Significance
Natural transformation is a major mechanism of horizontal gene transfer. Although the genes required for natural transformation are nearly ubiquitous in bacteria, it is commonly reported that some isolates of transformable species fail to transform. In Legionella pneumophila, we show that the inability of multiple clinical isolates to transform is caused by a conjugative element that shuts down expression of genes required for transformation. Diverse conjugative elements in the Legionella genus have adopted the same inhibition strategy. We propose that inhibition of natural transformation by episomal and integrated conjugative elements can explain the lack of transformability of isolates and also the apparent lack of natural transformation in some species.
Keywords: natural transformation, conjugative element, horizontal gene transfer, noncoding RNA, Legionella pneumophila
Abstract
Natural transformation (i.e., the uptake of DNA and its stable integration in the chromosome) is a major mechanism of horizontal gene transfer in bacteria. Although the vast majority of bacterial genomes carry the genes involved in natural transformation, close relatives of naturally transformable species often appear not competent for natural transformation. In addition, unexplained extensive variations in the natural transformation phenotype have been reported in several species. Here, we addressed this phenomenon by conducting a genome-wide association study (GWAS) on a panel of isolates of the opportunistic pathogen Legionella pneumophila. GWAS revealed that the absence of the transformation phenotype is associated with the conjugative plasmid pLPL. The plasmid inhibits transformation by simultaneously silencing the genes required for DNA uptake and recombination. We identified a small RNA (sRNA), RocRp, as the sole plasmid-encoded factor responsible for the silencing of natural transformation. RocRp is homologous to the highly conserved and chromosome-encoded sRNA RocR which controls the transient expression of the DNA uptake system. Assisted by the ProQ/FinO-domain RNA chaperone RocC, RocRp acts as a substitute of RocR, ensuring that the bacterial host of the conjugative plasmid does not become naturally transformable. Distinct homologs of this plasmid-encoded sRNA are found in diverse conjugative elements in other Legionella species. Their low to high prevalence may result in the lack of transformability of some isolates up to the apparent absence of natural transformation in the species. Generally, our work suggests that conjugative elements obscure the widespread occurrence of natural transformability in bacteria.
One of the remarkable features of bacteria is their ability to naturally undergo genetic transformation. This property is linked to the unique ability of bacteria to import exogenous DNA and integrate it in their chromosome through homologous recombination (1). Despite major difference in their cell-wall architecture, the process is generally conserved between gram-positive (gram+) and gram-negative (gram−) bacteria (2). Exogenous DNA is captured by an extracellular type IV pilus (3, 4). For gram−, retraction of the pilus conveys DNA through the outer membrane (3). Once at the cytoplasmic membrane, the captured DNA is converted into single-strand DNA (ssDNA) (5–7) and transported to the cytoplasm where it is brought to the chromosome for recombination. The entire process relies on the tightly regulated and concerted expression of the type IV pilus and of several proteins essential for importing the DNA and promoting its integration in the chromosome, a situation known as competence (8). Collectively forming a DNA uptake system, these proteins include the periplasmic ComEA protein, which interacts with double-stranded DNA and acts as a ratchet (9–11), and ComEC, a putative transmembrane channel for ssDNA (12). Crossing the inner membrane requires ComFA, a cytoplasmic ATPase unique to gram+ bacteria (13), and ComFC (ComF in gram−) (14). Both form a complex and interact with DprA (15), which protects the incoming ssDNA and promotes RecA-dependent recombination with the chromosome (16, 17). A DNA helicase (RadA in gram+, ComM in gram−) then facilitates recombination over long distance (18, 19).
The core components of the DNA uptake system are encoded by the vast majority of bacterial genomes, with, for instance, ComEC found in over 95% of bacterial genomes (20, 21). However, only a fraction of species are deemed naturally transformable (22). This is commonly explained by the inability of standard laboratory conditions to induce the competence state (i.e., the concerted expression of the type IV pilus and the DNA uptake system). This competence state is generally transient and triggered by signals that appear as species-specific (1, 23). Interestingly, and as illustrated with Pseudomonas stutzeri, closely related species sometimes share the ability to undergo natural transformation, while others, such as Pseudomonas aeruginosa, do not take up DNA and then become widely accepted as nontransformable (24). This apparent lack of transformability is also found at shorter phylogenetic distance. In well-established transformable species such as Streptococcus pneumoniae, P. stutzeri, and Haemophilus influenzae, from 30 to 60% of strains consistently fail to transform (25–29). The underlying cause of this phenotypic heterogeneity is not understood. We here sought to explore this phenomenon in Legionella pneumophila, a naturally transformable human pathogen whose genome is shaped by high rates of recombination (30–33).
We here show that a sparsely distributed mobile genetic element (MGE) conflicting with natural transformation contributes to instraspecific variations in natural transformability. Several other conjugative elements have adopted a similar strategy tailored to Legionella species. We propose that a widespread distribution of such elements may also cause the apparent lack of transformability of specific species.
Results
Large Instraspecific Variations in Natural Transformability Are Incongruent with the Phylogeny.
