Significance
Conventional metagenomic sequencing offers an unbiased approach to identify pathogens in clinical samples, but the presence of a pathogen is not synonymous with disease. Our assay simultaneously quantifies the abundance of a large range of viral and bacterial pathogens, and the degree of host tissue injury from host–microorganism interaction. This is accomplished with a genome-wide measurement of cell-free DNA methylation marks to trace the tissues of origin of cell-free DNA and quantify the degree of injury to different host tissues. We applied this assay to screen for infection of the urinary tract in kidney transplant patients and find excellent support for the utility of this assay to distinguish infection and infectious disease, and to assess the severity of disease.
Keywords: cell-free DNA, metagenomics, host injury
Abstract
High-throughput metagenomic sequencing offers an unbiased approach to identify pathogens in clinical samples. Conventional metagenomic sequencing, however, does not integrate information about the host, which is often critical to distinguish infection from infectious disease, and to assess the severity of disease. Here, we explore the utility of high-throughput sequencing of cell-free DNA (cfDNA) after bisulfite conversion to map the tissue and cell types of origin of host-derived cfDNA, and to profile the bacterial and viral metagenome. We applied this assay to 51 urinary cfDNA isolates collected from a cohort of kidney transplant recipients with and without bacterial and viral infection of the urinary tract. We find that the cell and tissue types of origin of urinary cfDNA can be derived from its genome-wide profile of methylation marks, and strongly depend on infection status. We find evidence of kidney and bladder tissue damage due to viral and bacterial infection, respectively, and of the recruitment of neutrophils to the urinary tract during infection. Through direct comparison to conventional metagenomic sequencing as well as clinical tests of infection, we find this assay accurately captures the bacterial and viral composition of the sample. The assay presented here is straightforward to implement, offers a systems view into bacterial and viral infections of the urinary tract, and can find future use as a tool for the differential diagnosis of infection.
Differential diagnosis of infectious disease in humans is complex. Metagenomic high-throughput DNA sequencing offers an unbiased approach for the detection of pathogens in clinical samples (1–4), but the presence of a pathogen is not necessarily synonymous with disease (5). Some microbes are commensals in all human hosts, some cause disease only in some hosts, and others cause disease in all hosts. To bring clarity to the lexicon of microbial pathogenesis, Casadevall and Pirofski (5) defined infectious disease as a clinical manifestation of damage to the host that results from host–microbe interaction. In this framework, the degree of host damage, mediated by the host response and/or by the pathogen, offers a quantifiable metric that can be used to distinguish between different outcomes of infection (5).
We report a high-throughput metagenomic sequencing assay that can both detect a diverse array of bacterial and viral pathogens and quantify damage to host tissues. The assay implements whole-genome bisulfite sequencing (WGBS) of cell-free DNA (cfDNA), small fragments of DNA released by host or microbial cells into blood, urine, and other bodily fluids, and brings together 2 previously reported concepts. First, the assay implements a genome-wide measurement of cytosine methylation marks comprised within cfDNA—marks that are highly cell, tissue, and organ-type specific—to determine the cell and tissue types that contribute to the mixture of host cfDNA in a sample. Several recent studies have shown that profiling CpG methylation marks in urinary or plasma cfDNA, via whole-genome sequencing, targeted sequencing, or PCR assays, can be used to determine their tissues of origin and to quantify tissue-specific injury in various diseased settings (6–8). Here, we explore this concept for the monitoring of injury due to infection. Second, the assay quantifies the relative abundance of microbes via WGBS of cfDNA. Several studies have investigated the utility of conventional, metagenomic sequencing of cfDNA for infection testing in clinical samples (2, 4, 9, 10). We show here that WGBS is compatible with such analyses.
We investigated the utility of this assay to monitor infectious complications of the urinary tract after kidney transplantation. More than 80,000 patients receive lifesaving kidney transplants worldwide each year (11). Immunosuppression after transplantation is required to manage the risk of rejection but leaves patients vulnerable to viral and bacterial infection. BK polyomavirus (BKV) infection has emerged as a serious risk factor for allograft survival. BKV reactivation occurs in up to 73% of kidney transplant recipients and leads to BK polyomavirus nephropathy (BKVN) in up to 8% of patients (12, 13). Renal biopsies are currently required to confirm BKVN and to distinguish BKVN from BKV reactivation without nephropathy (BKV+/N−). While BKVN histology is characterized by inflammation and necrosis of tissue, biopsies from BKV+/N− patients are similar to those without reactivation (12). It remains unclear whether BKV reactivation alone induces kidney damage. Bacterial urinary tract infection (UTI) affects ∼43% of kidney transplant recipients in the first 42 mo posttransplant (14). There is a disagreement in the literature regarding the appropriate balance between mitigating the risks of infectious complications and adverse effects of antimicrobial treatment for UTI. In this study, we describe a urinary cfDNA assay that can identify viral and bacterial infectious agents and can quantify the degree of host injury related to UTI.
