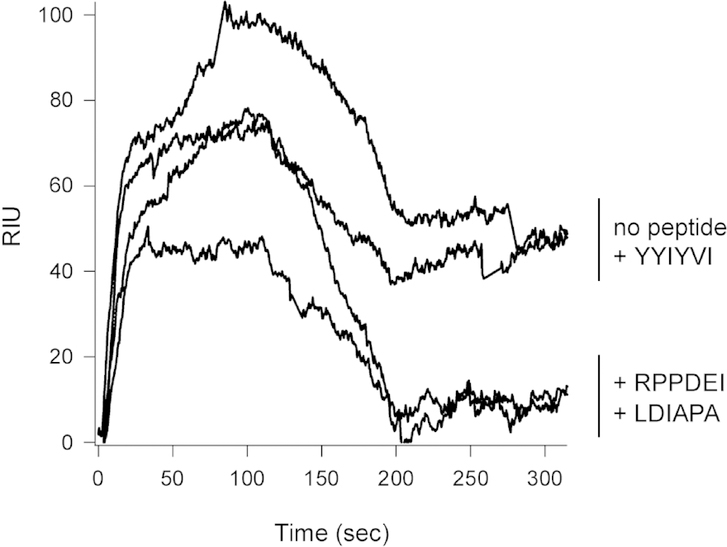
A competition assay further established the specificity of our identified interactions (Fig. 1). For this experiment, Hsp90 was perfused over an SPR sensor coated with CtxA1. Hsp90 bound to CtxA1 in the absence of peptide and in the presence of a peptide that contains the YYIYVI binding motif for Hsc70. In contrast, pre-incubation with the RPPDEI hexapeptide or RGYRDRYYSNLDIAPA peptide prevented Hsp90 from binding to CtxA1. Our collective observations thus identified two specific amino acid sequences from CtxA1 that are recognized by Hsp90: residues 11–16 (RPPDEI) and 153–158 (LDIAPA).
FIGURE 1.
Competitive inhibition of Hsp90 binding to CtxA1. Hsp90 was perfused over a CtxA1-coated SPR sensor at 37°C in the absence of peptide or in the presence of RPPDEI, RGYRDRYYSNLDIAPA, or GQTILSGHSTYYIYVI peptides. Hsp90 was removed from the perfusion buffer after 100 sec.
2.2. Both Hsp90 binding sites in CtxA1 are required for toxin translocation to the cytosol
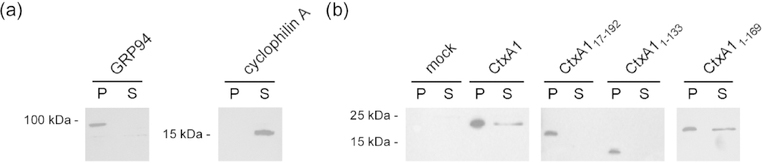
A plasmid-based expression system was used to determine whether one or both of the Hsp90 binding sites in CtxA1 is necessary for the translocation event. With this system, an N-terminal leader sequence directs CtxA1 for co-translational insertion into the ER lumen (Teter et al., 2002). The leader sequence is cleaved in the ER, and the mature CtxA1 polypeptide is subsequently exported from the ER to the cytosol. Translocation is monitored through the accumulation of CtxA1 in the cytosolic fraction from digitonin-permeabilized cells (Massey et al., 2009). As shown in Figure 2a, centrifugation of digitonin-permeabilized cells partitioned the cell extract into a cytosol-containing supernatant and a pellet containing intact ER membrane. Western blot analysis detected wild-type CtxA1 in both the membrane and cytosol fractions from transfected CHO cells (Fig. 2b). In contrast, CtxAl truncations lacking either the N-terminal (CtxA117–192) or C-terminal (CtxA11–133) Hsp90 binding site were not found in the cytosolic fraction. These deletion constructs were only found in the membrane fraction containing intact endomembrane organelles such as the ER. None of these constructs were detected in the extracellular medium of transfected cells (data not shown). A previous study reported the A13 subdomain of CtxAl, which encompasses amino acids 162–192, is not required for toxin translocation to the cytosol (Teter et al., 2006). We confirmed that observation here: transfected CtxA11–169 was detected in the cytosolic fraction, indicating ER-to-cytosol export had occurred. Neither Hsp90 binding site, nor the YYIYVI binding site for Hsc70, is located in the A13 subdomain. These collective results indicated both Hsp90 binding sites are required for the ER- to-cytosol export of CtxA1.
FIGURE 2.
Both Hsp90 binding sites are required for CtxA1 translocation to the cytosol. CHO cells were mock transfected or transfected with plasmids encoding wild-type CtxA1, an N-terminal deletion construct (CtxA117–192), or one of two C-terminal deletion construct (CtxA11.133, CtxA11–169). All constructs are co-translationally inserted into the ER by an N-terminal signal sequence that is proteolytically removed after delivery to the ER. At 24 h post-transfection, cells were partitioned into separate membrane (pellet; P) and cytosolic (supernatant; S) fractions by centrifugation after selective permeabilization of the plasma membrane with digitonin. (a) Both fractions were probed by Western blot to follow the distributions of ER lumenal protein GRP94 and cytosolic protein cyclophilin A. (b) Both fractions were probed by Western blot with a rabbit anti-Ctx primary antibody and HRP-conjugated goat anti-rabbit IgG secondary antibody. One of at least three representative results is shown for each construct.
2.3. An Hsp90 binding motif in ER-translocating ADPRTs
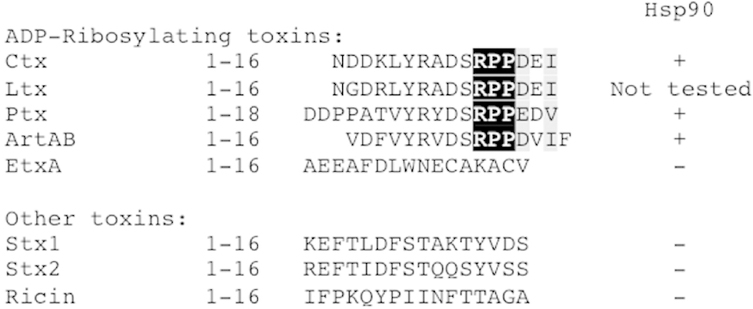
Our collective data support a model of CtxA1 translocation in which Hsp90 binds to the N-terminal RPPDEI motif as soon as the toxin emerges at the cytosolic face of the ER translocon pore. This allows Hsp90 to initiate the refolding process that extracts CtxA1 from the ER. Hsp90 is thought to recognize a conserved feature of ADPRTs (Barth, 2011; Ernst et al., 2016), so we looked for an N-terminal RPPDEI sequence in the A chains of other ER-translocating toxins. As shown in Figure 3, sequence alignments identified this motif in three additional toxins: Escherichia coli heat-labile toxin (Ltx), pertussis toxin (Ptx), and Salmonella enterica serovar Typhimurium ADPRT (ArtAB). The A chains from Pseudomonas aeruginosa exotoxin A (EtxA), Stx1, Stx2, and ricin do not contain an N-terminal RPPDEI-like motif. EtxA is an ADPRT, whereas Stx1, Stx2, and ricin have different catalytic mechanisms (Melton-Celsa, 2014; Michalska and Wolf, 2015; Spooner and Lord, 2015). An SPR-based assay documented the specific binding of Hsp90 to N-terminal toxin sequences with the RPPDEI motif (Fig. 3). Thus, a shared N-terminal binding site for Hsp90 is found in a subset of ADPRTs that move from the ER to the cytosol.
