Significance
The central metabolic pathways are remarkably conserved among diverse life forms, from bacteria to mammals. Distinct, alternative variants of these pathways have been identified, especially in prokaryotes. The Calvin–Benson–Bassham (CBB) cycle is a primary organic matter production pathway for assimilating CO2 to produce organic matter. Thus far, only a single version of this cycle, which functions in all plants and algae and many autotrophic bacteria, has been identified. Here, we show a distinct version of the CBB cycle in the chemolithoautotrophic bacterium Thermodesulfobium acidiphilum. This version has not been previously demonstrated experimentally and has been considered unlikely to occur.
Keywords: CBB cycle, form III RubisCO, transaldolase, Thermodesulfobium acidiphilum
Abstract
The Calvin–Benson–Bassham (CBB) cycle assimilates CO2 for the primary production of organic matter in all plants and algae, as well as in some autotrophic bacteria. The key enzyme of the CBB cycle, ribulose-bisphosphate carboxylase/oxygenase (RubisCO), is a main determinant of de novo organic matter production on Earth. Of the three carboxylating forms of RubisCO, forms I and II participate in autotrophy, and form III so far has been associated only with nucleotide and nucleoside metabolism. Here, we report that form III RubisCO functions in the CBB cycle in the thermophilic chemolithoautotrophic bacterium Thermodesulfobium acidiphilum, a phylum-level lineage representative. We further show that autotrophic CO2 fixation in T. acidiphilum is accomplished via the transaldolase variant of the CBB cycle, which has not been previously demonstrated experimentally and has been considered unlikely to occur. Thus, this work reveals a distinct form of the key pathway of CO2 fixation.
Life on our planet depends on the ability of autotrophic organisms to assimilate the inorganic carbon of CO2 and thus to build cellular material from de novo synthesized organic molecules. At present, seven pathways of autotrophic CO2 fixation are known (1–3). Six of these pathways are unique to prokaryotes, indicating the diversity of these mechanisms during the early stages of Earth’s evolution. The seventh pathway is the reductive pentose-phosphate cycle, also known as the Calvin–Benson–Bassham (CBB) cycle, following its discovery in the early 1950s by Melvin Calvin, Andrew Benson, and James Bassham. The CBB cycle occurs in all higher plants and algae, as well as in many photo- and chemoautotrophic bacteria. Thus, it is quantitatively the most important CO2 fixation mechanism in the modern biosphere.
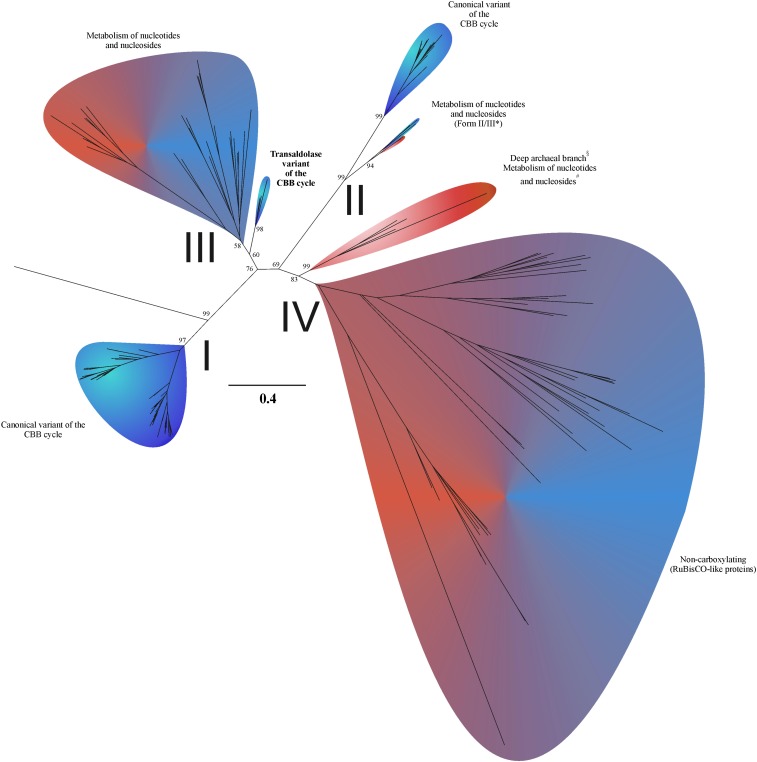
Ribulose-bisphosphate carboxylase/oxygenase (RubisCO) is the key enzyme of the CBB cycle. RubisCO catalyzes the formation of two molecules of 3-phosphoglycerate from ribulose-1,5-bisphosphate, CO2, and H2O. At present, three carboxylating forms (forms I, II, and III) of RubisCO are known (Fig. 1 and SI Appendix, Figs. S1–S5). Thus far, only forms I and II have been reported to participate in the CBB cycle (6, 7). The most widely distributed, form I of RubisCO is found in eukaryotes and some prokaryotes, whereas form II occurs only in prokaryotes, with the exception of the eukaryotic Alveolata (7). Aside from genetic engineering experiments (8–10), form III has never been implicated in autotrophic CO2 fixation. Instead, it has been demonstrated to be involved in the utilization of ribonucleotides and ribonucleosides via the pentose-bisphosphate pathway. Until recently, this pathway was found exclusively in archaea (11–13). It has since been posited to exist in several bacterial candidate divisions (14).
Fig. 1.
Phylogeny and functions of RubisCO proteins. Blue indicates that the host organisms are bacteria, and red shows archaea. The form III and form IV branches involve both bacteria and archaea as host organisms. For names of sequences and bootstrap values at nodes within colored petals, see the expanded subtrees in SI Appendix, Figs. S1–S5. Metabolic descriptions are generalizations based on available data and, in the case of the Calvin–Benson–Bassham cycle transaldolase variant, on this current work. *Termed form II/III by Wrighton et al. (4). §Labeled with an ambiguous term form III-a by Kono et al. (5). #Metabolism of nucleotides and nucleosides, likely overlapping in part with the reductive hexulose-phosphate pathway (5).
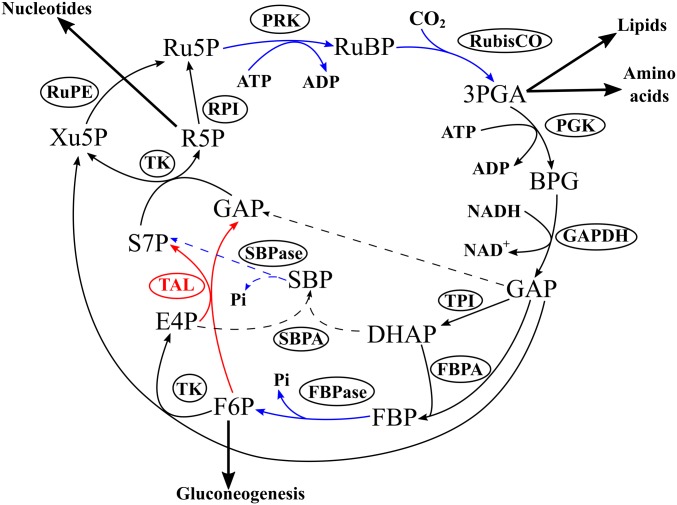
Most other enzymes of the CBB cycle are common with glycolysis, gluconeogenesis, or the nonoxidative branch of the pentose-phosphate pathway. These enzymes synthesize cycle products and regenerate ribulose-1,5-bisphosphate in rearrangement reactions (Fig. 2). The phosphoribulokinase (PRK) enzyme rarely occurs in organisms that lack the CBB cycle. Moreover, the sedoheptulose-1,7-bisphosphatase (SBPase) reaction is unique to the CBB cycle. This reaction renders the rearrangement reactions irreversible, directing carbon flow in the CBB cycle. However, an idea was put forward (15) that, despite its reversibility, the transaldolase reaction might substitute in the CBB cycle for the sedoheptulose-1,7-bisphosphate aldolase and SBPase reactions. Therefore, enzymes in the rearrangement reactions may be exactly the same as in the nonoxidative branch of the pentose-phosphate pathway (15). This hypothetical variant of the CBB cycle is termed the transaldolase variant. However, all biochemical results so far indicate that autotrophic organisms employ the canonical (i.e., SBPase) variant of the CBB cycle (16, 17). Recent proposals of the transaldolase variant of the CBB cycle in Thermithiobacillus tepidarius and Thiobacillus thioparus are based on genome analyses alone, lacking validation by any biochemical data (18, 19). Here, using multiomics and biochemical approaches, we present evidence of a form III RubisCO-mediated transaldolase variant of the CBB cycle.