Results
Methylation Marks Are Cell, Tissue, and Organ Type Specific.
We performed WGBS (Fig. 1A) on 51 urinary cfDNA isolates collected from a cohort of kidney transplant recipients (Fig. 1B) and used computational methods to quantify the burden of viral and bacterial cfDNA and the cell and tissue types of origin of host-derived cfDNA (15, 16) (Fig. 1C). We assayed urinary cfDNA isolates from patients who had a same-day corresponding bacterial culture (UTI positive, UTI group, n = 12; UTI negative, no-UTI group, n = 12) and from patients that were BKV positive in the blood and confirmed to have BKVN by biopsy (BKVN, n = 9), BKV positive in the blood without evidence of BKVN on biopsy (BKVN+/N−, n = 7), and negative for BKV in the blood and with normal surveillance biopsy (Normal, n = 6). In addition, we analyzed urinary cfDNA obtained from patients within the first 3 d after transplantation (Early group, n = 5). To obtain sequence information after bisulfite conversion of these molecules, we used a single-stranded sequencing library preparation (1, 17) (Fig. 1A). This library preparation employs ssDNA adapters and bead ligation to create diverse sequencing libraries from short, highly fragmented cfDNA (17, 18). We obtained 104.5 ± 43 million paired-end reads per sample, corresponding to a per-base human genome coverage of 1.4 to 4.1× (Materials and Methods).
Fig. 1.
Study methodology and cell type specificity of genome-wide methylation profiles. (A) Schematic of single-stranded library preparation method. cfDNA is denatured and treated with sodium bisulfite, which converts unmethylated cytosines (dark blue) into uracils (light blue) but not methylated cytosines (green). Bisulfite-treated DNA is first ligated to single-stranded adapters and bound to magnetic beads. Second-strand synthesis, and double-stranded adapter ligation are performed on the beads. The final step is a PCR, which converts uracils to thymines (red). (B) Pie chart with summary of samples included in this study, colored by pathology. (C) Schematic of WGBS analysis workflow. (D) PCA of reference whole-genome methylation profiles from human tissues. (E) UMAP of reference methylation profiles. Ellipses in D and E are normal 95% confidence ellipses (K means, 4 centers).
We sought to implement a reference-based approach for cell type deconvolution, thereby taking advantage of the large and growing number of genome-wide methylation profiles of tissues and cell types of interest that are available in public repositories. We downloaded 112 genome-wide methylation datasets representing 16 different tissue types (Dataset S1) (19–23) and determined tissue-specific differentially methylated regions (DMRs) using Metilene (24). We compared CpG methylation profiles of each tissue group in a 1-versus-1 approach and found 91,275 DMRs, with an average length of 453 bp. Principal-component analysis (PCA) of the methylation density measured across all DMRs revealed global tissue-specific clustering, with 3 heterogeneous clusters representing blood, gut, and a diverse group of other solid organ tissues (Fig. 1D and SI Appendix, Fig. S1). To identify both global and local structural features within reference methylomes, we applied uniform manifold approximation projection (UMAP) (25). We found that UMAP further resolves reference methylation profiles into clusters composed of specific cell types (Fig. 1 D and E). For example, among myeloid cells, cell types with similar lineages such as macrophages and monocytes clustered more closely than those from other lineages on the UMAP projection. These analyses confirm that genome-wide methylation profiles are cell, tissue, and organ-type specific, and can in principle inform its tissue of origin, as described previously (6, 7, 26–28).
Cell-Free DNA Origin Associated with Infection.
To determine methylation marks comprised within urinary cfDNA, we first aligned the WGBS reads to a human reference genome via bwa-meth (29). We quantified the efficiency of bisulfite conversion achieved in experiments from the fraction of reported methylated C[A/T/C] base pairs, which are rarely methylated in humans (30). We found a conversion efficiency greater or equal to 94.5% for all cfDNA isolates (Materials and Methods). We next projected the genome-wide cfDNA CpG methylation profiles onto the 2D feature spaces generated by PCA and UMAP for the 112 public references. We found that cfDNA profiles organized between the cluster that comprised kidney tissue and the white blood cell cluster on the PCA and UMAP 2D projections (Fig. 2 A and B). This observation provided a first indication that urinary cfDNA originates primarily from blood cell types and kidney tissue. Other cell types found in the urinary tract contributed less significantly to urinary cfDNA.
