Abstract
Multidrug resistance in Gram-negative bacteria can arise by the over-expression of multidrug efflux pumps, which can extrude a wide range of antibiotics. Here we describe the ancestral Haemophilus influenzae efflux pump AcrB (AcrB-Hi). We performed a phylogenetic analysis of hundreds of RND-type transporters. We found that AcrB-Hi is a relatively ancient efflux pump, which nonetheless can export the same range of antibiotics as its evolved colleague from Escherichia coli. AcrB-Hi was not inhibited by the efflux pump inhibitor ABI-PP, and could export bile salts weakly. This points to an environmental adaptation of RND transporters. We also explain the sensitivity of H. influenzae cells to β-lactams and novobiocin by the outer membrane porin OmpP2. This porin counterbalances the AcrB-Hi efflux by leaking the drugs back into the cells. We hypothesise that multidrug recognition by RND-type pumps is not an evolutionarily acquired ability, and has been present since ancient promiscuous transporters.
Subject terms: Biochemistry, Microbiology, Antimicrobial resistance
Zwama et al. demonstrate that the AcrB efflux pump from H. influenzae (AcrB-Hi) is phylogenetically far removed from AcrB in E. coli but show similar specificity. They also show that efflux is counteracted by leakage of some antibiotics back into the bacterial cell through the OmpP2 channel which can explain H. influenzae sensitivity to these compounds.
Introduction
Haemophilus influenzae Type b (Hib) is a clinically relevant pathogenic Gram-negative bacterium that causes several invasive diseases (especially in young children), including meningitis and pneumonia1,2. Although conjugate vaccinations significantly decreased the number of Hib-related infectious diseases, Hib still causes illnesses all around the world3. Today, antimicrobial-resistant pathogens form a great threat to modern health worldwide4. β-lactams (such as ampicillin or third-generation cephalosporins) are often used as first- and second-line drugs to treat Hib infections5, as until recently Hib did not possess β-lactamase genes. However, today, β-lactam resistance has been observed in clinical Hib isolates: BLNAR (Beta-Lactamase Non-producing Ampicillin Resistance, caused by TEM-1 and ROB-1 β-lactamases6) and BLPAR (Beta-Lactamase-Producing Ampicillin Resistance, caused by lower affinities in PBPs by amino acid substitutions7,8) have become serious threats in Hib chemotherapy9. Resistance to a single class of antibiotics can be caused by one resistance factor, such as the production of β-lactamases or altered PBPs10,11. On the other hand, multidrug resistance (MDR) in Gram-negative bacteria can be caused solely by the overexpression of antibiotic efflux pumps. These pumps render several or all classes of antibiotics commercially available today ineffective12. Such a multidrug efflux pump (called AcrB) belonging to the resistance-nodulation-division (RND) superfamily is expressed in H. influenzae cells. AcrB is part of the tripartite efflux system AcrAB-TolC that spans both the inner and outer membrane of the Gram-negative cells, directly facilitating the efflux of antibiotics from the cytoplasm or periplasm out of the cell. H. influenzae AcrB (AcrB-Hi) is an efflux pump that was shown to expel several antibiotics and dyes similar to Escherichia coli AcrB (AcrB-Ec)13. Although this efflux pump has been described previously to cause macrolide resistance14, relatively little is known about the role of AcrB in Hib drug resistance, in addition to its large outer membrane protein OmpP2.
Here we show that AcrB-Hi is a relatively ancient pump, phylogenetically far removed from AcrB-Ec. Despite their relatively low genetic relation and homology, AcrB-Hi can transport similar compounds as its evolved colleague AcrB-Ec. However, we also found differences in substrate specificities. We show that AcrB-Hi can export bile salts only weakly. Furthermore, AcrB-Hi is uninhibited by pyridopyrimidine-derived efflux pump inhibitor (EPI) ABI-PP, which completely inhibits AcrB-Ec. We observe that the defence mechanism of H. influenzae is counterbalanced by the leakage of antibiotics through the OmpP2 outer membrane channel back into the bacterial cells, providing a molecular explanation as to why Hib cells are sensitive to certain antibiotics. Our data suggest that ancestral less-efficient and nonspecific RND transporters have evolved to become efficient and more specific transporters, with physiologically relevant specificities. From our data, we hypothesise that multidrug recognition by multidrug efflux pumps did not increase during evolution and that the ability to export chemically different compounds is an intrinsic property since ancient transporters.
Results
Haemophilus influenzae AcrB is an ancestral RND efflux pump
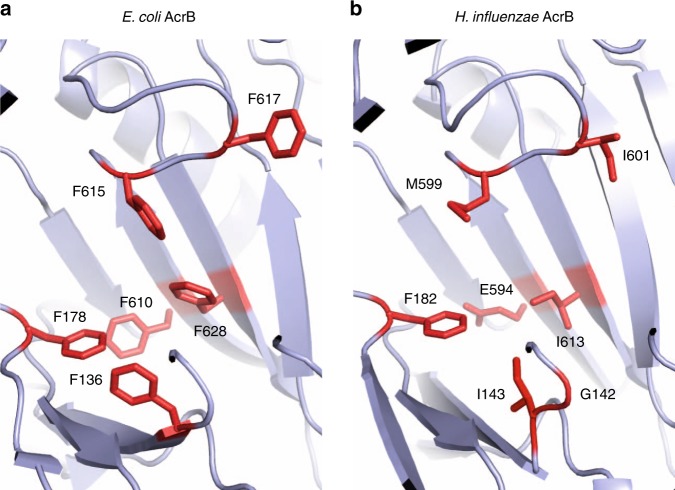
To investigate the RND efflux pump AcrB expressed in H. influenzae (AcrB-Hi), we decided to compare the pump to the probably most studied RND multidrug efflux pump AcrB from E. coli (AcrB-Ec). AcrB-Ec is known as a promiscuous efflux pump with an extremely wide efflux range of many structurally unrelated compounds, which include dyes, antibiotics, bile salts, and detergents15,16. Although the substrate specificity of RND transporters is generally wide, the pumps are not non-specific, as paralogues and orthologues (such as AcrB vs. AcrD, and MexB vs. MexY)17–20 differ in substrate recognition depending on the physiochemical properties of the compounds21–24. We found that acrB is the only RND transporter gene (HI0895) in the Haemophilus influenzae Rd KW20 genome with a translated length of 1032 amino acids (AcrB-Ec has 1049 amino acids). Initial amino acid sequence alignment shows the two proteins share 31% identity, 52% similarity and 4% gaps (Supplementary Fig. 1). The hydrophobic pit (or hydrophobic trap) of AcrB-Ec holds six phenylalanine residues25 (the Phe-rich pit), namely Phe136, Phe178, Phe610, Phe615, Phe617, and Phe628. These correspond to Gly142, Phe182, Glu594, Met599, Ile601, and Ile613 in AcrB-Hi. Our homology model suggests Phe136 may correspond to Ile143 (Fig. 1). Most of these residues are hydrophobic (Ile143, Met599, Ile601, and Ile613), one is aromatic (Phe182), and one is charged (Glu594). Between AcrB-Hi and AcrB-Ec, the transmembrane domain is mostly preserved (40% identity and 67% similarity), of which transmembrane helix 4 (TM4) is the most preserved (59% identity and 89% similarity).
Fig. 1.
