Significance
The mineralocorticoid receptor (MR), the receptor for aldosterone, appears in evolution well before the appearance of terrestrial vertebrates, yet aldosterone emerges in vertebrates only with terrestrial life. Curiously, in fish, the MR sees progesterone and spironolactone as agonists, whereas in terrestrial species, they are antagonist at the MR. We have identified a unique single amino acid difference between the fish MR and the other vertebrate MR that mediates this switch, agonist to antagonist. This striking evolutionary event was perhaps mandatory if the appearance of aldosterone as a specific mediator of the homeostatic salt retention required for terrestrial life was to be tolerated. The conformational changes also provide insights into the structural basis of agonism versus antagonism in steroid receptors.
Keywords: aldosterone, cortisol, mineralocorticoid receptor, sodium homeostasis, progesterone
Abstract
The mineralocorticoid receptor (MR) is highly conserved across vertebrate evolution. In terrestrial vertebrates, the MR mediates sodium homeostasis by aldosterone and also acts as a receptor for cortisol. Although the MR is present in fish, they lack aldosterone. The MR binds progesterone and spironolactone as antagonists in human MR but as agonists in zebrafish MR. We have defined the molecular basis of these divergent responses using MR chimeras between the zebrafish and human MR coupled with reciprocal site-directed mutagenesis and molecular dynamic (MD) simulation based on the crystal structures of the MR ligand-binding domain. Substitution of a leucine by threonine in helix 8 of the ligand-binding domain of the zebrafish MR confers the antagonist response. This leucine is conserved across fish species, whereas threonine (serine in rodents) is conserved in terrestrial vertebrate MR. MD identified an interaction of the leucine in helix 8 with a highly conserved leucine in helix 1 that stabilizes the agonist conformation including the interaction between helices 3 and 5, an interaction which has previously been characterized. This switch in the MR coincides with the evolution of terrestrial vertebrates and of aldosterone synthesis. It was perhaps mandatory if the appearance of aldosterone as a specific mediator of the homeostatic salt retention was to be tolerated. The conformational changes also provide insights into the structural basis of agonism versus antagonism in steroid receptors with potential implications for drug design in this important therapeutic target.
The mineralocorticoid receptor (MR) is a member of the nuclear receptor superfamily of ligand-dependent transcription factors; in vertebrates, it diverged with the glucocorticoid receptor (GR) by a gene duplication from the ancestral corticoid receptor >450 million years ago (1). MR has been highly conserved across vertebrate evolution (2). In terrestrial vertebrates, including humans, MR mediates the regulation of sodium homeostasis by aldosterone. The MR also binds cortisol and the less potent deoxycorticosterone (DOC), which is 21-hydroxyprogesterone. In most tetrapods (including human, rodent, alligator, and Xenopus), progesterone is an antagonist of MR, as is its derivative spironolactone (3). Although the MR is present in teleosts that lack aldosterone synthesis (4), the teleost MR responds to aldosterone, as well as to cortisol and DOC (2, 5, 6) that have been proposed as the “physiological” mineralocorticoids for the fish MR (5, 6). Characterization of various fish MR found that, in contrast to terrestrial animals, teleost MR were activated by both progesterone and spironolactone. These findings suggest that progesterone may be a physiological agonist for the MR in teleosts. The evolutionary adaptability of proteins is brought into sharp focus by divergent responses to the same ligands (progesterone and spironolactone) in such closely related receptors, while responses to other ligands are essentially equivalent. Understanding the structural basis of agonism versus antagonism in steroid receptors is also of considerable therapeutic importance (7).
In the present study, we demonstrate that a single residue change is responsible for the switch from an agonist response to progesterone for zebrafish MR (zMR) to antagonist for human MR (hMR). This change in the MR ligand-binding domain (LBD) is surprisingly distant from the ligand-binding site and has not previously been associated with agonist to antagonist switching in the MR.
Results
LBD Mediates the Agonist Response of the Zebrafish MR to Spironolactone.
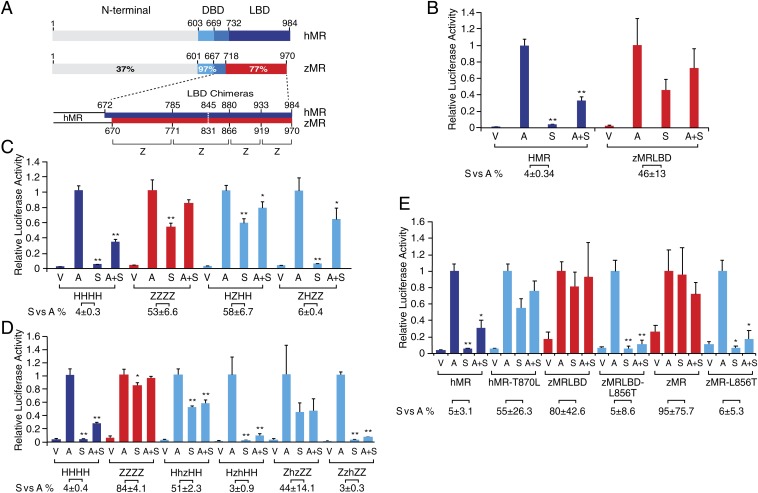
Previous studies using full-length MR in transactivation assays demonstrated that spironolactone and progesterone were agonists for fish MR (2, 5, 6); to formally demonstrate that the difference is a property of the LBD, the zMR LBD, amino acids 670 to 970, was substituted for the equivalent region (amino acids 672 to 984) in hMR (Fig. 1A). This initial chimera (zMR LBD) demonstrated the same robust agonist response to spironolactone (Fig. 1B) as for full-length zMR (6). Antagonism of the aldosterone response by spironolactone at the hMR demonstrates that there is a loss of receptor activation by spironolactone, not of ligand binding. The response of hMR to spironolactone alone is 4% that of aldosterone, while for the zMR LBD chimera it is 46% (Fig. 1B).