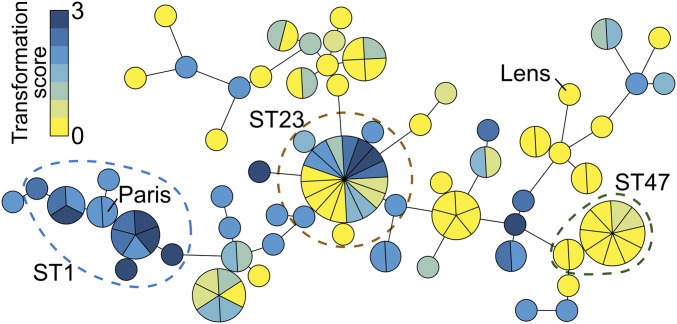
We initially determined the natural transformability of 25 clinical isolates, along with the Paris and Lens isolates, all belonging to 12 different sequence types (ST). As expected, the Paris strain and closely related ST1 isolates showed high transformation frequencies, ranging from 10−6 to 10−5 (SI Appendix, Fig. S1). In contrast, some strains were poorly transformable or even consistently unable to generate transformants, as for instance the Lens isolate. To gain insight into these extensive variations we used an overlapping and extended set of 113 isolates, encompassing 42 ST (SI Appendix, Table S1). For this extended set, transformability was scored from 0 (transformation frequency <10−8) to 3 (transformation frequency ∼10−5) using a higher-throughput assay. Phylogenetic analysis using core-genome polymorphism shows that variations in ability to transform do not always cluster according to phylogenetic distance/relatedness (Fig. 1 and SI Appendix, Table S1). For instance, all isolates of the ST1 and ST47 groups are consistently highly transformable and poorly transformable, respectively. In contrast, in other branches, nontransformable isolates are mixed with transformable isolates. This is particularly noticeable in the ST23 group which includes an equal number of nontransformable and highly transformable isolates. Overall, natural transformability is a conserved trait but shows extensive variations not associated with phylogenetic relationships.
Fig. 1.
Extensive variations of the trait of natural transformation are inconsistent with the phylogeny. Transformability of 113 isolates of L. pneumophila was determined using as transforming DNA a PCR product encompassing an rpsL allele conferring resistance to streptomycin. Following incubation with DNA for at least 24 h, 10 µL of the cultures were spotted on plates containing streptomycin. Transformation was scored from 0 to 3 as a function of the number of colonies that developed in the spot (Materials and Methods). The median score is calculated from 4 independent experiments (SI Appendix, Table S1). Genetic relationships were determined by cgMLST and visualized using a minimum spanning tree displaying transformation scores color-coded from 0 (yellow) to 3 (dark blue).
GWAS Associates the Sparsely Distributed Conjugative Plasmid pLPL with Nontransformability.
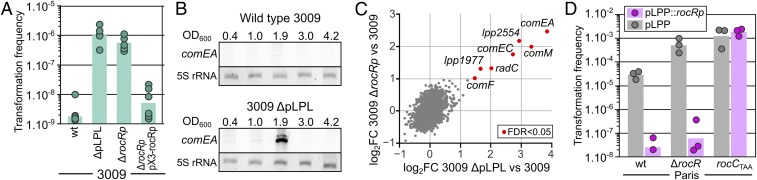
We hypothesize that genetic factors were responsible for the extensive variations of natural transformability. To reveal such factors, we converted the transformation scores into a binary phenotype (transformable T, score 1 to 3 or nontransformable NT, score 0 to 0.5) and conducted a genome-wide association study (GWAS) using DBGWAS (34). The output of DBGWAS was typical of a horizontal gene transfer (HGT) event, with the top 3 subgraphs (q-value <0.1) displaying nearly linear structures consisting of respectively 683, 51, and 93 unambiguous sequences that individually associate with the NT phenotype. The combined 827 sequences mapped onto the sequence of pLPL (SI Appendix, Fig. S2A), a 59.8-kb conjugative plasmid originally identified in the Lens strain (35) (SI Appendix, Fig. S2A). A pLPL variant is present in one-third (n = 14) of the NT isolates (SI Appendix, Figs. S2B and S3) and 5.4% of 537 available L. pneumophila genomes include a pLPL element. Consistent with the presence of a conjugative system, pLPL elements are sparsely distributed, suggesting that they are actively spreading by conjugation (SI Appendix, Fig. S2C). We thus tested the conjugative transfer of pLPL3009 of isolate HL-0640-3009, hereafter denoted strain 3009. In strain 3009, we generated pLPL3009KF which carries the selectable/counterselectable “kan-mazF” (KF) cassette in a pseudogene. In mixed cultures of strain 3009 and the nontransformable strain JR32, pLPL3009KF transfers at a frequency of 3.2 ± 1.18 × 10−6 (transconjugants per recipient). In 3 highly transformable isolates of the ST1 cluster, conjugative acquisition of pLPL3009KF decreased transformation frequencies to the detection limit (SI Appendix, Fig. S4C). The counterselectable KF cassette of pLPL3009KF allowed us to obtain a spontaneous pLPL-free clone of isolate 3009 (3009 ΔpLPL) (SI Appendix, Fig. S5). Rid of pLPL, the isolate now efficiently undergoes natural transformation (Fig. 2A), confirming the GWAS result that pLPL is a potent inhibitor of natural transformation.