The hydrophobic pits of AcrB-Ec and AcrB-Hi. a The phenylalanine residues in the hydrophobic trap in the crystal structure of AcrB-Ec (PDB identification code 3AOA). The six residues are Phe136, Phe178, Phe610, Phe615, Phe617, and Phe628 and this pit is therefore also described as the phenylalanine rich pit. b The corresponding residues in the hydrophobic trap of AcrB-Hi, based on the crystal structure of AcrB-Ec (PDB accession code 3AOA) according to homology modelling. These residues are Gly142 or Ile143, Phe182, Glu594, Met599, Ile601, and Ile613. The ABI-PP inhibitor-bound pit models can be found in Supplementary Fig. 6
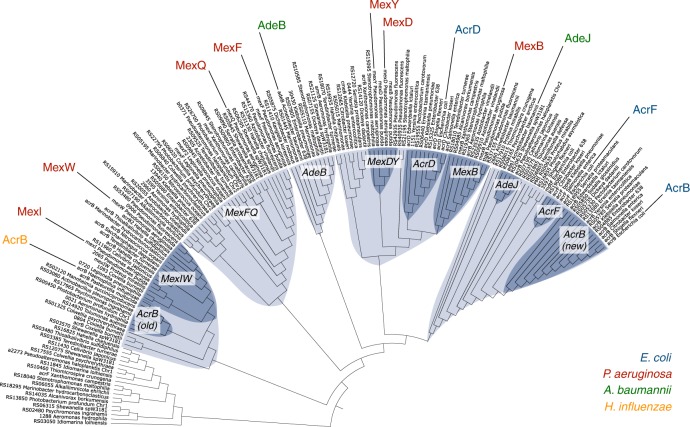
The next step in comparing AcrB-Hi with AcrB-Ec was to collect gene sequences from multiple Gram-negative bacteria homologous to the acrB-Hi and acrB-Ec genes. We collected 393 homologous genes from 51 gammaproteobacteria and analysed them by multiple sequence alignment (Supplementary Data 1). Genes could be classified into three major clusters: MDR (including clusters for mdtB and mdtC), heavy metal (separated into clusters for cusA and czcA) and other (including triclosan pump triC26) (Supplementary Figs. 2, 3). MDR transporters comprised about half (Supplementary Fig. 2) and are shown in Fig. 2 (with the exclusion of the heteromeric MdtBC-like systems). The phylogenetic tree (Fig. 2) shows that acrB-Hi is phylogenetically far removed from acrB-Ec. AcrB-Ec is one of the latest evolved pumps in the phylogenetic tree and is most closely related to homologue AcrF from E. coli and orthologue AdeJ from Acinetobacter baumannii. These transporters then share a branching point with a cluster containing E. coli AcrD and Pseudomonas aeruginosa MexB, MexD, and MexY. Further genetically distanced genes belong to other clusters; firstly harbouring A. baumannii AdeB, and then P. aeruginosa MexF27 and MexQ. Even further genetically distanced is a cluster separated into two genetic sub-clusters; one containing P. aeruginosa MexI28 and MexW29 and the other one AcrB-Hi. AcrB-Hi therefore appears to be a relatively ancestral efflux pump, closely related to the AcrB efflux pump from zoonotic pathogen Pasteurella multocida, and other RND pumps from e.g. pathogenic Q-fever causing Coxiella burnetii and marine bacterium Photobacterium profundum (Fig. 2).
Fig. 2.
Phylogenetic tree of a selection of RND transporters. Phylogenetic relationship between several homologous RND transporters (~160) from gammaproteobacteria analysed by multiple sequence alignment, branched into several clusters. One of the two latest branched clusters harbour E. coli AcrB-Ec (blue, right), AcrF (blue) and A. baumannii AdeJ (green), and the other cluster P. aeruginosa MexB, MexD, MexY (red) and E. coli AcrD (blue). Two genetically further distanced clusters contain A. baumannii AdeB (green) and P. aeruginosa MexB and MexD (red), respectively. Then H. influenzae AcrB (yellow, left) and P. aeruginosa MexI and MexW (red) are situated in genetically further removed phylogenetic clusters. Clusters and sub-clusters are highlighted in blue and dark blue, respectively. The complete tree of all 393 analysed RND genes can be found in Supplementary Fig. 2. Branch lengths can be seen in Supplementary Fig. 3
AcrB-Hi is an active promiscuous MDR efflux pump
As we found that AcrB-Hi is phylogenetically distant from AcrB-Ec, we wanted to test for its substrate specificity and compare it to AcrB-Ec. Table 1 shows the minimal inhibitory concentration (MIC) data for 15 different compounds, including planar aromatic cations (PACs), low-molecular-mass drugs (LMMDs), high-molecular-mass drugs (HMMDs) and β-lactams (including penicillins, cephalosporins, carbapenems, and monobactams). The acrRAB-cluster from the H. influenzae strain Rd KW20 genome was cloned into plasmids (pBAD33acrRABHiT, Supplementary Table 1) and transformed into E. coli MG1655∆acrAB cells. AcrB-Hi and adapter protein AcrA-Hi were functioning with E. coli TolC (TolC-Ec). AcrB-Ec and AcrB-Hi expression levels were similar (Supplementary Fig. 4). Compared to AcrB-Ec, AcrB-Hi is a relatively less-efficient transporter, with MIC values identical to or lower than AcrB-Ec. Nonetheless, it was able to transport the same compounds tested as AcrB-Ec.
Table 1.
Minimal inhibitory concentrations (MIC) for several antibiotics for AcrB-Ec and AcrB-Hi expressing E. coli MG1655 cells
| Genotype | Plasmid | Minimal inhibitory concentration (MIC, µg mL−1) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAC | LMMD | HMMD | β-lactam | |||||||||||||
| Penicillins | Cephalosporins | Carbapenem | Monobactam | |||||||||||||
| EtBr | R6G | CV | MINO | DEQ | KAN | EM | NOV | CLX | BPEN | MET | CEFP | CTX | DORI | AZT | ||
| ∆acrB | pBAD33 | 16 | 16 | 2 | 1 | 1.6 | 8 | 4 | 8 | 2 | 16 | 16 | 0.016 | 0.016 | 0.03125 | 0.0625 |
| ∆acrB | pBAD33acrBhis | 256 | >512 | 16 | 4 | 100 | 8 | 64 | 512 | 128 | 32 | >512 | 8 | 0.016 | 0.0625 | 0.0625 |
| ∆acrAB | pBAD33acrRABHiT | 128 | 256 | 16 | 2 | 50 | 8 | 32 | 64–128 | 64 | 16–32 | 128 | 1 | 0.016 | 0.0625 | 0.0625 |
Liquid MIC values determined by a twofold dilution method, where cells are grown in LB broth supplemented with drugs as described in Methods. Bold indicates a significant at least two dilutions increase in MIC compared to KO cells, italic indicates the same MIC as KO cells. Data based on at least two independent experiments. An overview of relative MICs between AcrB-Ec and AcrB-Hi can be seen in Supplementary Fig. 5
PAC planar aromatic cation, LMMD low-molecular-mass drug, HMMD high-molecular-mass drug, EtBr ethidium bromide, R6G rhodamine6G, CV crystal violet, MINO minocycline, DEQ dequalinium, KAN kanamycin, EM erythromycin, NOV novobiocin, CLX cloxacillin, BPEN benzylpenicillin, MET methicillin, CEFP cefcapene pivoxil, CTX ceftriaxone, DORI doripenem, AZT aztreonam, acrBhis his-tagged E. coli acrB, acrRABHiT H. influenzae acrR, acrA and his-tagged acrB
PACs, erythromycin (EM), antiseptic dequalinium (DEQ) and the penicillin cloxacillin (CLX) show a relatively high MIC compared to AcrB-Ec, showing only a lowering in MIC of up to one dilution (while also being significantly more active than acrB-deficient MG1655 cells). At the same time, HMMD aminocoumarin novobiocin (NOV), penicillin methicillin (MET) and cephalosporin cefcapene pivoxil (CEFP) show a somewhat lower MIC when AcrB-Hi was expressed, with a lowering of the MIC values by 2−3 dilutions compared to AcrB-Ec (although also still being significantly higher than acrB-deficient cells). On the other hand, compared to acrB knockout (KO) cells, CEFP shows the relatively highest MIC of six dilutions higher than KO cells. It appears that the antibiotic specificity of AcrB-Hi is very similar to AcrB-Ec, although depending on the compound, the MIC is either similar or significantly lower (Supplementary Fig. 5). For both AcrB-Ec and AcrB-Hi expressing cells, third-generation cephalosporin ceftriaxone (CTX), monobactam aztreonam (AZT) and aminoglycoside kanamycin (KAN) show identical MIC values as the acrB-deficient cells. LMMD tetracycline minocycline (MINO), carbapenem doripenem (DORI) and penicillin benzylpenicillin (BPEN) only show a small elevation of the MIC compared to acrB KO cells. (Supplementary Fig. 5).
