Significance
Th17 cells are a subset of T cells that produce interleukin 17 and other proinflammatory cytokines. They are involved in the development of autoimmune diseases such as multiple sclerosis and rheumatoid arthritis. Previous studies illustrated that RORγt is a driver for Th17 cell differentiation. Here, we reveal REV-ERBα as an antagonist of RORγt. REV-ERBα and RORγt share the same DNA binding motif. REV-ERB can inhibit the expression of RORγt target genes and suppress RORγt-driven Th17 cell differentiation. Treatment with a synthetic REV-ERB agonist delays the onset and impedes the progression of experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis. Taken together, our study suggests that modulating REV-ERBα activity may be used to manipulate Th17 cells in autoimmune diseases.
Keywords: transcriptional repressor, autoimmunity, nuclear receptors
Abstract
T helper 17 (Th17) cells produce interleukin-17 (IL-17) cytokines and drive inflammatory responses in autoimmune diseases such as multiple sclerosis. The differentiation of Th17 cells is dependent on the retinoic acid receptor-related orphan nuclear receptor RORγt. Here, we identify REV-ERBα (encoded by Nr1d1), a member of the nuclear hormone receptor family, as a transcriptional repressor that antagonizes RORγt function in Th17 cells. REV-ERBα binds to ROR response elements (RORE) in Th17 cells and inhibits the expression of RORγt-dependent genes including Il17a and Il17f. Furthermore, elevated REV-ERBα expression or treatment with a synthetic REV-ERB agonist significantly delays the onset and impedes the progression of experimental autoimmune encephalomyelitis (EAE). These results suggest that modulating REV-ERBα activity may be used to manipulate Th17 cells in autoimmune diseases.
T helper 17 (Th17) cells are the drivers of inflammatory responses in a large number of autoimmune diseases such as multiple sclerosis, rheumatoid arthritis, and psoriasis (1, 2). The orphan nuclear receptor RORγt is the lineage-specific transcription factor that regulates the differentiation of Th17 cells (3). RORγt expression is induced specifically under Th17 differentiation condition. Once expressed, RORγt in turn binds to the loci of Th17 signature genes Il17a and Il17f and up-regulates their expression (4). Several small-molecule RORγt antagonists were identified that can inhibit Th17 cell differentiation and effector function (5–8). These findings suggested that RORγt inhibitors could be developed for treatment of autoimmune diseases. However, RORγt is also known for its critical role in promoting survival of CD4+CD8+ double-positive (DP) thymocytes. A recent study showed that RORγt inhibitor treatment leads to not only reduced DP thymocyte numbers but also limited T cell repertoire diversity (9). Therefore, it is still a challenge to develop a safe strategy to inhibit RORγt activity in Th17 cells in vivo.
Beyond their critical roles in Th17 cell differentiation, members of the ROR family are known to be key players in the circadian regulatory machinery, where they function as transcriptional activators to turn on the expression of circadian genes (10, 11). In the circadian system, RORs’ transcriptional activity is opposed by a pair of repressors, REV-ERBα and REV-ERBβ. Like RORs, REV-ERBs are also members of the nuclear hormone receptor family and play critical roles in circadian and metabolic regulations (12). REV-ERBs recognize the same RORE DNA sequence as RORs and function as transcriptional repressors to suppress the expression of ROR target genes (13, 14). Although the antagonistic relationship between ROR and REV-ERB was well established in the circadian rhythm system, it is not clear if a similar interaction exists in the T cell lineage.
In this study, we show that REV-ERBα is also a key feedback regulator of RORγt in Th17 cells. REV-ERBα is specifically up-regulated during Th17 differentiation and plays a dual role in Th17 cells. When expressed at a low level, REV-ERBα promotes RORγt expression via the suppression of negative regulator NFIL3 as reported previously (15, 16). At high expression level, REV-ERBα directly competes with RORγt binding to the loci of Th17 signature genes and suppresses Th17 effector function. Elevated REV-ERBα activity also ameliorates Th17-driven autoimmune disease experimental autoimmune encephalomyelitis (EAE). Our results suggest that modulating REV-ERBα activity could provide a way to manipulate Th17 cells in autoimmune diseases.
Results
REV-ERBα Is Highly Expressed during Th17 Cell Differentiation.
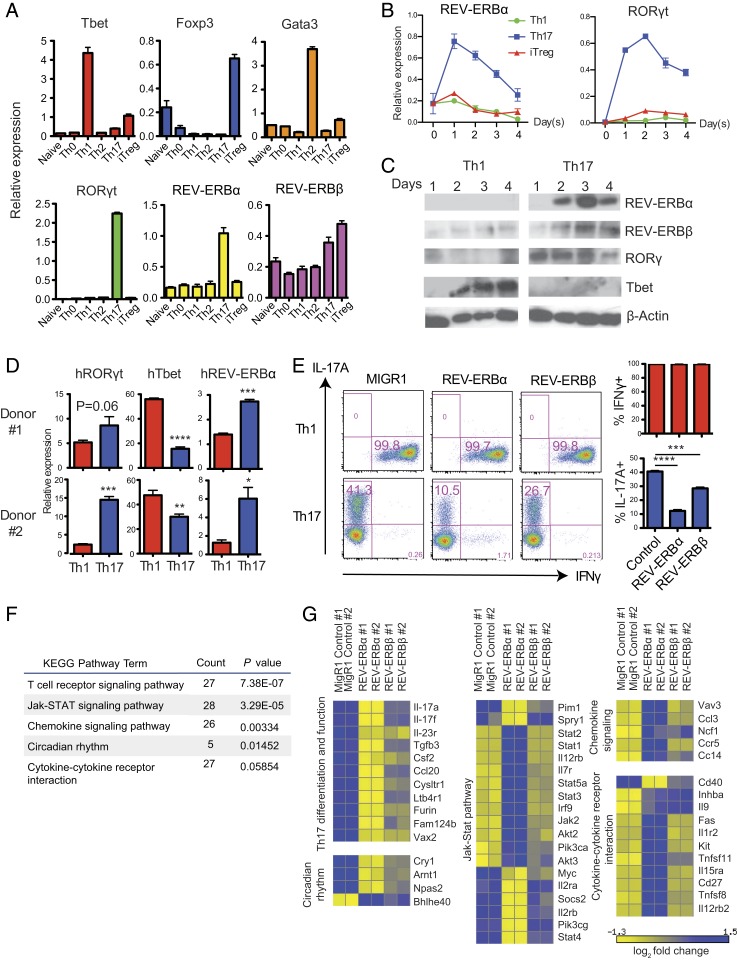
In an effort to identify novel players in the nuclear hormone receptor superfamily that are involved in T cell function, we conducted expression profiling of nuclear hormone receptors in different T helper cell subsets. We noticed that, similar to RORγt, REV-ERBα expression was uniquely up-regulated in Th17 cells at both mRNA and protein levels (Fig. 1 A–C). The differences in the kinetics of REV-ERBα mRNA and protein expression are likely due to a tightly regulated protein degradation pathway of REV-ERBα (17, 18). Furthermore, REV-ERBα expression was significantly higher in human Th17 cells relative to Th1 cells (Fig. 1D). The unique expression pattern of REV-ERBα suggested that it may play a role in the regulation of Th17 cells. Previous studies on circadian regulation demonstrated that, by binding to the same RORE motifs, RORs activate transcription of their target genes, whereas REV-ERBs act as repressors of the same targets (13, 14). We hypothesized that REV-ERBs may suppress Th17 cell differentiation and function by antagonizing RORγt.