Fig. 2.
Form III RubisCO-mediated transaldolase variant of the CBB cycle in T. acidiphilum. Black arrows indicate reversible reactions, and blue arrows show irreversible ones. Red arrows indicate the reversible transaldolase reaction, absent from the canonical CBB cycle. Dashed arrows show the missing steps of the canonical variant of the CBB cycle. 3PGA, 3-phosphoglycerate; BPG, 1,3-bisphosphoglycerate; DHAP, dihydroxyacetone phosphate; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; FBPA, fructose-1,6-bisphosphate aldolase; FBPase, fructose-1,6-bisphosphatase; GAP, glyceraldehyde-3-phosphate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; Pi, inorganic phosphate; PRK, phosphoribulokinase; R5P, ribose-5-phosphate; RPI, ribose-5-phosphate isomerase; Ru5P, ribulose-5-phosphate; RubisCO, ribulose-1,5-bisphosphate carboxylase (form III in T. acidiphilum); RuBP, ribulose-1,5-bisphosphate; RuPE, ribulose-5-phosphate 3-epimerase; S7P, sedoheptulose-7-phosphate; SBP, sedoheptulose-1,7-bisphosphate; SBPA, sedoheptulose-1,7-bisphosphate aldolase; SBPase, sedoheptulose-1,7-bisphosphatase; TAL, transaldolase; TK, transketolase; TPI, triose-phosphate isomerase; Xu5P, xylulose-5-phosphate.
Results and Discussion
Analysis of the Thermodesulfobium acidiphilum Genome.
In our recent study of sulfate reduction in acidic hot springs, an autotrophic sulfate-reducing bacterium, Thermodesulfobium acidiphilum strain 3127-1T, was isolated and characterized (20). This bacterium, together with Thermodesulfobium narugense Na82T, represents the family Thermodesulfobiaceae (21), which, according to the latest Bergey’s Manual (22), is assigned to the class Clostridia of the phylum Firmicutes. However, recent phylogenetic studies (23, 24), including those within the Genome Taxonomy Database project (25), show that the genus Thermodesulfobium represents a distinct phylum-level lineage.
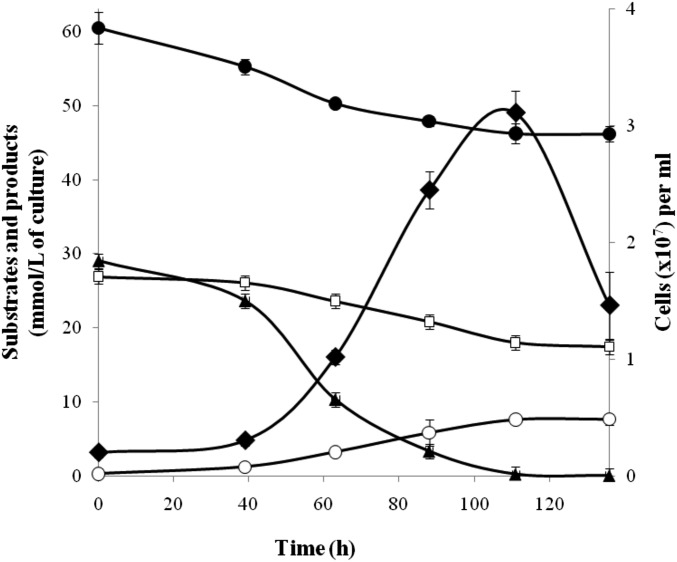
T. acidiphilum is an obligate autotroph growing on H2 or formate via sulfate or thiosulfate respiration (20), as shown in Fig. 3. The completely sequenced genome of T. acidiphilum (SI Appendix) was screened for the genes of key enzymes of CO2 fixation pathways. This analysis showed that the genomes of T. acidiphilum and its closest relative T. narugense lacked the genes encoding key enzymes of 6 of the 7 known autotrophic pathways (1–3): acetyl-CoA synthase (Wood–Ljungdahl pathway), ATP-citrate lyase and citryl-CoA lyase (two variants of the reductive tricarboxylic acid cycle), citrate synthase (reversible tricarboxylic acid cycle), 4-hydroxybutyryl-CoA dehydratase (3-hydroxypropionate/4-hydroxybutyrate and dicarboxylate/4-hydroxybutyrate cycles), and malonyl-CoA reductase (3-hydroxypropionate bicycle). However, the analysis revealed the presence of the cbbL gene encoding RubisCO that belonged to form III and, together with homologs from T. narugense, Ammonifex degensii, Ammonifex thiophilus, and Thermodesulfitimonas autotrophica, comprised a distinct subclade close to the root of the entire form III branch. Various treeing algorithms and sequence sets yielded the same branching order. Fig. 1 and SI Appendix, Figs. S1–S5 show the maximum-likelihood tree.
Fig. 3.
Growth (♦) and production of sulfide (○) by T. acidiphilum in medium with sulfate (☐) under H2 (▲)/CO2 (●) at 60 °C and pH 4.8. Error bars indicate SDs of biological replicates (n = 3).
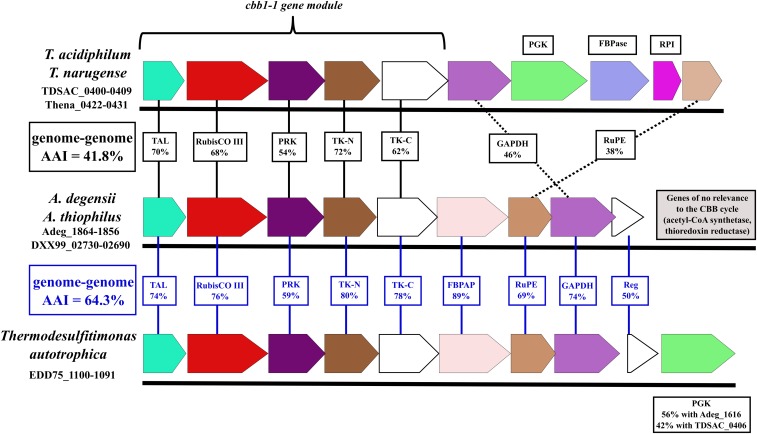
In the T. acidiphilum genome, the cbbL-III gene occurred within a cluster of 10 genes, referred to hereafter as cbb1, which we predicted to encode proteins involved in CO2 fixation. We also detected two additional groups of compactly positioned genes encoding proteins related to CO2 fixation. SI Appendix, Table S1 shows these gene clusters, termed hereafter cbb2 and cbb3, each one comprising 3 genes. The cbb1, cbb2, and cbb3 gene clusters contained all the necessary genes for enzymes of the CBB cycle. However, the presence of the genes for aldolase and two bisphosphatases, despite the known promiscuity of many of such enzymes (26, 27), could not guarantee that they could be active not only toward fructose-1,6-bisphosphate but also toward sedoheptulose-1,7-bisphosphate, which is crucial for the canonical variant of the CBB cycle.