We were particularly interested to check the AcrB-Hi efflux ability of β-lactams, as these antibiotics are often used as a first-line treatment against H. influenzae-induced infectious diseases5. AcrB-Hi can export penicillin (CLX, BPEN and MET), cephalosporin (CEFP) and slightly carbapenem (DORI) antibiotics (Table 1). Relatively to AcrB-Ec, from these antibiotics, AcrB-Hi-expressing cells showed a significantly high MIC for CLX and least significantly for MET (Supplementary Fig. 5). In absolute terms, for AcrB-Hi expressing cells, the highest MIC values were observed for MET (128 µg mL−1) and CLX (64 µg mL−1). It must be noted that AcrB-Hi was expressed within E. coli cells; it is therefore possible that the unnatural environment of E. coli played a role in the relatively lower MICs; however, AcrB-Ec and AcrB-Hi were expressed in similar abundance (Supplementary Fig. 4). The presence of AcrR-Hi and TolC-Ec is discussed in the Discussion section. Nonetheless, the MIC values for AcrB-Ec- and AcrB-Hi-expressing cells were either identical (for CV), just slightly lower (for DEQ, CLX, EtBr and EM) or significantly lower (for R6G, NOV, and MET) (Supplementary Fig. 5), pointing to a divergent resistance caused by intrinsic differences between these two transporters. Despite the relatively distant phylogenetic relationship with AcrB-Ec, AcrB-Hi too is a promiscuous MDR efflux pump.
AcrB-Hi is uninhibited and exports bile salts weakly
The phylogenetic tree (Fig. 2) shows that AcrB-Hi is a relatively ancestral MDR pump which—despite the low similarity with AcrB-Ec—could transport the same antibiotics and dyes (Table 1). We set out to investigate potential differences between AcrB-Ec and AcrB-Hi. As RND pumps have a physiological function in bacterial cells besides being MDR efflux systems30, and because AcrB-Ec seemed to be evolved significantly later, we wanted to investigate the efflux ability of AcrB-Hi for AcrB-Ec biologically relevant compounds: bile salts. These toxic compounds are present in the natural environment of enteric E. coli cells, but not of H. influenzae cells. Bile salts, including cholic acid (CHO) and deoxycholic acid (DEOX), are excellent substrates of AcrB-Ec, but not of all MDR RND transporters17. Table 2 shows the MIC results for bile salt (mixture), CHO, DEOX and indole for AcrB-Ec- and AcrB-Hi-expressing cells. AcrB-Hi could not export the bile salts efficiently, with MIC values of only one dilution higher than the acrB KO cells, while cells expressing AcrB-Ec had MIC values 16- to 32-fold higher than the acrB KO cells, for CHO, bile salts mixture, and DEOX, respectively. The MIC values for AcrB-Hi- and AcrB-Ec-expressing cells for bile salts and deoxycholic acid were 2000 vs. >16,000 µg mL−1 (>8-fold) and 1600 vs. >25,600 µg mL−1 (>16-fold), with acrB KO MICs as 1000 and 800 µg mL−1, respectively. The MIC values for AcrB-Ec could be even higher, as the tested compound concentrations reached its limit (Table 2, Supplementary Fig. 5). A recent crystal structure of efflux pump regulator RamR cocrystallised with bile salts shows that bile salts can bind in the voluminous binding pocket of the regulator protein by forming hydrogen bonds31 rather than by π−π interactions used to bind PACs32. The hydrophobic pit in AcrB-Hi does not comprise a Phe-rich cluster (Fig. 1, Supplementary Fig. 1); however, it is probably not the Phe-residues that recognise bile salts, because bile salt molecules do not contain aromatic moieties. AcrB-Hi also did not seem to export aromatic indole, with an identical MIC value as the acrB KO cells, although the MIC for AcrB-Ec was only (at least) one dilution higher compared to the KO cells (Table 2).
Table 2.
Minimal inhibitory concentrations (MIC) for bile salts for AcrB-Ec and AcrB-Hi expressing E. coli MG1655 cells
| Genotype | Plasmid | Minimal inhibitory concentration (MIC, µg mL−1) | |||
|---|---|---|---|---|---|
| Bile salts | Other | ||||
| Mix | CHO | DEOX | Indole | ||
| ∆acrB | pBAD33 | 1000 | 6400 | 800 | 512 |
| ∆acrB | pBAD33acrBhis | >16,000 | >25,600 | >25,600 | >512 |
| ∆acrAB | pBAD33acrRABHiT | 2000 | 12,800 | 1600 | 512 |
Liquid MIC values determined by a twofold dilution method, where cells are grown in LB broth supplemented with drugs as described in Methods. Bold shows an only one dilution higher MIC than KO cells, where bold underlined additionally indicates a significant decrease in MIC for AcrB-Hi compared to AcrB-Ec. Data based on at least two independent experiments. An overview of relative MICs between AcrB-Ec and AcrB-Hi can be seen in Supplementary Fig. 5
CHO cholic acid, DEOX deoxycholic acid, acrBhis his-tagged E. coli acrB, acrRABHiT H. influenzae acrR, acrA and his-tagged acrB
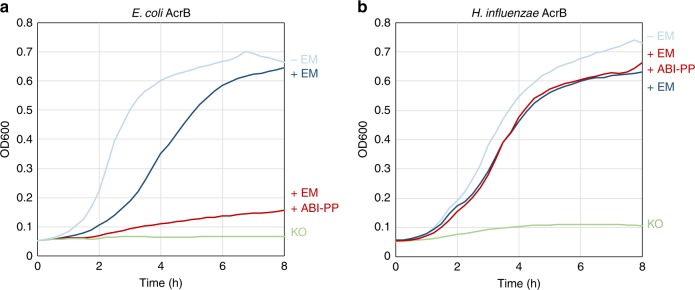
Because the Phe-lacking AcrB-Hi expressed in E. coli cells showed very low MICs for bile salts close to KO cells, we additionally wanted to know the potential implications of the deficiency of the Phe-rich hydrophobic trap. The first crystal structure of an RND transporter with an efflux pump inhibitor (EPI) was AcrB-Ec co-crystallised with pyridopyrimidine-derived EPI ABI-PP25. ABI-PP was located in the Phe-rich hydrophobic trap in the binding monomer of the trimer, blocking the transport ability of the pump. The Phe-residues were strongly interacting with the aromatic moieties of the inhibitor by π−π interactions, especially with Phe178 and Phe615. Figure 3 shows the growth ability of both AcrB-Hi- and AcrB-Ec-expressing cells in the presence or absence of EM and ABI-PP. AcrB-Ec was completely inhibited by the inhibitor ABI-PP as found before25; however, AcrB-Hi was completely unaffected by the EPI and cells showed full viability. When ABI-PP was added in even higher concentrations (double and triple), there was still no effect. Although the presence of the Phe cluster in AcrB-Ec may be important for efficient binding and export of some pit-binding compounds33,34 and inhibitors25,35, they seem not to be crucial for the export of antibiotics and dyes (Table 1), although they are important for the tight binding of and inhibition by pyranopyridine-derived EPIs (Figs. 1, 3, Supplementary Fig. 6)25,35. However, the inability to recognise and export bile salts effectively (Table 2) should perhaps be explained by other regions or differences in the transporter (possibly in the binding pockets36,37 or the entrance channels37,38).