Fig. 1.
REV-ERBα is up-regulated in Th17 cells and inhibits the expression of Th17 signature genes. (A) mRNA expression of REV-ERBα, REV-ERBβ, as well as T cell lineage specifying transcription factors T-bet, Gata3, RORγt, and Foxp3, in naïve T cells and Th0, Th1, Th2, Th17, and iTreg cells differentiated for 3 d in vitro. (B) REV-ERBα and RORγt mRNA expression in Th1, Th17, and iTreg cells over 4 d of in vitro differentiation. (C) Protein expression of REV-ERBα, REV-ERBβ, RORγt, and T-bet during in vitro differentiation of Th1 and Th17 cells over 4 d. (D) mRNA expression of RORγt, T-bet, and REV-ERBα in human CD4+ T cells activated under Th1 and Th17 polarizing conditions for 6 d. (E) FACS analysis of IL-17A and IFN-γ expression in mouse CD4+ T cells activated under Th1 and Th17 polarizing conditions and transduced with MIGR1, REV-ERBα or REV-ERBβ retroviral vectors. Data are representative of 3 independent experiments with triplicate wells for each condition. (F) KEGG pathway analysis of genes differentially expressed in REV-ERBα and MIGR1 retrovirally transduced Th17 cells. (G) Heat map of functional groups of differentially expressed genes in Th17 cells transduced with MIGR1, REV-ERBα, or REV-ERBβ retroviral vectors. Relative fold change was normalized to the average of each row in the matrix. Data represents mean ± SEM. Statistical analyses were performed using unpaired 2-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Ectopic Expression of REV-ERBα Inhibits the Expression of Th17 Signature Genes.
To assess the role of REV-ERBs in Th17 cells, we examined the effects of ectopic expression of REV-ERBs on Th1 and Th17 cell differentiation. Retroviral expression of REV-ERBα during Th17 differentiation significantly suppressed interleukin-17A (IL-17A) production compared to T cells transduced with control vector MIGR1 (Fig. 1E). The inhibitory effect of REV-ERBα is specific to Th17 cells, as it did not suppress interferon (IFN)-γ expression in Th1 cells. Ectopic expression of REV-ERBβ showed a modest negative impact on Th17 cells (Fig. 1E). Th17 differentiation can also be driven by ectopic expression of RORγt in T cells cultured without Th17 polarizing cytokines (3). We found that coexpression of REV-ERBα along with RORγt also led to significant decrease of IL-17A expression (SI Appendix, Fig. S1), suggesting that REV-ERBα can suppress RORγt-dependent IL-17A expression. To evaluate the genome-wide effects of REV-ERBs’ ectopic expression in Th17 cells, we performed RNA-sequencing (RNA-seq) analysis of Th17 cells retrovirally transduced with REV-ERBα, REV-ERBβ, or MIGR1 control vector (19). KEGG pathway analysis of the differentially expressed genes indicated that REV-ERBα regulates genes involved in T cell receptor signaling, cytokine/chemokine signaling, as well as circadian rhythm regulation (Fig. 1F). REV-ERBα–transduced cells differentially expressed a number of Th17 cell signature genes compared with MIGR1-transduced cells, which include Il17a, Il17f, Il23r, Csf2, and Tgfb3. Interestingly, most of these genes were significantly down-regulated by REV-ERBα (Fig. 1G). REV-ERBβ expression also suppressed most Th17 signature genes, but its impact was modest compared to REV-ERBα (Fig. 1G). Therefore, we decided to focus on the role of REV-ERBα in suppressing Th17 cell differentiation and the expression of Th17 signature genes.
REV-ERBα Directly Competes with RORγt and Represses Th17 Signature Gene Expression.