At the same time, we found two transaldolase-encoding genes, tal1 and tal2, in the cbb1 and cbb2 gene clusters, respectively (SI Appendix, Table S1). Both T. acidiphilum transaldolases belonged to the subfamily of small transaldolases (28), as shown in SI Appendix, Fig. S6. Transaldolase activity of some representatives of this subfamily was demonstrated for Bacillus subtilis and Thermotoga maritima (29) and for Methanocaldococcus jannaschii (30). Notably, the cbb1 gene cluster was apparently an operon (Fig. 4), with intergenic distances of −9 to +64 bp and the tal1 gene immediately upstream (+34 bp) of the RubisCO gene. Based on these observations, we hypothesized a form III RubisCO-mediated transaldolase variant of the CBB cycle for CO2 fixation in T. acidiphilum (Fig. 2).
Fig. 4.
Comparison of the cbb1 gene cluster of Thermodesulfobium spp. with the gene clusters in Ammonifex spp. and T. autotrophica in terms of gene order and amino acid sequence identity of the encoded proteins. Solid vertical lines denote high identity values and dotted lines denote lower identity values. FBPAP, fructose-1,6-bisphosphate aldolase/phosphatase; RubisCO III, ribulose-1,5-bisphosphate carboxylase, form III; TK-C, transketolase, C-terminal section; TK-N, transketolase, N-terminal section. Reg is annotated as a transcriptional regulator, LytTR family.
Analyses of the T. acidiphilum Proteome and CBB Cycle Enzyme Activity.
To substantiate the advanced hypothesis, we performed a proteomic analysis of T. acidiphilum cells grown autotrophically by oxidation of H2 with sulfate (Fig. 5). The proteomic results supported our genomic reconstructions: All CBB cycle enzymes (including the two transaldolases) were among the most represented proteins in the total proteome (Fig. 5). Moreover, the activities of the key enzymes (including the transaldolase activity) were detected in extracts of autotrophically grown cells of T. acidiphilum (Table 1). The specific activities of these enzymes upon heterologous expression in Escherichia coli and partial purification (Table 1) confirmed the attribution of the activities toward particular genes. Both candidate transaldolases possessed the conserved catalytic amino acid residues described in ref. 28; however, only the protein encoded by tal1 (TDSAC_0400) within the cbb1 gene cluster exhibited transaldolase activity after heterologous expression.
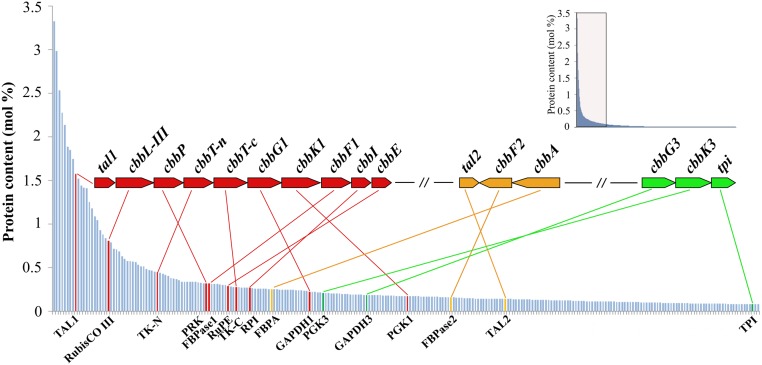
Fig. 5.
Overview of the genomic and proteomic analysis results for T. acidiphilum. The proteins are sorted according to their relative abundances in cells, and the 260 most abundant proteins are shown. The data for the 1,300 proteins are outlined in the small nested box. Proteins encoded in the cbb1 gene cluster are indicated in red, those encoded in the cbb2 gene cluster are in orange, and those in the cbb3 gene cluster are in green. The genes tal1 and tal2 encode transaldolases TAL1 and TAL2; cbbL-III, ribulose-1,5-bisphosphate carboxylase, RubisCO form III; cbbP, phosphoribulokinase; cbbT-n, transketolase, N-terminal section; cbbT-c, transketolase, C-terminal section; cbbG1 and cbbG3, glyceraldehyde-3-phosphate dehydrogenases (GAPDH1 and GAPDH3); cbbK1 and cbbK3, phosphoglycerate kinases (PGK1 and PGK3); cbbF1 and cbbF2, fructose-1,6-bisphosphatases (FBPase1 and FBPase2); cbbI, ribose 5-phosphate isomerase; cbbE, ribulose-phosphate 3-epimerase; cbbA, fructose-1,6-bisphosphate aldolase; and tpi, triose-phosphate isomerase.
Table 1.
Activities of some enzymes of the form III RubisCO-mediated transaldolase variant of the CBB cycle in T. acidiphilum
| Enzyme | Assay temperature, °C* | Specific activity, nmol⋅min−1⋅mg protein−1 | Gene in T. acidiphilum | |
| T. acidiphilum cell extract | Partially purified cloned enzyme | |||
| Phosphoribulokinase | 45 | 35 ± 2 | — | cbbP (TDSAC_0402) |
| Ribulose-bisphosphate carboxylase | 60 | 72 ± 6 | (23 ± 1) × 103 | cbbL-III (TDSAC_0401) |
| Aldolase class II | 45 | 3 ± 1 | (11 ± 3) × 103 | cbbA (TDSAC_1156) |
| Transaldolase | 45 | 51 ± 1 | (9 ± 0.4) × 103 | tal1 (TDSAC_0400) |
| n.d. | tal2 (TDSAC_1154) | |||
n.d., not detected: Cloned TAL2 (TDSAC_1154) exhibited no detectable transaldolase activity.
Due to the necessity of adding mesophilic coupling enzymes, some of the assay incubation temperatures had to be 45 °C (the T. acidiphilum cultivation temperature was 60 °C).
Fructose-1,6-Bisphosphate Aldolase Step of the T. acidiphilum CBB Cycle.
Two proteins were encoded in the genome that might catalyze the fructose-1,6-bisphosphate aldolase (FBPA) reaction, and both were atypical. The first one, encoded by TDSAC_0449 beyond the cbb gene clusters, was a defective homolog of the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase (FBPAP) (31). The defectiveness (at least with respect to the discussed functions) of the homolog in T. acidiphilum followed from its length, which was 287 aa compared with the usual 360 to 385 aa, and, more importantly, from the lack in it of the crucial amino acid residues of the active sites (31, 32): Tyr-229, Lys-232, and Asp-233, essential for aldolase activity, and Tyr-348, essential for phosphatase activity [SI Appendix, Fig. S7; numbering is by the Sulfolobus tokadaii enzyme, for which the crystal structure has been solved (33)]. Notably, the same defects are inherent in the Thena_0472–encoded FBPAP homolog in T. narugense (SI Appendix, Fig. S7), showing that the defectiveness of the T. acidiphilum homolog did not arise from mutations that occurred during isolation and maintenance of the culture.
