Abstract
The zebrafish (Danio rerio) is a popular vertebrate model organism used in a wide range of research fields. Mycobacteriosis, caused by Mycobacterium species, is particularly concerning because it is a common disease associated with chronic infections in these fish. Infections are also a source of uncontrolled experimental variance that may influence research results. Live feeds for zebrafish are common and include paramecia (Paramecium caudatum), brine shrimp (Artemia franciscana), and rotifers (Branchionus spp.). Although nutritionally beneficial, live feeds may pose a biosecurity risk. In this study, we investigate transmission of Mycobacterium chelonae and Mycobacterium marinum through these three live feeds. We show that all three live feeds ingest both M. marinum and M. chelonae and can transmit mycobacterial infections to zebrafish. This observation emphasizes the need for live feeds to be included in the consideration of potential biosecurity risks. This study is of importance to other beyond the zebrafish community, including those of additional aquatic models and those using live feeds for other types of aquaculture.
Keywords: Mycobacterium chelonae, Mycobacterium marinum, zebrafish, transmission, live feeds, aquaculture
1. Introduction
Mycobacteriosis is a current problem in laboratory zebrafish, occurring in approximately 40% of facilities submitting cases to the Zebrafish International Resource Center diagnostic service (Kent, Spitsbergen et al. 2012). Disease management recommendations highlight the importance of disease prevention, because although eradication can be effective, it is intensive and requires re-derivation of fish strains and complete facility disinfection (Whipps et al., 2012). Despite the common reporting of mycobacteriosis in zebrafish, further understanding of its disease ecology including reservoirs and transmission is required. Additionally, live feeds are used commonly during the rearing of larval and juvenile fish, a time at which the adaptive immune system of zebrafish is still developing with immunocompetence occurring between 2–6 weeks post-fertilization (Lam et al. 2004).
The natural route of transmission of mycobacteria in zebrafish hypothesized to be predominantly through ingestion. This is due, in part, to inference from transmission in other fishes. In salmonids, outbreaks in the 1950s and 1960s were correlated to unpasteurized fish feed derived from carcasses of returning salmon (Ross 1970, Belas, Faloon et al. 1995). Mutoji (2011) also showed a significant increase in infection of Mycobacterium marinum by cannibalism in Medaka (Oryzias latipes). Transmission through ingestion has also been demonstrated in zebrafish (Chang et al., 2019), and the intestinal tract identified as a route of mycobacterial entry (Harriff, Bermudez et al. 2007). Other route of exposure may be possible (Chang et al., 2019), but ingestion it likely the most important source of mycobacterial transmission in zebrafish, this highlights feed as a potential risk factor.
Live feeds for zebrafish are both common and recommended by husbandry experts (reviewed in Lawrence 2007). These feeds include the ciliated protozoan Paramecium caudatum, brine shrimp Artemia spp., as well as rotifers Branchionus spp. Despite nutritional benefits gained from the usage of these live feeds, they pose a potential risk for pathogen transmission. In aquaculture, bacteria present in brine shrimp and rotifers have been transmitted to larvae where survival and growth of fishes was negatively impacted (Gatesoupe 1989). In a study of mycobacteriosis of ornamental fish, artemia feeds have been shown to be contaminated with Mycobacterium spp. (Beran, Matlova et al. 2006). Transmission within another organism may also enhance infectivity in fish. For example, mosquito larvae infected with Mycobacterium spp. were more infective to Japanese medaka (O. latipes) than cultured mycobacteria (Mutoji 2011) and Mycobacterium spp. transmitted to zebrafish through the ciliated protozoan Paramecium caudatum resulted in a higher prevalence of infections than bacteria transmitted from culture plates alone (Peterson, Ferguson et al. 2013). Mycobacterium spp. infected Acanthamoeba castellanii resulted in higher bacterial load in zebrafish relative to fish fed bacteria directly (Harriff, Bermudez et al. 2007).