Fig. 2.
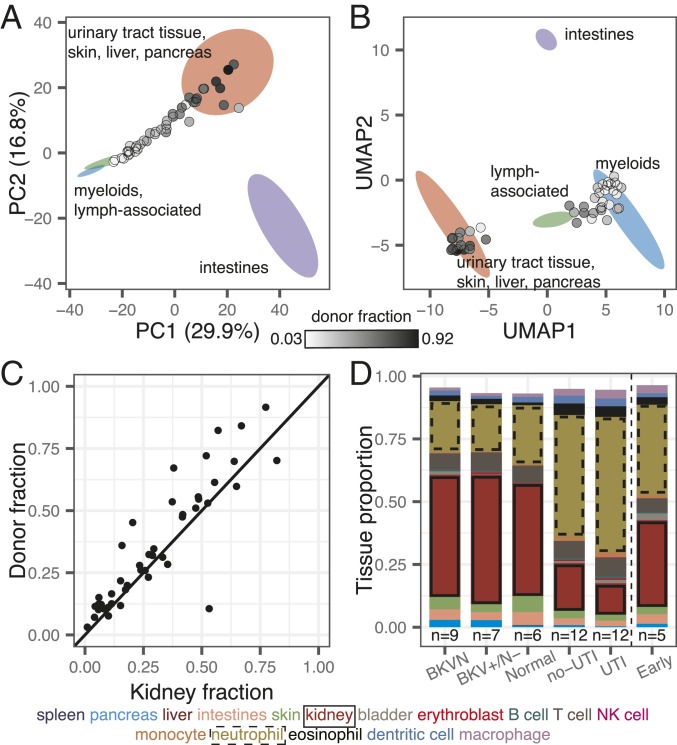
Methylation marks within cfDNA inform its tissues of origin. (A and B) Projection of cfDNA genome-wide methylation profiles onto PCA and UMAP feature space (from Fig. 1 D and E). Data points colored according to donor fraction (n = 44). (C) Comparison of the proportion of kidney-derived cfDNA and donor-derived cfDNA measured for sex-mismatched donor–recipient pairs (n = 44). Spearman’s rho = 0.88; P < 2.2 × 10−16. Samples from dual bone marrow and kidney transplants are excluded. (D) Barplot of cfDNA cell and tissue type composition measured for clinical groups.
We next computed the proportion of transplant donor-derived cell-free DNA (ddcfDNA) in these samples. We and others have identified ddcfDNA as a noninvasive, quantitative marker of graft injury in solid-organ transplantation (2, 4, 31, 32). For sex-mismatched donor–recipient pairs, the proportion of ddcfDNA can be estimated by evaluating the coverage of the Y chromosome relative to the autosomal chromosomes (Materials and Methods) (2, 33). We verified that the proportion of ddcfDNA measured by sequencing of bisulfite-treated cfDNA matched the proportion of ddcfDNA measured using conventional sequencing (n = 36 matched samples, Spearman’s rho = 0.97, P < 2.2 × 10−16; Materials and Methods and SI Appendix, Fig. S2), and then quantified the proportion of ddcfDNA in urine for all sex-mismatched donor–recipient transplant pairs (n = 46). We observed a very large range of ddcfDNA values across all samples (3 to 99%). Superimposing the ddcfDNA proportion on the PCA and UMAP projections revealed that samples with a higher proportion of ddcfDNA landed closer to the reference cluster composed of kidney tissue (Fig. 2 A and B; n = 44; 2 samples from a patient that received both a kidney and bone marrow transplant were excluded from this analysis, because the donor fraction represents the summation of kidney DNA and engrafted bone marrow-derived cells for this case). This observation provided a second line of evidence that urinary cfDNA is a mixture of cfDNA derived from blood cell types and kidney tissue.
To quantify the contributions of different tissues to the mixture of cfDNA in urine, we implemented quadratic programming. Quadratic programming retrieves the fractional contribution of each tissue, πi, from the ensemble cfDNA methylation profile Y, and the public reference methylation profile for each tissue, Xi: Y = πiXi + ε, where ε is an error term (Materials and Methods) (34). Using this approach, we found an excellent quantitative agreement (Spearman’s rho = 0.88, P < 2.2 × 10−16; Fig. 2C) between the proportion of kidney-specific cfDNA (determined from methylation marks) and ddcfDNA (determined from genetic marks in cfDNA) for sex-mismatched donor–recipient transplant pairs (n = 44). This analysis provided support for the use of our bioinformatic and molecular approaches to quantify the tissue and cell types of origin of cfDNA in urine.