The second candidate to catalyze the FBPA step of the CBB cycle was a class II aldolase encoded by the cbbA (TDSAC_1156) gene from the cbb2 cluster (SI Appendix, Table S1). Our phylogenetic analysis showed that this enzyme belonged to a class II aldolase subclade for which activity toward fructose-bisphosphate has not been previously shown (SI Appendix, Fig. S8). Proteome analysis of cells grown autotrophically on H2 and sulfate (Fig. 5) revealed overrepresentation of the class II aldolase but not of the defective homolog of bifunctional FBPAP (the latter ranked as low as 807th among the 1,323 totally revealed proteins).
On these grounds, we proposed that the class II aldolase participates in the CO2 fixation pathway in T. acidiphilum, catalyzing the formation of fructose-1,6-bisphosphate from two triose-phosphates. The activity of the FBPA reaction in the T. acidiphilum cell extract was 3 ± 1 nmol⋅min−1⋅mg protein−1. The FBPA activity of the class II aldolase (TDSAC_1156) after cloning and expression in E. coli and partial purification was (11 ± 3) × 103 nmol⋅min−1⋅mg protein−1(Table 1), supporting our genomic predictions and proteomic analysis results.
Summing Up Arguments for the Operation of the Transaldolase Variant of the CBB Cycle in T. acidiphilum.
According to our in silico reconstructions and in vitro experiments, all enzymes necessary for the transaldolase variant of the CBB cycle were present in T. acidiphilum, encoded by genes assembled in clusters, and highly expressed during autotrophic growth. Given that key enzymes of other known autotrophic pathways are not encoded in the T. acidiphilum genome, these results indicate that the CBB cycle that involves form III RubisCO does function in this bacterium, and that it is, most likely, a transaldolase variant of the cycle. Aside from genetic engineering experiments (8–10, 34), no evidence has previously been reported either of the participation of form III RubisCO in autotrophic CO2 fixation or of the operation of the CBB cycle transaldolase variant. The involvement of transaldolase is evidenced by the clustering of the tal1 gene in an apparent operon with the genes of key enzymes of the CBB cycle (RubisCO and PRK) and by the strong parallel expression of these genes, demonstrated by proteomics and activity measurements. Transaldolase activity was attributed to the product of the tal1 gene via its heterologous expression.
An argument against the transaldolase mechanism of the operation of the T. acidiphilum CBB cycle may be the well-known promiscuity of many prokaryotic FBPAs (26) and fructose-1,6-bisphosphatases (FBPases) (27). In prokaryotes, the sedoheptulose-1,7-bisphosphate aldolase and SBPase reactions, essential for the canonical variant of the CBB cycle, may be catalyzed by the same proteins that fulfill the FBPA and FBPase functions. Therefore, the same assumption could be made for the FBPA and FBPase(s) of T. acidiphilum. However, we believe this contention to be invalid, at least in vivo. Our main counterargument derives from the above-substantiated involvement of transaldolase in the CBB cycle in T. acidiphilum. Presence of an enzyme with SBPase activity along with transaldolase would make transaldolase operate in the direction opposite that leading to sedoheptulose-7-phosphate production, and it would deplete the sedoheptulose-7-phosphate pool instead of replenishing it. It has been argued on a number of occasions that transaldolase activity is hardly compatible with the activity of SBPase in the context of the CBB cycle (17, 35–37). All this strongly suggests the absence of the SBPase reaction and the functioning of the transaldolase variant of the CBB cycle in T. acidiphilum.
These arguments in favor of the transaldolase variant of the CBB cycle in T. acidiphilum are not applicable to the aforementioned T. tepidarius and T. thioparus. There are no proteomic or enzyme activity data for these species. Moreover, our analysis showed no clustering of transaldolase and RubisCO genes in their genomes and the presence of genes of possibly promiscuous aldolases and phosphatases.
As for T. acidiphilum, the ultimate proof of the transaldolase variant should be demonstration of the lack of SBPase activity in the cell extract and the lack of the activity in purified candidate enzymes, whose properties should be studied in detail.
Occurrence and Distribution of the cbb1 Gene Cluster and Its Key Module in Available Genomes.
The occurrence of RubisCO, phosphoribulokinase, and transaldolase genes in a common gene cluster, such as the cbb1 gene cluster in the T. acidiphilum genome (Fig. 4 and SI Appendix, Table S1), indicates tight cooperation among these genes. We screened available genomes for the occurrence of similar organization patterns of these three genes, which we believe to comprise the cbb1 cluster key module, indicative of the CBB cycle transaldolase variant. The T. narugense Na82T genome harbored close homologs of all genes determining the transaldolase variant of the CBB cycle in T. acidiphilum (SI Appendix, Table S1). These genes were arranged in gene clusters that shared synteny with the cbb1, cbb2, and cbb3 clusters of T. acidiphilum.
The chemoautotrophic thermophilic Firmicutes representative A. degensii has been mentioned to possess genes encoding key enzymes of the reductive acetyl-CoA pathway and also genes for form III RubisCO and phosphoribulokinase and, thus, possibly, the CBB cycle (1). We found the A. degensii KC4T and A. thiophilus SRT genomes to contain a gene cluster whose 5 upstream genes were identical in gene organization and quite similar in amino acid sequence of the encoded proteins to the 5 upstream genes of the cbb1 gene cluster of Thermodesulfobium spp. (hereafter, cbb1-1 gene module), as seen from Fig. 4 and SI Appendix, Table S1. In the recently sequenced genome of the chemoautotrophic thermophilic firmicute T. autotrophica, representing a genus related to Ammonifex (38), we found a gene cluster (Fig. 4) very similar to the previously mentioned Ammonifex spp. cluster. According to our genomic analysis, Ammonifex spp. and T. autotrophica have the CBB cycle, along with another autotrophic mechanism, the reductive acetyl-CoA pathway. We assume that if the CBB cycle operates in these bacteria under certain conditions, then it must be the transaldolase variant of the cycle. Using Integrated Microbial Genomes and Microbiomes Expert Review (IMG/MER) cassette search tools, we failed to find gene cassettes similar in gene organization to the cbb1-1 gene module in any genomes other than those of Thermodesulfobium spp., Ammonifex spp., and T. autotrophica. Moreover, we could not find other genomes with adjacent transaldolase and RubisCO genes.
While Ammonifex and Thermodesulfitimonas are closely related genera, Thermodesulfobium belongs to a different phylum-level lineage, as evidenced, in particular, by the average amino acid identity (AAI) values between deduced proteomes: 64.3% in the former case and 41.8% in the latter (Fig. 4); our interpretation of the AAI values relies on Rodriguez-R et al. (39). We compared identity percentages between the proteins encoded by the cbb1-1 gene modules of Ammonifex spp., T. autotrophica, and Thermodesulfobium spp. with the AAI values between their deduced proteomes (Fig. 4). The results suggest vertical inheritance of cbb1-1 in the Ammonifex–Thermodesulfitimonas lineage (59 to 80% identity of proteins encoded by cbb1-1 vs. an AAI of 64.3%) and horizontal transfer of cbb1-1 between this lineage and the Thermodesulfobium lineage (54 to 72% identity of proteins encoded by the cbb1-1 modules of the lineages vs. an AAI of 41.8%). The trees for the five proteins encoded by the cbb1-1 gene modules showed coherent clustering of the proteins from Thermodesulfobium spp. and proteins from the Ammonifex–Thermodesulfitimonas lineage (see the RubisCO and transaldolase trees in Fig. 1 and SI Appendix, Figs. S2 and S6), supporting the above assumption of the cbb1-1 gene module transfer event.
Further Considerations and Conclusion.