Fig. 3.
Inhibitory effect of ABI-PP on AcrB-Ec and AcrB-Hi. a Efflux pump inhibitor ABI-PP was able to inhibit the efflux activity of AcrB-Ec (red). b ABI-PP was unable to inhibit AcrB-Hi (red). Colours light blue and dark blue show AcrB-expressing cells without or with the addition of erythromycin, respectively, and green shows acrB knockout cells in the presence of erythromycin. Red shows the growth ability of AcrB-expressing E. coli cells with the addition of both erythromycin and ABI-PP. All erythromycin concentrations were 32 µg mL−1 and ABI-PP concentrations were 64 µg mL−1 (a, b). EM erythromycin, KO acrB knockout. Data shown are one of the results, repeats (at least four) showed similar results
As we found that AcrB-Hi was not inhibited by ABI-PP and that for certain compounds AcrB-Hi-expressing cells show a divergent MIC pattern compared to AcrB-Ec-expressing cells, we tried to partly mimic the AcrB-Hi pit residues in AcrB-Ec (Supplementary Table 2). We made several single and double Phe-substitutions and checked the MIC for several compounds. Most single and double mutations had a negative effect on the MIC of all tested compounds, which may explain the higher MIC for certain compounds by AcrB-Ec. For example, replacing the Phe136 and Phe628 with Ile or Gly had relatively the biggest effect on R6G and to a lesser extent on EtBr and MET (Supplementary Table 2). Probably multiple differences in residues in both the binding pocket and the hydrophobic pit cause the divergent MIC differences between AcrB-Hi- and AcrB-Ec-expressing cells. Other factors also play a role in substrate recognition. As also seen in Supplementary Table 2, the mutation Phe610Glu in AcrB-Ec (mimicking Glu594 in AcrB-Hi) had little effect on the MIC values, despite the significant change from a hydrophobic aromatic side chain to a negatively charged side chain. Interestingly, the similar mutation Phe610Ala, described too by others39 in AcrB-Ec, had a significant negative effect on the MIC values. It is therefore difficult to interpret mutations in the pockets of AcrB, and binding efficiencies alone are not the only determinant of the export of the compounds by AcrB39. Additionally, we made mutations in AcrB-Hi to include Phe-residues in the hydrophobic pit to mimic AcrB-Ec to see if the transporter would become sensitive to ABI-PP. In the crystal structure of ABI-PP bound to AcrB, the residues binding most strongly are Phe178, Phe615 and Phe628, and slightly Phe610. Supplementary Fig. 7 shows the results. Phe178 is conserved in AcrB-Hi (Phe182). E594F (resulting in double Phe178 and Phe610) had no effect. M599F (resulting in double Phe178 and Phe615, which strongly interact with ABI-PP in AcrB-Ec) had no effect on ABI-PP inhibition. The introduction of I613F (corresponding to AcrB-Ec Phe628) resulted in a completely inactive transporter. For this reason, we are cautious in interpreting the MIC findings presented in Supplementary Table 2. A more thorough analysis of the binding pockets is needed (by molecular dynamic or chimeric experiments replacing the whole pit or the pockets) in order to better understand the differences in substrate export efficiency and EPI inhibition.
Antibiotics leak through outer membrane porin OmpP2
AcrB-Hi can actively transport β-lactams (Table 1). Previous research showed that MICs for a β-lactam (carbenicillin), fusidic acid, ciprofloxacin, tetracycline (TET), chloramphenicol (CHL) and norfloxacin (NOR) for Hib cells (expressing AcrB-Hi) were identical to acrB KO cells40. It was also found that TET, CHL, and NOR were substrates of AcrB-Hi13, and here we show that AcrB-Hi can export many other compounds, including a variety of classes of β-lactams (Table 1). β-lactams, including penicillins, are often used as first-line drugs to treat Hib infections5, despite that these compounds are actively being effluxed by the H. influenzae AcrAB-TolC system (Table 1). The outer membrane (OM) of Hib cells is very permeable41, including for β-lactams42. The outer membrane of H. influenzae contains a wide outer membrane porin OmpP2. We wanted to directly see the possible effect of this large porin on the efflux by AcrB-Hi. We therefore cloned the ompP2 gene from the H. influenzae strain Rd KW20 genome into plasmids (creating pET26b(+)ompP2, Supplementary Table 1). Table 3 shows the MICs for ten different compounds. Basically no effect was seen for DEQ, EM and most tested PACs, for both AcrB-Ec- and AcrB-Hi-expressing cells when OmpP2 was introduced. The MICs were identical (for EtBr, DEQ and EM) or decreased one dilution (for R6G). For the cationic CV, however, the MIC values (for AcrB-Ec expressing cells) was decreased by two dilutions. Additionally, for the large molecule NOV the MIC was affected significantly by the expression of OmpP2, as the MIC values (for both AcrB-Ec- and AcrB-Hi-expressing cells) were decreased a significant two dilutions. It must also be noted that for NOV, the expression of OmpP2 had a big effect for all tested concentrations and that therefore the MIC values are within a range and difficult to determine; the significant effect on growth ability under NOV can be observed in Supplementary Fig. 8. Furthermore, a significant decrease in MIC was seen for all the β-lactams CLX, BPEN and MET, with a decrease of MIC ranging between one and three dilutions. Penicillin MET was affected mostly with a decrease in MIC from 128 µg mL−1 for AcrB-Hi-expressing cells, to 16 µg mL−1 when AcrB-Hi and OmpP2 were both expressed. CLX was also significantly affected, with an MIC decrease from 128 or 64 µg mL−1 to 16 µg mL−1 when OmpP2 was expressed. A previous study showed that the MICs for other LMMDs (such as TET and CHL) for Hib cells were the same with or without AcrB-Hi40 (indicating an influx of these LMMDs through the OM), while the MIC for the LMMD DEQ in our study showed to be unaffected by OmpP2, perhaps because this compound is positively charged. The PACs tested here are also positively charged and also seem to be unaffected by the expression of OmpP2 (Table 3). We did not test for CHL, as this was the selection marker on our pBAD33 constructs, and the tetracycline MINO is not a very good substrate of both AcrB-Ec and AcrB-Hi (Table 1).
Table 3.