The ability for mycobacteria to survive, and possibly thrive, within these invertebrates is likely reflects their natural environmental interactions and adaptations for intracellular survival. Free-living aquatic protozoans (e.g. Acanthamoeba and Dictyostelium) have evolved as phagocytic cells, actively preying on bacteria within the air water interface of biofilms (Barker and Brown 1994). These aquatic protozoans inhabit the same environmental niches as mycobacteria, and they digest mycobacteria in food vacuoles, where mycobacteria are processed for energy and eliminated via excretory vacuoles. However in some instances, mycobacteria persist. This mycobacterial persistence within a free-living aquatic protozoan host has been examined in the amoebae host where an endosymbiotic relationship between the bacteria and the amoebae has been hypothesized (Winiecka-Krusnell and Linder 2001). More than 25 species of Mycobacterium have been shown to have some form of endosymbiotic relationship with free-living amoebae (Steinert, Birkness et al. 1998). Many bacterial traits co-evolved with amoebae in order to resist predation and digestion, including: bacterial encapsulation, toxin secretion, blockage of phagolysosomal fusion, and intracellular replication within the host (Cosson and Soldati 2008). The amoebae are thought to act as surrogates for macrophages causing Mycobacterium spp. to upregulate virulence genes in the intracellular environment (Harriff, Bermudez et al. 2007). A similar mechanism is thought to be responsible for the increase in virulence observed in paramecium and mosquito larvae (Peterson et al., 2013; Mutoji 2011). More work investigating the virulence of Mycobacterium spp. when transmitted through a live feed vector is required to determine if live feeds used in zebrafish culture pose a risk factor for enhanced mycobacterial transmission.
The goal of this study was to investigate the uptake and transmission of Mycobacterium chelonae and Mycobacterium marinum by three different live feeds (paramecia, brine shrimp, and rotifers), compared to transmission by ingestion through no live feed as a control.
2. Methods
2.1. Fish
The fish used for this study were bred and maintained in the zebrafish facility at the SUNY-ESF Center for Integrated Teaching and Research in Aquatic Science. Adult casper (nacrew2/w2;roya9/a9) zebrafish (n=396; 198 male and 198 female; age = 6 months) lines, originally obtained from the SARL at Oregon State University (Corvallis, OR) and bred for two generations at SUNY-ESF, were utilized in this study. Animals were housed at a density of 6–10 fish/liter in 1.8 L tanks on a timed, flow-through housing system where tanks received an approximately 25% water change 6–8 times between 6:00 am to 6:00 pm (Aquaneering, San Diego, CA). The housing system included ultraviolet disinfection of dechlorinated (carbon filter) municipal tap water maintained to pH 7.6, a conductivity of 600–700 us/cm2, and a temperature of 28.5°C, and ammonia levels ranging from 0–0.25 ppm. The zebrafish facility maintained a 14:10 light:dark photoperiod. Fish were fed a commercial feed for zebrafish (Gemma 500, Gemma, Skretting, Maine, USA) twice daily on weekdays and once daily on weekends during periods where they were not being fed a treatment feed. All animal work was approved by the SUNY-ESF Institutional Animal Care and Use Committee, protocol #151001. Fish were randomly divided into 33, 1.8L tanks (n=12, 6 male and 6 female). Groups of three tanks were randomly designated to one of the following treatments, M. chelonae: Gelly Belly, Artemia, Paramecium, Rotifers; M. marinum: Gelly Belly, Artemia, Paramecium, Rotifers; Sham: Artemia, Paramecium, Rotifers.
2.2. Bacterial culture and growth media
For this study, a Mycobacterium chelonae (H1E2) transgenic strain expressing green fluorescence protein (GFP) provided by M.L. Kent, and Mycobacterium marinum (OSU214) were used. The M. chelonae was maintained at 28–30°C on solid-phase Middlebrook 7H10 agar supplemented with oleic albumin dextrose catalase (OADC) and 50 μg/ml kanamycin for 5 days. Mycobacterium marinum was cultured at 28–30°C on solid-phase Middlebrook 7H10 agar supplemented with OADC and 60 μM hemin for 10–14 days.