Fig. 1.
Identification of the critical amino acid switch that determines agonism versus antagonism for spironolactone in the MR. (A) Schematic representation of the comparison of the hMR and zMR. The MR is divided into three principal domains: the N-terminal domain, the DNA-binding domain (DBD), and the ligand-binding domain (LBD). The amino acid numbers and the percentage amino acid identity are shown (6, 24). The architecture of the LBD chimeras is shown below with the amino acid numbers of the breakpoints shown above (hMR) and below (zMR). (B) Transactivation responses of hMR and a chimera of hMR N terminus and DBD with the zMR LBD (zMRLBD) to aldosterone and spironolactone. CV-1 cells were transiently transfected with 250 ng of pRShMR or pRSzMRLBD together with 250 ng of MMTV-LUC reporter gene and 50 ng of pREN-LUC. The cells were treated with vehicle (V), 10 nM aldosterone (A), 1 µM spironolactone (S), or aldosterone plus spironolactone (A+S). Corrected luciferase activity is expressed relative to the response of the hMR to 10 nM aldosterone (mean ± SEM) derived from two independent experiments with treatment groups aldosterone and aldosterone plus spironolactone being greater than vehicle alone (P < 0.05). Spironolactone and spironolactone plus aldosterone were less than aldosterone alone for hMR (**P < 0.0001) but not for zMR LBD. The activation with spironolactone alone as a percentage of that for aldosterone (S vs. A) for each MR is shown below the graph and significantly differs between the MR (P < 0.05). (C) Transactivation responses of hMR:zMR LBD chimeras to aldosterone and spironolactone highlighting the critical role of the second region in reciprocal chimeras. The four regions of the LBD are shown in A. Analysis is as in B with aldosterone and aldosterone plus spironolactone being greater than vehicle alone (P < 0.001). Spironolactone and spironolactone plus aldosterone are less than aldosterone alone (*P < 0.05, **P < 0.001) where indicated. The relative response, S vs. A, differs significantly (P < 0.001) between the wild type and its chimeric MR. (D) Transactivation responses with the second region of the LBD further subdivided as indicated by lowercase h and z. Spironolactone and spironolactone plus aldosterone are less than aldosterone alone (*P < 0.05, **P < 0.001) where indicated. The relative response S vs. A differs significantly (P < 0.001) for HhzHH but not HzhHH from HHHH and for ZzhZZ but not ZhzZZ from ZZZZ. (E) Transactivation responses of hMR 870 leucine and zMR 856 threonine. The response of WT hMR (hMR) and hMR threonine 870 leucine (hMR-T870L), and both the chimera, zMRLBD, and WT zMR (zMR) with leucine 856 threonine (zMR-L856T and zMRLBD-L856T, respectively) to aldosterone (A) and spironolactone (S) was examined as described above in three independent experiments. Spironolactone and spironolactone plus aldosterone are less than aldosterone alone (mean ± SEM, *P < 0.01, **P < 0.001) where indicated. The relative response, S vs. A, significantly differs between the intact MR LBD and the corresponding mutant MR LBD (P < 0.01).
Amino Acids in the hMR Known to Interact with Spironolactone Are Conserved in the zMR.
The 49 amino acid differences in the LBD between the hMR and the zMR (5) do not include amino acids reported to be of importance in the interaction of spironolactone with the MR. Alanine 773 in the hMR has been associated with activation by 11β-substituted spironolactones in the hMR (8), but it is conserved in the zMR, as is hMR methionine 852, which has been characterized as playing a key role in the interaction with the C7 substituents on spironolactones (9). hMR serine 810 in helix 5 was identified as being mutated to leucine (Ser810Leu) in a kindred with early-onset low renin hypertension; this mutation creates an agonist response to a range of ligands including progesterone and spironolactone (10, 11). Analyses of the conformational changes induced by the Ser810Leu mutation in the hMR has highlighted the importance of an interaction between helix 3 and helix 5 (11). Ser810 is found at the equivalent position in the hMR across species (2). While there is a serine at zMR position 770 in helix 5, with the equivalent amino acid in the hMR being alanine 813, reciprocal mutagenesis of these residues showed that this difference was without impact (6).
Functional Analyses of zMR/hMR LBD Chimeras.
Of the differences in the LBD between the hMR and the zMR, we reasoned that the critical residue(s) were likely to be one or more of the 39 amino acid pairs conserved in the rainbow trout and zebrafish but which differed from the equivalent residues in mouse, rat, and human (6). Chimeras based on the two LBD were designed, an approach previously used with the MR and GR to define the determinants of aldosterone selectivity in the MR (12). Three break points were chosen to divide the LBD into four sections (Fig. 1A) fused with the N terminus and DBD derived from the hMR. Each chimera (SI Appendix, Fig. S1) was examined in transactivation assays for their response to aldosterone, spironolactone, and the combination of both. The initial screen (Fig. 1C and SI Appendix, Fig. S1) suggested that the critical region was within residues 785 to 880 of the hMR (zMR 771 to 866). Four subchimeras for this region (Fig. 1D) containing 10 of the 39 species-specific amino acid differences, demonstrated the region hMR 844 to 880/zMR 830 to 866 to be critical.