Minimal inhibitory concentrations (MIC) for several compounds for AcrB-Ec, AcrB-Hi and OmpP2 expressing E. coli C43(DE3) cells
| Genotype | Plasmid(s) | Minimal inhibitory concentration (MIC, µg mL-1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PACs | LMMD | HMMD | β-lactams | |||||||||||
| Proteins expressed | Penicillins | |||||||||||||
| AcrB-Ec | AcrB-Hi | OmpP2 | EtBr | CV | R6G | DEQ | EM | NOV | CLX | BPEN | MET | |||
| ∆acrB∆ompF | pBAD33 + pET26b(+) | <4 | 2 | 8 | 0.4 | 2 | 2 | <4 | 1 | 2 | ||||
| AcrB-Ec | ∆acrB | pBAD33acrBhis | ✓ | 32 | 32 | >512 | 50 | 64 | 512 | 128 | 4 | 256 | ||
| ∆acrB∆ompF | pBAD33acrBhis | ✓ | 32 | 32 | >512 | 50 | 32 | 256–512 | 64 | 4 | 256 | |||
| ∆acrB∆ompF | pBAD33acrBhis + pET26b(+)ompP2 | ✓ | ✓ | 32 | 8 | >512 | 25 | 32 | 128 | 16 | 2 | 64 | ||
| AcrB-Hi | ∆acrB | pBAD33acrRABHiT | ✓ | 32 | 16 | 256 | 50 | 64 | 256–512 | 64 | 8 | 128 | ||
| ∆acrAB | pBAD33acrRABHiT | ✓ | 32 | 8 | 128–256 | 50 | 64 | 256–512 | 64 | 8 | 128 | |||
| ∆acrB∆ompF | pBAD33acrRABHiT + pET26b(+)ompP2 | ✓ | ✓ | 32 | 4 | 128–256 | 50 | 64 | <64–128 | 16 | 2 | 16 | ||
Liquid MIC values determined by a twofold dilution method, where cells are grown in LB broth supplemented with drugs as described in Methods. Italic indicates no change in MIC when OmpP2 is expressed, and bold underlined indicates 2 or 3 dilutions decrease in MIC. A check mark (✓) indicates the presence by expression of the respective protein. Data based on at least two independent reproduced experiments
PAC planar aromatic cation, LMMD low-molecular-mass drug, HMMD high-molecular-mass drug, EtBr ethidium bromide, CV crystal violet, R6G rhodamine6G, DEQ dequalinium, EM erythromycin, NOV novobiocin, CLX cloxacillin, BPEN benzylpenicillin, MET methicillin, acrBhis his-tagged E. coli acrB, acrRABHiT H. influenzae acrR, acrA and his-tagged acrB
We show that OmpP2 is a porin with high permeability, while maintaining a semi-specificity: one cationic compound (CV) and one HMMD (NOV) was affected, while the others were not, and all tested β-lactams seemed to be affected by OmpP2. The effect of OmpP2 on the MICs for our tested compounds can be summarised as β-lactams > NOV > CV> PACs ≧ EM. Large antibiotic EM may also traverse through OmpP2, but at a much slower rate, hence largely not affecting the MIC. The high permeability by the influx through OmpP2 can explain why Hib cells are sensitive to β-lactams and novobiocin, despite the fact that these are actively transported by the AcrB-Hi efflux transporter (Table 1).
Discussion
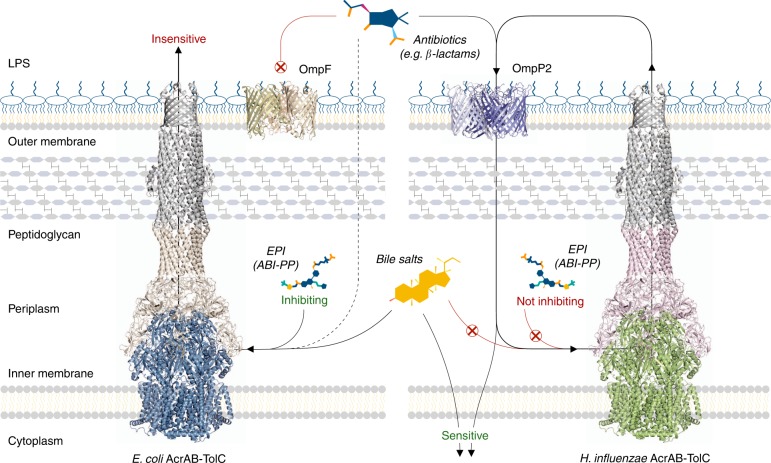
We showed that AcrB-Hi is a relatively ancient efflux pump, with a similar antibiotic efflux range as its evolved colleague AcrB-Ec. Despite the similar drug range, differences were additionally observed: AcrB-Hi is a significantly weak bile salt exporter compared to AcrB-Ec. In addition, AcrB-Hi is not inhibited by the efflux pump inhibitor ABI-PP. Furthermore, we showed that outer membrane porin OmpP2 leaks β-lactams and novobiocin, providing a molecular explanation for the high sensitivity of H. influenzae to these antibiotics. The results are summarised in Fig. 4.
Fig. 4.
The interplay between efflux pumps and porins from E. coli and H. influenzae. AcrB efflux pumps can actively transport antibiotics from the periplasm and cytoplasm of bacterial cells, rendering them ineffective. AcrB-Ec is able to export bile salts, which is present in the enteric environment of E. coli cells. On the other hand, AcrB-Hi can export bile salts only weakly. Additionally, efflux pump inhibitor ABI-PP cannot inhibit AcrB-Hi, while it inhibits the efflux ability of AcrB-Ec completely. The wide OmpP2 outer membrane porin from H. influenzae leaks small and elongated antibiotics such as β-lactams and novobiocin back into the cells, making Hib cells sensitive to these antibiotics. OmpF was rendered from a crystal structure (PDB identification code 3POX) and the structure of OmpP2 is based on a homology model. EPI efflux pump inhibitor
We found that ancestral RND transporter AcrB-Hi is relatively closely related to P. aeruginosa MexI and MexW. The MexGHI-OpmD system can export vanadium28 and several classes of antibiotics43. MexVW-OprM can export a wide range of antibiotics, including erythromycin and ethidium bromide29. As H. influenzae cells harbour merely one RND transporter, it can be argued that AcrB-Hi plays a physiological role besides being a MDR efflux pump. Being closely related to MexW, AcrB-Hi may be involved in the production of quorum sensing signal molecules43, indirectly accounting for pathogenicity and promotion of persistence and antibiotic resistance of other bacteria44. Additionally, the transporter causes a decreased susceptibility for macrolide antibiotics in clinical strains14.
We also found that AcrB-Hi has a similar antibiotic efflux spectrum as AcrB-Ec, despite these transporters being significantly phylogenetically unrelated within the analysed 393 RND efflux genes (Fig. 2, Supplementary Fig. 2, 3, Supplementary Data 1). In a previous study, the highest MIC values for H. influenzae cells were observed for crystal violet and erythromycin (4 and 2.5 µg mL−1, respectively)40, consistent to the high efflux activity of these compounds by AcrB-Hi (Table 1). Interestingly, in the same study, the MIC values for novobiocin were significantly lower (0.13 µg mL−1, and only two dilutions higher than acrB KO cells). Additionally, in the same study, the MIC for β-lactam carbenicillin was unaffected (0.08 µg mL−1) within H. influenzae cells when the acrB gene was disrupted40. Furthermore, MIC values for other β-lactams (e.g. for ampicillin and benzylpenicillin) are significantly low (0.02 and 0.06 µg mL−1, respectively) for H. influenzae cells41. The quantitative MIC order for these compounds for intrinsically AcrB-Hi-expressing H. influenzae cells is therefore CV > EM≫NOV > β-lactams. On the other hand, in our study we observe high MIC values for novobiocin and β-lactams when AcrB-Hi was expressed in E. coli cells, which were even higher than for erythromycin and crystal violet (Table 1). For these compounds the MIC order is reversed to β-lactams = NOV > EM > CV. We provide a molecular explanation by the presence of OmpP2 in the OM of H. influenzae cells. This porin is naturally not present in E. coli cells. When we expressed OmpP2 in ompF-deficient E. coli cells, we saw a significant decrease in MICs for β-lactams (including cloxacillin and methicillin) and novobiocin (Table 3, Supplementary Fig. 8). At the same time, we saw no effect for erythromycin, and somewhat effect for crystal violet (Table 3). Therefore, we can directly explain the sensitivity of the Hib cells to β-lactams and novobiocin, as these compounds traverse through the porin channel back into the cells. Similarly, mutations in the OmpC porin in E. coli caused a higher sensitivity to β-lactams, as these mutations increased the width of the pore45. As the MICs were unaffected for high-molecular-mass drug erythromycin, it is reasonable to argue that novobiocin despite its high-molecular mass is a rod-like elongated molecule, while erythromycin is a bulkier sphere-like molecule46, hence a slow influx rate. In a previous study41, the OmpP2 molecular weight exclusion limit was estimated to be about 1400 Da. To compare, E. coli OmpF has an exclusion limit estimated to be 600 Da47. Interestingly, despite the high exclusion limit of OmpP2, small cationic compounds such as ethidium bromide were unaffected by the expression of OmpP2 (Table 3), which corroborates the high MIC of ethidium bromide for Hib cells (1.25 µg mL−1, and three dilutions higher than the acrB KO cells)40. OmpP2 seems to be a semi-selective channel for neutral compounds; however, it must be noted that the porin also impacted the MIC for cationic crystal violet (Table 3).
