Many organisms have established symbiotic relationships with acquired mobile genetic elements (MGEs) integrated in their genomes (1). MGEs spread among genomes within and across microbial species through horizontal gene transfer and, once integrated into host chromosome, are disseminated vertically to the progeny, causing rapid evolution of drug resistance, pathogenicity, and virulence traits (2, 3). The MGEs that integrates into the host bacterial chromosomes (IMGEs) either carry their own DNA integration machineries or exploit machineries already existing in the host organisms for integration (4). The latter elements are of interest, in part, because of their contribution to pathogenesis, antimicrobial resistance, and other medically relevant properties (5, 6). One of the host site-specific recombination systems frequently exploited by IMGEs is the widely distributed bacterial chromosome dimer-resolving Xer recombination system, a system that recombines chromosomes at dif site located near where DNA replication terminates (7). As in all organisms, during DNA replication of bacteria, many DNA damages need to be repaired by homologous recombination reaction. For bacteria having circular chromosomes, this often generates circular dimer chromosome, causing problems when the cell divides. Hence, when a pair of unresolved chromosome dimer junctions get trapped at the closing cell division septum, the pair of dif sites with XerC and XerD recombinases bound across the recombination junction encounter FtsK DNA translocation pump, a component of the closing septum complex, whose job is to clear trapped DNA out of the septum. This encounter triggers initiation of recombination by activating XerD to carry out the first strand exchange, generating a Holliday junction recombination intermediate, which is resolved by XerC-mediated second pair of strand exchange (8). XerC is an efficient resolver of the recombination intermediate but a poor recombination initiator. Without FtsK activation, Xer remains essentially silent, avoiding formation of chromosome dimer out of 2 separable replicated chromosome copies. IMGEs that exploit the Xer machinery (IMEXs) harbor a short attachment sequence (attP) that mimics the dif site, XerC- and XerD-binding DNA sequences separated by a cross-over spacer region at the border of which sequential strand exchanges occur (9). However, above-mentioned control of Xer recombination by FtsK, which serves well for the bacteria, would prevent Xer-mediated IMGE integration into chromosome at dif, and these elements apparently evolved a number of ways to circumvent this problem.
Vibrio cholerae, the etiological agent of the disease cholera, has acquired several fitness traits, including metabolic functions, antimicrobial resistance, colonization factors, cholera toxin, and several signaling pathways through horizontal gene transfer involving Xer recombination system (5, 10). Horizontally acquired IMEXs in clinical and environmental V. cholerae isolates can be categorized into 3 classes, which follow a distinct mode of integration by exploiting the Vibrio Xer machinery (Fig. 1). The replicative double-stranded DNA (dsDNA) genome of CTXΦ, the representative phage of IMEX category 1, harbors 2 distinct XerC and XerD binding site pairs, attP1 and attP2, arranged in inverted orientations and separated by ∼90-bp intervening sequence (11). In the (+) single-stranded DNA (ssDNA) genome of CTXΦ, attP1 and attP2 can form intrastrand base pairs and generate a functional phage attachment site attP(+) recognized by the XerC and XerD, which mediate integration at dif site (12, 13). In CTXΦ integration, the unique attP structure tricks XerC, which does not need activation by FtsK, to carry out the first strand exchanges, generating a pseudo-Holliday junction intermediate (Fig. 1). Integration of CTXΦ is completed with the help of host DNA replication, which resolved the recombination intermediate into 1 daughter chromosome with integrated copy of the phage genome and the other without.
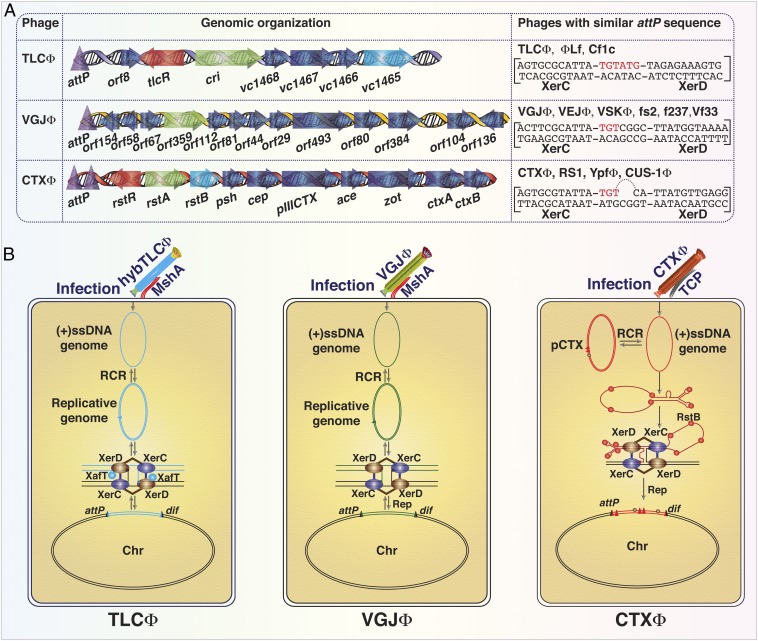
Fig. 1.