Recent experiments by Antonovsky et al. (34) demonstrate that when RubisCO and PRK are heterologously expressed in E. coli and selective pressure is applied, a functional and reproducible CBB cycle (the transaldolase variant of the CBB cycle) can be quickly established. Thus, the CBB cycle could have arisen repeatedly during evolution, either based on preexisting ancestor RubisCO forms or via horizontal gene transfer in various phylogenetic lineages, as illustrated by the previously mentioned E. coli laboratory experiment and the likely natural transfer of the cbb1-1 gene module between the Ammonifex–Thermodesulfitimonas and Thermodesulfobium lineages.
The direction of the cbb1-1 gene module transfer and origin of the CBB cycle of T. acidiphilum will be subjects of further study. Regardless of the future choice among the conceivable evolutionary scenarios, the current work emphasizes the importance of investigating deep prokaryotic lineages to broaden knowledge of the biochemical diversity of life on Earth. In conclusion, our study demonstrates an operative form III RubisCO-mediated transaldolase variant of the CBB cycle and thus provides grounds for insights into the evolution of the CBB cycle.
Materials and Methods
Bacterial Strain and Growth Conditions.
T. acidiphilum 3127-1T (=DSM 102892T =VKM B-3043T) was isolated from geothermally heated soil sampled at the Oil Site, Uzon Caldera, Kronotsky Nature Reserve, Kamchatka, Russia (N54° 30.023′ E160° 00.088′, elevation 654 m) in August 2014. T. acidiphilum was grown under anaerobic conditions on a chemically defined medium (20) with CO2 as the carbon source, H2 as the electron donor, and sulfate as the electron acceptor.
Analytical Methods.
Sulfide was measured colorimetrically with N,N-dimethyl-p-phenylenediamine (40). Sulfate was analyzed with a Stayer ion chromatograph (Aquilon) equipped with an IonPack AS4-ASC column (Dionex) and conductivity detector; the eluent was bicarbonate (1.36 mM)/carbonate (1.44 mM) buffer at a flow rate of 1.5 mL⋅min−1. H2 and CO2 were analyzed with a Crystal 5000.2 chromatograph (Chromatec) equipped with an NaX zeolite 60/80–mesh 3-m × 2-mm column (Chromatec) for H2 and a Hayesep Q 80/100–mesh 3-m × 2-mm column (Chromatec) for CO2.
Genome Sequencing and Assembly.
Sequencing of the T. acidiphilum genome, briefly mentioned by us earlier (20), is described in detail below. For sequencing of the genome of T. acidiphilum, a standard fragment DNA library was used. The fragment library was prepared from 0.5 μg of genomic DNA with an NEBNext Ultra DNA Library Preparation Kit (New England Biolabs) according to the manufacturer’s instructions to obtain a mean library size of 700 bp. The resulting library was sequenced on a MiSeq Personal Sequencing System (Illumina) using a 2 × 250-bp paired-end read cartridge. After sequencing, all reads were subjected to stringent quality trimming and filtering with CLC Genomics Workbench 9.5 (Qiagen). Finally, 984,791 read pairs were used for de novo assembly.
Reads were assembled by SPAdes 3.10.0 (41), resulting in 126 genomic scaffolds with an N50 of 403,920 bp. After filtering of scaffolds by coverage, only 13 scaffolds longer than 1,000 bp were obtained. To improve the quality of the assembly, two alternative de novo assembly algorithms were used: the MIRA 4.0 assembler (42) and CLC de novo assembler, provided as a part of the CLC Genomics Workbench Package (Qiagen). Manual comparison of the assemblies guided by the T. narugense reference genome sequence (NC_015499) allowed the sequence of the circular chromosome of T. acidiphilum to be obtained. The quality of the assembly was checked by analysis of read mappings back to the obtained T. acidiphilum chromosome. Neither regions with 0 coverage nor regions enriched with unaligned read termini were detected. Average chromosome coverage for T. acidiphilum was 245-fold.
Genome sequence, as well as related project information and sample details, have been deposited in the NCBI database under accession numbers CP020921, PRJNA353274, and SAMN06013554, respectively.
For sequencing of the genome of A. thiophilus SRT, a standard fragment DNA library was used. The fragment library was prepared from 50 ng of genomic DNA with a Nextera Library Preparation Kit (Illumina) according to the manufacturer’s instructions. The resulting library was sequenced on a MiSeq Personal Sequencing System (Illumina) with a 2 × 250-bp paired-end read cartridge. After sequencing, all reads were subjected to stringent quality trimming and filtering with CLC Genomics Workbench 9.5 (Qiagen); 574,698 read pairs were used for the assembly.
Reads were assembled by SPAdes 3.10.0 (41), resulting in a 2,114,402-bp assembly consisting of 125 genomic scaffolds with an N50 of 87,296 bp. After filtering of scaffolds by coverage, 111 genomic scaffolds longer than 200 bp were obtained and submitted to the NCBI. Average assembly coverage for A. thiophilus was 106-fold.
Genomic assembly, as well as related project information and sample details, has been deposited in the NCBI database under accession numbers QSLN00000000, PRJNA484335, and SAMN09763181, respectively.
Genome Annotation.
Gene prediction and primary annotation of the T. acidiphilum genomic assembly were performed with an IMG/MER System (43). Refining of the automated annotations and other predictions was done manually according to a genome annotation protocol (44). After the refinement, annotation files were adapted to NCBI genome annotation requirements with the tbl2asn tool set (https://www.ncbi.nlm.nih.gov/genbank/tbl2asn2/) and submitted to the NCBI. Prediction of mobile elements was performed with the ISsaga online server (45), and identification and prediction of phage-related genes were done with the PHASTER server (46). Prediction of genomic islands was performed by three independent tools: SeqWord Gene Island Sniffer (47), IslandViewer 4 (48), and AlienHunter (49). Tetranucleotide composition biases were estimated as described by Daims et al. (50). Genome visualization was performed by Circos (51).
Annotation of the A. thiophilus genomic assembly was performed by the automatic NCBI Prokaryotic Genome Annotation Pipeline (52) with manual curation of key metabolic genes.
Phylogenetic Analysis.
Protein phylogeny studies were performed as follows: Respective Pfam domain sequences were downloaded in July to September 2017 from http://pfam.xfam.org/. The “SEED” and “rp15” datasets were downloaded for each domain: PF00016 for RubisCO large, PF01116 for aldolase class II, and PF00923 for transaldolase/fructose-6-phosphate aldolase. Additionally, the 100 best BLAST hits of the respective T. acidiphilum proteins were added to the corresponding datasets. All sequences shorter than 200 amino acids in the RubisCO dataset were discarded. Sequences of the fructose-bisphosphate aldolase and transaldolase datasets were clustered using CD-HIT utility and 50% identity cutoff (53) to eliminate duplicates and reduce redundancy. Upon addition of the respective T. acidiphilum sequences the datasets were aligned using the MAFFT version 7 (54) web server using the L-INS-I algorithm. The evolutionary history was inferred in MEGA6 (55) by using the maximum-likelihood method based on the Le and Gascuel matrix (56) +G +I (4 gamma categories). All positions with less than 95% site coverage were eliminated. The percentages of trees in which the associated taxa clustered together (bootstrap values, 1,000 replicates) are shown next to the branches. There were a total of 186 amino acid sequences and 228 positions in the RubisCO final dataset, 240 amino acid sequences and 164 positions in the transaldolase dataset, and 179 amino acid sequences and 249 positions in the fructose-bisphosphate aldolase dataset. The trees are drawn to scale, with branch lengths measured as the number of substitutions per site.