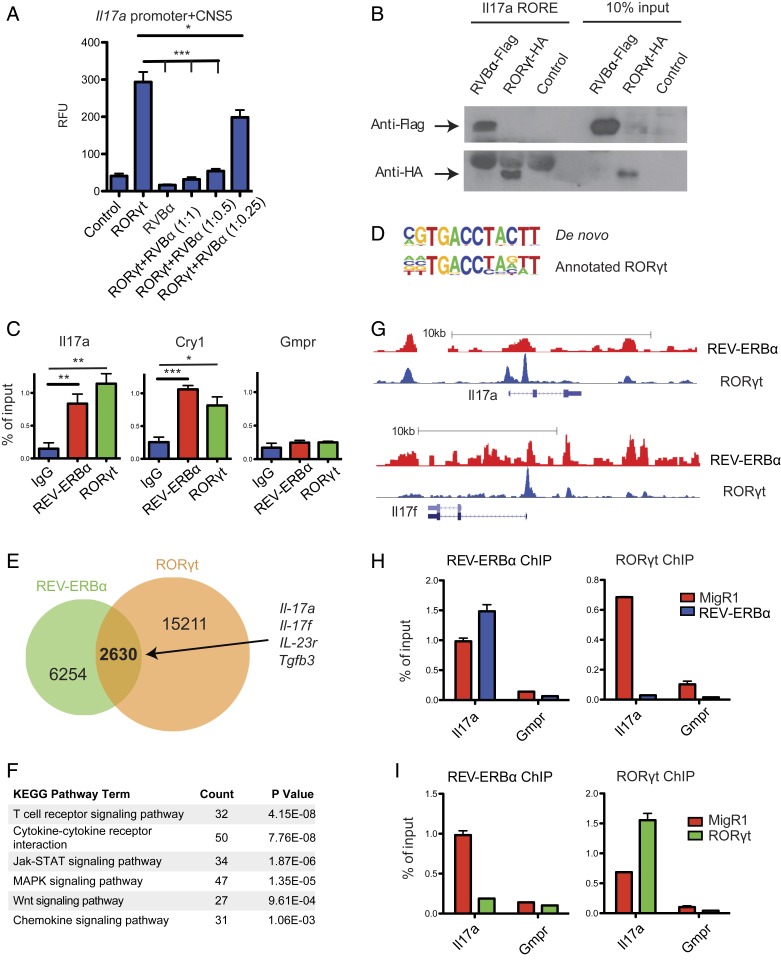
Since REV-ERBs and RORs both recognize ROREs, we hypothesized that REV-ERBα could directly interact with the Il17a locus and repress its transcription. RORE motifs located in CNS5 (also named CNS2), an enhancer 5 kb upstream of the Il17a locus, are critical for optimal expression of Il17a (20–22). Using a reporter driven by the Il17a promoter and CNS5 (20), we measured luciferase activity after transfecting RORγt with or without REV-ERBα. Cotransfection of REV-ERBα inhibited RORγt-dependent Il17a reporter activity in a dose-dependent manner (Fig. 2A). To investigate whether REV-ERBα can directly bind to ROREs located at the Il17a locus, we performed an in vitro DNA binding assay. Biotinylated oligonucleotides containing the RORE motif derived from the Il17a CNS5 enhancer were incubated with nuclear extracts from mouse CD4 T cells transduced with either REV-ERBα− or RORγt-expressing retroviral vectors. The DNA:protein complexes were then precipitated with streptavidin beads, and Western blots were performed to detect precipitated REV-ERBα and RORγt. As shown in Fig. 2B, both REV-ERBα and RORγt bind to the RORE motif. To determine if the REV-ERBα:CNS5 interaction occurs in vivo, we performed chromatin immunoprecipitation (ChIP) experiments in Th17 cells with anti-REV-ERBα and anti-RORγt antibodies. Indeed, both REV-ERBα and RORγt bound to the CNS5 region in Th17 cells (Fig. 2C). These findings suggest that REV-ERBα can directly repress Il17a expression by binding to the Il17a CNS5 enhancer.
Fig. 2.
REV-ERBα directly competes with RORγt and represses Th17 signature gene expression. (A) Luciferase assays of EL4 T cells cotransfected with an Il17a luciferase reporter, and combinations of RORγt and REV-ERBα at various ratios, with the amount of RORγt transfected remaining constant. Renilla luciferase activity was used as internal control. (B) Western blot to detect the binding of HA-tagged RORγt and Flag-tagged REV-ERBα expressed in CD4 T cells to biotinylated oligonucleotides containing RORE motif derived from the Il17a CNS5 enhancer. (C) ChIP-qPCR to detect the binding of Il17a CNS5 enhancer, Cry1 (positive control) and Gmpr (negative control) by REV-ERBα and RORγt in Th17 cells. (D–G) Analysis of Th17 cell REV-ERBα ChIP-seq data along with published RORγt ChIP-seq data. (D) Alignment of de novo generated REV-ERBα binding sequence to annotated RORγt binding sequence. (E) Venn diagram depicting the numbers of unique and shared genes bound by REV-ERBα and RORγt. (F) KEGG pathway analysis of REV-ERBα bound genes. (G) Trace analysis of ChIP-seq data visualized on the UCSC genome browser showing overlapping binding sites of REV-ERBα and RORγt at Il17a and Il17f loci. (H and I) ChIP-qPCR to detect changes in REV-ERBα and RORγt binding to the Il17a locus in response to ectopic expression of REV-ERBα (H) or RORγt (I). Data represents mean ± SEM. Statistical analyses were performed using unpaired 2-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
To identify genome-wide REV-ERBα target genes in Th17 cells, we performed REV-ERBα ChIP-seq assays (23). As expected, the de novo REV-ERBα binding motif is highly similar to the established RORγt binding motif (Fig. 2D). When compared to previously published RORγt ChIP-seq data, about 30% of the 8,884 REV-ERBα binding sites in Th17 cells are also RORγt binding sites (Fig. 2E) (4). It is expected that a large proportion of REV-ERBα binding peaks are different from RORγt binding peaks despite their shared binding motif. This is partly due to the finding that REV-ERBα can target DNA indirectly by interacting with other transcription factors (24). KEGG pathway analysis of genes bound by both REV-ERBα and RORγt revealed that they are enriched with genes involved in T cell signal and cytokine/chemokine pathways (Fig. 2F). In addition to Il17a, several other Th17 cell signature genes, including Il17f, Il23r, and Tgfb3, were also identified as direct targets of REV-ERBα (Fig. 2 E and G and SI Appendix, Fig. S2). The finding that REV-ERBα binds to a large number of RORγt target genes suggests that REV-ERBα inhibits Th17 cell differentiation through direct suppression of the expression of key Th17 cell signature genes. To further test this hypothesis, we examined whether ectopic REV-ERBα expression decreases the binding of RORγt to Il17a by ChIP-qPCR. Indeed, RORγt binding at the Il17a locus decreased significantly in cells transduced with a REV-ERBα expressing retroviral vector (Fig. 2H). Conversely, ectopic expression of RORγt diminished REV-ERBα binding at the Il17a locus (Fig. 2I).