Fig. 2.
Plasmid pLPL silences the DNA uptake system by expressing the sRNA RocRp. (A) Natural transformation frequencies of the clinical isolate 3009 and derivatives. Transformation was tested with a nonreplicative plasmid (pGEM-ihfB::kan) carrying a kanamycin resistance gene in a 4-kb chromosomal region encompassing the ihfB gene. Transformation frequencies represent the ratio of CFUs determined by plating serial dilutions on selective vs. on nonselective solid media. (B) Northern blot analysis of comEA expression during growth of strains 3009 and 3009 ΔpLPL in AYE medium at 30 °C. Total RNA was extracted at the indicated ODs (measured at 600 nm, OD600) of the culture, and corresponding to the exponential growth phase (0.4 to 1.0), the transition phase (1.9), and the early (3.0) and late stationary phases (4.2). Total RNA was separated on an 8% denaturing polyacrylamide gel and comEA transcripts were revealed with a biotinylated probe. The 5S ribosomal RNA (rRNA) was used as a loading control. (C) RNAseq transcriptional profiling of strains 3009 ΔpLPL and 3009 ΔrocRp compared to 3009. Total RNA was extracted from 3 independent cultures (n = 3) collected at an OD of 1.8 to 2.0 (at which expression of comEA was observed). Following rRNA depletion, RNAs were reverse-transcribed and sequenced. Normalized read counts are reported as log2-transformed fold change (log2FC). Benjamini–Hochberg correction was applied to P values (false discovery rate, FDR). Individual genes (gray dots) were considered differentially expressed if log2FC was >1 or <−1 and if FDR < 0.05 (red dots). (D) Natural transformability of the Paris strain and isogenic mutants rocCTAA and ΔrocR in which the rocRp gene was introduced in the pLPP plasmid (pink) or not (gray). Transformation data are from at least 3 independently performed experiments and bars represent the geometric mean of the transformation frequencies.
Plasmid pLPL Inhibits Natural Transformation by Silencing the DNA Uptake System.
Natural transformability is a transient phenotype in L. pneumophila, occurring at the onset of the stationary phase. Transformability correlates with expression of the genes encoding the DNA uptake system, notably comEA, whose expression is a marker of natural transformability (36–38). In the Paris strain, expression of comEA is repressed in the exponential phase, expressed at the transition between the exponential to stationary phase, and then repressed again in the stationary phase (36). Northern blot analyses show that comEA is never expressed during growth of the original isolate 3009 (Fig. 2B). However, curing pLPL restored a transient expression pattern of comEA (Fig. 2B). In addition to inhibiting expression of comEA, RNA-sequencing (RNAseq) transcriptional profiling shows that pLPL3009 also prevents expression of comF, comEC, comM, and radC (Fig. 2C, x axis and SI Appendix, Table S2). The concerted targeted inhibition of these 5 genes is reminiscent of the action of RocR, a Legionella-specific but highly conserved 66-nt-long small noncoding RNA (sRNA) that silences these genes in the exponential phase (36). RocR depends on the RocC RNA chaperone to form a duplex with the 5′ untranslated region of the genes encoding the DNA uptake system, preventing translation and exposing the targeted mRNA to cellular nucleases (36). Absence of either RocR or RocC results in a constitutively transformable phenotype (36). A BLAST search revealed a sequence with 80% identity to RocR and located in an intergenic region of pLPL (red bar between 35 and 40 kb, SI Appendix, Fig. S2A). Supporting a role in inhibition of transformation, this sequence is missing in the pLPL of isolates HL-0637-4025 and HL-0703-5020 that appeared transformable (SI Appendix, Fig. S3). Rapid amplification of cDNA ends (RACE) confirmed transcription initiating at 3 positions, generating 3 sRNA variants of 65, 68, and 70 nt (SI Appendix, Fig. S6). We termed this gene rocRp for “rocR plasmidic.” RNAseq transcriptional profiling of a deletion mutant 3009 ΔrocRp shows overexpression of the same genes induced in the 3009 ΔpLPL strain (Fig. 2C, y axis and SI Appendix, Table S2). Thus, rocRp is responsible for the repression of the DNA uptake system caused by pLPL. Accordingly, and similarly to strain 3009 ΔpLPL, strain 3009 carrying pLPL deleted of rocRp (3009 ΔrocRp) can undergo natural transformation (Fig. 2A), while reintroduction rocRp on a nonnative plasmid (pX3-rocRp) restored inhibition (Fig. 2A).
To test the hypothesis that rocRp can inhibit transformation when carried by other native plasmids, we artificially introduced the rocRp gene in pLPP, a conjugative plasmid of 131.8 kb distinct from pLPL. Initially reported in the Paris strain (35), pLPP plasmids are commonly found in the ST1 cluster (SI Appendix, Fig. S4A). When the rocRp gene is inserted in pLPP (pLPP::rocRp), the Paris strain is no longer transformable (Fig. 2D). In addition, rocRp can also fully repress the constitutive and elevated transformability of a ΔrocR mutant, indicating that RocRp can fulfill the same function as RocR. Consistent with this result, and just like RocR, the RocRp RNA appears to require the RNA chaperone RocC as it cannot repress natural transformation of the rocCTAA mutant (Fig. 2D). In conclusion, pLPL inhibits transformation by encoding an sRNA that mimics the chromosome-encoded sRNA RocR and silences the genes encoding the DNA uptake system.