Average amino acid identity values of the deduced proteomes were determined with the AAI calculator (http://enve-omics.ce.gatech.edu/aai/) (57).
Preparation of Cell Extracts.
Cell extracts of T. acidiphilum were prepared under anoxic conditions. Cells were suspended in 50 mM Tris⋅HCl buffer (pH 7.8) containing 120 mM KCl. The lysate was obtained using the ultrasonic disintegrator Soniprep 150 Plus (150 W, 23 kHz). The lysate was centrifuged for 30 min (12,100 × g; 4 °C), and the supernatant (cell extract) was stored anaerobically at +4 °C until use.
Enzyme Assays.
Carboxylase activity of RubisCO.
The carboxylase activity of RubisCO was measured using a previously described radiometric assay (58), with modifications to create anaerobic conditions. All buffers and solutions were autoclaved (121 °C, 30 min), cooled, and bubbled under an N2 atmosphere free of CO2 and O2. Enzymes (cell extract or partially purified cloned enzyme) were preincubated for 1 h at 60 °C in an assay buffer containing 50 mM Tris⋅HCl (pH 8.0), 10 mM MgCl2, 50 mM NaH14CO3, and 5 mM DTT for enzyme activation. Reactions were initiated by the addition of 1 M ribulose-1,5-bisphosphate (RuBP). All assays contained 2 µCi H14CO3−. Reactions, carried out at 60 °C, were stopped after 15, 30, 60, 120, and 240 min by the injection of aerobic 1 M HCl. Vials were opened to the aerobic environment and allowed to dry overnight at 80 °C. Samples were resuspended in 200 µL 1.0 N HCl and placed in a scintillation mixture. Radiolabeled 14C was detected using a Tri-Carb 2800TR PerkinElmer scintillation counter. As controls, assays lacking enzyme (cell extract or partially purified cloned enzyme) or substrate were performed.
Phosphoribulokinase activity.
Phosphoribulokinase activity was assayed at 45 °C with a procedure for a cyanobacterial PRK reported previously (59), with modifications. The reaction mixture (1 mL) contained 50 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 0.2 mM NADH, 2 mM ATP, 2.5 mM phosphoenolpyruvate, 2 mM ribulose-5-phosphate, 2.5 U l-lactate dehydrogenase (from human, recombinant, expressed in E. coli; Sigma-Aldrich), 1 U pyruvate kinase (rabbit muscle; Sigma-Aldrich) and 0.05 mL cell extract. The reaction was initiated by adding the cell extract, and the absorbance at 340 nm was monitored with a Specord spectrophotometer. Activity was calculated from the molecular extinction coefficient of NADH (6.22 × 103 M−1⋅cm−1 at 340 nm). As controls, assays lacking cell extract or substrate were performed.
Transaldolase activity.
Transaldolase activity was measured at 45 °C using a previously described assay (30), with modifications. The reaction mixture (1 mL) contained 50 mM Tris⋅HCl (pH 7.6), 5 mM fructose-6-phosphate, 0.2 mM erythrose-4-phosphate, 0.2 mM NADH, 10 U triose-phosphate isomerase (baker’s yeast; Sigma-Aldrich), 5 U α-glycerolphosphate dehydrogenase (rabbit muscle; Sigma-Aldrich), and 0.05 mL cell extract (or partially purified cloned enzyme). The reaction was initiated by adding the cell extract, and the absorbance at 340 nm was monitored with a Specord spectrophotometer. Activity was calculated from the molecular extinction coefficient of NADH (6.22 × 103 M−1⋅cm−1 at 340 nm). As controls, assays lacking enzyme (cell extract or partially purified cloned enzyme) or substrates were performed.
FBP aldolase activity.
FBP aldolase (class II aldolase) activity was measured at 45 °C using a previously described assay (31), with modifications. The reaction mixture (1 mL) contained 50 mM Tris⋅HCl (pH 7.6), 10 mM MgCl2, 10 mM DTT, 0.2 mM NADH, 10 U triose-phosphate isomerase (baker’s yeast; Sigma-Aldrich), 5 U α-glycerolphosphate dehydrogenase (rabbit muscle; Sigma-Aldrich), and 0.05 mL cell extract (or partially purified cloned enzyme). The reaction was started by the addition of FBP (5 mM), and the absorbance at 340 nm was monitored with a Specord spectrophotometer. Activity was calculated from the molecular extinction coefficient of NADH (6.22 × 103 M−1⋅cm−1 at 340 nm). As controls, assays lacking enzyme (cell extract or partially purified cloned enzyme) or substrate were performed.
Proteomic Analyses.
The following chemicals were used: sequencing-grade modified trypsin (Promega); 2-chloroacetamide (2-CAA), formic acid (FA), sodium deoxycholate (SDC), trifluoroacetic acid (TFA), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and Empore SPE disks SDB-RPS, SCX, and C18 (Sigma-Aldrich); Tris(hydroxymethyl)aminomethane (Tris⋅HCl) (Panreac); ammonium acetate (Fluka); hypergrade-quality acetonitrile (ACN) for LC-MS (LiChrosolv), acetone for liquid chromatography (LiChrosolv), gradient-grade methanol for liquid chromatography (LiChrosolv), and HPLC-grade water (Merck).
Cell lysis and reduction, alkylation, and digestion of proteins.
Cell lysis was done as follows: Frozen cell residue was suspended in 500 µL SDC reduction and alkylation buffer (4% SDC, 100 mM Tris, pH 8.5, 10 mM TCEP, 40 mM 2-CAA) and sonicated for 20 s using an ultrasonic cell disrupter VirSonic 100 (VirTis). The solution obtained was boiled for 10 min and, after cooling to room temperature, centrifuged at 15,000 × g for 15 min. Protein concentration in the cell lysate was determined using Bradford reagent. An aliquot of cell lysate (100 µg of protein) was diluted 8 times with 100 mM Tris (pH 8.5), and 1 µg of trypsin was added. Digestion was performed at 37 °C overnight. Peptides were desalted using SDB-RPS StageTips as described previously (60) with minor modifications: Peptides were acidified to a final concentration of 0.1% TFA, and 20 µg was loaded on three 14-gauge StageTip plugs. Ethyl acetate/1% TFA (125 µL) was added, and the StageTips were centrifuged at 300 × g. After washing the StageTips using two wash steps of 100 µL ethyl acetate/1% TFA and one of 100 µL 0.2% TFA consecutively, peptides were eluted by 60 µL elution buffer (80% acetonitrile, 5% ammonia). The collected material was completely dried using a SpeedVac centrifuge (Savant) and stored at −80 °C before analysis.
SCX StageTip fractionation.
Before LC-MS/MS analysis, peptides were fractionated using SCX as described previously (60), with minor modifications. In brief, peptides (40 µg) were dissolved in 100 µL 1% TFA in water and loaded on two 10-gauge SCX StageTip plugs, washed with 100 µL 0.2% TFA, and successively eluted by a step gradient of ammonium acetate (100, 150, 200, 300, 400, 600 mM) in 20% (vol/vol) ACN, 0.5% (vol/vol) formic acid, with final elution by 5% (vol/vol) ammonium hydroxide, 80% (vol/vol) ACN. All fractions obtained were desalted on three 14-gauge SCX StageTip plugs, completely dried using a SpeedVac centrifuge (Savant), and stored at −80 °C before analysis.
NanoLC-MS/MS analysis was performed as described previously (61).
Data analysis.