Insights into the IMEXs for their integration into chromosome dimer resolution site (dif). (A) Schematic diagram showing attachment sequences (attP) and the genomic organization of IMEXs associated with clinical and environmental isolates of V. cholerae. IMEXs are classified into 3 distinct classes based on the sequence and structure of their attP. Bases at the cross-over (CO) region that are important for strand exchanges are labeled in red. XerC and XerD binding sequences in the attP site of TLCΦ, VJGΦ, and CTXΦ are indicated within the parentheses. Arrows indicate different open reading frames (ORFs). Orientations of arrows specify transcription direction. Green arrows indicate ORF essential for phage replication. Red arrows indicate functions important for transcription regulation of phage genes. Sky blue arrows depict functions that are important for phage integration. (B) Schematic representation of the integration mechanisms of 3 classes of IMEXs. Each class of IMEX harbors distinct attachment sequences and follows a distinct mode of integration. While integration of IMEXs belonging to CTXΦ class is irreversible, integration of IMEXs belonging to the other 2 classes is reversible. The single-stranded genome (ssDNA) of TLCΦ used morphogenesis proteins of helper phage like fs2 and infected mannose-sensitive hemagglutinin (MshA) pili-positive V. cholerae. In the host cytoplasm, the ssDNA genome of TLCΦ converted into dsDNA and produced ssDNA phage genome by rolling circle replication (RCR). XerC and XerD recombinases recognize 28-bp attPTLC site of TLCΦ and mediate its integration with the help of XerD activating factor XafT. The microhomology at the overlap region between attPTLC and dif sites determines integration compatibility and strand exchanges. XerD mediates the first pair of strand exchanges and generates a Holliday junction. After isomerization, XerC mediates the second pair of strand exchanges and enables TLCΦ integration. TLCΦ integration is reversible, and excision probably follows the same sequences of strand exchanges as described for integration. The attP sequence in the dsDNA replicative form of V. cholerae Gillermo Javier filamentous phage (VGJΦ) harbors dif like XerC- and XerD-binding sites separated by a 7-bp CO region. Three base pairs of the CO region immediately adjacent to the XerC-binding site are identical to those of the dif1 sequence. The pseudo-Holliday junction resulting from the strand exchanges between attPVGJ and dif1 catalyzed by XerC is converted into product by chromosomal replication. The cholera toxin encoding CTXΦ recognizes host-encoded toxin coregulated pilus (TCP) as receptor and introduce its (+)ssDNA genome into host cytoplasm. The (+)ssDNA genome CTXΦ is either converted to replicative dsDNA or directly integrated into the host chromosome. For integration, the ∼150-nucleotide-long attachment site folds into a fork hairpin structure where inversely oriented attP1 and attP2 form intrastrand base pairing and generates functional attP(+) site. XerC mediates 1 pair of strand exchanges and generates a pseudo-Holliday junction. Host DNA replication resolves the pseudo-Holliday junction and completes the irreversible integration of CTXΦ.
The V. cholerae Gillermo Javier filamentous phage (VGJΦ) represents a large number of phages that belong to the second category of IMEXs. The replicative genome of VGJΦ harbors an attP site (attPVGJ), sequence of which, like that of CTXΦ, tricks XerC to carry out the first strand exchanges. The resulting Holliday recombination intermediate is resolved by chromosome replication producing integrated progeny. However, unlike stable CTXΦ lysogens, integration of VGJΦ is reversible and integrated copies of VGJΦ are unstable (14).