Additionally, we found that AcrB-Hi was able to export bile salts only weakly, while AcrB-Ec is a strong bile salt pump17 (Table 2, Supplementary Fig. 5). As E. coli is an enteric bacterium, it is essential for the organism to export bile salts48; however, H. influenzae is naturally not residing in a bile salts-rich environment. We also found that aminoglycoside kanamycin, third-generation cephalosporin ceftriaxone, and monobactam aztreonam were not exported. Third-generation cephalosporins such as ceftriaxone or cefotaxime are recommended antibiotics to treat H. influenzae infections—even when β-lactamases are identified in the strain5—and the inability of AcrB-Hi to export these drugs (Table 1) in addition to the high permeability of OmpP2 (Table 3) explains the effectiveness of these β-lactams. Monobactam aztreonam (used to treat E. coli and P. aeruginosa infections5) is exported by AcrD-Ec, but not by AcrB-Ec nor AcrB-Hi22. Figure 2 shows that AcrD-Ec is located in a different cluster than AcrB-Ec, which include MexB and MexY. Aminoglycosides are exported by AcrD-Ec and MexY, but not by AcrB-Ec, MexB and AcrB-Hi49,50. Figure 2 shows that MexB, MexY, and AcrD-Ec share the same cluster; however, MexB is located in a separate sub-cluster. A. baumannii AdeB from another earlier distinct cluster (Fig. 2) is also involved in aminoglycoside resistance51. It is interesting that AcrB-Hi from an ancestral cluster cannot export aminoglycosides, while some RND transporters from adapted clusters (MexY, AcrD-Ec) can, while other RND transporters (MexB, AcrB-Ec) cannot.
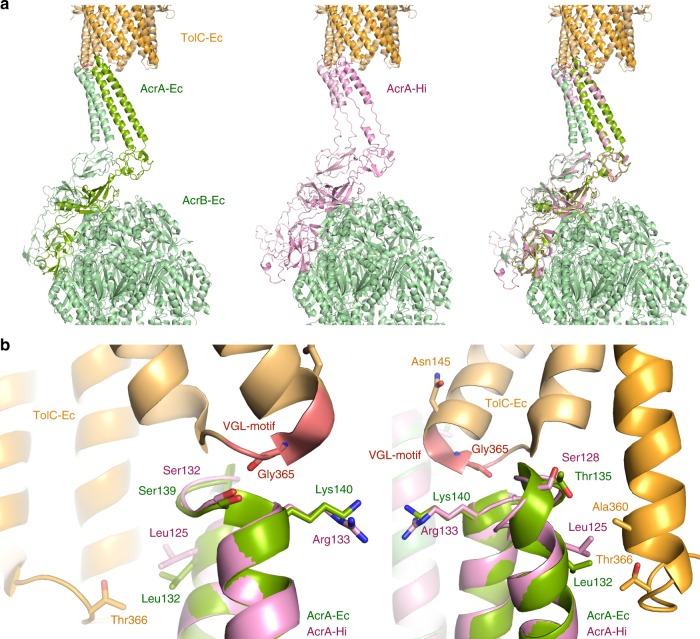
We were only able to express functional AcrAB-Hi when the upstream AcrR-Hi gene was added to the construct. The presence of AcrR-Hi could have influenced the MIC data, as the repressor would have bound to the DNA if no AcrR substrate was present. However, we expressed AcrAB-Hi in the presence of bile salts (Supplementary Fig. 4) and found that the expression level of AcrB-Hi was similar to that of AcrB-Ec. We are not sure why the removal of acrR-Hi from the construct resulted in non-functional acrAB-constructs; however, the same phenomenon was observed previously13. In addition, AcrAB-Hi was functional with TolC-Ec. We investigated the AcrA/TolC interactions by homology modelling based on the tripartite structure of AcrAB-TolC52. Initial alignment of AcrA-Hi (382 amino acids) with AcrA-Ec (397 amino acids) results in 24% identity, 43% similarity and 15% gaps. Figure 5 shows the homology model of AcrA-Hi modelled on the tripartite structure of E. coli AcrAB-TolC52. In the model, the helices of the arms of AcrA-Hi (about 26 residues) are significantly shorter than AcrA-Ec (about 35 residues). However, residues interacting with TolC-Ec are conserved (Leu125 and Ser132) or similar (Arg133 and Ser128) in AcrA-Hi compared to AcrA-Ec. These residues can explain by tip−tip interactions the functional AcrAB-Hi when coupled with TolC-Ec. We cannot be completely sure whether or not the substitution of TolC-Hi with TolC-Ec slightly altered the drug susceptibility. However, the active pumping of the drugs by AcrB-Hi resulted in the elevated MICs. This indicates the substrate specificity to these tested compounds of the efflux pump AcrB-Hi itself.
Fig. 5.
AcrA-Hi modelled in the tripartite structure of AcrAB-TolC. The homology model of AcrA-Hi is shown in pink and the actual AcrAB-Ec cryo-EM structure is shown in green. TolC-Ec is shown in orange. The TolC conserved VGL-motif is shown in red. a The AcrAB-TolC complex (shown are two out of the total six AcrA chains). The left image is the AcrAB-TolC-Ec structure (PDB identification code 5O66). The centre image is the homology model of AcrA-Hi modelled on AcrB-Ec and TolC-Ec. AcrA-Hi is depicted in pink. Visible are the elongated chains, rather than a helix conformation. In AcrA-Ec, the two helices are Pro98-Leu133 (36 residues) and Ser139- Ala172 (34 residues). In the AcrA-Hi model, the helices are Ser99-Leu125 (27 residues) and Gln134-Ile158 (25 residues), a significantly shorter region than for AcrA-Ec. The shorter AcrA-Hi is functional with TolC-Ec, despite the shorter arms. b Interactions of AcrA-Ec (green) or AcrA-Hi (pink) with TolC-Ec (orange). The AcrA-Ec residues interacting with TolC-Ec are depicted as sticks. The backbone of Gly365 in TolC-Ec is interacting with AcrA-Ec Ser139 and the backbone of Lys140, or with the corresponding AcrA-Hi Ser132 and the backbone of Arg133. Conserved interacting residues among AcrA-Ec and AcrA-Hi are Leu132/Leu125 and Ser139/Ser132, respectively, and similar residues are Lys140/Arg133 (positively charged) and Thr135/Ser128 (polar). These conserved residues can explain why AcrAB-Hi is functional with TolC-Ec
Furthermore, AcrB-Hi was completely uninhibited by ABI-PP (Fig. 3). ABI-PP strongly inhibits AcrB-Ec and MexB25. ABI-PP cannot inhibit MexY, as MexY has a bulky tryptophan in the Phe-rich pit of the distal binding pocket, sterically hindering the binding of ABI-PP25. As for AcrB-Hi, the lack of a Phe-rich pit (Fig. 1, Supplementary Fig. 1) probably prevents the strong required binding to the inhibitor’s aromatic moieties. Homology modelling of the AcrB-Hi hydrophobic trap shows no steric hindrance (Supplementary Fig. 6). However, we cannot be certain how the hydrophobic pit of AcrB-Hi actually is. The addition of the Phe-residues by evolution may have contributed to the increase of export efficiency by AcrB-Ec, as aromatic compounds such as novobiocin and rhodamine 6G are significantly more efficiently exported by AcrB-Ec than AcrB-Hi (Table 1, Supplementary Fig. 5), and Phe-substitutions in AcrB-Ec significantly decrease the MIC for rhodamine 6G (Supplementary Table 2). The Phe-rich pit is nonetheless a disadvantage of these improved RND efflux pumps, since they can be inhibited by current EPIs53. It additionally points to a potential weakness in these presently developed EPIs25,35,54, as genomic point mutations causing Phe-substitution may render the EPIs ineffective.