RocRp Acts as a Substitute for RocR.
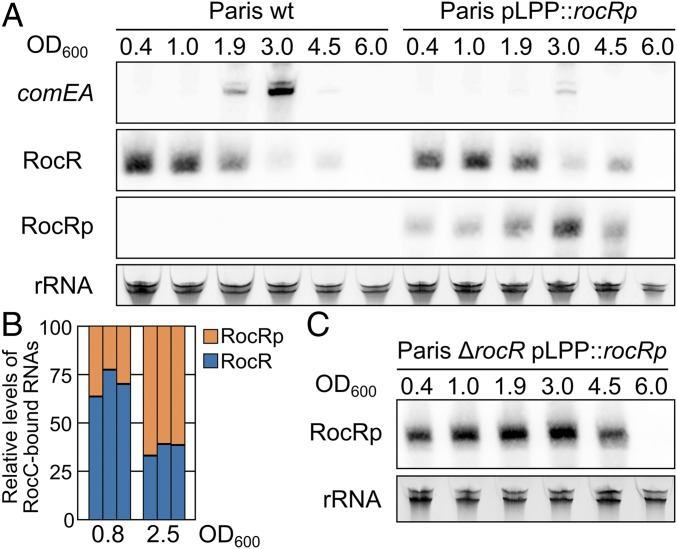
How does RocRp exert a dominant effect over the chromosome-encoded RocR? We investigated this situation by looking at the expression of one of their targets, comEA. We previously found that during the transition phase the decreased expression level of RocR relieves silencing, allowing for the transient expression of comEA (Fig. 3A and ref. 36). However, in the strain Paris pLPP::rocRp, comEA is silenced in the transition phase, suggesting that RocRp may be expressed differently than RocR. Indeed, expression of RocRp from the pLPP plasmid shows a pattern opposite of that of RocR (Fig. 3A). Expression of RocRp is low in the exponential phase but increases to reach its maximal level at the transition phase (Fig. 3A). This expression pattern is also observed when RocRp is expressed from the original pLPL plasmid (SI Appendix, Fig. S7A). Both RocR and RocRp are undetectable in the late stationary phase (Fig. 3A), possibly because of the absence of RocC in the stationary phase (optical density [OD] >4.5) (SI Appendix, Fig. S7B), which is needed to stabilize RocR (36). The Northern blot analysis indicates that RocRp levels in the transition phase compensate for the decreased levels of RocR. Yet, Northern blot analyses are not suited to determine the relative levels of transcripts. To determine the relative levels of RocR and RocRp engaged in silencing of the DNA uptake system, we immunopurified RocC from cultures at OD 0.9 to 1 and OD 2.5 to 3. RNA bound to RocC was purified and the relative abundance of RocR and RocRp was determined using a restriction analysis following a reverse-transcription step (Materials and Methods). In the exponential phase (OD 0.9 to 1), RocC binds 3 times more RocR than RocRp, but in the transition phase this ratio shifts in favor of RocRp, which becomes the most abundant sRNA bound to RocC (Fig. 3B). Interestingly, the half-life of RocR and RocRp correlates with their relative association with RocC. In the exponential phase, when RocC mainly binds RocR, RocR shows a long half-life of 330 ± 22 min (n = 3), which decreases by 3-fold to 109 ± 65 min in the transition phase. RocRp seems less stable than RocR but shows the opposite pattern with a half-life of 36 ± 7 min in the exponential phase that increases 3-fold to 107 ± 39 min. when it is bound to RocC in the transition phase (SI Appendix, Fig. S7C). The results are consistent with our previous finding that RocC stabilizes the bound sRNA (36) and suggest that the engagement of RocRp in RocC-assisted silencing increases its stability.
Fig. 3.
RocRp acts as a substitute for RocR. (A) Northern blot analysis of comEA mRNA, RocR, and RocRp expression during growth of the wild-type Paris strain or carrying the rocRp gene in pLPP (Paris pLPP::rocRp) in AYE medium at 30 °C. Total RNA was extracted at the indicated ODs (OD600) during the exponential growth phase (0.4 to 1.0), the transition phase (1.9 and 3.0), and the early (4.5) and late stationary phases (6.0). (B) Relative quantification of RocR and RocRp bound to RocC in the exponential phase (OD = 0.8) and transition phase (OD = 2.5). For each OD, 3 independent cultures of the strain Paris pLPP::rocRp were obtained. RNAs that copurified with RocC were reverse-transcribed with a mix of primers specific for RocR and RocRp, each bearing the same 5′ end extension. The resulting cDNAs were amplified by PCR using primers corresponding to the 5′ extension of the reverse transcription primer and a reverse primer that matches both RocR and RocRp. Amplification of either RocR or RocRp produces an 85-bp DNA. PCR products originating from RocR and RocRp were distinguished and quantified by restriction with NruI, which cuts only the RocRp PCR product into 2 fragments. (C) Northern blot analysis of RocRp expression during growth in AYE medium at 30 °C of the strain Paris pLPP::rocRp deleted of rocR. Ribosomal RNAs were used as loading controls.