In Vivo Induction of REV-ERBα Expression in T Cells Suppresses EAE Disease Progression.
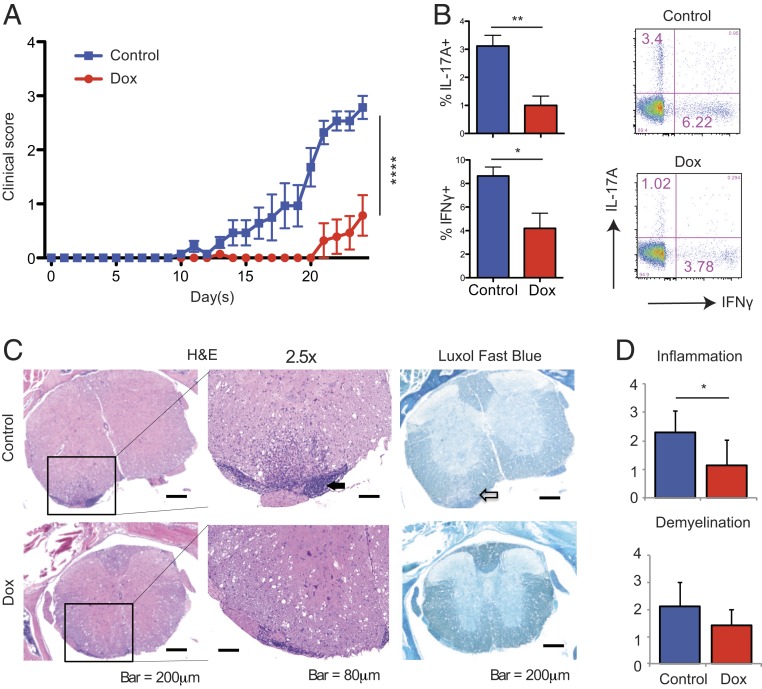
Since high levels of REV-ERBα expression are inhibitory to Th17 cell differentiation in vitro, we explored whether constitutive REV-ERBα expression in T cells could ameliorate Th17 cell-mediated autoimmune diseases in vivo. We tested this hypothesis in EAE, a mouse model for multiple sclerosis, as Th17 cells play a critical role in EAE disease development. A tetracycline-inducible REV-ERBα transgenic mouse (TRE-REV-ERBα/Rosa-M2rtTA) (25) was crossed with a 2D2 TCR transgenic mouse, which carries a TCR that recognizes the MOG (myelin oligodendrocyte glycoprotein) peptide (26), to generate a triple transgenic mouse strain (TTg). T cells from the TTg mice express REV-ERBα constitutively under doxycycline treatment in vivo (SI Appendix, Fig. S3A). CD4+ T cells from the TTg mice were activated in vitro under Th17 conditions for 4 d before being adoptively transferred into WT recipient mice to induce EAE. Mice were given doxycycline water to induce REV-ERBα expression in transferred 2D2 T cells or normal water. The doxycycline-treated group showed delayed EAE disease onset as well as slower disease progression compared to mice that were given normal water (Fig. 3A). Elevated REV-ERBα expression did not affect homing, proliferation, or survival of the transferred CD4+ T cells (SI Appendix, Fig. S3B). However, consistent with milder disease progression observed in mice treated with doxycycline, the frequency of IL-17A producing 2D2 T cells were significantly reduced in the CNS tissues of these mice compared to controls (Fig. 3B). Histopathology analysis showed that doxycycline treatment significantly reduced inflammation levels with a decreasing trend for demyelination in the spinal cord of these mice (Fig. 3 C and D). These differences were dependent on REV-ERBα induction because the same doxycycline treatment of mice transferred with WT 2D2 T cells did not delay or ameliorate EAE disease progression (SI Appendix, Fig. S3C). Thus, elevated expression of REV-ERBα in T cells can attenuate Th17 cell-mediated EAE.
Fig. 3.
In vivo induction of REV-ERBα expression in Th17 cells suppresses EAE disease progression. EAE was induced in C57/BL6 mice by adoptive transfer of in vitro differentiated Rosa-M2rtTAxTRE-REV-ERBax2D2 transgenic Th17 cells. Recipient mice were treated with or without Doxycycline water (n = 7 per group) starting 2 d before Th17 cell adoptive transfer and were monitored for EAE disease progression. Mice were analyzed on day 24 after transfer. (A) Clinical scores of mice induced with EAE. (B) FACS analysis of IL-17A and IFN-γ production of transferred 2D2 CD4+ T cells infiltrating in the CNS tissues. (C) Representative H&E and Luxol Fast Blue staining of the spinal cords to show the sites of immune cell infiltration (filled arrow) and demyelination (open arrow). Data represents mean ± SEM. Statistical analyses were performed using 2-way analysis of variance (ANOVA) for EAE clinical score analysis and 2-tailed unpaired Student’s t test for other analysis, comparing the indicated groups (*P < 0.05, **P < 0.01).
REV-ERBα Deficiency Impairs Th17 Cell Differentiation.