2.3. Live Feed Culture
2.3.1. Artemia
For artemia experiments, Grade A Brine Shrimp Eggs were used (Brine Shrimp Direct, Utah, USA). Manufacturer instructions were used to prepare artemia hatching water from dechlorinated system water with a salinity of 25 parts per thousand and a pH of 8. Following preparation, the hatching water was autoclaved and cooled to room temperature. Prior to hatching, artemia cysts were disinfected and decapsulated by bleaching. For bleaching, cysts were placed in a mesh bottom container, nested within a glass beaker on ice. Cysts were bleached in a 1:1 ratio of Clorox® to hatching water under constant aeration. Bleaching continued until cysts were decapsulated and orange in color. Following bleaching, cysts were rinsed three times with hatching water. Next, rinsed cysts were placed in hatching cones containing hatching water (Aquaneering). Hatching cones were maintained on a bench top with constant overhead lighting at room temperature (25°C). Experiments involving uptake of bacteria by artemia were carried out 24h post-hatching.
2.3.2. Paramecium
Starter cultures of Paramecium caudatum were obtained from Carolina Biological Supply (Burlington, NC). Paramecia were cultured following previously described directions using autoclaved distilled water (Westerfield, 2000). Wheat berries and brewer’s yeast were purchased in the bulk section of a local health food store. Briefly, autoclaved and dried wheat berries were boiled for 10 minutes in previously autoclaved distilled water. Following boiling, wheat berries were cooled to room temperature on the bench top in a sterile petri plate. Several sterile 150×20 mm petri plates were prepared with autoclaved distilled water, 0.01 g of powdered brewer’s yeast and 5 boiled wheat berries. Prepared plates were inoculated with an approximately 1:3 ratio of starter paramecia culture and autoclaved distilled water. Inoculated plates were incubated at 26°C for approximately 2 weeks before being used for feeding. Plates were checked bi-weekly for contamination and paramecia growth.
2.3.3. Rotifers
A starter culture of 1 million live marine L-type rotifers was obtained from Reed Mariculture Inc. (Campbell, CA), along with a Compact Culture System (CCS) for benchtop-scale culture of the rotifers. Rotifers were cultured following directions from the manufacturer with a 20% daily harvest and daily feeding of RGComplete (Reed Mariculture, https://reedmariculture.com/support_rotifers_culturing.php). Dechlorinated, UV filtered, system water was used as a water source.
2.4. Vector Uptake of M. chelonae GFP and M. marinum
To gain insight into the uptake of Mycobacterium spp. by vectors, initial bacterial incubations were conducted using the GFP expressing M. chelonae. For artemia and paramecia, several incubation time-points (10 min, 30 min, 1 hour, and 24 hour) were chosen to determine the optimal time required for feeds to incubate with bacteria prior to zebrafish infections. Artemia and paramecia were concentrated by filtration through a mesh-bottom cylinder and incubated at a 3:1 ratio with M. chelonae GFP diluted in autoclaved dH2O to a concentration of 2.7 × 108 cfu/ml using a nephelometer (Sensititre/Thermo Fisher Scientific, MA, USA) where 1 ml of diluted M. chelonae was used per 3 ml of concentrated artemia or paramecium. Control incubations with no bacteria were also conducted. For each incubation time point, a sample of artemia and paramecia were filtered and rinsed using a mesh-bottom cylinder and resuspended in autoclaved hatching water. For rotifer incubation with M chelonae GFP, the same procedures were followed for the 24-hour time-point. This 24 h infection procedure was also repeated for all live feeds and M. marinum, however, no imaging was done as the M. marinum did not express GFP.
For imaging, a sterile 6-well plate was used to immobilize and image all vectors. Mycobacterium chelonae-exposed artemia were immobilized using carbonated water (Wegmans), paramecia were immobilized using MS222 (0.1g/L was dropped into the petri dish containing paramecia until movement ceased), and rotifers were immobilized using magnesium chloride. Imaging was conducted using a Leica inverted compound microscope (Leica Microsystems, Buffalo Grove, IL, USA) under both brightfield and fluorescence.