FIGURE 3.
An RPPDEI-like motif is present at the N-termini of A chains from a subset of ER-translocating toxins. N-terminal sequences from the A chains of ER-translocating toxins are shown. Black and grey columns denote amino acid identity and similarity, respectively, within the RPPDEI binding motif. Peptides corresponding to the listed sequences were screened by SPR for binding to Hsp90; plus signs represent a positive interaction. Hsp90 did not bind to any peptide in the absence of ATP.
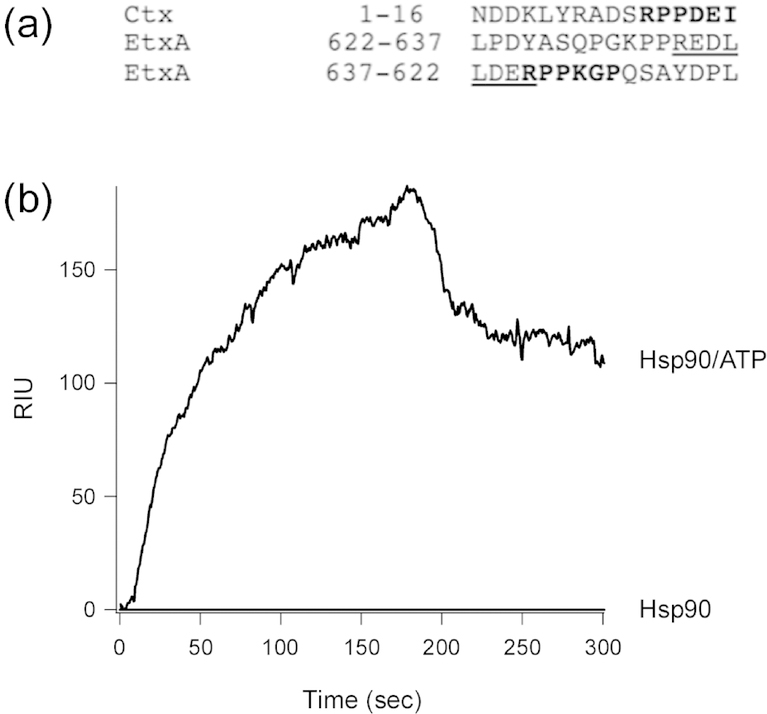
We next examined each toxin A chain from Figure 3 for additional Hsp90 binding sites along the full-length protein. LtxA1, which is highly homologous to CtxA1 (Heggelund et al., 2015), contains an LNIAPA sequence spanning residues 153–158 of the 192 amino acid protein. No LDIAPA motif was identified for any other toxin. No internal RPPDEI motif was found in an N-to-C-terminal orientation for any toxin, but the C-terminus of EtxA contains an RPPDEI-like sequence in reverse order: PGKPPREDL (Fig. 4a). Using our SPR-based system, we confirmed the C-terminus of EtxA interacts with Hsp90/ATP. As seen with our SPR screens of other toxin-derived peptides, Hsp90 did not bind to EtxA in the absence of ATP (Fig. 4b). These observations suggest EtxA exits the ER in a C-terminal to N-terminal direction, which would expose its C-terminus to the cytosol in the LDERPPKGP orientation.
FIGURE 4.
The C-terminus of the A chain from EtxA contains an Hsp90 binding motif. (a) Comparison of the N-terminal sequence from CtxAl with the C-terminal sequence of the A chain from EtxA, shown in both N-to-C-terminal (622–637) and C-to-N-terminal (637–622) orientations. Amino acid residues in the RPPDEI-like motif are highlighted in black, while the REDL tetrapeptide tag for EtxA transport to the ER is underlined. (b) Hsp90 in the absence or presence of ATP was perfused over an SPR sensor coated with the LPDYASQPGKPPREDL peptide from EtxA. Hsp90 was removed from the perfusion buffer after 180 sec.
Surprisingly, the A chains of endosome-translocating toxins do not contain an RPPDEI motif. This includes several ADPRTs known to interact with Hsp90 (Table 3). The LDIAPA binding motif for Hsp90 was likewise absent from endosome-translocating toxins. The recognition of endosome-translocating ADPRTs by Hsp90 thus appears to be distinct from the process involving ER-translocating ADPRTs.
TABLE 3.
Endosome-translocating toxins that lack the A chain RPPDEI binding motif for Hsp90.
| Toxin | ADPRT | Hsp90-dependent | References |
|---|---|---|---|
| Diphtheria toxin | + | + | (Ratts et al., 2003; Dmochewitz et al., 2011; Schuster et al., 2017) |
| Clostridium perfringens iota toxin | + | + | (Haug et al., 2004; Kaiser et al., 2011) |
| Clostridium difficile CDT toxin | + | + | (Haug et al., 2004; Kaiser et al., 2011) |
| Photorhabdus luminescens PTC3 | + | + | (Lang et al., 2014) |
| Clostridium botulinum C2 toxin | + | + | (Haug et al., 2003) |
| Botulinum and tetanus neurotoxins | − | + | (Azarnia Tehran et al., 2017) |
| C. difficile TcdA and TcdB toxins | − | − | (Steinemann et al., 2018) |
| Bacillus antrhacis lethal factor | − | − | (Dmochewitz et al., 2011) |
2.4. A functional role for Hsp90 in Ptx intoxication
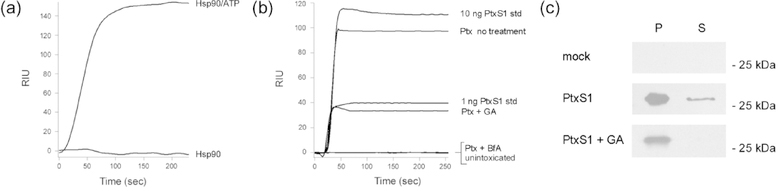
The shared RPPDEI motif in ER-translocating ADPRTs suggested a common role for Hsp90 in the intoxication process for each of these toxins. The role of Hsp90 in CtxA1 translocation is well-established (Taylor et al., 2010; Burress et al., 2014), and we have published preliminary data linking Hsp90 function to the cytotoxic activity of EtxA (Taylor et al., 2015). Additional experiments established an active role for Hsp90 in the Ptx intoxication process (Fig. 5). As expected, Hsp90 directly bound to disordered PtxSl in an ATP-dependent manner (Fig. 5a). To detect a cellular role for Hsp90 in Ptx intoxication, CHO cells were incubated with Ptx for 3 h in the absence or presence of the Hsp90 inhibitor geldanamycin (GA). Control cells were either unintoxicated or intoxicated in the presence of brefeldin A (BfA), a drug that blocks Ptx trafficking to the ER translocation site (Xu and Barbieri, 1995; el Baya et al., 1997; Plaut and Carbonetti, 2008; Banerjee et al., 2016). Cell extracts were collected and partitioned into separate organelle and cytosolic fractions. An SPR sensor coated with a PtxS1 antibody was then used to detect the cytosolic pool of toxin. GA-treated cells contained substantially less cytosolic PtxS1 than untreated cells, thus demonstrating the efficiency of PtxS1 delivery to the cytosol is linked to Hsp90 function (Fig. 5b).