MS raw files were analyzed by MaxQuant software version 1.5.6.5 (62), and peptide lists were searched against the T. acidiphilum genome and a common contaminants database by the Andromeda search engine (63) with cysteine carbamidomethylation as a fixed modification and N-terminal acetylation and methionine oxidations as variable modifications. The false discovery rate was set to 0.01 for both proteins and peptides with a minimum length of 7 amino acids and was determined by searching a reverse database. Enzyme specificity was set to trypsin, and a maximum of 2 missed cleavages was allowed in the database search. Peptide identification was performed with an allowed initial precursor mass deviation up to 20 ppm and an allowed fragment mass deviation of 20 ppm. Matching between runs was performed with 6 SCX fractions serving as a library. Proteins matching the reverse database were filtered out.
Cloning and Heterologous Expression of T. acidiphilum Genes in E. coli.
Standard protocols were used for molecular cloning (64–66). Plasmid DNA was isolated with the Plasmid Miniprep Kit (Evrogen). Insert verification for all recombinant plasmids was performed by Sanger sequencing. (i) cbbL-III was amplified from T. acidiphilum chromosomal DNA using the forward primer (5′-GGCGCTAGCAATAAATTAAATATTAAATATACAAATTTTTTGGATTTTGACTACG-3′) introducing an NheI site (underlined) and reverse primer (5′-AGACCTCGAGTTATGCACCTCCCCATTTTGATAGG-3′) introducing an XhoI site (underlined). The PCR product was isolated and cloned into the vector pET-6×His-(-30)GFP (Addgene), generating pET_His_cbbL-III. The His-tagged recombinant enzyme was produced in E. coli BL21(DE3) transformed with pET_His_cbbL-III. (ii) tal1 was amplified from T. acidiphilum chromosomal DNA using the forward primer (5′-GGCGCTAGCATGCAAATTTTTTTAGATACTGCGAAAATTGAC-3′) introducing an NheI site (underlined) and the reverse primer (5′-GTGCTCGAGCTATTTTGATTCCTGAAGCCCTTTCCAATC-3′) introducing an XhoI site (underlined). The PCR product was isolated and cloned into pET-6×His-(-30)GFP (Addgene), generating pET_His_tal1. The His-tagged recombinant enzyme was produced in E. coli BL21(DE3) transformed with pET_His_tal1. (iii) tal2 was amplified from T. acidiphilum chromosomal DNA using the forward primer (5′-GGCGCTAGCTTGGAACTATATTTAGATACTGCAAAAATAGAAGAAATAGAAGAAGGC-3′) introducing an NheI site (underlined) and the reverse primer (5′-CACCTCGAGCTAAGATACTATAGAAATACCCTTTGAATCACTTAGAAACTTTGC-3′) introducing an XhoI site (underlined). The PCR product was isolated and cloned into pET-6×His-(-30)GFP (Addgene), generating pET_His_tal2. The His-tagged recombinant enzyme was produced in E. coli BL21(DE3) transformed with pET_His_tal2. (iv) cbbA was amplified from T. acidiphilum chromosomal DNA using the forward primer (5′-GGCGCTAGCTTGTTTAGGACAAAAGAGGAAGTTGCAAAAATTC-3′) introducing an NheI site (underlined) and the reverse primer (5′-AGACTCGAGTCAGTCAGCCCCCTCTTCAAATTCC-3′) introducing an XhoI site (underlined). The PCR product was isolated and cloned into pET-6×His-(-30)GFP (Addgene), generating pET_His_cbbA. The His-tagged recombinant enzyme was produced in E. coli BL21(DE3) transformed with pET_His_cbbA.
The transformed cells were grown at 37 °C in 2 L LB medium with 100 μg⋅mL−1 ampicillin. Expression was induced at an OD578 of 0.5 with 1 mM isopropylthiogalactopyranoside, and the temperature was lowered to 22 °C.
Purification of Recombinant Enzymes.
Frozen cells (1 to 2 g) from the respective overexpression were suspended in 25 mM sodium phosphate buffer (pH 7.4) containing 500 mM NaCl and 25 mM imidazole. The lysate obtained using the ultrasonic disintegrator Soniprep 150 Plus (150 W, 23 kHz) was centrifuged for 30 min (12,100 × g; 4 °C). His-tagged proteins were purified by gravity-flow chromatography using 1 mL nickel agarose (Ni-NTA agarose; Qiagen) equilibrated with 25 mM sodium phosphate buffer (pH 7.4) containing 500 mM NaCl and 25 mM imidazole. The column was washed with the same buffer containing 50 mM imidazole. Thereafter, the recombinant His-tagged enzymes were eluted with the same buffer containing 500 mM imidazole. The enzymes were stored at −20 °C.
Denaturing Polyacrylamide Gel Electrophoresis.
Denaturing polyacrylamide gel electrophoresis in the presence of SDS, used to monitor purification of the heterologously expressed enzymes, was performed on a 10% polyacrylamide gel according to Laemmli (67). Enzyme aliquots were added to the sample buffer (100 mM Tris⋅HCl, pH 6.8, 25 °C, 10% [wt/vol] SDS, 5% [vol/vol] 2-mercaptoethanol, 1% [wt/vol] bromophenol blue) and heated before loading. A mixture of standard marker proteins with molecular masses of 6.5 to 200 kDa (Sigma) was used. Proteins in the gel were stained using Coomassie brilliant blue G-250 (Sigma).
Data Availability.
All data for understanding and assessing the conclusions of this research are available in the main text, SI Appendix, GenBank database (https://www.ncbi.nlm.nih.gov/genbank/), and ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org). The genome sequences of T. acidiphilum and A. thiophilus are deposited under GenBank accession numbers CP020921.1 and QSLN00000000.1, respectively. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (68) with the dataset identifier PXD014169.
Supplementary Material
Acknowledgments
We are grateful to I. A. Berg (Institute for Molecular Microbiology and Biotechnology, University of Münster) for helpful discussions. This work was supported by the Russian Science Foundation (Grant 17-74-30025; genome sequencing and assembly, phylogenetic analysis, proteomic analyses, cloning and heterologous expression of genes), Russian Foundation for Basic Research (Grant 18-34-00258; purification of recombinant enzymes and enzyme assays), and Ministry of Science and Higher Education of the Russian Federation (growth product analysis).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome sequences of Thermodesulfobium acidiphilum and Ammonifex thiophilus reported in this paper have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank (accession nos. CP020921.1 and QSLN00000000.1, respectively). The mass spectrometry proteomics data reported in this paper have been deposited in the ProteomeXchange Consortium, http://proteomecentral.proteomexchange.org, via the PRIDE partner repository (dataset identifier PXD014169).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904225116/-/DCSupplemental.