To test this hypothesis we analyzed the expression and stability of RocRp in a ΔrocR mutant. RocRp can effectively silence natural transformation in the absence of RocR (Fig. 2D). In the absence of RocR, RocRp remains expressed in the transition phase but now shows high expression levels in the exponential phase, behaving like RocR (Fig. 3C). Indeed, in the exponential phase RocRp now shows a half-life of 412 ± 91 min, similar to that of RocR (SI Appendix, Fig. S7D). This indicates that the stability of RocRp is not only dictated by its intrinsic properties but rather also by its involvement in RocC-dependent silencing. If RocRp levels are affected by the absence of RocR, in contrast, the presence of RocRp does not significantly alter the expression levels of RocR (Fig. 3A). This suggests that RocRp cannot displace RocR from RocC. Altogether the data suggest that RocRp does not compete with RocR. Rather, RocRp fulfills the same function as RocR, but only when the latter is missing. Thus, RocRp acts as a substitute, taking over the function of RocR if its levels were to decrease.
Diverse Conjugative Elements in the Legionella Genus Carry RocRp Homologs.
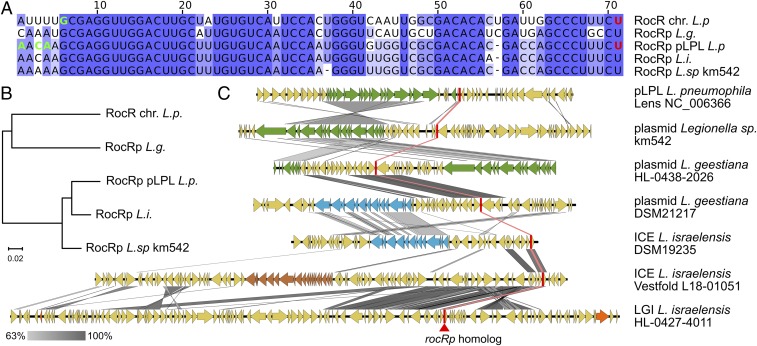
RocR is highly conserved in the Legionella genus. We found RocR in 833 out of 835 Legionella/Fluoribacter/Tatlockia genome assemblies. One assembly in which RocR is missing is incomplete, and the other one belongs to a symbiont with a drastically reduced genome (one-fifth of a Legionella genome). The RocR sequence is also uniquely invariant, showing no single-nucleotide polymorphism (SNP) in 100% L. pneumophila isolates and in 70% of non-pneumophila isolates. Only 38 (out of 138) non-pneumophila species isolates showed 1 SNP and 3 showed 2 SNPs. The ubiquity of RocR in the Legionella genus suggests that in all species natural transformation is regulated by this sRNA. Consequently, other MGEs could also use a RocRp homolog to interfere with natural transformation. In publicly available genomes 3 distinct RocRp homologs were found in an unclassified Legionella species (km542) and in the type strains of Legionella geestiana (DSM21217) and Legionella israelensis (DSM19235) (Fig. 4A). The RocRp homologs of Legionella sp. km542 and L. israelensis are closely related to the RocRp from pLPL, whereas the RocRp from L. geestiana is more closely related to the chromosomal RocR (Fig. 4B). Both the Legionella sp. km542 and L. geestiana RocRp homologs are carried by episomal conjugative elements (of 66.4 and 60.9 kb, respectively), while the L. israelensis RocRp homolog is part of a 46.6-kb conjugative element integrated in chromosome at the transfer-messenger RNA-encoding gene. In gram− bacteria, conjugative systems fall into 6 phylogenetically distinct types (B, C, F, G, I, and T) (39). Like pLPL, the plasmid of Legionella sp. km542 encodes a type-F conjugative system. In contrast, the integrated conjugative element (ICE) of L. geestiana DSM21217 and the plasmid of L. israelensis DSM19235 encode type-T systems (39). All 4 elements share no other homology (Fig. 4B). The L. geestiana and L. israelensis type strains are their only representatives, isolated in England and Israel, respectively. We could retrieve 1 more L. geestiana isolate from France and 2 isolates of L. israelensis from France and Norway. The genomes of all 3 isolates revealed RocRp homologs, but again on distinct MGEs. In L. geestiana HL-0438-2026, the RocRp homolog is found again on a 53.3-kb episomal element but this time encoding a type-F conjugative system (Fig. 4B) (39). In L. israelensis strain Vestfold L18-01051 from Norway, the RocRp homolog belongs to an ICE with an unclassified conjugative system previously reported in the L. pneumophila genomic islands (LGI) LpcGI-2 and LppGI-2 of L. pneumophila (40). In L. israelensis HL-0427-4011, the RocR homolog is in another LGI inserted at a transfer RNA gene (40). Except for TraC, this element shows no known conjugative system but encodes a TraK homolog and an origin of transfer. Thus, this LGI may be mobilized by other conjugative systems of plasmids or ICE. Interestingly, the RocRp homologs and the proximal sequences were identical between the MGEs found in each species, suggesting intraspecific transfers. In conclusion, the strategy to inhibit transformation by encoding a RocRp homolog has been adopted by a diverse set of conjugative and mobilizable elements in Legionella species. Importantly, both known isolates of L. geestiana and all 3 known isolates L. israelensis are infected with distinct conjugative elements carrying RocRp homologs.