FIGURE 5.
Hsp90 is required for PtxSl delivery to the cytosol. (a) Hsp90 was perfused over a PtxSl-coated SPR sensor at 37°C in the absence or presence of ATP. (b) CHO cells pulse-loaded with 1 μg/mL of Ptx at 4°C were chased at 37°C for 3 h in toxin-free medium. The cytosolic extracts from digitonin-permeabilized cells were then perfused over an SPR sensor appended with an anti-PtxSl antibody. Cells were untreated (no treatment), treated with the Hsp90 inhibitor geldanamycin (GA), or treated with a drug that blocks toxin transport to the ER (brefeldin A; BfA). A cell extract from unintoxicated CHO cells was also perfused over the sensor, as were 1 ng/mL and 10 ng/mL PtxSl standards. (c) CHO cells mock transfected or transfected with an ssPtxSl expression plasmid were chased for 24 h in the absence or presence of the Hsp90 inhibitor GA. Separate organelle (pellet; P) and cytosolic (supernatant; S) fractions generated from digitonin-permeabilized cells were then probed by Western blot with a rabbit anti-PtxS1 primary antibody and HRP-conjugated goat anti-rabbit IgG secondary antibody. One of two representative experiments is shown.
The loss of Hsp90 function could affect Ptx trafficking to the ER as well as PtxS1 translocation to the cytosol. The experiment of Figure 5b could not distinguish between these possibilities, so we ran an additional assay in which PtxS1 was expressed directly in the ER of transfected CHO cells. This approach has been used by other investigators to follow the ER-to-cytosol export of PtxS1 (Veithen et al., 2000; Castro et al., 2001), is similar to the strategy employed for ssCtxA1 (Fig. 2), and bypasses the upstream toxin trafficking events that complicate data interpretation. For this experiment, CHO cells transfected with ssPtxS1 expression plasmid were incubated in the absence or presence of GA. The membrane and cytosolic fractions from digitonin-permeabilized cells were then probed by Western blot to track the intracellular distribution of PtxS1. As shown in Figure 5c, PtxS1 was found in the cytosol of untreated but not GA-treated cells. Hsp90 is therefore required for the ER-to-cytosol export of PtxSl.
2.5. Identification of key residues in the RPPDEI binding motif
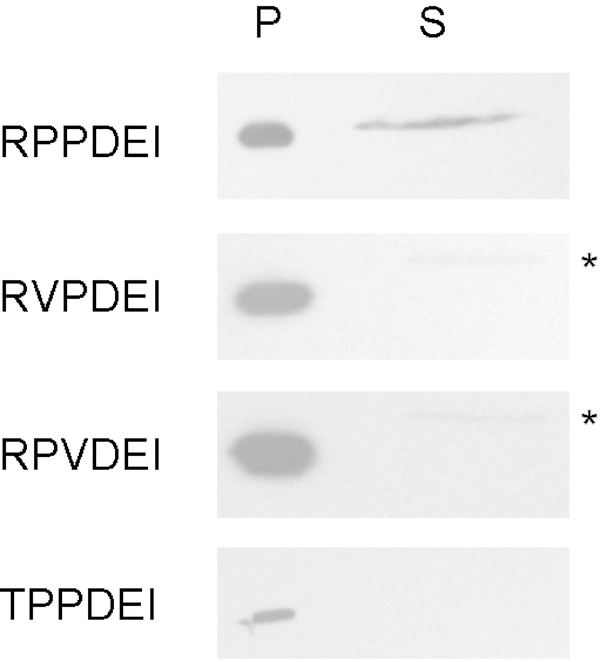
The RPP residues in the RPPDEI motif are conserved in the subset of Hsp90-dependent ADPRTs that move from the ER to the cytosol (Fig. 3). To determine whether these residues play an essential role in toxin translocation, each individual residue was altered in a mutated ssCtxAl construct. We then used our transfection/translocation system to assess the impact of each mutation on toxin export to the cytosol. As shown in Figure 6, wild-type CtxAl could move from the ER to the cytosol and was accordingly detected in both membrane and cytosol fractions. In contrast, CtxAl constructs with RVPDEI or RPVDEI mutations were detected in the membrane fraction but were absent from the cytosolic fraction. Point mutations that disrupt either of the core RPP proline residues therefore block CtxAl translocation to the cytosol. We were unable to express CtxAl with a VPPDEI mutation, but a TPPDEI variant of CtxAl was expressed and found exclusively in the membrane fraction. All three residues in the RPP sequence are thus required for CtxAl export to the cytosol. Interestingly, the catalytic pertussis-like toxin A subunit from typhoid toxin contains an N-terminal TPPDVI sequence (residues l0 - l5 of the mature polypeptide) (Spano et al., 2008). Our results with the TPPDEI variant of CtxAl suggest the pertussis-like toxin A subunit will not use an Hsp90-dependent mechanism for its predicted ER-to-cytosol translocation pathway (Fowler et al., 2017).
FIGURE 6.
The RPP residues of the RPPDEI motif are required for CtxA1 translocation to the cytosol. CHO cells were transfected with plasmids encoding ER-localized variants of wild-type CtxA1 or CtxA1 variants with point mutations in one of the RPP amino acids. At 24 h post-transfection, digitonin-permeabilized cells were partitioned into separate organelle (pellet; P) and cytosolic (supernatant; S) fractions. Both fractions were probed by Western blot with a rabbit anti-Ctx primary antibody and HRP-conjugated goat anti-rabbit IgG secondary antibody. Asterisks denote a faint background band that was also detected in the mock supernatant from Figure 2. One of at least three representative results is shown for each construct.