Reciprocal Substitution of hMR T870 with zMR L856 Reversed the Response to Spironolactone.
Extensive site-directed mutagenesis (SI Appendix, Fig. S1) eliminated all of the species-specific amino acids in the critical hMR 844 to 880/zMR 830 to 866 regions. We next looked at the differences between the zMR and hMR, without regard to the trout or rodent MR sequences, identifying an additional 11 differences in the LBD. Within a residue pair that differed between the hMR LBD (threonine 870: isoleucine 871) and the zMR LBD (leucine 856: alanine 857), substitution of hMR threonine 870 with leucine in zMR, and leucine 856 substituted with threonine in hMR, reciprocally reversed the response. hMR LBD T870L showed a relative response to spironolactone as 55% of aldosterone, whereas with zMRLBD-L856T spironolactone was antagonist with a relative response of 5% (Fig. 1E). The antagonist response to spironolactone was also observed when this single residue substitution was introduced into the full-length zMR (Fig. 1E).
Progesterone Is Also an Agonist with hMR LBD T870L.
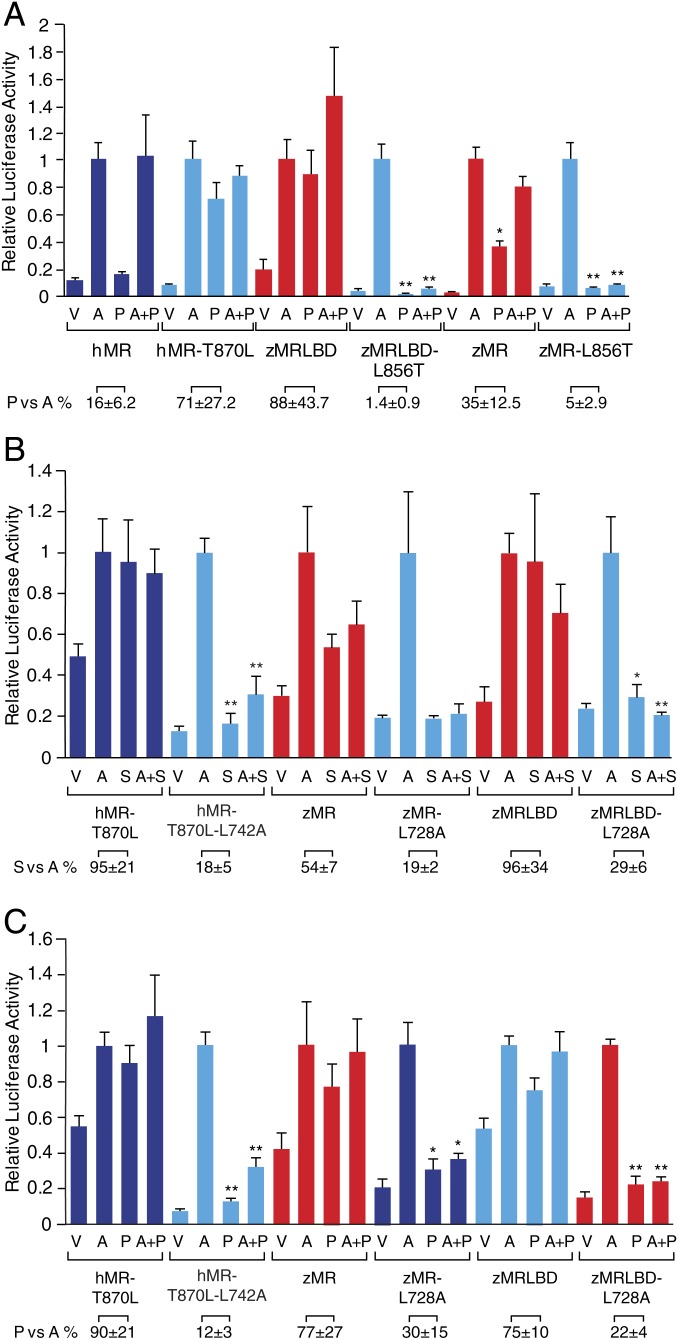
The analyses in Fig. 1 use spironolactone as the hMR antagonist/zMR agonist; the equivalent results were observed with progesterone (Fig. 2A). Eplerenone, which is derived from spironolactone and is a weak agonist at the zMR (6), also exhibited clear evidence of the antagonist/agonist switch (SI Appendix, Fig. S2). The pattern of responses across both the wild-type and mutant MR for both species was confirmed using two other physiological MR agonists, cortisol and DOC, both of which, in contrast to aldosterone, are present in fish (SI Appendix, Fig. S3). DOC exhibits a marked right shift in its activation in the presence of the mutations, e.g., hMR leucine 870 or zMR threonine 856 but not between the zMR and the hMR (SI Appendix, Fig. S3), suggesting that coincident with the evolutionary transition of the zMR leucine 856 to a threonine, other changes occurred elsewhere to ensure that DOC retained its potency at the MR, reinforcing a previous observation that the interaction of DOC with the MR is not equivalent to that of aldosterone (13).