References
- 1.Berg I. A., Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77, 1925–1936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunoura T., et al. , A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 359, 559–563 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mall A., et al. , Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359, 563–567 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Wrighton K. C., et al. , Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Kono T., et al. , A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat. Commun. 8, 14007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabita F. R., et al. , Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71, 576–599 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabita F. R., Satagopan S., Hanson T. E., Kreel N. E., Scott S. S., Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Finn M. W., Tabita F. R., Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185, 3049–3059 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida S., et al. , Phototrophic growth of a Rubisco-deficient mesophilic purple nonsulfur bacterium harboring a type III Rubisco from a hyperthermophilic archaeon. J. Biotechnol. 124, 532–544 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Yoshida S., Atomi H., Imanaka T., Engineering of a type III Rubisco from a hyperthermophilic archaeon in order to enhance catalytic performance in mesophilic host cells. Appl. Environ. Microbiol. 73, 6254–6261 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T., Atomi H., Imanaka T., Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315, 1003–1006 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Aono R., et al. , Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. J. Bacteriol. 194, 6847–6855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aono R., Sato T., Imanaka T., Atomi H., A pentose bisphosphate pathway for nucleoside degradation in Archaea. Nat. Chem. Biol. 11, 355–360 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Wrighton K. C., et al. , RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 10, 2702–2714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs M., “Intermediary metabolism and pathology” in Plant Physiology, Steward F. C., Ed. (Academic Press, New York, 1966), vol. IVB, pp. 3–115. [Google Scholar]
- 16.Shively J. M., van Keulen G., Meijer W. G., Something from almost nothing: Carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52, 191–230 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Sharkey T. D., Weise S. E., The glucose 6-phosphate shunt around the Calvin-Benson cycle. J. Exp. Bot. 67, 4067–4077 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Boden R., et al. , Permanent draft genome of Thermithiobacillus tepidarius DSM 3134T, a moderately thermophilic, obligately chemolithoautotrophic member of the Acidithiobacillia. Stand. Genomic Sci. 11, 74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutt L. P., et al. , Permanent draft genome of Thiobacillus thioparus DSM 505T, an obligately chemolithoautotrophic member of the Betaproteobacteria. Stand. Genomic Sci. 12, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frolov E. N., et al. , Thermodesulfobium acidiphilum sp. nov., a thermoacidophilic, sulfate-reducing, chemoautotrophic bacterium from a thermal site. Int. J. Syst. Evol. Microbiol. 67, 1482–1485 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Mori K., Kim H., Kakegawa T., Hanada S., A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles 7, 283–290 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Ludwig W., Schleifer K.-H., Whitman W. B., “Taxonomic outline of the phylum Firmicutes” in Bergey’s Manual of Systematics of Archaea and Bacteria, Whitman W. B., Ed. (Wiley, 2015), pp. 1–4. [Google Scholar]
- 23.Zhang W., Lu Z., Phylogenomic evaluation of members above the species level within the phylum Firmicutes based on conserved proteins. Environ. Microbiol. Rep. 7, 273–281 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Kunisawa T., Evolutionary relationships of completely sequenced Clostridia species and close relatives. Int. J. Syst. Evol. Microbiol. 65, 4276–4283 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Parks D. H., et al. , A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Flechner A., Gross W., Martin W. F., Schnarrenberger C., Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 447, 200–202 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y. H., Wang D. Y., Wen J. F., The independent prokaryotic origins of eukaryotic fructose-1, 6-bisphosphatase and sedoheptulose-1, 7-bisphosphatase and the implications of their origins for the evolution of eukaryotic Calvin cycle. BMC Evol. Biol. 12, 208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samland A. K., Sprenger G. A., Transaldolase: From biochemistry to human disease. Int. J. Biochem. Cell Biol. 41, 1482–1494 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Schürmann M., Sprenger G. A., Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 276, 11055–11061 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Soderberg T., Alver R. C., Transaldolase of Methanocaldococcus jannaschii. Archaea 1, 255–262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Say R. F., Fuchs G., Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464, 1077–1081 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Du J., Say R. F., Lü W., Fuchs G., Einsle O., Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase. Nature 478, 534–537 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Nishimasu H., Fushinobu S., Shoun H., Wakagi T., The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea. Structure 12, 949–959 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Antonovsky N., et al. , Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinser E. R., et al. , Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One 4, e5135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson L. R., et al. , Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U.S.A. 108, E757–E764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caillau M., Paul Quick W., New insights into plant transaldolase. Plant J. 43, 1–16 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Slobodkina G. B., Baslerov R. V., Novikov A. A., Bonch-Osmolovskaya E. A., Slobodkin A. I., Thermodesulfitimonas autotrophica gen. nov., sp. nov., a thermophilic, obligate sulfite-reducing bacterium isolated from a terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 67, 301–305 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-R L. M., et al. ; The Microbial Genomes Atlas , The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 46, W282–W288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trueper H. G., Schlegel H. G., Sulfur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek 30, 225–238 (1964). [DOI] [PubMed] [Google Scholar]
- 41.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chevreux B., Wetter T., Suhai S., “Genome sequence assembly using trace signals and additional sequence information” in Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB), Wingender E., Ed. (Department of Bioinformatics, GBF-Braunschweig, 1999), vol. 99, pp. 45–56. [Google Scholar]
- 43.Chen I. A., et al. , IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toshchakov S. V., Kublanov I. V., Messina E., Yakimov M. M., Golyshin P. N., “Genomic analysis of pure cultures and communities” in Hydrocarbon and Lipid Microbiology Protocols. Genetic, Genomic and System Analyses of Communities, McGenity T. J., Timmis K. N., Nogales B., Eds. (Springer, Berlin, 2015), pp. 5–27. [Google Scholar]
- 45.Varani A. M., Siguier P., Gourbeyre E., Charneau V., Chandler M., ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12, R30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arndt D., et al. , PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bezuidt O., Lima-Mendez G., Reva O. N., SeqWord Gene Island Sniffer: A program to study the lateral genetic exchange among bacteria. World Acad. Sci. Eng. Technol. 58, 1169–1174 (2009). [Google Scholar]
- 48.Bertelli C., et al. ; Simon Fraser University Research Computing Group , IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernikos G. S., Parkhill J., Interpolated variable order motifs for identification of horizontally acquired DNA: Revisiting the Salmonella pathogenicity islands. Bioinformatics 22, 2196–2203 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Daims H., et al. , Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krzywinski M., et al. , Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatusova T., et al. , NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 44, 6614–6624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y., Niu B., Gao Y., Fu L., Li W., CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le S. Q., Gascuel O., An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez-R L. M., Konstantinidis R. T., Bypassing cultivation to identify bacterial species. Microbe 9, 111–118 (2014). [Google Scholar]
- 58.Tabita F. R., Caruso P., Whitman W., Facile assay of enzymes unique to the Calvin cycle in intact cells, with special reference to ribulose 1,5-bisphosphate carboxylase. Anal. Biochem. 84, 462–472 (1978). [DOI] [PubMed] [Google Scholar]
- 59.Kobayashi D., Tamoi M., Iwaki T., Shigeoka S., Wadano A., Molecular characterization and redox regulation of phosphoribulokinase from the cyanobacterium Synechococcus sp. PCC 7942. Plant Cell Physiol. 44, 269–276 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Nezametdinova V. Z., et al. , Species-specific serine-threonine protein kinase Pkb2 of Bifidobacterium longum subsp. longum: Genetic environment and substrate specificity. Anaerobe 51, 26–35 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Cox J., Mann M., Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J., Fritsch E. F., Maniatis T., Eds., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1989). [Google Scholar]
- 65.Studier F. W., Moffatt B. A., Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 (1986). [DOI] [PubMed] [Google Scholar]
- 66.Ausubel F. M., et al., Eds., Current Protocols in Molecular Biology (Wiley, New York, 1987). [Google Scholar]
- 67.Laemmli U. K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- 68.Vizcaíno J. A., et al. , ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for understanding and assessing the conclusions of this research are available in the main text, SI Appendix, GenBank database (https://www.ncbi.nlm.nih.gov/genbank/), and ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org). The genome sequences of T. acidiphilum and A. thiophilus are deposited under GenBank accession numbers CP020921.1 and QSLN00000000.1, respectively. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (68) with the dataset identifier PXD014169.