3. DISCUSSION
3.1. Defined amino acid sequences as binding motifs for Hsp90
No specific binding motif for Hsp90 has been identified until now. An interaction with co-chaperones is thought to dictate Hsp90 substrate specificity (Li et al., 2012; Karagoz and Rudiger, 2015; Schopf et al., 2017), but we have shown that Hsp90 can directly bind to the RPPDEI and LDIAPA sequences in the absence of co-chaperones. Furthermore, peptides containing the RPPDEI or LDIAPA sequences disrupted Hsp90 binding to full-length CtxA1. This competitive inhibition indicated the RPPDEI and LDIAPA sequences occupy the same general binding site within Hsp90 - otherwise, peptide occlusion of one binding site in Hsp90 would not prevent CtxA1 from interacting with Hsp90 at a second, distinct binding site within Hsp90.
3.2. A shared Hsp90 binding motif in ER-translocating ADPRTs
Hsp90 is functionally linked to the cellular activity of several ER-translocating ADPRTs (Taylor et al., 2015) (Fig. 5). These toxins have an N- or C-terminal RPPDEI-like motif that is recognized by Hsp90. In the case of CtxA1, we have shown the RPPDEI motif and Hsp90 are required for toxin translocation to the cytosol (Taylor et al., 2010; Burress et al., 2014) (Fig. 6). Hsp90 is also required for PtxS1 translocation to the cytosol, and it directly binds to disordered PtxS1 (Fig. 5). An active role for Hsp90 in the intoxication process for EtxA has been established as well (Taylor et al., 2015). The A chains from Ptx and EtxA likely utilize their RPPDEI-like motif for Hsp90-driven toxin extraction from the ER, although this prediction has not yet been tested. Likewise, we predict the currently unknown ER-to-cytosol export mechanism for ArtAB (Herdman et al., 2017) will require an interaction between Hsp90 and the N-terminal RPPDVI sequence in ArtA.
In addition to the core RPPDEI motif, four additional N-terminal residues are conserved in the A chains of Ctx, Ltx, Ptx, and ArtAB (Fig. 3). The YRxDS residues (6–10 in CtxA1) could represent an extended YRxDSRPPDEI recognition sequence for Hsp90, but some of these residues are also involved with the toxin-catalyzed ADP-ribosylation reaction. For example, the enzymatic functions of both CtxA1 and PtxS1 require the YRxDS arginine and aspartic acid residues (Barbieri and Cortina, 1988; Burnette et al., 1988; Burnette et al., 1991; Domenighini et al., 1994; Vadheim et al., 1994). PtxS1 activity also requires the RPPDEI arginine residue (Barbieri and Cortina, 1988). Thus, the N-terminal sequences could be conserved for enzymatic activity, Hsp90 binding, or both. Furthermore, the conserved N-terminal YR residues in a potential extended consensus sequence are absent from the C-terminal Hsp90 binding site in EtxA (Fig. 4a). These considerations do not negate the possibility of a larger consensus sequence for Hsp90 binding, but our work suggests the RPPDEI residues represent a core binding motif: an RPPDEI hexapeptide was recognized by Hsp90 and could act as a competitive inhibitor of Hsp90 binding to full-length CtxA1, so additional YRxDS residues are not required for the interaction with Hsp90.
3.3. Dual functions for the C-terminal Hsp90 binding motif of EtxA
EtxA contains a C-terminal REDL sequence that enhances toxin transport to the ER by serving as an imperfect binding motif for the host KDEL receptor (Chaudhary et al., 1990). The arginine residue in the C-terminal REDL sequence is also a component of its Hsp90 binding motif (Fig. 4a), and the aspartic acid could be part of an extended DERPPKGP recognition sequence for Hsp90. The need to maintain a high-affinity interaction between Hsp90 and EtxA may have therefore selected the REDL sequence over the related and more common KDEL sequence. Interestingly, the REDL sequence is required for EtxA activity against cultured cells (Chaudhary et al., 1990) even though EtxA can follow a retrograde transport route to the ER that does not involve the KDEL receptor (Smith et al., 2006). Ctx, in contrast, does not require the KDEL sequence at the C-terminus of its A2 subunit to elicit a cytopathic effect (Lencer et al., 1995). These collective observations suggest the REDL sequence is linked to both the intracellular trafficking and Hsp90-driven translocation of EtxA.
3.4. Distinct Hsp90-dependent export mechanisms for ER-translocating toxins and endosome-translocating toxins
The Barth lab has proposed all ADPRTs utilize Hsp90 for A chain translocation to the cytosol. It was further suggested that Hsp90 recognizes a common structural feature of ADPRTs (Barth, 2011; Ernst et al., 2016). This model was based upon extensive studies with endosome-translocating toxins, but our work has provided further support for the model and expanded its scope to include ER-translocating ADPRTs (Taylor et al., 2010; Burress et al., 2014; Taylor et al., 2015). In addition, we have identified a shared Hsp90 binding motif in the A chains of ER- translocating ADPRTs. This RPPDEI sequence is absent from ER-translocating toxins that are not ADPRTs and do not utilize an Hsp90-dependent export mechanism. Surprisingly, the sequence is also absent from endosome-translocating ADPRTs. Hsp90 thus recognizes a common feature of ER-translocating ADPRTs, but it must interact with a different feature of endosome-translocating ADPRTs. The molecular details of Hsp90-driven toxin export therefore differ to some extent for endosome-translocating ADPRTs and ER-translocating ADPRTs. Another example of this difference is the influence of peptidyl prolyl cis-trans isomerases on the intoxication process: these enzymes are required for the activity of endosome-translocating ADPRTs (Barth, 2011; Ernst et al., 2016) but are dispensable for the cellular activity of Ctx1. Interestingly, peptidyl prolyl cis-trans isomerases do appear to be involved with Ptx intoxication (Ernst et al., 2018). These collective observations support a general role, with toxin-to-toxin differences, for Hsp90, Hsc70, and peptidyl prolyl cis-trans isomerases in A chain translocation for the family of ADPRTs.
3.5. Potential for enhanced cytosolic delivery of exogenous cargo with an RPPDEI tag
Both endosome-translocating and ER-translocating AB toxins have been modified to act as vehicles for the delivery of therapeutic cargo to the cytosol of a target cell (Guillard et al., 2015; Beilhartz et al., 2017). Other toxin variants have been designed as vaccine agents to deliver immunogenic proteins into the cytosol for MHC class I antigen presentation (Moron et al., 2004). These strategies replace the toxin A subunit with a recombinant protein that is shuttled to the intracellular translocation site through its association with the toxin B subunit. Yet delivery to the translocation site does not ensure cargo export to the cytosol, which is likely inefficient in comparison to toxin A chains that actively exploit host mechanisms for passage into the cytosol. Our work suggests the addition of an N- or C-terminal RPPDEI motif to the recombinant protein in a chimeric toxin could, at least for the ER translocation site, facilitate an Hsp90-driven export mechanism and enhanced cargo delivery to the cytosol.