Fig. 4.
Diverse conjugative elements carry RocRp homologs. (A) Sequence alignment of RocR and RocRp homologs found in Legionella species. Experimentally determined transcription start and termination sites are highlighted with bold green and red nucleotides, respectively. (B) Parsimony-based unrooted phylogenetic tree of the genes encoding RocRp homologs. (C) Schematic representation of MGEs carrying a RocRp homolog-encoding gene. Conjugative systems of the type F and T are colored in green and blue, respectively. Incomplete and unclassified conjugative systems are colored in orange and brown, respectively. RocRp homologs are represented as a red bar. Pairwise nucleotide sequence identity is represented by a gradient from light to dark gray.
Discussion
The transformability phenotype is generally conserved in L. pneumophila, yet not homogeneously distributed. The ST1 clade groups a majority of transformable isolates, consistent with high recombination rates in this clade (31, 32). This contrasts with the absence of transformable isolates in the ST47 clade in which no recombination events could be detected (32). Although not a demonstration, this correlation suggests that natural transformation is a major contributor to genome recombination in L. pneumophila. The occurrence of mixed phenotypes in the ST23 clade provided the strongest signal for GWAS and allowed for the identification of pLPL as an inhibitor of natural transformation. Plasmid pLPL is found in one-third of nontransformable isolates and GWAS could not identify other genetic determinants associating with the remaining nontransformable isolates. This may be because different polymorphisms (or diverse MGEs) can result in the loss of transformability. GWAS also could not identify the underlying reason for the lack of transformability of ST47 isolates, which do not carry pLPL. The ST47 cluster is homogeneously nontransformable and shows too little genetic polymorphism for a successful GWAS approach.
It is striking that a single conjugative element could be responsible for the lack of transformability of one-third of the nontransformable isolates in our panel of isolates. On a more global scale of over 500 genomes, at least 5% of L. pneumophila isolates are likely not transformable due to this single conjugative element. This provides evidence that conjugative elements significantly contribute to intraspecific variation in natural transformability, a situation observed in other species but so far unexplained (25–28). The same phenomenon is likely at play in other species, as cases of transformation inhibition by plasmids and conjugative elements have been sporadically reported. For instance, in Bacillus subtilis the 84-kb plasmid pBS32 encodes a 30-amino acid protein which seems to interfere with the activity the DNA uptake system without altering its expression (41). In contrast, still in B. subtilis, the native conjugative plasmid pLS20 does interfere with expression of the DNA uptake system by encoding a homolog of Rok, the repressor of the master activator of competence ComK (42). In Vibrio cholerae, isolates from the Haiti outbreak are poorly transformable because the VchInd5 ICE encodes a secreted DNase that degrades exogenous DNA (43). It was proposed that the inhibition of natural transformation by a secreted DNase was coincidental, resulting from the serendipitous acquisition of a “stowaway” gene (43). Far from fortuitous, all these active interferences now designate natural transformation as a common target of conjugative elements but also of other MGEs. Indeed, a repressor of the ComRS system, which controls competence in Streptococcus pyogenes, was recently identified on a prophage (44). Also, several ICE and prophages have been reported to insert and disrupt genes involved in natural transformation (refs. 45 and 46 and references therein). These observations support the proposal that natural transformation is a mechanism to cure the genome of parasitic MGEs (45). In this model, bacteria whose chromosomes carry an integrated MGEs could import DNA from uninfected cells. Homologous recombination would restore the original chromosomal segment and delete the integrated MGE (45). MGEs inserting in genes required for natural transformation would block chromosome curing. Thus, the insertional inactivation of transformation genes by MGEs can be viewed as an “anticuring strategy” that would favor the persistence of MGEs in the bacterial community. For a chromosomal MGE, carrying a transformation-interfering genetic cargo would similarly function as an anticuring strategy. However, this hardly explains the inhibition of transformation by nonintegrative plasmids (such as pLPL, pBS32, and pLS20). Alternatively, blocking transformation could prevent import of competing plasmids, yet this process is poorly efficient and plasmids can spread at high rates by conjugation. However, blocking DNA uptake could limit recombination and rearrangements that could otherwise jeopardize the plasmid stability or impair its propagation.
From only a handful of cases, it is already apparent that MGEs have evolved diverse mechanisms to interfere with natural transformation, targeting several stages of the process. We speculate that a plethora of diverse transformation-inhibiting factors (TIF), frequent genetic cargo of MGEs, are obscuring the widespread occurrence of natural transformability in bacteria. Conversely, once identified, they may also represent evidence of natural transformation activities. This is exemplified here by L. geestiana and L. israelensis, in which the presence of TIF cargo on distinct conjugative elements is a strong indication that natural transformation is active in these species. Revealing the intricacies of the conflict between MGEs and natural transformation represents a unique opportunity to gain insight into a mechanism of HGT, which has captivated scientists for nearly a century.
Materials and Methods
Bacterial Strains, Growth Conditions, and Mutant Construction.