We hypothesise based on the phylogenetic analysis and drug export ability data that multidrug recognition is not an evolutionarily acquired ability present only in the latest evolved efflux pumps. Supplementary Table 3 shows the residues in the pockets and the hydrophobic trap of several RND transporters interacting with substrates (minocycline, doxorubicin, and erythromycin) according to the AcrB-Ec crystal structures36,37. Some residues are conserved or similar between transporters (but not for all transporters) and certain residues within the same phylogenetic cluster tend to be more similar. AcrB-Ec and AcrB-Hi also show conserved residues in the binding pockets; however, there are also several differences (Supplementary Table 3). We hypothesised before that drugs may be occluded in the distal binding pocket, and not bound strongly to specific residues, oscillating and moving within the voluminous binding pocket, before being pushed out by peristaltic motion16,55. Multidrug recognition may not be dependent on recognition within the distal binding pocket. We hypothesise that compounds are initially selected at the entrances at the lipid bilayer, periplasm, and central cavity, based on their physicochemical properties. Supplementary Table 3 suggests that the Phe-rich pit is present in all three AcrB-Ec, AcrF-Ec and AdeJ (from Acinetobacter baumannii) belonging to the same cluster. Transporters in clusters genetically further distanced show fewer Phe-residues. We hypothesise that the Phe-rich hydrophobic trap is acquired by evolution. The inhibition by ABI-PP might be an unexpected evolutionary result for the AcrB-Ec efflux pump. We note that this is a hypothesis and it would be interesting for future research to study the differences between other RND transporters in more detail.
To conclude, a recent serious problem in Hib chemotherapy are β-lactam-resistant strains, which are accounted for by β-lactamases (in BLPAR strains) or mutations in PBPs (in BLNAR strains). However, we emphasise that the overexpression of the AcrB-Hi pump in clinical strains is a potential risk for the development of MDR Hib pathogenic strains, as we show the wide substrate specificity of this pump. Furthermore, the loss of porin expression or porin mutations are well-known factors in acquired drug resistance in many organisms including E. coli and P. aeruginosa56,57. Mutations causing a loss in expression of OmpP2 or a narrowing of the porin could potentially also turn sensitive Hib strains into drug-resistant clinically relevant strains. In addition, we suggest that ancestral efflux pumps extrude similar hydrophobic and amphiphilic toxic compounds and that these nonspecific and less-efficient transporters have evolved into specific, more efficient transporters, and that multidrug recognition itself is not an evolutionarily acquired ability. Our knowledge on pharmacokinetics and the molecular defence mechanism by multidrug efflux systems including its evolution is crucial for the development of novel antibiotics.
Methods
Bacterial strains and growth conditions
The E. coli MG1655 strain58 was used as wild-type strain and ∆acrB (NKE96)59 and ∆acrAB were derived from MG1655. In addition, the E. coli C43(DE3) strain was used for the OmpP2-related minimal inhibition concentration experiments. Gene deletions (∆acrB, ∆acrAB and ∆acrB∆ompF) were performed according to the method of Datsenko and Wanner, with recombination between short homologous DNA regions catalysed by phage λ Red recombinase60. The drug resistance markers were eliminated using plasmid pCP2060. The bacterial strains were grown at 37 °C in Luria−Bertani broth61.
Protein analysis and phylogenetic tree
Homologous RND transporters genes were identified by homology search of acrB-Hi and acrB-Ec genes within the genome of interest from the NCBI genome database (GenBank) by GENETYX software. An overview of the analysed genes and their sequences can be found in Supplementary Data 1. Multiple sequence alignment was executed by Clustal Omega from EMBL-EBI62 and phylogenetic trees were visualised with iTOL63. Homology models were created using SWISS-MODEL64.
Amplification and cloning
Three adjacent genes HI0893 (acrR), HI0894 (acrA) and HI0895 (acrB) from the Haemophilus influenzae strain Rd KW20 (ATCC 51907) genome were amplified together using primers, including a C-terminal 6-histidine tag. pBAD33 vectors were digested by SalI and HindIII restriction enzymes (NEB) and the insert was cloned into pBAD33 vectors by InFusion (TaKaRa), resulting in pBAD33acrRABHiT (10.3 kbp). The ompP2 gene (HI0139) was amplified using primers and cloned by InFusion (TaKaRa) into XbaI- and HindIII-digested pET26b(+) vectors (Novagen, Merck), resulting in pET26b(+)ompP2 (6.4 kbp). All constructs were confirmed by agarose gel and nucleotide sequencing (FASMAC, Japan). Amplification primers can be found in Supplementary Table 1.
Site-directed mutagenesis
The plasmid pBAD33acrB (pBAD33 carrying the acrB-gene from E. coli MG1655, including a C-terminal hexahistidine-tag) was used for the site-directed mutagenesis. Primers were used to introduce the mutations by polymerase chain reaction (PCR). The mutations were confirmed by nucleotide sequencing (FASMAC, Japan). Plasmids were transformed in acrB-deficient MG1655 E. coli cells.
Drug susceptibility by MIC
The MIC values were determined using growth ability in liquid LB or on LB agar plates supplemented with substrates, in a series of dilutions. Cell cultures of MG1655∆acrB, MG1655∆acrAB, C43(DE3)∆acrB, C43(DE3)∆acrAB and C43(DE3)∆acrB∆ompF cells harbouring the plasmid of interest (pBAD33acrRABHiT, pBAD33acrBhis and/or pET26b(+)ompP2) were grown overnight, and liquid cultures (supplemented with 10 mM arabinose, or both 10 mM arabinose and 0.1 mM IPTG) were incubated, shaken at 37 °C and OD600 nm readings were done using the Infinite M Nano (200 PRO series, Tecan). ABI-PP concentrations were 64 µg mL−1. For LB agar MIC determination experiments, overnight grown cells were stamped on LB agar plates (supplemented with 10 mM arabinose) and incubated at 37 °C overnight. MIC values were defined as the lowest drug concentrations at which the cells were no longer viable. AcrB expression levels were measured by western blotting using an iBind western system (ThermoFisher), with an anti-his-tag mAb (MBL) as the primary and anti-mouse IgG HRP-linked whole Ab from sheep (GE Healthcare) as the secondary antibody. AcrB-Ec and AcrB-Hi were expressed in E. coli cells in LB medium supplemented with 10 mM arabinose, with and without the addition of 1000 µg mL−1 bile salts.
Statistics and reproducibility
MIC experiments and the inhibition ability experiments were repeated multiple times to validate the reproducibility. Phylogenetic analysis and clustering was based on 393 different homologous RND-type transporters and multiple sequence alignment was performed several times.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by CREST and the Center of Innovation Program (COI) from the Japan Science and Technology Agency (JST), the Program for the Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, Grants-in-Aid, Network Joint Research Center for Materials and Devices, Dynamic Alliance for Open Innovation Bridging Human, Environment and Materials from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), Grant-in-Aid Research Activity Start-up (Kakenhi 18H06103 and 19K21223) from MEXT, Grant-in-Aid for Scientific Research (B) (Kakenhi 17H03983) and for Challenging Research (Exploratory) (Kakenhi 18K19451) from Japan Society for the Promotion of Science (JSPS), and the Japan Agency for Medical Research and Development (AMED). We would like to thank Manami Tanaka, Norihiro Kawashita, Teruo Yasunaga, and Tatsuya Takagi for the initial explorative comparison of several RND-type efflux pumps. We would also like to thank Chihana Hayashi and Kyoka Tada for helping with the plate MIC experiments.
Author contributions
M.Z. performed the biochemical experiments and the gene analysis. M.Z., A.Y. and K.N. designed the research and wrote the manuscript.