Fig. 2.
Characterization of the role of leucine at position 856 in the zMR in the agonist response to progesterone and spironolactone. The approach is described for Fig. 1E with each data point representing the mean ± SEM derived from three independent experiments. (A) Transactivation responses of hMR 870 leucine and zMR 856 threonine. The analysis in Fig. 1E was repeated with 1 μM progesterone (P). Progesterone and progesterone plus aldosterone are less than aldosterone alone (*P < 0.01; P < 0.001) where indicated. The relative response, progesterone versus aldosterone (P vs. A), significantly differs between the intact MR LBD and the corresponding mutant MR LBD (P < 0.01). (B) Transactivation responses when the leucine in helix 1 (hMR 742 and zMR 728) is converted to alanine. The response of hMR threonine 870 leucine (hMR-T870L), hMR-T870L containing an alanine at position 742 (hMR-T870L-L742A), WT zMR (zMR), WT zMR (zMR) containing an alanine at position 728 (zMR-L728A), zMRLBD, and zMRLBD containing an alanine at position 728 (zMRLBD-L728A) to ligand (10 nM aldosterone [A] and 1 μM spironolactone [S]) were analyzed as for Fig. 1E. Spironolactone and spironolactone plus aldosterone are less than aldosterone alone (*P < 0.05; **P < 0.001) where indicated. The relative response, S vs. A, differs between the intact MR LBD and the corresponding mutant MR LBD (P < 0.001). (C) Transactivation responses when the leucine in helix 1 (hMR 742 and zMR 728) is converted to alanine as in C but with progesterone (P). All treatment groups (A, P, and A+P) are greater than vehicle alone (P < 0.05). Progesterone and progesterone plus aldosterone are less than aldosterone alone (*P < 0.05; **P < 0.001) where indicated. The relative response, P vs. A, significantly differs between the intact MR LBD and the corresponding mutant MR LBD (P < 0.01).
Spironolactone Is Agonist for the zMR under Physiological Conditions.
These analyses were conducted in a heterologous system; to address this potential confounder, we examined the responses in the zebrafish embryonic fibroblast-derived cell line, ZF4, which is cultured at 28 °C (14). The ligand-discriminate responses to spironolactone for the zMR versus the hMR were again observed (SI Appendix, Fig. S4). The magnitude of the response to aldosterone of the full-length hMR versus zMR differed, being relatively higher for the hMR in the CV-1 cells and conversely higher for the zMR in the ZF4 cells; this species-specific difference appears to be a property of the N terminus. Western blot analysis of hMR, zMRLBD, and the mutated MR showed no evidence of an effect on receptor stability (SI Appendix, Fig. S5).
The Antagonist Response Mediated by hMR Threonine 870 Is Not Modulated by Phosphorylation.
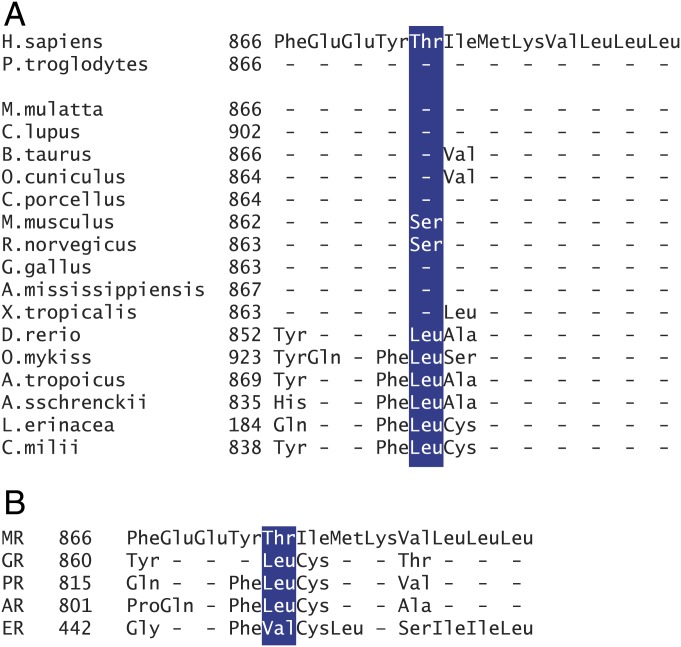
Threonine 870 as the critical residue in the hMR was unexpected as its location is distant from the ligand binding pocket. Leucine at position 856 in the zMR is conserved across all fish species whose MR has been shown to respond to progesterone with an agonist response (Fig. 3A); leucine is also present at the equivalent position on the human GR, progesterone, and androgen receptors (Fig. 3B). Conversely, the threonine at position 870 in the hMR is conserved across the other vertebrate species with the exception of the rat and mouse MR where this residue is a serine (Fig. 3A). When serine is substituted for leucine at position 856 in both the zMRLBD chimera and the full-length zMR, the response to spironolactone was the same as seen with the threonine (SI Appendix, Fig. S6). That either a threonine or serine can replace the leucine suggested that phosphorylation might be involved in the switch; however, substitution of the threonine 870 in the hMR by the phosphomimetic residues aspartate or glutamate, or the neutral residue alanine, did not alter the antagonist response to spironolactone. This suggests that leucine at this position plays a specific role (SI Appendix, Fig. S7).