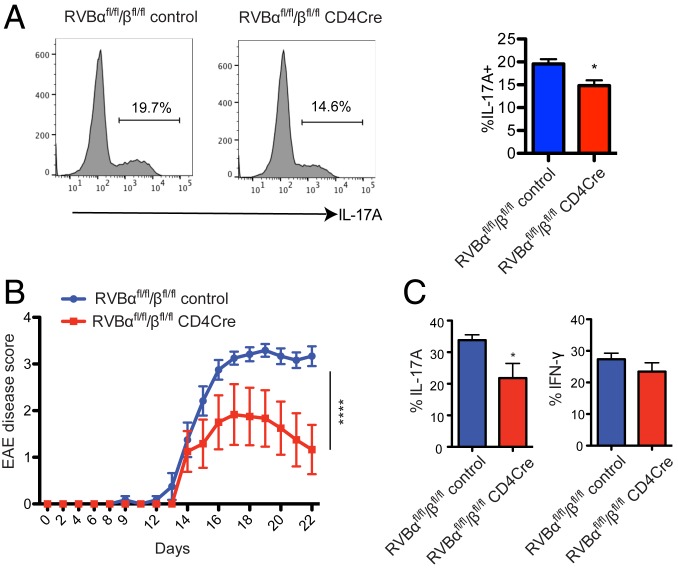
Despite the suppressive role of REV-ERBα on the expression of Th17 signature genes, a previous study by Hooper and coworkers (15) showed that REV-ERBα–deficient T cells were also defective in Th17 differentiation. It was proposed that in the absence of REV-ERBα, expression of NFIL3 increases, which, in turn, suppresses Th17 cell development by directly binding to the RORγt promoter and repressing its expression. We assessed the effect of REV-ERB ablation in Th17 cell differentiation and function. The REV-ERBα/β knockout T cells showed a moderate reduction in Th17 differentiation measured by IL-17A expression (Fig. 4A), consistent with the findings by Hooper’s group. To test the impact of REV-ERB deletion in vivo, we utilized T cell-specific REV-ERBα/β conditional knockout mice (REV-ERBαfl/fl/βfl/fl CD4Cre) (14). The REV-ERB conditional knockout mice and WT controls were immunized with MOG/CFA to induce EAE. Disease development was monitored for 3 wk and followed by analysis of T cell composition in CNS tissues. Mice carrying REV-ERB null T cells developed milder EAE accompanied with reduced IL-17A–producing T cells in the CNS tissues (Fig. 4 B and C). These results suggest that REV-ERBα expression needs to be tightly regulated for robust Th17 cell differentiation. Insufficient REV-ERBα activity leads to decreased Th17 cell differentiation due to increased levels of NFIL3, which suppress RORγt expression, whereas at high levels REV-ERBα outcompetes RORγt for regulatory binding sites in Th17 signature genes such as Il17a and Il17f, also resulting in the suppression of Th17 cell differentiation. It is worth noting that overexpression of REV-ERBα exerts a much stronger inhibitory effect on Th17 cells than absence of REV-ERBα expression (Figs. 1E and 4A).
Fig. 4.
REV-ERBα deficiency impairs Th17 cell differentiation. (A) CD4+ T cells from REV-ERBαfl/fl/βfl/fl control and CD4Cre REV-ERBαfl/fl/βfl/fl mice were activated under Th17 polarizing condition. After 3 d of culturing, IL-17A production was analyzed by flow cytometry. FACS plots shown were representative of 3 independent experiments. (B and C) REV-ERBαfl/fl/βfl/fl control and CD4Cre REV-ERBαfl/fl/βfl/fl mice were immunized with MOG/CFA to induce EAE. (B) EAE disease progression scores. (C) IL-17A and IFN-γ production in the CNS infiltrating CD4 T cell population at endpoint. Data represents mean ± SEM. Statistical analyses were performed using unpaired 2-tailed Student’s t test or 2-way ANOVA for EAE disease scores (*P < 0.05, ****P < 0.0001).
A Synthetic Agonist of REV-ERBα Can Inhibit Th17 Cell Differentiation In Vitro and Ameliorate EAE In Vivo.
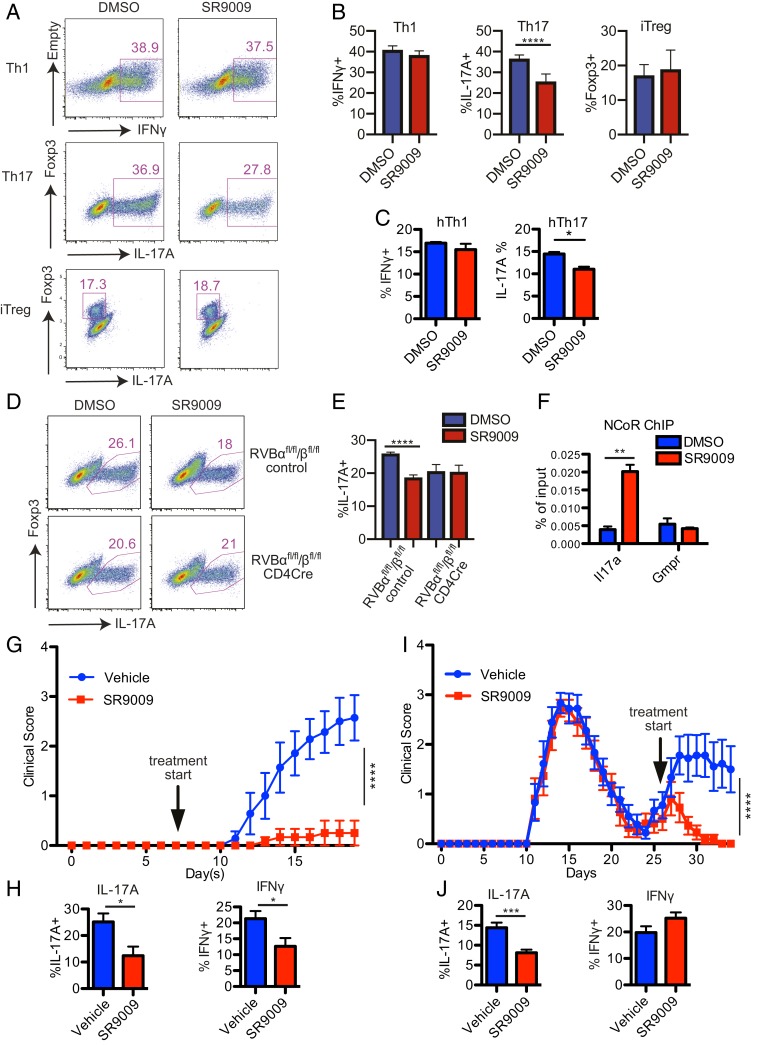
Structural studies have shown that REV-ERBs contain a ligand binding domain, and their activity can be modulated by specific ligands (27). Heme, the prosthetic group in hemoglobin, was identified as an endogenous agonist that binds to REV-ERBs and potentiates their activity (28, 29). Efforts have also been made to generate synthetic REV-ERB agonists with higher specificity and fewer side effects. Two chemical compounds, SR9009 and SR9011, bind specifically to REV-ERBs and modulate their activity, and exhibited favorable pharmacokinetic properties when tested in mice (30–32). Since increased REV-ERBα expression suppresses Th17 cell differentiation and function, we tested if potentiating REV-ERB activity via agonist treatment could have a similar effect on Th17 cells. First, we cultured naïve mouse CD4 T cells under Th1, Th17, or iTreg differentiation conditions with or without SR9009. SR9009 treatment significantly inhibited Th17 cell differentiation but did not affect Th1 or iTreg differentiation (Fig. 5 A and B). Similarly, Th17, but not Th1, differentiation of human CD4 T cells was significantly inhibited by SR9009 treatment (Fig. 5C). To test the specificity of SR9009, we cultured CD4 T cells isolated from REV-ERBα/β conditional knockout and WT control mice under Th17 differentiation condition with or without SR9009 and measured their IL-17A expression. While Th17 differentiation of WT T cells was suppressed by SR9009 treatment, IL-17A expression in REV-ERB null T cells was not affected (Fig. 5 D and E). These results suggest that SR9009 can inhibit Th17 differentiation by modulating REV-ERB activity.