3.6. Identification of chaperone binding sites in CtxA1 defines the directionality of toxin translocation
ERAD substrates emerge at the cytosolic face of the translocon pore in an unfolded or partially unfolded conformation. The use of peptide sequences to screen for Hsp90 binding sites in CtxA1 therefore mimicked the probable linearized state of CtxA1 during its ER-to-cytosol export. Our data strongly suggest Hsp90 is the first chaperone recruited to CtxA1 as the toxin exits the ER translocon pore. The refolding of CtxA1 by Hsp90 (Burress et al., 2014) could then initiate a ratchet mechanism for toxin extraction from the ER: the folded N-terminal domain of CtxA1 would no longer fit into the translocon pore, thus preventing back-sliding of CtxA1 into the ER. This, in turn, would ensure the unidirectional movement of CtxA1 from the ER to the cytosol. Subsequent, sequential binding of Hsc70 to an internal segment of CtxA1 (residues 83–88) and Hsp90 binding to a C-terminal region of CtxA1 (residues 153–158) would continue the refolding process for unidirectional ER-to-cytosol extraction from the translocon pore.
Our data support a model for toxin translocation involving an N-terminal to C-terminal direction for CtxA1 passage through the ER translocon pore. Consistent with this model, we have shown that the C-terminal region of CtxA1 (amino acid residues 169–192) is not required for toxin translocation to the cytosol (Teter et al., 2006) (Fig. 2). None of the Hsp90 or Hsc70 binding sites were missing from this translocation-competent CtxA11–169 deletion construct. Further truncation of the C-terminal domain generated a CtxA11–132 construct that lacked the LDIAPA binding site for Hsp90 and could not move from the ER to the cytosol (Fig. 2). A CtxA117–192 construct that lacked the N-terminal RPPDEI binding motif for Hsp90 was also unable to exit the ER. These results demonstrated both Hsp90 binding sites in CtxA1 are required for toxin translocation to the cytosol.
Recombinant CtxA1 constructs with N-terminal extensions derived from the E. coli heat-stable enterotoxin have slightly reduced in vitro activities (0–30 fold less than wild-type toxin) but greatly attenuated cellular activities (1,000–100,000 fold less than wild-type toxin) (Sanchez et al., 2002). The dramatic reduction of cellular activity was directly related to the length of the N-terminal extension: recombinant toxins with longer extensions were less active than recombinant toxins with shorter extensions. Hsp90 must contact the N-terminal RPPDEI sequence of CtxA1 to initiate the coupled refolding/translocation process, so N-terminal extensions to CtxA1 would inhibit toxin export to the cytosol and the resulting toxicity. As the length of the N-terminal extension increases, it would become increasingly unlikely that the Brownian back-and-forth motion of CtxA1 within the translocon pore would allow the RPPDEI sequence to appear at the cytosolic face of the ER membrane for recognition by Hsp90. It should be noted, however, that other recombinant CtxA1 constructs appended with unstructured peptides at the N-terminus were exported to the cytosol and rapidly degraded by the ubiquitin-proteasome system (Wernick et al., 2010). These unstructured N-terminal extensions may have diverted CtxA1 to a different, Hsp90-independent translocation route just as they diverted CtxA1 to the ubiquitin-proteasome pathway. Our work has thus generated new insight into the CtxA1 translocation event and provided additional context to previous studies on toxin export to the cytosol.
4. EXPERIMENTAL PROCEDURES
4.1. CtxA1 expression and purification
Escherichia coli strain BL21(DE3)pLysS harboring expression plasmid CtxA1-Hisx6/pET-22(b) was grown overnight at 37°C in Luria broth supplemented with 100 μg/mL of ampicillin. The culture was used to inoculate 1 L of Luria broth supplemented with ampicillin and was incubated at 37°C until an O.D.600 of 0.6–0.8 was achieved. A final concentration of 1 mM IPTG was used to induce protein expression for 18 h at 25°C. The culture was centrifuged (6,000 × g for 30 min at 4°C), and the resulting cell pellet was resuspended in lysis buffer (20 mM Tris-HCl pH 7.0, 300 mM NaCl, 8 M urea, 0.1% Triton-X-100, 1% sodium deoxycholate, 100 μg/mL lysozyme). Lysis was allowed to proceed at 37°C for 20–30 minutes before the lysate was sonicated for 3 minutes. The lysate was subjected to a second centrifugation step (16,000 × g for 30 min at 4°C), and the resulting supernatant was transferred to equilibrated TALON (Takara Bio, Mountain View, CA) resin. The resin and lysate were incubated together overnight at 4°C in the presence of EDTA-free protease inhibitor (Pierce Biotechnology, Rockford, IL). The TALON resin was then washed 3 times with wash buffer (20 mM Tris-HCl pH 7.0, 600 mM NaCl, 8 M urea, 0.1% Triton-X100) before it was loaded into a gravity column. After an additional wash with extraction buffer (20 mM Tris-HCl pH 7.0, 600 mM NaCl, 8 M urea) CtxA1 was eluted from the column using an imidazole gradient. Fractions were analyzed for purity by Coomassie staining. Fractions containing pure protein were pooled and refolded by step-wise dialysis against 20 mM sodium phosphate buffer pH 7.0 with 150 mM NaCl. Final protein concentration was assessed using a BCA kit (Pierce Biotechnology). CtxA1 samples were lyophilized and stored at −80°C until use.
4.2. SPR
All SPR experiments were performed using a Reichert (Depew, NY) SR7000 SPR refractometer at 37°C with a flow rate of 41 μL/min. Full-length CtxA1, full-length PtxS1 (List Biologicals, Campbell, CA), or peptide (Peptide 2.0, Chantilly, VA) was amide-coupled to an SPR sensor slide with a mixed-assembled monolayer as previously described (Massey et al., 2009). The slide was then washed with phosphate-buffered saline with 0.1% Tween 20 (Medicago AB, Uppsala, Sweden) until a stable baseline signal was achieved. Hsp90 with 1 mM ATP was then added to the perfusion buffer for 100–180 sec. After the removal of Hsp90/ATP, the slide was washed with phosphate-buffered saline with 0.1% Tween 20. Hsp90 without ATP was used as a negative control for all experiments. For competition assays, Hsp90 was first incubated with the indicated peptide in the presence of ATP for 10 min at room temperature before addition to the perfusion buffer. SPR traces were processed using Reichert software, BioLogic (Campbell, Australia) Scrubber2 software, and WaveMetrics (Lake Oswego, OR) Igor Pro software.