Fig. 3.
Conservation of the amino acids in helix 8: Alignment of the amino acid sequences of helix 8 of the MR (27) across species (A). The sequences are from Sugimoto et al. (2), where their derivation is described in detail; with additional mammalian sequences: Pan troglodytes (XP_001150516.1), Macaca mulatta (XP_001099855.2), Canis lupus (XP_003639585.2), and Bos taurus (NP_001178278.1), Oryctolagus cuniculus (XP_008265594), and C. porcellus (XP_003476911). (B) Alignment of the amino acid sequences of the helix 8 region of the human MR, GR, PR, androgen receptor (AR), and estrogen receptor (ER) LBD (31).
Molecular Modeling of the Leucine for Threonine Substitution.
To identify the structural basis of how the substitution of threonine for leucine mediates this evolutionary switch in ligand activity from agonist to antagonist, we performed molecular dynamic (MD) simulations on the native and T870L mutant hMR LBD in complex with spironolactone. Since there exists no experimental structure of either, we created models of both LBD based on the triple mutant crystal structure (Protein Data Bank [PDB] ID code: 4PF3; C808S, S810L, A976V) (15); all three mutations were reverted to WT residues. Unrestrained MD simulations showed both structures maintained the typical and stable NR LBD’s three-layer sandwich conformation along the 2-μs trajectories (SI Appendix, Fig. S8). In the template, and therefore the starting structures of MD simulations, the guanidinium group of R817 forms a hydrogen bond with the A-ring ketone of the ligand. In native hMR this interaction is lost early in the simulation; release of R817 ultimately results in it forming an interaction with the loop preceding helix 3 with an accompanying rotation through about 15.5° of the C terminus of helix 5 (Fig. 4A). In comparison, in hMR-T870L, helix 5 retains its original position preserving the interaction between R817 and ligand. Another consequence of the change in position of helix 5 is a relocation of the loop preceding the first β-sheet (Fig. 4A). In hMR-T870L, this loop retained interactions with the residues in helix 7; for instance, S824 in the loop forms a hydrogen bond with H853. In the following, we refer to this state as the “closed” state for the loop. In native hMR, the hydrogen bond between R817 and the ligand ketone was not observed to form due to the corresponding movement of the loop following the tilting of helix 5; this state of the loop is referred as the “open” state.
Fig. 4.
Structural differences between agonist and antagonist state of MR. (A) Relative dispositions of H5, H6, and H7 between native (gray) and T870L mutant (cyan) hMR LBD, with H5–H6 loop in either open or closed conformation, respectively. R817 disengages from the A-ring ketone carbonyl of spironolactone (black dotted line) in native hMR during MD. Axes of H5 are showed as slender cylinders showing relative tilt. Side-chain carbon atoms of R817 in each model are colored green, nitrogen blue. Ligands carbon atoms are colored coral, oxygen red, sulfur yellow. (B) Relative dispositions of H5, H7, and H8 between native and T870L mutant hMR LBD. Helices of native hMR are also showed as half-transparent cartoon on the right for comparison purposes. Packing of bulky residues F866 and W816 is maintained in both conformations.
In native hMR, the side chain hydroxyl group of T870 forms an intrahelix hydrogen bond, extending helix 8 by stabilizing the helix hydrogen bond between the carbonyl of E867 and the amide of I871 (Fig. 5A and SI Appendix, Fig. S9). The absence of this extension in hMR-T870L is complemented by a displacement of helices 5, 7, and 8 relative to native hMR (Fig. 4B); the disposition of these helices characterizes the open and closed configurations. Packing of key bulky residues W816 and F866 is preserved in both configurations and provides the conduit of structural differences in helix 8 through helix 5, on to helix 7, and therein to the distant ligand binding pocket. We could demonstrate that this intrahelix hydrogen bond is necessary to stabilize the closed state, and these observations were not simple artifacts of the initial geometry used in the MD simulations.
Fig. 5.
Structural differences between agonist and antagonist state of MR. (A) Intrahelix hydrogen bond network within H8 of native (gray) and T870L mutant (cyan) hMR LBD. An extra hydrogen bond is formed between the hydroxyl of T870 and amide carbonyl of F866. Asterisk indicates the backbone carbonyl group of the residue (i) that forms a hydrogen bond with the residue (i+4) in native LBD, which is absent in the T870L mutant. (B) Hydrophobic network mediated through L742 in hMR-T870L. H1 remains helical in hMR-T870L H1 but uncoils in native LBD. Hydrophobic interactions are showed as dotted lines. (C) The missing hydrophobic network in both native (gray) and L742A-T870L double mutant (dark cyan). In both cases, due to the absence of effective mediators L870 or L742, the hydrophobic network is not maintained.