The bacterial load of either M. chelonae or M. marinum was evaluated byqPCR. Using sterile technique, triplicate 200 μl samples from each incubation time-point were taken from the rinsed and concentrated artemia, paramecia, and rotifers. An additional 200 μl sample was taken and set aside so that the number of organisms in the sample could be counted. Samples were homogenized using a sterile plastic tissue pestle. DNA was extracted using the MO BIO Laboratories, Inc. UltraClean® Microbial DNA Isolation Kit following manufacturer protocol. Following DNA extraction, a real-time qPCR was performed with technical triplicates for each triplicate sample using primer, probes, and protocol previously published (Meritet, Mulrooney et al. 2017). Ct values of serial dilutions of M. chelonae were used to create a standard curve and calculate the colony forming units for each sample (Excel 2010, Microsoft). From the qPCR assays, it was determined that 24 hour incubation was sufficient for all vectors to obtain the desired bacterial dose.
2.5. Vector Feed
Vectors (artemia, paramecium, rotifers) were incubated with either M. chelonae, M. marinum, or no bacteria for 24 hours as described above. Following incubations, vectors were filtered and rinse. Triplicate 200 μl samples were taken while the remaining concentrated vectors were stored at 4°C. A sample of M. chelonae or M. marinum broth culture was also taken for the bacteria-only treatments. From these samples a DNA extraction followed by qPCR assay was performed as described above to verify the amount of vector required to reach the desired infective dose. This dose of concentrated vector needed for each treatment based on each fish receiving 1.0 × 104 cfu per feed each day for five days at a daily feeding amount of 5% body weight based on an average fish size of 0.4 g.
To determine the effect of oral infection using bacterial culture only (no vector), gelatin feed (Gelly Belly, Florida Aqua Farms) was prepared as previously described (Sciarra, Tyler et al. 2014) with 10 g of Gelly Belly being dissolved in 20 ml (minus volume of inoculant) of clam juice (grocery store brand) warmed to approximately 50°C. Once the gelatin was mixed using a flame sterilized metal spatula, the inoculant was then added to the warmed mixture. Next, the entire mixture was spread on a sterile plastic petri dish and allowed to set at 4°C. After setting, the gelatin was minced using a flame-sterilized razor blade and aliquoted into the mass of feed needed for one tank’s (N=12) daily feed. Aliquots were stored at −20°C until feeding time. Treatment tanks were fed the treatment feeds for five consecutive days. Aliquoted treatment feeds were briefly thawed from storage in the freezer and then emptied into treatment tanks. Following 30 min of feeding tanks were observed for any excess food, none was noted. Following the 5 treatment feeds, fish were returned to regular feeding and care as listed in the details above.
2.6. End Analyses
After 8 weeks post-infection all fish were euthanized by submersion in 0.3 g/L MS222 buffered to a pH of 7.5 for a minimum of 10 minutes following the cessation of opercular movement. Using sterile technique, euthanized fish were dissected and samples of liver and spleen were removed and stored individually in 200 μl of sterile dH2O in a 2.0 ml sterile tube. These samples were further processed by homogenization using a sterile plastic tissue pestle. Next, 900 μl of autoclaved dH2O was added, followed by vortexing. The vortexed tubes were pelleted and subsequently cultured on MB agar plates as described above. Resulting growth was recorded. Finally, real-time qPCR of the homogenate was conducted using M. chelonae (M. chelonae treatments), M. marinum (M. marinum treatments), or both (sham infections) specific primers as previously described (Meritet, Mulrooney et al. 2017). Finally, euthanized fish were fixed in Davidson’s solution for 48h and then transferred to 70% EtOH for storage.
2.7. Statistics
The quantity of Mycobacterium spp. that were taken up by each vector was compared following the collection of real-time qPCR data. Prior to statistical analyses the resulting quantity of bacteria following qPCR was calculated by both sample (50 μl DNA extraction product) and also by organism based on counts of organisms in each sample. For each treatment, descriptive statistics were obtained. Data were also checked for normality equal variances. Because all data sets were found to have non-normal distributions (p<0.05) and unequal variances (p<0.05) the non-parametric Kruskal-Wallis rank sum test was used to compare bacterial count values. In the case of a significant result, indicating differences between vector treatments, post-hoc tests for pairwise multiple comparisons of the ranked data were performed. Data were then visualized as a clustered bar graph using GraphPad Prism v5.0.