Fig. 5.
REV-ERB agonist SR9009 inhibits Th17 differentiation and suppresses EAE. (A and B) Mouse CD4+ T cells were activated under Th1, Th17, and iTreg polarizing conditions and treated with DMSO or SR9009. IFN-γ, IL-17A, and Foxp3 expression in Th1, Th17, and iTreg cells, respectively, were analyzed by flow cytometry (n = 3). (C) IFN-γ and IL-17A production in human Th1 and Th17 polarized cells treated with either DMSO or SR9009. (D and E) CD4+ T cells from REV-ERBαfl/fl/βfl/fl control and CD4Cre REV-ERBαfl/fl/βfl/fl mice were activated under Th17 polarizing condition and treated with DMSO or SR9009. IL-17A expression in Th17 cells was analyzed by flow cytometry (n = 5). (F) ChIP-qPCR to detect enhanced NCoR recruitment to the Il17a locus in response to SR9009. (G) EAE disease progression of C57/BL6 mice that were immunized with MOG/CFA and treated with vehicle control or SR9009 via i.p. injections starting on day 7 after immunization (n = 5–6 per group). (H) IL-17A and IFN-γ production in the CNS infiltrating CD4 T cell population at endpoint. (I) EAE disease progression of SJL mice immunized with PLP/CFA and treated with vehicle control or SR9009 daily starting at the beginning of the relapsing phase of EAE as indicated by the arrow (n = 9 to 10 per group). (J) IL-17A and IFN-γ production in the CNS infiltrating CD4 T cell population at endpoint. Data represents mean ± SEM. Statistical analyses were performed using 2-way ANOVA for EAE clinical score analysis and 2-tailed unpaired Student’s t test for other analysis, comparing the indicated groups (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
To investigate the molecular mechanism of SR9009’s effect in Th17 cells, we examined the recruitment of NCoR, a corepressor that binds to REV-ERB and represses its target gene expression (24). ChIP-qPCR assay showed that NCoR recruitment to the Il17a CNS5 enhancer increased significantly in the presence of SR9009, indicating that SR9009 can target the IL17A pathway directly via enhancing NCoR:REV-ERB interaction at the Il17a locus (Fig. 5F).
The inhibitory effect of SR9009 on Th17 differentiation in vitro compelled us to test if it could also exert similar modulating effect on Th17-induced autoimmune diseases in vivo. Unlike RORγt antagonists, SR9009 treatment did not skew T cell development in the thymus or grossly affect T cell activation state in the periphery (SI Appendix, Fig. S4).
To test the effect of SR9009 treatment in mouse EAE models, we immunized C57/BL6 mice with MOG/CFA and started injection of SR9009 or vehicle control 7 d after initial immunization. Mice treated with SR9009 showed significantly delayed onset and slower progression of EAE compared to vehicle-treated control group (Fig. 5G). SR9009 treatment also reduced the frequency of IL-17A–producing CD4 T cells infiltrating the CNS tissues during EAE (Fig. 5H). We next explored the efficacy of REV-ERB agonist treatment on mice that have already developed EAE. We induced EAE in SJL mice, which upon PLP (myelin proteolipid protein) peptide immunization, exhibit disease progression in remitting and relapsing patterns mimicking the development of multiple sclerosis in humans. SR9009 treatment of SJL mice during the primary phase of EAE showed inhibitory effects similar to its effects in C57/BL6 mice (data not shown). When SR9009 was administered in the remitting phase, SR9009-treated mice maintained their remitting state, whereas vehicle-treated control mice developed additional episodes of EAE symptoms (Fig. 5I). Furthermore, the frequency of IL-17A–producing CD4 T cells was also significantly reduced in the CNS tissues of SR9009-treated mice compared to controls (Fig. 5J). These results demonstrate that modulating in vivo REV-ERB activity by its agonist SR9009 effectively suppresses development and progression of Th17 cell-mediated EAE.
Discussion
In this study, we demonstrated a role for REV-ERBα in the regulation of Th17 cell differentiation and function in addition to its established roles in circadian rhythm and metabolism. REV-ERBα is induced during Th17 cell differentiation and directly competes with RORγt by binding to the RORE sites to repress the expression of key Th17 cell signature genes such as Il17a and Il17f. At the same time, normal RORγt induction is also dependent on repression of Nfil3 by REV-ERBα (15). This is substantiated by reduction of IL-17A production in vitro and milder EAE phenotype in vivo as a result of T cell-specific REV-ERB ablation. These observations suggest that REV-ERBα serves as a feedback regulator for RORγt in T cells, and its expression needs to stay at the right level for optimal Th17 differentiation.