The modeling also suggested a possible interaction between this leucine at position 870 and a highly conserved leucine in helix 1 (hMR742 and zMR728). Helix 1 uncoiled in native hMR with MD, however, remained stable throughout the 2-μs simulation in hMR-T870L. In the simulation of double mutant hMR-L742A-T870L, helix 1 again uncoiled. In hMR-T870L, where helix 1 was stable, leucine residues 742 and 870 formed a stable compact hydrophobic network that involved several highly conserved residues among the MR of different species, including L787 (helix 3) and V874 (helix 8), while in native hMR or the L742A-T870L mutant, the network was absent (Fig. 5 B and C). In all crystal structures of hMR (in an agonist-stabilized form), helix 1 is coiled with L742 packing against T870 maintaining the hydrophobic network, likely due to stabilization of helix 3 through mutations. To test our hypothesis, a series of double mutants for both hMR and zMR were made and tested in transcriptional assays. When this helix 1 leucine was replaced with alanine in either zMR or hMR-T870L, the agonist responses to spironolactone (Fig. 2B) and progesterone (Fig. 2C) were switched back to an antagonist responses with no loss of the agonist response to aldosterone Thus, this compact arrangement of helices 1 and 8 also defines the agonist form of the receptor. A previous study in the peroxisome proliferator activated receptor gamma, demonstrated a critical role for an equivalent leucine–leucine interaction between helices 1 and 8 in maintaining a stable LBD agonist conformation (16), indicating a shared activation and regulation mechanism across the nuclear receptor superfamily.
Stablization of the Helix 3–Helix 5 Interaction.
Previous studies have shown that the ketone of the ligand is responsible for bridging the interaction between helix 3 and helix 5, which is vital for receptor activation (17). Our MD simulations show that in native hMR, leucine at position 870 will drive a series of conformational changes that sees R817 disengage from the ligand and, consequently, dislodge the bridge between helix 3 and helix 5. Notably, it has been shown previously that in the Ser810Leu mutation, the interaction between these helices is enhanced and is able to switch the ligand behavior from antagonist to agonist (11). Likewise, we propose that the weakened interaction between helix 3 and helix 5 in native hMR is also not able to maintain the receptor LBD in an active conformation and, consequently, spironolactone behaves as an antagonist. In contrast, stabilizing helix 5 in hMR-T870L renders an agonist conformation, which allows spironolactone to present the MR in an agonist conformation.
Confirming the Role of T870.
To test the role of T870 in dictating the LBD structural form, we examined the outcome of MD simulations where the hydroxyl group was rendered incapable of forming a hydrogen bond (Fig. 5A). After 1 μs of MD simulation on this form of native hMR, R817 remained engaged with the ligand ketone and the loop preceding the first two β-strands remained in the closed state, reminiscent of the agonist form of the LDB (Fig. 4A). Thus, hydrogen bonding of the threonine at position 870 can determine the state of the LBD.
Ensuring the Structure of Initial Model Template Did Not Influence the Observed Structural Changes.
To ensure the structural changes observed were not simple artifacts of simulations conducted with the template that harbors several amino acid substitutions that stabilize the agonist form of the receptor (C808S, S810L, A976V: SI Appendix), T870 in the structure resulting from 2 μs of native hMR simulation was mutated to leucine, and MD continued from this model. Helix 5 was observed to relocate to the position observed in the agonist form, stabilizing the interaction with H3, and allowing R817 to reengage with the ligand ketone (Fig. 4A). At the end of this simulation, the LBD had reversed most of the structural changes observed in the final hMR model; the root mean square derivative (rmsd) across C⍺ pairs of residues from helix 3 to helix 8 (encompassing the ligand binding pocket) between this model and the initial hMR-T870L was 1.88 Å, whereas the rmsd between this model and the model of hMR was 5.65 Å. Thus, we are confident the models of native hMR and hMR-T870L are representative of the antagonist and agonist forms of MR LBD.
Discussion
The MR is an important therapeutic target in the treatment of cardiovascular disease (7) and hypertension (18). The MR antagonists currently in clinical use, spironolactone and eplerenone, are limited by hyperkalaemia, an “on-target” consequence of blocking the renal MR (19). Spironolactone is also used widely both as an experimental tool despite it being an agonist in fish MR (20). In this study, we identified a single amino acid difference between the fish MR and terrestrial vertebrate MR that mediates the switch, agonist to antagonist, in the response of the MR to spironolactone and to the physiological ligand, progesterone. Molecular modeling reveals that the agonist response seen in fish is mediated by a stabilizing leucine–leucine interaction between helices 1 and 8 in the MR LBD, helices not previously identified as contributing to the agonist response, not the least because neither is in direct contact with the ligand. The conformational differences resulting from this switch impacts other helices, including an interaction between helices 3 and 5, that has previously been characterized in the context of an unrelated mutation in one human kindred (10).
The MR may be seen as being unique among steroid hormone receptors in that it is a receptor for both mineralocorticoids and glucocorticoids. In mammalian physiology, in the MR-expressing cells of the kidney and colon, MR acts predominantly as a receptor for the “mineralocorticoid” aldosterone; cortisol (or corticosterone in rodents) is precluded from binding the MR due to metabolism by 11β-hydroxysteroid dehydrogenase type 2 (HSD2). In other tissues where HSD2 is absent, including inflammatory cells, heart, and brain, the MR predominantly binds the physiological glucocorticoids. In fish, cortisol has been shown to be involved in a number of osmoregulatory processes including MR-mediated adaptation to ion-deficient water (21). Aldosterone itself first appears as an active steroid in amphibians (4). It has been postulated that the MR originated in fish to regulate ion balance under the control of glucocorticoids and that amphibians evolved mineralocorticoids to appropriate the MR for this function. Possibly the requirement for glucocorticoids in other crucial aspects of homeostasis, particularly those involved in adaptation to terrestrial life, were no longer compatible with their having a continuing role in ion balance. These considerations take little account of the ability of progesterone to act as an agonist of the fish MR. The physiological significance of the MR as a progesterone receptor or perhaps as a receptor for a progesterone metabolite, has not been characterized (22). There is also an underappreciated caution, that spironolactone should not be used in fish as the MR antagonist (20). Similarly, the observation that the evolution of terrestrial species has been associated with the appearance of aldosterone synthase and with a switch in the response of the MR to progesterone from agonist to antagonist has received little attention.