All Legionella (SI Appendix, Table S1 and S3) were grown in liquid medium ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract (AYE) or on solid media ACES-buffered charcoal yeast extract (CYE) plates. Oligonucleotides and plasmids are listed in SI Appendix, Table S4. DNA manipulation and construction of the mutants can be found in SI Appendix.
Transformation Assay of Clinical Isolates and GWAS.
Clinical isolates (SI Appendix, Table S1) were tested for natural transformation in 100 µL of AYE without or with 2 µg/mL of a transforming PCR product in 96-well plates at 30 °C. After 48 h, 10 µL of cultures were then spotted on CYE plates containing streptomycin. Transformation was scored as a function of the approximate number of colonies that developed in the spot (0 colony, score 0; 1 to 9, score 1; 10 to 50, score 2; >50, score 3). Transformation scores were determined 4 times independently. A score was retained only if it was superior to the score determined in the no-DNA condition. GWAS were carried out using DBGWAS 0.5.2 (34) on a matrix of NT (score 0 to 0.5) and T (score 1 to 3) phenotypes.
Quantitative Transformation Assays.
Strains were inoculated at OD = 0.1 to 0.2 in 3 mL of AYE in 13-mL tubes with 1 µg of transforming DNA and incubated at 30 °C with shaking for 24 h. A PCR product encompassing the rpsL gene from the Paris_S strain was used for streptomycin-sensitive strains. Alternatively, transformation was tested with a nonreplicative circular DNA carrying the kanamycin resistance gene inserted in a nonessential gene. Transformation frequencies correspond to the ratio of transformants colony-forming units (CFU) over total CFU counts. All assays were performed at least 3 times independently, several days or weeks apart.
Gene Expression Analysis by Northern Blot, RNAseq, and RNA Decay.
Total RNA extraction and Northen blot analysis were performed as previously described (36). For RNAseq analysis, RNAs from cultures grown to OD600 = 2 at 30 °C were treated with DNase I and purified. Following ribosomal RNA depletion, strand-specific cDNA libraries were sequenced (Illumina) and enriched transcripts were determined using DESeq2 (47). Half-lives of RocR and RocRp were determined by RNA decay experiments. Bacterial cultures were treated with 100 μg/mL of rifampicin to stop transcription. RocR and RocRp levels were detected by Northern blot analysis and quantitated by densitometry with ImageJ and fit to a first-order exponential decay.
The 5′/3′ RACE of RocR and RocRp.
Total RNA was treated with RppH for 1 h at 37 °C and circularized with T4 RNA ligase. RocR and RocRp were then reverse-transcribed with a specific primer (LA124). RocR (primers LA124/LA125) and/or RocRp (primers LA124/LA126) were PCR-amplified. The PCR products were cloned and sequenced.
Purification and Relative Quantification of RocC-Bound RNAs.
RocC-bound RNAs were immunopurified with rabbit-raised affinity-purified antibodies directed against RocC as previously described (36). RNAs were reverse-transcribed in a single reaction with a mix of 2 primers specific for RocRp and RocR (primers RT19-rocRlike and RT20-rocR). Both primers carry a 5′ extension which was then used for the simultaneous PCR amplification of RocR and RocRp with primers RRl17F and RRl20R. The resulting PCR products, a mix of RocR (86 bp) and RocRp (85 bp), were digested with NruI, which cleaves only RocRp, giving 2 fragments of 42 and 43 bp. The restriction digests were quantified by capillary electrophoresis.
Accession Numbers.
Genome assemblies have been deposited in GenBank (Bioproject PRJNA528641). RNAseq reads have been deposited to the European Nucleotide Archive (PRJEB31835). Sequence data of all clinical isolates are available from the European Nucleotide Archive (PRJEB15241) (48).
Supplementary Material
Acknowledgments
This work was supported in part by a grant from Agence Nationale de la Recherche to L.A. (Project RNAchap, ANR-17-CE11-0009-01). This work was performed within the framework of the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon, within the program “Investissements d’Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency. We thank Magali Jaillard and Laurent Jacob for introducing us to the DBGWAS tool and for all subsequent discussions about its use in our project, Louise Kindingstad (Stavanger University Hospital, Norway) for providing the L. israelensis isolate Vestfold L18-01051, and Olga Soutourina (I2BC, France) for RACE protocols.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome assemblies have been deposited in the GenBank database (Bioproject PRJNA528641). RNA-sequencing reads have been deposited in the European Nucleotide Archive (PRJEB31835).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909374116/-/DCSupplemental.