A recent study by Amir et al. reported similar results showing reduced Th17 activity when REV-ERB expression is increased (33). However, the same study also showed Th17 differentiation was enhanced in REV-ERBα knockout T cells, which differs from our results and the study performed by Yu et al. (15). One primary difference between the 2 studies is that T cell-specific REV-ERB conditional knockout mice were used in our study, while REV-ERBα germline knockout mice were used in the study by Amir et al. Since REV-ERBα germline knockout mice carried severe defects in circadian and metabolic regulation, it is possible that these perturbations originated outside of the immune system rendered T cells more inflammatory under Th17-inducing conditions. Additionally, differences in gut microbiota between mouse facilities might also contribute to the contradictory results.
Given the key role REV-ERBα plays in Th17 cells, we explored if tuning REV-ERBα activity can influence Th17 differentiation and function. Our results showed that elevated REV-ERBα expression in T cells or treatment with REV-ERB ligand SR9009 suppresses Th17 cell differentiation in vitro and inhibits the development of EAE in vivo. Although specific REV-ERBα induction in T cells is sufficient to ameliorate EAE, SR9009 treatment in mice might also impact non-Th17 cells. A previous study demonstrated that REV-ERBα could suppress macrophage expression of IL-6, a key cytokine for Th17 cell differentiation (34). We also observed that subsets of gamma/delta T cells and regulatory T cells could express high levels of REV-ERBα, although the significance of these cell subsets in EAE pathogenesis is currently unclear and requires further characterization. A recent study raised concern on the specificity of SR9009 by demonstrating that SR9009 could exert REV-ERB independent effects in certain tissues, such as mouse embryonic stem cells and hepatocytes (35). In our experiments, SR9009 treatment only affects Th17 differentiation in WT T cells, not REV-ERBα/β double knockout T cells (Fig. 5 D and E), suggesting that SR9009s inhibitory effects on Th17 cells is REV-ERB dependent.
A concerted effort has been made to identify RORα/γ antagonists for treatment of Th17-related autoimmune diseases (5–8). In fact, a recent clinical trial on an RORγ antagonist showed encouraging results in psoriasis patients (36). In addition to Th17 cells, RORγt is also highly expressed in developing T cells in the thymus. A recent report showed that RORγ antagonist treatment leads to DP thymocyte apoptosis and reshapes the T cell repertoire by skewing TCRα rearrangement (9). Although limiting the diversity of the T cell repertoire could be beneficial in some autoimmune disease settings, its long-term effect could also increase the risk to cancer and certain infections. In contrast to RORγt, the low expression levels of REV-ERBα and REV-ERBβ in thymocytes and our own results (SI Appendix, Fig. S4) suggest that REV-ERB agonists will not likely have the same impact on thymocytes and the T cell repertoire as RORγ antagonists (37). Therefore, a strategy of targeting REV-ERB alone or in combination with RORγ may provide a unique advantage in developing treatments for Th17 cell-mediated autoimmune diseases.
Materials and Methods
Mice.
Rosa-M2rtTA, TRE-REV-ERBα, and 2D2 transgenic mice were purchased from Jackson Laboratory. The 3 transgenic lines were crossed to generate Rosa-M2rtTAxTRE-RVBx2D2 triple transgenic mice. REV-ERBαfl/fl/βfl/fl mice were generated previously (14). CD4Cre transgenic, C57BL/6, SJL/J, and Ly5.1+ congenic mice were purchased from the Jackson Laboratory. All mice were maintained in the Salk Institute specific pathogen free (SPF) animal facility in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the Salk Institute.
Reverse Transcription and Quantitative PCR.
Total RNA was isolated from CD4 T cells using TRIzol reagent (Life Technologies). cDNA was synthesized with iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad), followed by qPCR using SYBR Green PCR Master Mix (Applied Biosystems). Quantitative PCR was performed on an Applied Biosystems ViiA 7 Real-Time PCR System with gene specific primers listed in SI Appendix, Table S1.
Retroviral Transduction.
HEK 293T cells were transfected via FuGENE6 reagent (Promega), which contained 0.8 μg of pCL-Eco retroviral packaging plasmid and 1.2 μg of expression plasmid. pCL-Eco was a gift from Inder Verma (Salk Institute, La Jolla, CA) (38). Viral supernatant was harvested 48 and 72 h after transfection. CD4+ T cells were cultured in Th17 polarizing condition and retroviral transduction was performed 24 and 48 h after activation by incubating cells with viral supernatant in the presence of polybrene (4 μg/mL; Millipore) and centrifuged at 2,500 rpm for 90 min at 32 °C.
ChIP.
Naive CD4+ T cells were activated and polarized in Th17 condition for 3 d for ChIP experiments as described previously (14). Mouse IgG control antibody was purchased from Santa Cruz Biotechnology. RORγt ChIP was performed with a combination of antibodies from BioLegend and Santa Cruz Biotechnologies. REV-ERBα antibody was generated as previously described (14). NCoR1 antibody was purchased from Cell Signaling. Primers spanning the regulatory regions of Il17a, Cry1, and Gmpr are described in SI Appendix, Table S2.
ChIP-Seq and Data Analysis.
ChIP-seq libraries were constructed and sequenced as described previously (14). Reads were aligned against the mouse mm9 reference genome using the Bowtie2 aligner with standard parameters that allow up to 2 mismatches in the read. Peak calling, motif analyses, and other data analysis were performed using HOMER, a software suite for ChIP-seq analysis as described previously (14). Visualization of ChIP-Seq results was achieved by uploading custom tracks onto the University of California, Santa Cruz (UCSC) genome browser. ChIP-seq data can be accessed in the National Center for Biotechnology Information (NCBI) GEO database under the accession no. GSE72271.