Remarkably, the work presented here shows this amino acid switch in the MR LBD involves a single residue change from a leucine in fish to a threonine (or serine in rodents) in terrestrial vertebtrates. The precursor of the MR, which first appears in the evolution in cartilaginous fish, is an ancestral corticoid receptor (CR) that is found in lamprey and hagfish (jawless fish). Lamprey CR have a partial agonist response to progesterone, whereas the hagfish CR does not exhibit an agonist response (1). The lamprey has a leucine at the relevant position, whereas this is an arginine in the hagfish CR (23). Lungfish, which are the closest extant ancestors of tetrapods, have aldosterone synthesis (23) but retain a leucine at the equivalent position to zMR 865 (ALN70157: Noeoceratodus fosteri). The response of the lungfish MR to progesterone has not been reported. Although all of the terrestrial vertebrate MR have a threonine (or serine) at the equivalent of hMR870 (Fig. 3A) and progesterone is an antagonist at amphibian, reptile, and mammalian MR (24), the chicken MR exhibits a predominant agonist response to progesterone and spironolactone (25); this suggests divergent evolution with a yet-to-be-identified change involving neither helix 8 (Fig. 3A) nor helices 3 and 5 (24). The evolutionary relationship of the various species is shown in SI Appendix, Fig. S10.
Although our studies primarily used spironolactone, it is clear that the observations apply equally to progesterone (and also to eplerenone). A further feature of this switch is that across this evolutionary landmark, the MR retains its agonist responses to cortisol and DOC. This switch in the MR response to progesterone and related compounds is a striking evolutionary event that was perhaps mandatory if the appearance of aldosterone as a specific mediator of the homeostatic salt retention required for terrestrial life was to be tolerated during this transition to terrestrial life. Our findings provide the structural basis for this unique evolutionary switch. The conformational changes also provide insights into the structural basis of agonism versus antagonism in steroid receptors, perhaps with implications for drug design in this important therapeutic target.
Materials and Methods
Construction of MR Chimeras and Point Mutations.
The previously described hMR expression vector pRShMR (6) was used as the starting point with the LBD being replaced by a series of hMR:zMR chimeras created (SI Appendix) between amino acids 672 and 984 of the hMR LBD and the corresponding amino acids 670 to 970 of the zMR LBD (SI Appendix, Fig. S1). These chimeras were given a four-letter name based on the sequence in each of four sections of the LBD, where H is hMR sequence and Z is zMR sequence (Fig. 1A). Single, double, and multiple reciprocal point mutations in the chimeras (SI Appendix, Fig. S1).
Transactivation Assays.
The MMTV-LUC reporter and Ren-LUC control plasmids have been described previously (6). CV-1 African green monkey cells at 37 °C or ZF-4 zebrafish embryonic fibroblast cells (14) at 28 °C were transiently transfected, incubated with steroid for 24 h, and harvested for measurement of luciferase activity as detailed in SI Appendix.
Western Blot Analysis.
Western blotting with mouse monoclonal antibody MR1-18 (1:1,000) (a gift from Celso Gomez-Sanchez, Department of Internal Medicine, University of Mississippi, Jackson, MS) was as detailed in SI Appendix, Fig. S4.
Statistical Analyses.
The response relative to that of aldosterone for each construct was compared using a one-way ANOVA followed by Tukey’s post hoc test with correction for multiple comparisons using Prism Version 7.0b (GraphPad Software). The relative agonist/antagonist response for spironolactone or progesterone between MR is based on the percent agonist response as described in SI Appendix.
Molecular Modeling and MD Simulations.
Models of native hMR and hMR-T870L LBD were generated using the MODELER software (version 9.14) (26); the X-ray crystal structure of hMR with the highest resolution (PDB ID code: 4PF3) was used as the template for these models (27) as detailed in SI Appendix.
MD Simulations.
All MD simulations were performed using Gromacs software package (version 5.1.2) (28) with the GROMOS 54A7 United-Atom force field (29) and the G54A7FF United-Atom topology for spironolactone obtained using the ATB website (30).
Supplementary Material
Acknowledgments
We thank Dr. Maria-Cristina Keightley for the gift of the ZF-4 cells, Jeana Thomas and Karen O’Keefe for preparation of the manuscript, and Sue Panckridge for preparation of the figures. This work was supported by the National Health & Medical Research Council of Australia through Project Grant 1058336 and Senior Principal Research Fellowship 1002559 (to P.J.F.). The Hudson Institute is supported by the Victorian Government’s Operational Infrastructure Scheme. R.J. and S.H. acknowledge receipt of Australian Research Training Scholarships. Part of this work was undertaken using resources from the National Computational Infrastructure, which is supported by the Australian Government and provided through Intersect Australia Ltd., and through the HPC-GPGPU Facility, which was established with the assistance of LIEF Grant LE170100200.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903172116/-/DCSupplemental.