References
- 1.Johnston C., Martin B., Fichant G., Polard P., Claverys J.-P., Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Chen I., Dubnau D., DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Ellison C. K., et al. , Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 3, 773–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurenceau R., et al. , A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 9, e1003473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vos W. M., Venema G., Canosi U., Trautner T. A., Plasmid transformation in Bacillus subtilis: Fate of plasmid DNA. Mol. Gen. Genet. 181, 424–433 (1981). [DOI] [PubMed] [Google Scholar]
- 6.Chaussee M. S., Hill S. A., Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J. Bacteriol. 180, 5117–5122 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacks S., Greenberg B., Carlson K., Fate of donor DNA in pneumococcal transformation. J. Mol. Biol. 29, 327–347 (1967). [Google Scholar]
- 8.Claverys J.-P., Martin B., Polard P., The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol. Rev. 33, 643–656 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Provvedi R., Dubnau D., ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol. Microbiol. 31, 271–280 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Seitz P., et al. , ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet. 10, e1004066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hepp C., Maier B., Kinetics of DNA uptake during transformation provide evidence for a translocation ratchet mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, 12467–12472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draskovic I., Dubnau D., Biogenesis of a putative channel protein, ComEC, required for DNA uptake: Membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55, 881–896 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Londoño-Vallejo J. A., Dubnau D., comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol. Microbiol. 9, 119–131 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Sinha S., Mell J. C., Redfield R. J., Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J. Bacteriol. 194, 5245–5254 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diallo A., et al. , Bacterial transformation: ComFA is a DNA-dependent ATPase that forms complexes with ComFC and DprA. Mol. Microbiol. 105, 741–754 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Mortier-Barrière I., et al. , A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130, 824–836 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Bergé M., Mortier-Barrière I., Martin B., Claverys J.-P., Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50, 527–536 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Marie L., et al. , Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension. Nat. Commun. 8, 15638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nero T. M., et al. , ComM is a hexameric helicase that promotes branch migration during natural transformation in diverse Gram-negative species. Nucleic Acids Res. 46, 6099–6111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pimentel Z. T., Zhang Y., Evolution of the natural transformation protein, ComEC, in bacteria. Front. Microbiol. 9, 2980 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendler K., et al. , AnnoTree: Visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 47, 4442–4448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnsborg O., Eldholm V., Håvarstein L. S., Natural genetic transformation: Prevalence, mechanisms and function. Res. Microbiol. 158, 767–778 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Seitz P., Blokesch M., Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 37, 336–363 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Carlson C. A., Pierson L. S., Rosen J. J., Ingraham J. L., Pseudomonas stutzeri and related species undergo natural transformation. J. Bacteriol. 153, 93–99 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maughan H., Redfield R. J., Extensive variation in natural competence in Haemophilus influenzae. Evolution 63, 1852–1866 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Li G., et al. , Addiction of hypertransformable pneumococcal isolates to natural transformation for in vivo fitness and virulence. Infect. Immun. 84, 1887–1901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans B. A., Rozen D. E., Significant variation in transformation frequency in Streptococcus pneumoniae. ISME J. 7, 791–799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski J., Teschner N., Wackernagel W., Highly different levels of natural transformation are associated with genomic subgroups within a local population of Pseudomonas stutzeri from soil. Appl. Environ. Microbiol. 68, 865–873 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godeux A.-S., et al. , Fluorescence-based detection of natural transformation in drug-resistant Acinetobacter baumannii. J. Bacteriol. 200, e00181-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Valero L., et al. , Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12, 536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David S., et al. , Dynamics and impact of homologous recombination on the evolution of Legionella pneumophila. PLoS Genet. 13, e1006855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David S., et al. , Multiple major disease-associated clones of Legionella pneumophila have emerged recently and independently. Genome Res. 26, 1555–1564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Busó L., Comas I., Jorques G., González-Candelas F., Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat. Genet. 46, 1205–1211 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Jaillard M., et al. , A fast and agnostic method for bacterial genome-wide association studies: Bridging the gap between k-mers and genetic events. PLoS Genet. 14, e1007758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cazalet C., et al. , Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Attaiech L., et al. , Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc. Natl. Acad. Sci. U.S.A. 113, 8813–8818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charpentier X., Kay E., Schneider D., Shuman H. A., Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J. Bacteriol. 193, 1114–1121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juan P.-A., Attaiech L., Charpentier X., Natural transformation occurs independently of the essential actin-like MreB cytoskeleton in Legionella pneumophila. Sci. Rep. 5, 16033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cury J., Touchon M., Rocha E. P. C., Integrative and conjugative elements and their hosts: Composition, distribution and organization. Nucleic Acids Res. 45, 8943–8956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lautner M., Schunder E., Herrmann V., Heuner K., Regulation, integrase-dependent excision, and horizontal transfer of genomic islands in Legionella pneumophila. J. Bacteriol. 195, 1583–1597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konkol M. A., Blair K. M., Kearns D. B., Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195, 4085–4093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh P. K., et al. , Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ. Microbiol. 14, 2812–2825 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Dalia A. B., Seed K. D., Calderwood S. B., Camilli A., A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 112, 10485–10490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mashburn-Warren L., Goodman S. D., Federle M. J., Prehna G., The conserved mosaic prophage protein paratox inhibits the natural competence regulator ComR in Streptococcus. Sci. Rep. 8, 16535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croucher N. J., et al. , Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. 14, e1002394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blokesch M., In and out-contribution of natural transformation to the shuffling of large genomic regions. Curr. Opin. Microbiol. 38, 22–29 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandewalle-Capo M., et al. , Minimum inhibitory concentration (MIC) distribution among wild-type strains of Legionella pneumophila identifies a subpopulation with reduced susceptibility to macrolides owing to efflux pump genes. Int. J. Antimicrob. Agents 50, 684–689 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.