In PNAS, Midonet et al. (15) report yet another integration mechanism of toxin-linked cryptic satellite phages (TLCΦs), a third category of IMEXs. In this study, a XerD activation function XafT, which takes over the role of FtsK, was identified in the genome of TLCΦ. Almost all pandemic V. cholerae strains harbor single or multiple copies of TLCΦ in chromosome 1 (16, 17). However, it is notably absent in the genomes of nontoxigenic environmental isolates (18, 19). The work of Midonet et al. (9) began with the observation that integration of TLCΦ relies on the XerD-initiated Holliday junction formation, which is then converted into fully integrated prophage by the XerC-mediated second strand exchange. Thus, the TLCΦ integration bypasses the FtsK control that limits Xer recombination to chromosome dimer resolution at the substrate site pair trapped at the closing cell division septum (Fig. 1). It was a long-standing question how XerD recombinase can initiate recombination from a degenerate dif-like sequence in the absence of FtsK stimulation. Midonet et al. (15) demonstrate that TLCΦs encode their own Xer recombination activation factor, XafT, which activates XerD catalysis. The yeast 2-hybrid assay and in vitro pull-down experiments demonstrate that XafT directly interacts with XerD recombinase. This perhaps explains how the XerD catalysis initiates recombination from a degenerate dif-like attachment site in the absence of FtsK stimulation. It is important to note that CTXΦ and RS1 encode a DNA-binding protein RstB, which is also important for their efficient integration (13). But RstB has no direct interaction with any of the Xer recombinases. Finally, in silico analysis of Midonet et al. (15) shows that the genomes of different human, animal, and plant bacterial pathogens harbor IMEXs, which also encode XafT homologs. Thus, the integration strategy of TLCΦs discovered here appears to be widely employed, and XafT homologs are expected to be important for many IMEX dynamics. A deeper understanding may help to induce IMEX instability in the genome of bacterial pathogens that can reverse the pathogenicity by removing IMEX-associated toxin or other virulence factors.
Acknowledgments
The work was supported by the Intramural Research funding of Translational Health Science and Technology Institute and Department of Biotechnology, Government of India (Grant BT/MB/THSTI/HMC-SFC/2011).
Footnotes
The author declares no conflict of interest.
See companion article on page 18391.
References
- 1.Frost L. S., Leplae R., Summers A. O., Toussaint A., Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Soucy S. M., Huang J., Gogarten J. P., Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 16, 472–482 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Brookfield J. F., The ecology of the genome — Mobile DNA elements and their hosts. Nat. Rev. Genet. 6, 128–136 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Partridge S. R., Kwong S. M., Firth N., Jensen S. O., Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das B., et al. , Molecular evolution and functional divergence of Vibrio cholerae. Curr. Opin. Infect. Dis. 29, 520–527 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hassan F., Kamruzzaman M., Mekalanos J. J., Faruque S. M., Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature 467, 982–985 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo F., Benmohamed A., Szatmari G., Xer site specific recombination: Double and single recombinase systems. Front. Microbiol. 8, 453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigot S., Sivanathan V., Possoz C., Barre F. X., Cornet F., FtsK, a literate chromosome segregation machine. Mol. Microbiol. 64, 1434–1441 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Midonet C., Das B., Paly E., Barre F. X., XerD-mediated FtsK-independent integration of TLCϕ into the Vibrio cholerae genome. Proc. Natl. Acad. Sci. U.S.A. 111, 16848–16853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazen T. H., Pan L., Gu J. D., Sobecky P. A., The contribution of mobile genetic elements to the evolution and ecology of Vibrios. FEMS Microbiol. Ecol. 74, 485–499 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Heidelberg J. F., et al. , DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Val M. E., et al. , The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19, 559–566 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Das B., Bischerour J., Val M. E., Barre F. X., Molecular keys of the tropism of integration of the cholera toxin phage. Proc. Natl. Acad. Sci. U.S.A. 107, 4377–4382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das B., Bischerour J., Barre F. X., VGJphi integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc. Natl. Acad. Sci. U.S.A. 108, 2516–2521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Midonet C., Miele S., Paly E., Guerois R., Barre F.-X.. The TLCΦ satellite phage harbors a Xer recombination activation factor. Proc. Natl. Acad. Sci. U.S.A. 116, 18391–18396 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weill F. X., et al. , Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785–789 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Weill F. X., et al. , Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature 565, 230–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin E. J., Lin W., Mekalanos J. J., Waldor M. K., Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28, 1247–1254 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Faruque S. M., Mekalanos J. J., Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 3, 556–565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]