4.3. Round-the-horn mutagenesis
The mammalian expression plasmid ssCtxAl/ pcDNA3.1, which encodes a CtxAl subunit that is co-translationally inserted into the ER via an N-terminal signal sequence (Teter et al., 2004), was used to generate mutations according to the OpenWetWare (openwetware.org/wiki/Main_Page) protocol. Briefly, primers containing the desired CtxAl mutation (Table 4) were phosphorylated with T4 polynucleotide kinase following the manufacturer protocol (Thermo Fisher Scientific, Waltham, MA) and were then used to PCR amplify the entire ssCtxA1/pcDNA3.1 plasmid. The resulting linear mutant ssCtxA1/pcDNA3.1 plasmids were ligated overnight using T4 DNA ligase. To remove the wild-type ssCtxA1/pcDNA3.1 template, DpnI was added to the ligation reaction. Ligated plasmids were transformed into chemically-competent DH5a cells. Mutations were confirmed by DNA sequencing (Genewiz, South Plainfield, NJ).
TABLE 4.
Primers used for CtxA1 mutagenesis.
| Construct | Primer | Sequence |
|---|---|---|
| TPPDEI | Forward | ACACCTCCTGATGAAATAAAGCAGTCAG |
| Reverse | AGAATCTGCCCGATATAACTTATCATCATT | |
| RVPDEI | Forward | GTCCCTGATGAAATAAAGCAGTCAGGTG |
| Reverse | TCTAGAATCTGCCCGATATAACTTATCATCATT | |
| RPVDEI | Forward | GTCGATGAAATAAAGCAGTCAGGTGG |
| Reverse | AGGTCTAGAATCTGCCCGATATAACTTATCATC | |
| CtxA117–192 | Forward | AAGCAGTCAGGTGGTCTTATGCC |
| Reverse | TGCATATGAAAATGATGATAAGAAAATAAAAAACACAAATATTATCTTTAC | |
| CtxA11–133 | Forward | TTAGCTGCAGGTCGACTCTAGAGG |
| Reverse | CCAAAATGAACTCGATACC | |
| CtxA11–169 | Forward | TAGCTGCAGGTCGACTCTAGAGG |
| Reverse | ACCATCTGCTGCTGGAGCAATATC |
4.4. Toxin transfection/translocation assay
CHO cells were seeded to 60–70% confluency in 6-well plates in Ham’s F-12 media supplemented with 10% fetal bovine serum and antibiotic-antimycotic. All cell culture reagents were from Gibco (Gaithersburg, MD), and all incubations were at 37°C with 5% CO2. Cells were first washed with PBS and then transfected with 1 μg of the indicated DNA using Lipofectamine as the transfection reagent. The ssPtxS1/pCMV/myc/ER expression vector (Castro et al., 2001) was generously provided by Dr. Nicholas Carbonetti (Department of Microbiology and Immunology, University of Maryland School of Medicine). After 3 h, the transfection mix was removed and replaced with Ham’s F-12 media supplemented with 10% fetal bovine serum and incubated at 37°C. At 24 h post-transfection, cells were lifted using PBS supplemented with 0.5 mM EDTA. The contents of 3 wells for each condition were pooled together. Samples were then centrifuged at 5,200 × g for 2 min at 4°C, and the resulting pellets were chilled on ice for 10 min. The pellets were then resuspended in HCN buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM CaCl2, 10 mM N-ethylmaleimide, protease inhibitor cocktail) containing 0.04% digitonin and incubated on ice for an additional 10 min. Cellular components were then separated by centrifugation (14,000 × g for 10 min at 4°C). Cytosolic fractions (i.e., supernatants) were collected and transferred to new tubes for lyophilization. Both pellet (i.e., intact membranes) and supernatant samples were resuspended in 2× sample buffer and separated by SDS-PAGE with 15% polyacrylamide gels, using 5-fold more supernatant sample than pellet sample. Gels were transferred to PVDF membranes and blocked with 4% milk in Tris-buffered saline with 0.5% Tween 20 (TBS-T). The membranes were incubated overnight at 4°C with either rabbit anti-CT antibody at 1:20,000 dilution (Sigma-Aldrich, St. Louis, MO) or rabbit anti-Ptx antibody at 1:30,000 dilution (Abcam, Cambridge, United Kingdom). Membranes were then washed 3 times with TBS-T and incubated for 1 h at room temperature with a HRP-conjugated goat anti-rabbit IgG antibody at 1:20,000 dilution (Jackson ImmunoResearch, West Grove, PA). Following 3 additional washes with TBS-T, the blots were developed using ECL.
To demonstrate the fidelity of our cellular fractionation, the pellet and supernatant fractions were also probed with a rabbit antibody against cytosolic marker cyclophilin A (1:10,000 dilution; Abcam) or a rabbit antibody against the ER lumenal maker GRP94 (1:1,000 dilution; Stressgen, Victoria, British Columbia) as described above.
4.5. Ptx translocation assay
As previously described (Banerjee et al., 2016), CHO cells grown to 80% confluency in 6-well plates were incubated with 1 μg of Ptx (List Biologicals) in serum-free Ham’s F-12 medium for 30 min at 4°C. Unbound toxin was removed by washing, and the temperature was raised to 37°C in order to permit endocytosis and retrograde transport of the surface-bound toxin. The Ham’s F-12 chase medium contained 1 μM GA (Enzo Life Sciences) or 5 μg/mL of BfA as indicated. After 3 h at 37°C, separate membrane and cytosolic fractions were generated from digitonin-permeabilized cells as described above for the transfection/translocation assay. The cytosolic fraction was brought to a final volume of 1 mL in HCN buffer and then perfused over an SPR sensor coated with an anti-PtxS1 antibody (Santa Cruz Biotechnology, Dallas, TX).
ACKNOWLEDGEMENTS
We thank Neyda VanBennekom for technical assistance with the preliminary screen of Hsp90 binding to CtxA1-derived peptides and Dr. Nicholas Carbonetti (Department of Microbiology and Immunology, University of Maryland School of Medicine) for providing the ssPtxS1/pCMV/myc/ER expression vector. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI099493 to KT. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors have no conflict of interest to declare.
Footnotes
Helen Burress, Alisha Kellner, Jessica Guyette, Suren A. Tatulian, and Ken Teter. “HSC70 and HSP90 Chaperones Perform Complementary Roles in Translocation of the Cholera Toxin A1 Subunit from the Endoplasmic Reticulum to the Cytosol”. under revision.