RNA-Seq and Data Analysis.
RNA-seq libraries were prepared from 100 ng of total RNA (TrueSeq v2, Illumina) and single-ended sequencing was performed on the Illumina HiSeq 2500. Read alignment and junction finding was accomplished using STAR (39) and differential gene expression with Cuffdiff 2 (40). Student’s t test was performed to generate a list of differentially expressed genes (P < 0.05), which was then run through KEGG pathway analysis on DAVID (41, 42) to examine enriched functional groups. Heatmaps were generated on Matrix2png (43). RNA-seq data can be accessed in the NCBI Sequence Read Archive under accession no. SRP062715.
EAE Models.
For active EAE, mice were immunized s.c. with 200 ng of MOG (35–55) peptide (BL6 mice) or PLP (139–151) peptide (SJL mice) in CFA and received 200 ng of Pertussis toxin intraperitoneally on days 0 and 2. Mice were monitored daily for disease progression. At the end point, the brain and spinal cord were harvested for histology and immune cell profiling. For passive EAE, CD4 T cells from Rosa-M2rtTAxTRE-RVBx2D2 mice were activated under Th17 condition for 3 d, then restimulated overnight in the presence of IL-18 (20 ng/mL; Fisher Scientific). Two to 3 million T cells were adoptively transferred into WT recipient mice, which were given normal water or Doxycycline water to induce REV-ERBα expression. EAE disease progression was monitored as in the active EAE model.
Supplementary Material
Acknowledgments
We thank A. Cheng, Y. Zhang, and C. Gordon for mouse colony management and Y. Dai, M. Ku, S. Heinz, and C. Benner for assistance in RNA-seq experiments. C.C. is supported by the H.A. and Mary K. Chapman Charitable Trust. C.-S.L. is supported by the Albert G. and Olive H. Schlink Foundation. S.P.B. is supported by NIH Grants DK096828 and T32 GM007198. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology and is supported by NIH Grants HL088093 and HL105278, Leona M. and Harry B. Helmsley Charitable Trust Grant 2017PG-MED001, Ipsen/Biomeasure, and the Fondation Leducq. Y.Z. is supported by the NOMIS Foundation, the Rita Allen Foundation, National Multiple Sclerosis Society Grant RG4978-A-2, and NIH Grants AI107027 and OD023689. This work was also supported by National Cancer Institute-funded Salk Institute Cancer Center core facilities Grant CA014195. Research reported in this publication was also supported by the National Institute of Environmental Health Sciences of the NIH under Award P42ES010337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-Seq data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. PRJNA293472). ChIP-Seq data have been deposited in Gene Expression Omnibus (GEO), www.ncbi.nlm.nih.gov/geo (accession no. GSE72271).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907563116/-/DCSupplemental.
References
- 1.Weaver C. T., Hatton R. D., Mangan P. R., Harrington L. E., IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Korn T., Bettelli E., Oukka M., Kuchroo V. K., IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Ivanov I. I., et al. , The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Ciofani M., et al. , A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh J. R., et al. , Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature 472, 486–490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solt L. A., et al. , Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472, 491–494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao S., et al. , Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity 40, 477–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheepstra M., et al. , Identification of an allosteric binding site for RORγt inhibition. Nat. Commun. 6, 8833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y., et al. , Inhibition of RORγT skews TCRα gene rearrangement and limits T cell repertoire diversity. Cell Rep. 17, 3206–3218 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Sato T. K., et al. , A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Jetten A. M., Retinoid-related orphan receptors (RORs): Critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 7, e003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin L., Wu N., Lazar M. A., Nuclear receptor Rev-erbalpha: A heme receptor that coordinates circadian rhythm and metabolism. Nucl. Recept. Signal. 8, e001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda H. R., et al. , A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Cho H., et al. , Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X., et al. , TH17 cell differentiation is regulated by the circadian clock. Science 342, 727–730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farez M. F., et al. , Melatonin contributes to the seasonality of multiple sclerosis relapses. Cell 162, 1338–1352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin L., Wang J., Klein P. S., Lazar M. A., Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311, 1002–1005 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Zhao X., et al. , Circadian amplitude regulation via FBXW7-targeted REV-ERBα degradation. Cell 165, 1644–1657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu R., Th17 cells transcriptome. National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database. http://www.ncbi.nlm.nih.gov/sra/?term=PRJNA293472. Deposited 20 August 2015.
- 20.Zhang F., Meng G., Strober W., Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9, 1297–1306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X. O., et al. , T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28, 29–39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., et al. , Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity 36, 23–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu R. T., Zheng Y., The nuclear receptor REV-ERBa modulates Th17 cell differentiation and function by competing with RORgt. NCBI Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72271. Deposited 21 August 2015.
- 24.Zhang Y., et al. , GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U., System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettelli E., et al. , Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197, 1073–1081 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo E.-J., et al. , Structural insight into the constitutive repression function of the nuclear receptor Rev-erbbeta. J. Mol. Biol. 373, 735–744 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Raghuram S., et al. , Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 14, 1207–1213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin L., et al. , Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Solt L. A., et al. , Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woldt E., et al. , Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulli G., et al. , Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553, 351–355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amir M., et al. , REV-ERBα regulates TH17 cell development and autoimmunity. Cell Rep. 25, 3733–3749.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibbs J. E., et al. , The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. U.S.A. 109, 582–587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dierickx P., et al. , SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc. Natl. Acad. Sci. U.S.A. 116, 12147–12152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gege C., RORγt inhibitors as potential back-ups for the phase II candidate VTP-43742 from Vitae Pharmaceuticals: Patent evaluation of WO2016061160 and US20160122345. Expert Opin. Ther. Pat. 27, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Heng T. S. P., Painter M. W.; Immunological Genome Project Consortium , The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Naviaux R. K., Costanzi E., Haas M., Verma I. M., The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70, 5701–5705 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C., et al. , Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavlidis P., Noble W. S., Matrix2png: A utility for visualizing matrix data. Bioinformatics 19, 295–296 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.