For M. chelonae and M. marinum treatment groups, with qPCR prevalence data, treatment groups were compared. Sham infection prevalence values were not included as they were zero. To compare treatments, an ANOVA test and Tukey post-hoc analyses were performed using GraphPad PRISM v 5.0. Data were then visualized as a clustered bar graph using GraphPad Prism v5.0
3. Results
3.1. Feed Analyses
Both M. chelonae and M. marinum were taken up in all vectors. This was first visualized through observation of the M. chelonae GFP expression in all live feeds (Fig. 1). In the artemia, the M. chelonae was observed to be collected in the organism’s gut, in the paramecia food vacuoles, and in the stomach of the rotifers. Based on real-time qPCR analysis, the amount of both M. chelonae and M. marinum taken up by each vector could be quantified and compared between organisms.
Figure 1.
Expression of M. chelonae GFP transgenic bacteria in zebrafish feed vectors. (A) Artemia is shown in bright field (inset) and under fluorescence where GFP is observed in the gut of the artemia. (B) GFP expression is observed in the paramecium vacuoles. (C) GFP expression is observed in the stomach of the rotifer. Scale = 100μm.
The amount of M. chelonae quantified by qPCR (Fig. 2A) showed no significant difference between vectors [F(5)=1.07, p=0.3870]. When comparing incubation time points for each vector individually, there was also no significant difference in uptake of M. chelonae at different time points [F(15)=1.07, p=0.4041].
Figure 2.
(A) Quantity of M. chelonae in live feeds based on real-time qPCR analyses. Paramecia (white bars), artemia (grey bars), and rotifers (black bars) are shown. (B) Quantity of M. marinum in live feed vectors based on real-time qPCR analyses following 24 hours of incubation. Paramecia (white bars), artemia (grey bars), and rotifers (black bars) are shown. Significant differences (p<0.01) between vectors at the same time-point is shown (*) and significant difference between the same vector and time-point for different bacterial species is shown (++).
For M. marinum (Fig. 2B), the amount quantified from the DNA extraction differed significantly between vector organisms at the 24-hour incubation time-point, with paramecia having significantly more bacteria than both artemia and rotifers [F(5)=1.07, p <0.01)]. Additionally, the uptake of M. marinum by paramecium was significantly greater at 24- hours than all vector M. chelonae 24 hour incubations (p<0.01) (Compare Fig. 2A to B).
3.2. End Prevalence Analyses
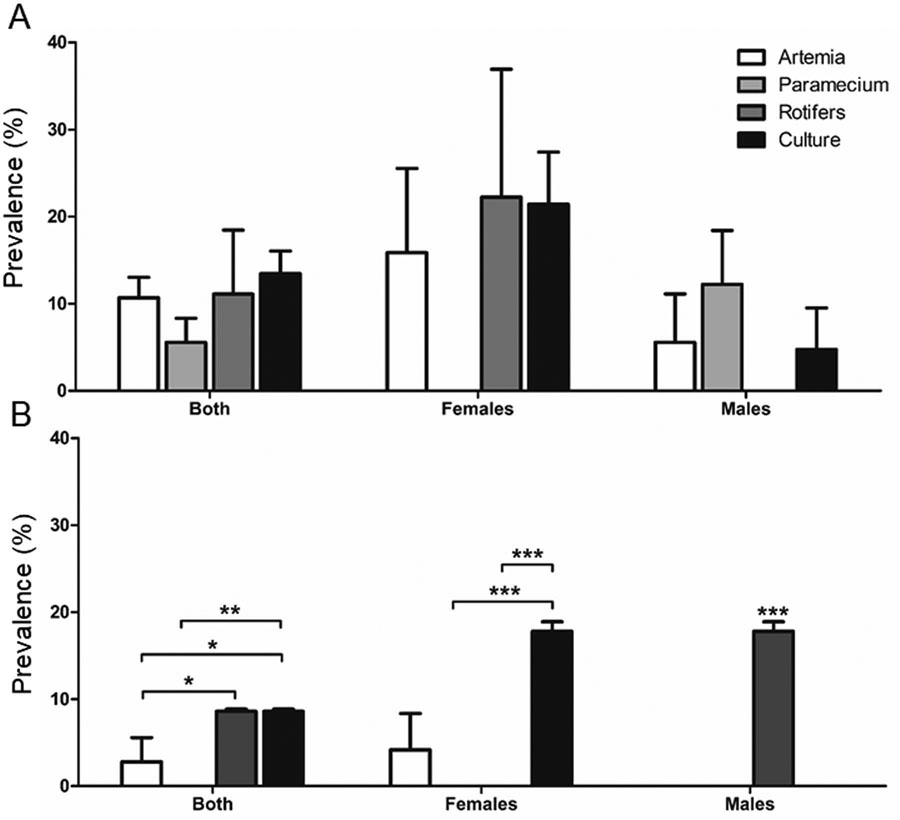
End prevalence values for M. chelonae and M. marinum treatments are shown based on real-time qPCR results indicating bacterial prevalence in liver and spleen samples of treated fish (Fig. 3). Based on qPCR testing, there was no prevalence of either M. chelonae or M. marinum in any sham treatment fish. For M. chelonae-exposed vectors, feeding all three invertebrates, as well as the Gelly Belly diet (Culture), resulted in transmission (Fig. 3A). Overall, there was no significant difference in resulting prevalence amongst the invertebrate vectors based on real-time qPCR testing. For M. marinum (Fig. 3B) transmission occurred with the Gelly Belly diet, artemia, and rotifers, but