REFERENCES
- Azarnia Tehran D, Pirazzini M, Leka O, Mattarel A, Lista F, Binz T, Rossetto O & Montecucco C (2017). Hsp90 is involved in the entry of clostridial neurotoxins into the cytosol of nerve terminals. Cell Microbiol, 19, e12647. [DOI] [PubMed] [Google Scholar]
- Banerjee T, Cilenti L, Taylor M, Showman A, Tatulian SA & Teter K (2016). Thermal unfolding of the pertussis toxin S1 subunit facilitates toxin translocation to the cytosol by the mechanism of endoplasmic reticulum-associated degradation. InfectImmun, 84, 3388–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Taylor M, Jobling MG, Burress H, Yang Z, Serrano A, Holmes RK, Tatulian SA & Teter K (2014). ADP-ribosylation factor 6 acts as an allosteric activator for the folded but not disordered cholera toxin A1 polypeptide. Mol Microbiol, 94, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri JT & Cortina G (1988). ADP-ribosyltransferase mutations in the catalytic S-1 subunit of pertussis toxin. Infect Immun, 56, 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H (2011). Exploring the role of host cell chaperones/PPIases during cellular up-take of bacterial ADP-ribosylating toxins as basis for novel pharmacological strategies to protect mammalian cells against these virulence factors. Naunyn Schmiedebergs Arch Pharmacol, 383, 237–245. [DOI] [PubMed] [Google Scholar]
- Barth H, Aktories K, Popoff MR & Stiles BG (2004). Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol Mol Biol Rev, 68, 373–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilhartz GL, Sugiman-Marangos SN & Melnyk RA (2017). Repurposing bacterial toxins for intracellular delivery of therapeutic proteins. Biochem Pharmacol, 142, 13–20. [DOI] [PubMed] [Google Scholar]
- Burnette WN, Cieplak W, Mar VL, Kaljot KT, Sato H & Keith JM (1988). Pertussis toxin S1 mutant with reduced enzyme activity and a conserved protective epitope. Science, 242, 72–74. [DOI] [PubMed] [Google Scholar]
- Burnette WN, Mar VL, Platler BW, Schlotterbeck JD, McGinley MD, Stoney KS, Rohde MF & Kaslow HR (1991). Site-specific mutagenesis of the catalytic subunit of cholera toxin: substituting lysine for arginine 7 causes loss of activity. Infect Immun, 59, 4266–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burress H, Taylor M, Banerjee T, Tatulian SA & Teter K (2014). Co- and posttranslocation roles for Hsp90 in cholera intoxication. J Biol Chem, 289, 33644–33654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MG, McNamara U & Carbonetti NH (2001). Expression, activity and cytotoxicity of pertussis toxin S1 subunit in transfected mammalian cells. Cell Microbiol, 3, 45–54. [DOI] [PubMed] [Google Scholar]
- Chaudhary VK, Jinno Y, FitzGerald D & Pastan I (1990). Pseudomonas exotoxin contains a specific sequence at the carboxyl terminus that is required for cytotoxicity. Proc Natl Acad Sci USA, 87, 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmochewitz L, Lillich M, Kaiser E, Jennings LD, Lang AE, Buchner J, Fischer G, Aktories K, Collier RJ & Barth H (2011). Role of CypA and Hsp90 in membrane translocation mediated by anthrax protective antigen. Cell Microbiol, 13, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenighini M, Magagnoli C, Pizza M & Rappuoli R (1994). Common features of the NAD-binding and catalytic site of ADP-ribosylating toxins. Mol Microbiol, 14, 41–50. [DOI] [PubMed] [Google Scholar]
- el Baya A, Linnemann R, von Olleschik-Elbheim L, Robenek H & Schmidt MA (1997). Endocytosis and retrograde transport of pertussis toxin to the Golgi complex as a prerequisite for cellular intoxication. Eur J Cell Biol, 73, 40–48. [PubMed] [Google Scholar]
- Ernst K, Schnell L & Barth H (2016). Host cell chaperones Hsp70/Hsp90 and peptidyl-prolyl cis/trans isomerases are required for the membrane translocation of bacterial ADP-ribosylating toxins. Curr Top Microbiol Immunol, doi: 10.1007/1082_2016_1014. [DOI] [PubMed] [Google Scholar]
- Ernst K, Eberhardt N, Mittler AK, Sonnabend M, Anastasia A, Freisinger S, SchieneFischer C, Malesevic M & Barth H (2018). Pharmacological cyclophilin inhibitors prevent intoxication of mammalian cells with Bordetella pertussis toxin. Toxins, 10, pii: E181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CC, Chang SJ, Gao X, Geiger T, Stack G & Galán JE (2017). Emerging insights into the biology of typhoid toxin. Curr OpinMicrobiol, 35, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard S, Minter RR & Jackson RH (2015). Engineering therapeutic proteins for cell entry: the natural approach. Trends Biotechnol, 33, 163–171. [DOI] [PubMed] [Google Scholar]
- Haug G, Aktories K & Barth H (2004). The host cell chaperone Hsp90 is necessary for cytotoxic action of the binary iota-like toxins. Infect Immun, 72, 3066–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug G, Leemhuis J, Tiemann D, Meyer DK, Aktories K & Barth H (2003). The host cell chaperone Hsp90 is essential for translocation of the binary Clostridium botulinum C2 toxin into the cytosol. J Biol Chem, 278, 32266–32274. [DOI] [PubMed] [Google Scholar]
- Hazes B & Read RJ (1997). Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry, 36, 11051–11054. [DOI] [PubMed] [Google Scholar]
- Heggelund JE, Bjornestad VA & Krengel U (2015) Vibrio cholerae and Escherichia coli heat-labile enterotoxins and beyond In The Comprehensive Sourcebook of Bacterial Protein Toxins, 4th edition. Alouf JE, Ladant D, and Popoff MR (eds.) Waltham, MA: Elsevier, pp. 195–229. [Google Scholar]
- Herdman BP, Paton JC, Wang H, Beddoe T & Paton AW (2017). Vacuolation activity and intracellular trafficking of ArtB, the binding subunit of an AB5 toxin produced by Salmonella enterica serovar Typhi. Infect Immun, 85, e00214–00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser E, Kroll C, Ernst K, Schwan C, Popoff M, Fischer G, Buchner J, Aktories K & Barth H (2011). Membrane translocation of binary actin-ADP-ribosylating toxins from Clostridium difficile and Clostridium perfringens is facilitated by cyclophilin A and Hsp90. Infect Immun, 79, 3913–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagoz GE & Rudiger SGD (2015). Hsp90 interaction with clients. Trends Biochem Sci, 40, 117–125. [DOI] [PubMed] [Google Scholar]
- Lang AE, Ernst K, Lee H, Papatheodorou P, Schwan C, Barth H & Aktories K (2014). The chaperone Hsp90 and PPIases of the cyclophilin and FKBP families facilitate membrane translocation of Photorhabdus luminescens ADP-ribosyltransferases. Cell Microbiol, 16, 490–503. [DOI] [PubMed] [Google Scholar]
- Lencer WI, Constable C, Moe S, Jobling MG, Webb HM, Ruston S, Madara JL, Hirst TR & Holmes RK (1995). Targeting of cholera toxin and Escherichia coli heat labile toxin in polarized epithelia: role of COOH-terminal KDEL. J Cell Biol, 131, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Soroka J & Buchner J (2012). The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta, 1823, 624–635. [DOI] [PubMed] [Google Scholar]