Data availability
Data are available in this published article itself and the supporting figures and tables are available as Supplementary Information files. The table with the homologous RND genes can be found as Supplementary Data 1. Other data that support the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0564-6.
References
- 1.Peltola H. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 2000;13:302–317. doi: 10.1128/CMR.13.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watt JP, et al. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years: global estimates. Lancet. 2009;374:903–911. doi: 10.1016/S0140-6736(09)61203-4. [DOI] [PubMed] [Google Scholar]
- 3.Morris SK, Moss WJ, Halsey N. Haemophilus influenzae type b conjugate vaccine use and effectiveness. Lancet Infect. Dis. 2008;8:435–443. doi: 10.1016/S1473-3099(08)70152-X. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. Antimicrobial Resistance Global Report on Surveillance: 2014 Summary (World Health Organization, Geneva, 2014).
- 5.Tunkel AR, et al. Practice guidelines for the management of bacterial meningitis. Clin. Infect. Dis. 2004;39:1267–1284. doi: 10.1086/425368. [DOI] [PubMed] [Google Scholar]
- 6.Farrell DJ, Morrissey I, Bakker S, Buckridge S, Felmingham D. Global distribution of TEM-1 and ROB-1 β-lactamases in Haemophilus influenzae. J. Antimicrob. Chemother. 2005;56:773–776. doi: 10.1093/jac/dki281. [DOI] [PubMed] [Google Scholar]
- 7.Ubukata K, et al. Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 2001;45:1693–1699. doi: 10.1128/AAC.45.6.1693-1699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark C, et al. In vitro selection of resistance in Haemophilus influenzae by amoxicillin-clavulanate, cefpodoxime, cefprozil, azithromycin, and clarithromycin. Antimicrob. Agents Chemother. 2002;46:2956–2962. doi: 10.1128/AAC.46.9.2956-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tristram S, Jacobs MR, Appelbaum PC. Antimicrobial resistance in Haemophilus influenzae. Clin. Microbiol. Rev. 2007;20:368–389. doi: 10.1128/CMR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikaido H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 12.Blair JMA, Richmond GE, Piddock LJV. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9:1165–1177. doi: 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 13.Dastidar V, Mao W, Lomovskaya O, Zgurskaya HI. Drug-induced conformational changes in multidrug efflux transporter AcrB from Haemophilus influenzae. J. Bacteriol. 2007;189:5550–5558. doi: 10.1128/JB.00471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peric M, Bozdogan B, Jacobs MR, Appelbaum PC. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob. Agents Chemother. 2003;47:1017–1022. doi: 10.1128/AAC.47.3.1017-1022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:1–19. doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwama M, Yamaguchi A. Molecular mechanisms of AcrB-mediated multidrug export. Res. Microbiol. 2018;169:372–383. doi: 10.1016/j.resmic.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishino K, Yamada J, Hirakawa H, Hirata T, Yamaguchi A. Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to beta-lactams. Antimicrob. Agents Chemother. 2003;47:3030–3033. doi: 10.1128/AAC.47.9.3030-3033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aires JR, Kohler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 1999;43:2624–2628. doi: 10.1128/AAC.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elkins CA, Nikaido H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 2002;184:6490–6498. doi: 10.1128/JB.184.23.6490-6499.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi N, Tamura N, van Veen HW, Yamaguchi A, Murakami S. β-Lactam selectivity of multidrug transporters AcrB and AcrD resides in the proximal binding pocket. J. Biol. Chem. 2014;289:10680–10690. doi: 10.1074/jbc.M114.547794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramaswamy VK, Vargiu AV, Malloci G, Dreier J, Ruggerone P. Molecular rationale behind the differential substrate specificity of bacterial RND multi-drug transporters. Sci. Rep. 2017;7:8075. doi: 10.1038/s41598-017-08747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramaswamy VK, Vargiu AV, Malloci G, Dreier J, Ruggerone P. Molecular determinants of the promiscuity of MexB and MexY multidrug transporters of Pseudomonas aeruginosa. Front. Microbiol. 2018;9:19–17. doi: 10.3389/fmicb.2018.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakashima R, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 26.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J. Bacteriol. 2007;189:7600–7609. doi: 10.1128/JB.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler T, et al. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 28.Aendekerk S, Ghysels B, Cornelis P, Baysse C. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology. 2002;148:2371–2381. doi: 10.1099/00221287-148-8-2371. [DOI] [PubMed] [Google Scholar]
- 29.Li Y. A new member of the tripartite multidrug efflux pumps,MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2003;52:572–575. doi: 10.1093/jac/dkg390. [DOI] [PubMed] [Google Scholar]
- 30.Piddock LJV. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki S, et al. Crystal structure of the multidrug resistance regulator RamR complexed with bile acids. Sci. Rep. 2019;9:177. doi: 10.1038/s41598-018-36025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamasaki S, et al. The crystal structure of multidrug-resistance regulator RamR with multiple drugs. Nat. Commun. 2013;4:2078. doi: 10.1038/ncomms3078. [DOI] [PubMed] [Google Scholar]
- 33.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2012;109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinana AD, Vargiu AV, Nikaido H. Effect of site-directed mutations in multidrug efflux pump AcrB examined by quantitative efflux assays. Biochem. Biophys. Res. Commun. 2016;480:552–557. doi: 10.1016/j.bbrc.2016.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sjuts H, et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA. 2016;113:3509–3514. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 37.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 38.Zwama M, et al. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 2018;9:124. doi: 10.1038/s41467-017-02493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vargiu AV, et al. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J. Am. Chem. Soc. 2011;133:10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez L, Pan W, Vinas M, Nikaido H. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 1997;179:6855–6857. doi: 10.1128/jb.179.21.6855-6857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vachon V, Lyew DJ, Coulton JW. Transmembrane permeability channels across the outer membrane of Haemophilus influenzae type b. J. Bacteriol. 1985;162:918–924. doi: 10.1128/jb.162.3.918-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coulton JW, Mason P, Dorrance D. The permeability barrier of Haemophilus influenzae type b against beta-lactam antibiotics. J. Antimicrob. Chemother. 1983;12:435–449. doi: 10.1093/jac/12.5.435. [DOI] [PubMed] [Google Scholar]
- 43.Aendekerk S. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005;151:1113–1125. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 44.Armbruster CE, et al. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio. 2010;1:S2–S9. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misra R, Benson SA. Isolation and characterization of OmpC porin mutants with altered pore properties. J. Bacteriol. 1988;170:528–533. doi: 10.1128/jb.170.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakurai K, et al. Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds. Sci. Rep. 2019;9:4359. doi: 10.1038/s41598-019-40232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 48.Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenberg EY, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 2000;182:1754–1756. doi: 10.1128/JB.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikaido H, Pagès JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife. 2017;6:e24905. doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aron Z, Opperman TJ. The hydrophobic trap-the Achilles heel of RND efflux pumps. Res. Microbiol. 2017;169:1–8. doi: 10.1016/j.resmic.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen ST, et al. Structure−activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorg. Med. Chem. 2015;23:2024–2034. doi: 10.1016/j.bmc.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 56.Delcour AH. Outer membrane permeability and antibiotic resistance. BBA-Proteins Proteome. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernández L, Hancock REW. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 59.Senda Y, Yamaguchi A, Nishino K. The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J. Infect. Chemother. 2008;14:23–29. doi: 10.1007/s10156-007-0575-Y. [DOI] [PubMed] [Google Scholar]
- 60.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd edn (Cold Spring Laboratory Press, New York, 1989).
- 62.McWilliam H, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data are available in this published article itself and the supporting figures and tables are available as Supplementary Information files. The table with the homologous RND genes can be found as Supplementary Data 1. Other data that support the findings of this study are available from the corresponding author upon request.