no prevalence was observed with the paramecia vector. When comparing M. marinum-treatment groups consisting of both sexes significant differences between treatments were identified [F(3)=26.42, p<0.0001], artemia prevalence was significantly lower than rotifers and Gelly Belly (p<0.05) as was paramecium prevalence (p<0.01). For females only, Gelly Belly was significantly higher (p< 0.001) than paramecium and rotifers. For males only, rotifers was higher than all other treatments (p<0.001).
Figure 3.
End real-time qPCR prevalence values for the detection of (A) M. chelonae and (B) M. marinum in liver and spleen samples are shown for each treatment group. Differences between treatment groups consisting of both sexes only males and only females are shown (* = p<0.05, ** = p<0.01, *** = p<0.001). Note sham controls were all negative and are not shown in this figure. Also treatments with no prevalence bar showing represent a prevalence of zero.
4. Discussion
All feeds readily took up mycobacteria, which was not surprising because the uptake of bacteria in live feeds has been demonstrated for other pathogens (Rombaut, Dhert et al. 1999, Sahul Hameed and Balasubramanian 2000) and in previous studies looking at Mycobacterium spp. (Harriff, Bermudez et al. 2007, Peterson, Ferguson et al. 2013). No significant difference was observed between feeds in terms of the quantity of bacteria taken up, with the exception of paramecia which had a significantly higher load of M. marinum after 24-hours of incubation. However, this study incubated feed with a high dose of mycobacterial inoculant, and differential uptake of bacteria between vectors may occur with a lower dose of inoculant. We also only looked at uptake over a maximum period of 24 hours, and continued bacterial uptake or elimination cannot be ruled out with extended exposure times. In terms of management, it is important to note that artemia do not take up bacteria until feeding by mouth begins at approximately 8 hours post-hatch (Sorgeloos, et al., 2001). Thus, the use of younger artemia may help reduce the risk of pathogen ingestion and subsequent transmission. We also observed that in paramecia, M. chelonae was taken up by the feeding vacuoles compared to artemia and rotifers taking bacteria up in the gut/stomach regions. Further investigation into differences between these forms of uptake between single-celled versus multi-celled organisms should be carried out.
Transmission of M. chelonae from live feeds to zebrafish was detected using our most specific test, qPCR. End real-time qPCR analyses of M. marinum treated fish resulted in M. marinum-paramecia treated fish having the lowest prevalence among M. marinum treatments (no infected fish) for groups of both sexes. Mycobacterium marinum- rotifer treatment resulted in the highest prevalence among males, and the M. marinum – Gelly Belly treatment resulted in the highest prevalence among females. Thus no clear trend in virulence between vector feed treatment was observed. Previous studies (Harriff, Bermudez et al. 2007, Peterson, Ferguson et al. 2013) have evaluated the influence of feed vectors separately, and this is the first study that compared transmission following treatment of multiple live-feed types.