- Massey S, Banerjee T, Pande AH, Taylor M, Tatulian SA & Teter K (2009). Stabilization of the tertiary structure of the cholera toxin A1 subunit inhibits toxin dislocation and cellular intoxication. J Mol Biol, 393, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton-Celsa AR (2014). Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol Spectr, 2, EHEC-0024–2013. doi: 0010.1128/microbiolspec.EHEC-0024-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska M & Wolf P (2015). Pseudomonas Exotoxin A: optimized by evolution for effective killing. Front Microbiol, 6, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moron G, Dadaglio G & Leclerc C (2004). New tools for antigen delivery to the MHC class I pathway. Trends Immunol, 25, 92–97. [DOI] [PubMed] [Google Scholar]
- Orlandi PA (1997). Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. JBiol Chem, 272, 4591–4599. [PubMed] [Google Scholar]
- Pande AH, Scaglione P, Taylor M, Nemec KN, Tuthill S, Moe D, Holmes RK, Tatulian SA & Teter K (2007). Conformational instability of the cholera toxin A1 polypeptide. J Mol Biol, 374, 1114–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaut RD & Carbonetti NH (2008). Retrograde transport of pertussis toxin in the mammalian cell. Cell Microbiol, 10, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Ratts R, Zeng H, Berg EA, Blue C, McComb ME, Costello CE, vanderSpek JC & Murphy JR (2003). The cytosolic entry of diphtheria toxin catalytic domain requires a host cell cytosolic translocation factor complex. J Cell Biol, 160, 1139–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodighiero C, Tsai B, Rapoport TA & Lencer WI (2002). Role of ubiquitination in retro- translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep, 3, 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J, Wallerstrom G, Fredriksson M, Angstrom J & Holmgren J (2002). Detoxification of cholera toxin without removal of its immunoadjuvanticity by the addition of (STa-related) peptides to the catalytic subunit. A potential new strategy to generate immunostimulants for vaccination. J Biol Chem, 277, 33369–33377. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM & Buchner J (2017). The HSP90 chaperone machinery. Nat Rev Mol Cell Biol, 18, 345–360. [DOI] [PubMed] [Google Scholar]
- Schuster M, Schnell L, Feigl P, Birkhofer C, Mohr K, Roeder M, Carle S, Langer S, Tippel F, Buchner J, Fischer G, Hausch F, Frick M, Schwan C, Aktories K, SchieneFischer C & Barth H (2017). The Hsp90 machinery facilitates the transport of diphtheria toxin into human cells. Sci Rep, 7, 613. doi: 610.1038/s41598-41017-00780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DC, Spooner RA, Watson PD, Murray JL, Hodge TW, Amessou M, Johannes L, Lord JM & Roberts LM (2006). Internalized Pseudomonas exotoxin A can exploit multiple pathways to reach the endoplasmic reticulum. Traffic, 7, 379–393. [DOI] [PubMed] [Google Scholar]
- Spano S, Ugalde JE & Galan JE (2008). Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe, 3, 30–38. [DOI] [PubMed] [Google Scholar]
- Spooner RA & Lord JM (2015). Ricin trafficking in cells. Toxins (Basel), 7, 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner RA, Hart PJ, Cook JP, Pietroni P, Rogon C, Hohfeld J, Roberts LM & Lord JM (2008). Cytosolic chaperones influence the fate of a toxin dislocated from the endoplasmic reticulum. Proc Natl Acad Sci U S A, 105, 17408–17413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann M, Schlosser A, Jank T & Aktories K (2018). The chaperonin TRiC/CCT is essential for the action of bacterial glycosylating protein toxins like Clostridium difficile toxins A and B. Proc Natl Acad Sci U S A, 115, 9580–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Britt CBT, Fundora J & Teter K (2015) Modulation of cholera toxin structure/function by Hsp90. In Proceedings from the Physical Biology of Proteins and Peptides Conference Olivares-Quiroz L, Guzman-Lopez O, and Jardon-Valadez E (eds.) New York: Springer, pp. 67–80. [Google Scholar]
- Taylor M, Navarro-Garcia F, Huerta J, Burress H, Massey S, Ireton K & Teter K (2010). Hsp90 is required for transfer of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J Biol Chem, 285, 31261–31267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Banerjee T, Navarro-Garcia F, Huerta J, Massey S, Burlingame M, Pande AH, Tatulian SA & Teter K (2011). A therapeutic chemical chaperone inhibits cholera intoxication and unfolding/translocation of the cholera toxin A1 subunit. PLoS ONE, 6, e18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K (2013). Toxin instability and its role in toxin translocation from the endoplasmic reticulum to the cytosol. Biomolecules, 3, 997–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K & Holmes RK (2002). Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect Immun, 70, 6172–6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K, Jobling MG & Holmes RK (2003). A class of mutant CHO cells resistant to cholera toxin rapidly degrades the catalytic polypeptide of cholera toxin and exhibits increased endoplasmic reticulum-associated degradation. Traffic, 4, 232–242. [DOI] [PubMed] [Google Scholar]
- Teter K, Jobling MG & Holmes RK (2004). Vesicular transport is not required for the cytoplasmic pool of cholera toxin to interact with the stimulatory alpha subunit of the heterotrimeric G protein. Infect Immun, 72, 6826–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K, Allyn RL, Jobling MG & Holmes RK (2002). Transfer of the cholera toxin A1 polypeptide from the endoplasmic reticulum to the cytosol is a rapid process facilitated by the endoplasmic reticulum-associated degradation pathway. Infect Immun, 70, 6166–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter K, Jobling MG, Sentz D & Holmes RK (2006). The cholera toxin A13 subdomain is essential for interaction with ADP-ribosylation factor 6 and full toxic activity but is not required for translocation from the endoplasmic reticulum to the cytosol. Infect Immun, 74, 2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI & Rapoport TA (2001). Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell, 104, 937–948. [DOI] [PubMed] [Google Scholar]
- Vadheim KL, Singh Y & Keith JM (1994). Expression and mutagenesis of recombinant cholera toxin A subunit. Microb Pathog, 17, 339–346. [DOI] [PubMed] [Google Scholar]
- Veithen A, Raze D & Locht C (2000). Intracellular trafficking and membrane translocation of pertussis toxin into host cells. Int J Med Microbiol, 290, 409–413. [DOI] [PubMed] [Google Scholar]
- Wernick NL, De Luca H, Kam WR & Lencer WI (2010). N-terminal extension of the cholera toxin A1-chain causes rapid degradation after retrotranslocation from endoplasmic reticulum to cytosol. J Biol Chem, 285, 6145–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y & Barbieri JT (1995). Pertussis toxin-mediated ADP-ribosylation of target proteins in Chinese hamster ovary cells involves a vesicle trafficking mechanism. Infect Immun, 63, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuverink M & Barbieri JT (2018). Protein toxins that utilize gangliosides as host receptors. Prog Mol Biol Transl Sci, 156, 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]