References
- 1.Bridgham J. T., Carroll S. M., Thornton J. W., Evolution of hormone-receptor complexity by molecular exploitation. Science 312, 97–101 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Sugimoto A., et al. , Corticosteroid and progesterone transactivation of mineralocorticoid receptors from Amur sturgeon and tropical gar. Biochem. J. 473, 3655–3665 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Claire M., Rafestin-Oblin M.-E., Michaud A., Roth-Meyer C., Corvol P., Mechanism of action of a new antialdosterone compound, prorenone. Endocrinology 104, 1194–1200 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Jiang J. Q., Young G., Kobayashi T., Nagahama Y., Eel (Anguilla japonica) testis 11beta-hydroxylase gene is expressed in interrenal tissue and its product lacks aldosterone synthesizing activity. Mol. Cell. Endocrinol. 146, 207–211 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Sturm A., et al. , 11-deoxycorticosterone is a potent agonist of the rainbow trout (Oncorhynchus mykiss) mineralocorticoid receptor. Endocrinology 146, 47–55 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Pippal J. B., Cheung C. M., Yao Y.-Z., Brennan F. E., Fuller P. J., Characterization of the zebrafish (Danio rerio) mineralocorticoid receptor. Mol. Cell. Endocrinol. 332, 58–66 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Pitt B., Pedro Ferreira J., Zannad F., Mineralocorticoid receptor antagonists in patients with heart failure: Current experience and future perspectives. Eur. Heart J. Cardiovasc. Pharmacother. 3, 48–57 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Auzou G., et al. , A single amino acid mutation of ala-773 in the mineralocorticoid receptor confers agonist properties to 11beta-substituted spirolactones. Mol. Pharmacol. 58, 684–691 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Fagart J., Seguin C., Pinon G. M., Rafestin-Oblin M. E., The Met852 residue is a key organizer of the ligand-binding cavity of the human mineralocorticoid receptor. Mol. Pharmacol. 67, 1714–1722 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Geller D. S., et al. , Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science 289, 119–123 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Simisky J., Tsai F. T., Geller D. S., A critical role of helix 3-helix 5 interaction in steroid hormone receptor function. Proc. Natl. Acad. Sci. U.S.A. 102, 2707–2712 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogerson F. M., et al. , A critical region in the mineralocorticoid receptor for aldosterone binding and activation by cortisol: Evidence for a common mechanism governing ligand binding specificity in steroid hormone receptors. Mol. Endocrinol. 21, 817–828 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Pippal J. B., Yao Y., Rogerson F. M., Fuller P. J., Structural and functional characterization of the interdomain interaction in the mineralocorticoid receptor. Mol. Endocrinol. 23, 1360–1370 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsachaki M., et al. , Absence of 11-keto reduction of cortisone and 11-ketotestosterone in the model organism zebrafish. J. Endocrinol. 232, 323–335 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Bledsoe R. K., et al. , A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J. Biol. Chem. 280, 31283–31293 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Holt J. A., et al. , Helix 1/8 interactions influence the activity of nuclear receptor ligand-binding domains. Mol. Endocrinol. 17, 1704–1714 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Fagart J., et al. , Crystal structure of a mutant mineralocorticoid receptor responsible for hypertension. Nat. Struct. Mol. Biol. 12, 554–555 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Kolkhof P., Bärfacker L., 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Mineralocorticoid receptor antagonists: 60 years of research and development. J. Endocrinol. 234, T125–T140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juurlink D. N., et al. , Rates of hyperkalemia after publication of the randomized aldactone evaluation study. N. Engl. J. Med. 351, 543–551 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Katsu Y., Baker M. E., Progesterone activation of zebrafish mineralocorticoid receptor may influence growth of some transplanted tumors. Proc. Natl. Acad. Sci. U.S.A. 115, E2908–E2909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiilerich P., et al. , Implication of the mineralocorticoid axis in rainbow trout osmoregulation during salinity acclimation. J. Endocrinol. 209, 221–235 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Kiilerich P., Geffroy B., Valotaire C., Prunet P., Endogenous regulation of 11-deoxycorticosterone (DOC) and corticosteroid receptors (CRs) during rainbow trout early development and the effects of corticosteroids on hatching. Gen. Comp. Endocrinol. 265, 22–30 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Baker M. E., Katsu Y., 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Evolution of the mineralocorticoid receptor: Sequence, structure and function. J. Endocrinol. 234, T1–T16 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Katsu Y., Oka K., Baker M. E., Evolution of human, chicken, alligator, frog, and zebrafish mineralocorticoid receptors: Allosteric influence on steroid specificity. Sci. Signal. 11, 537 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Proszkowiec-Weglarz M., Porter T. E., Functional characterization of chicken glucocorticoid and mineralocorticoid receptors. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1257–R1268 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Šali A., Blundell T. L., Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Hasui T., et al. , Discovery of 6-[5-(4-fluorophenyl)-3-methyl-pyrazol-4-yl]-benzoxazin-3-one derivatives as novel selective nonsteroidal mineralocorticoid receptor antagonists. Bioorg. Med. Chem. 22, 5428–5445 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Van Der Spoel D., et al. , GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Schmid N., et al. , Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Malde A. K., et al. , An automated force field topology builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 7, 4026–4037 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Suino K., Daugherty J., Xu H. E., Structural and biochemical mechanisms for the specificity of hormone binding and coactivator assembly by mineralocorticoid receptor. Mol. Cell 19, 367–380 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.