One of these studies (Peterson, Ferguson et al. 2013) found transmission through a live feed vector resulted in increased virulence compared to treatment with culture (no live feed). However, the dosage in this previous study was determined by visually counting the number of acid-fast bacilli in food vacuoles of paramecia. In contrast, Harriff et al (2007) only reported an increase in overall bacterial load, but not pathogenicity when comparing M. marinum infections using an amoebae vector compared to culture. They estimated a dose of 1 × 107 bacteria based on previous knowledge of amoebae phagocytosis of bacteria and dosage was later verified by plating. It was also noted that differences in the original treatment inoculum amounts prevented them from interpreting any increase in virulence based on vector transmission.
Based on the differing results of these previous studies, standardizing inoculum dosage for each treatment was a priority for this study so that results could be compared across different treatments. In preparation of treatment inoculums, qPCR assays were performed to verify the amount of infected vector needed to reach our desired, standardized dosage of 1.0 × 104 cfu per feed. By ensuring a standardized dosage of bacteria across all treatments we are able to compare prevalence following each treatment. We did not observe an increased prevalence when mycobacteria were transmitted in live feeds, and this finding could be due to several possible reasons. First, with regard to dose and exposure, adult zebrafish in a previous study (Peterson, Ferguson et al. 2013) were fed an experimental dose of M. chelonae and M. marinum for 14 days compared the 5 days of feeding used in this study. A longer treatment period or higher dose may result in more severe infections and greater difference in prevalence between feed vectors. Second, in Peterson et al. (2013), end point analyses consisted of histological analyses, whereas we used culture and qPCR analyses of dissected spleen and liver tissue, thus future histological analyses may help to further inform mycobacterial prevalence and infection severity in these fish. Third, we used exclusively adult casper (nacrew2/w2;roya9/a9) zebrafish. Differences may occur between wild-type and mutant fish, as well as zebrafish at different developmental time-points.
In this study, we demonstrated mycobacterial transmission through live feed vectors commonly used in zebrafish facilities. This observation emphasizes the need for consideration of live feeds as a potential biosecurity risk. However, steps such as routine testing of live feed cultures for pathogens, and routine disinfection of equipment may mitigate this posed risk. Future studies should aim to increase our understanding of the role live feeds play in mycobacterial transmission within zebrafish facilities, as live feeds have been shown to be a beneficial nutrition for zebrafish culture. These future studies, in addition to this one, will help not only zebrafish research facilities, but also those of additional aquatic models (i.e. Japanese medaka, swordtails, killifish, and Mexican cavefish).
Supplementary Material
Acknowledgements
This research was funded in part by the Office of Research Infrastructure Programs of the National Institutes of Health (NIH) under award number R24OD010998. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We also thank the Natural Sciences and Engineering Research Council of Canada for a postgraduate scholarship-doctorate to CTC. Thanks are extended to Michael L. Kent at Oregon State University for providing the transgenic strain of M. chelonae expressing GFP. We thank members of the Whipps Fish and Wildlife Disease Lab for their ongoing support, especially: Jet’aime Lewis, Julia Williamson, and K. Alice Lindsay.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
6 References
- Barker J and Brown MR (1994). Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140 (Pt 6): 1253–1259. [DOI] [PubMed] [Google Scholar]
- Belas R, Faloon R and Hannaford A (1995). Potential applications of molecular biology to the study of fish mycobacteriosis. Annual Review of Fish Diseases 5, 133–173. [Google Scholar]
- Beran V, Matlova L, Dvorska L, Svastova P and Pavlik I (2006). Distribution of mycobacteria in clinically healthy ornamental fish and their aquarium environment. Journal of Fish Diseases 29(7), 383–393. [DOI] [PubMed] [Google Scholar]
- Chang CT, Lewis J, and Whipps CM. (2019) Source or Sink: Examining the Role of Biofilms in Transmission of Mycobacterium spp. in Laboratory Zebrafish. Zebrafish. April 2019 10.1089/zeb.2018.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P and Soldati T (2008). Eat, kill or die: when amoeba meets bacteria. Current Opinion in Microbiology 11(3), 271–276. [DOI] [PubMed] [Google Scholar]
- Gatesoupe FJ (1989). Further advances in the nutritional and antibacterial treatments of rotifers as food for turbot larvae, Scophthalmus maximus L., in: De Pauw N et al. (Ed.) Aquaculture: a biotechnology in progress: ume 1 pp. 721–730 [Google Scholar]
- Harriff MJ, Bermudez LE and Kent ML (2007). Experimental exposure of zebrafish, Danio rerio (Hamilton), to Mycobacterium marinum and Mycobacterium peregrinum reveals the gastrointestinal tract as the primary route of infection: a potential model for environmental mycobacterial infection. Journal of Fish Diseases 30(10), 587–600. [DOI] [PubMed] [Google Scholar]
- Kent ML, Spitsbergen JM, Matthews JM, Fournie JW, Murray KN and Westerfield M. (2012). Diseases of zebrafish in research facilities. Available online at http://zebrafish.org/zirc/health/diseaseManual.php.Accessed 10/25/16. [Google Scholar]
- Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM (2004). Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Developmental Comparative Immunology 28 (1), 9–28. [DOI] [PubMed] [Google Scholar]
- Lawrence C (2007). The husbandry of zebrafish (Danio rerio): A review. Aquaculture 269(1–4), 1–20. [Google Scholar]
- Meritet DM, Mulrooney DM, Kent ML and Lohr CV (2017). Development of Quantitative Real-Time PCR Assays for Postmortem Detection of Mycobacterium spp. Common in Zebrafish (Danio rerio) Research Colonies. Journal of the American Association for Laboratory Animal Science 56(2), 131–141. [PMC free article] [PubMed] [Google Scholar]
- Mutoji KN (2011). Investigation into Mechanisms of Mycobacterial Transmission Between Fish. Doctor of Philosophy, University of Louisiana at Lafayette. Available from ProQuest Dissertations & Theses A&I. (876198923). Retrieved from https://search.proquest.com/docview/876198923?accountid=12154 [Google Scholar]
- Peterson TS, Ferguson JA, Watral VG, Mutoji KN, Ennis DG and Kent ML (2013). Paramecium caudatum enhances transmission and infectivity of Mycobacterium marinum and Mycobacterium chelonae in zebrafish Danio rerio. Diseases of Aquatic Organisms 106(3), 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombaut G, Dhert P, Vandenberghe J, Verschuere L, Sorgeloos P and Verstraete W (1999). Selection of bacteria enhancing the growth rate of axenically hatched rotifers (Branchionus plicatilis). Aquaculture 176(3–4), 195–207. [Google Scholar]
- Ross AJ (1970). Mycobacteriosis among Pacific salmonid fishes A Symposium on Diseases of Fishes and Shellfishes. Sniesko SF. Washington, D.C., American Fisheries Society, 279–283. [Google Scholar]
- Sahul Hameed AS and Balasubramanian G (2000). Antibiotic resistance in bacteria isolated from Artemia nauplii and efficacy of formaldehyde to control bacterial load. Aquaculture 183, 195–205. [Google Scholar]
- Sciarra JB, Tyler AT and Kolb A (2014). A gelatin-based diet for oral dosing juvenile to adult zebrafish (Danio rerio). Laboratory Animal Science Professional June, 32–35. [Google Scholar]
- Sorgeloos P, Ghert P, and Candreva P. (2001). Use of the brine shrimp, Artemia spp., in marine fish larviculture. Aquaculture 200 (1–2) pp 147–159. [Google Scholar]
- Steinert M, Birkness K, White E, Fields B and Quinn F (1998). Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Applied and Environmental Microbiology 64(6), 2256–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (2000). The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed, Univ. of Oregon Press, Eugene. [Google Scholar]
- Whipps CM, Lieggi C and Wagner R (2012). “Mycobacteriosis in Zebrafish Colonies.” ILAR Journal 53 (2): 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winiecka-Krusnell J and Linder E (2001). Bacterial infections of free-living amoebae. Research in Microbiology 152(7), 613–619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.