We proceeded to analyze the relative contributions of all cell and tissue types comprised within the pure cell and tissue references against clinical tests of infection. We found that the cfDNA cell and tissue composition was associated with infection status (Fig. 2D). For example, the relative contribution of kidney-derived cfDNA was elevated in samples from patients with BKV infection compared to patients diagnosed with bacterial UTI (P = 2.0 × 10−4; mean of 48.6% and mean of 12.5%, respectively; Fig. 2D). We further found that leukocytes were enriched in samples from patients diagnosed with bacterial UTI by conventional culture, and that neutrophils are the main contributors to the differences in white blood cell content, as expected from their role as first responders to infection (Fig. 2D).
The relative proportion of cfDNA from a specific tissue is a function of the proportion of DNA released from all other cell types or tissues and may therefore be a convoluted measure of tissue-specific injury. To overcome this limitation, we computed the absolute concentration of tissue-specific cfDNA by multiplying the proportion of tissue and cell type-specific DNA obtained using the approaches above with the concentration of total host-derived cfDNA in the sample (Materials and Methods). We observed marked temporal dynamics of the concentration of cfDNA from different cell and tissue types in absence of infection (no-UTI, Normal and Early groups, Fig. 3A). Recovery of postoperative stress in the first 3 d after transplantation was associated with a marked increase in cfDNA from most tissues. This signal of early postoperative injury decayed to a low baseline within 10 d after transplantation, and after 3 mo posttransplantation quiescence was observed with markedly low amount of cfDNA from all cell and tissue types in absence of infection.
Fig. 3.
Cell type deconvolution of urinary cfDNA reveals host response to infection. (A) Concentration of tissue-specific cfDNA for samples with no clinical indication of infection. Triangles represent mean of multiple measurements at the same time point; black circles indicate time points with a single measurement. Error bars indicate the uncertainty of the mean (SE). (B and C) Kidney (B) and leukocyte (C) cfDNA concentration for BKVN, BKV+/N−, Normal and Early groups. (D) Bladder cfDNA concentration for no-UTI and UTI groups. Inset shows receiver operating characteristic analysis of the performance of bladder cfDNA in distinguishing no-UTI from UTI groups (blue) and in distinguishing hematuria (RBC per HPF > 4) from no hematuria (RBC per RBC ≤ 4) in samples from the UTI group. TPR: true positive rate; FPR: false positive rate. (E) Serum creatinine versus kidney urinary cfDNA concentration (n = 50). Spearman’s rho = 0.51; P = 1.4 × 10−4. (F) Mean host cfDNA fragment length versus leukocyte proportion (n = 51). Spearman’s rho = −0.45; P = 1.1 × 10−3. Inset shows boxplots of fragment lengths measured for the UTI and no-UTI groups. Boxplot features detailed in Materials and Methods. *P < 0.05, **P < 0.01, and ***P < 0.001.
We examined whether the cfDNA concentration of certain tissues was associated with infection pathology. We first examined all samples from patients that were screened for BKVN via needle biopsy (all samples collected after day 100). We observed marked differences in the concentration of kidney-derived cfDNA for samples from patients diagnosed with BKVN (BKVN vs. Normal; mean kidney cfDNA, 21.9 and 1.5 ng/mL, respectively; P = 4.0 × 10−4; Fig. 3B), indicating significant tissue injury associated with BKVN. In addition, we found that this cfDNA measurement can distinguish BKVN from BKV reactivation without nephropathy (BKVN vs. BKV+/N−, mean kidney cfDNA, 21.9 and 6.9 ng/mL, respectively; P = 7.9 × 10−3), and BKV+/N− from Normal (mean kidney cfDNA, 6.9 and 1.5 ng/mL, respectively; P = 1.2 × 10−3). The concentration of leukocyte cfDNA was elevated in urine from patients diagnosed with BKVN (mean, 19.0 vs. 1.9 ng/mL in Normal; P = 2.8 × 10−3) and BKV+/N− (mean, 4.0 vs. 1.9 ng/mL; P = 3.5 × 10−2) but could not distinguish BKVN from BKV reactivation without nephropathy (P = 5.5 × 10−2) (Fig. 3C). Together, these experiments point to the utility of the assay presented here to noninvasively distinguish nephropathy and inflammation due to BK virus infection. The kidney cfDNA concentration in samples collected from patients within the first 3 d after transplantation, a period during which we observed significant postoperative injury (Fig. 3A), was also elevated (12.4 ng/mL) and could not be differentiated from the concentration measured in samples from patients with BK disease (P = 0.30 and 0.20 when compared to BKVN and BKV+/N−, respectively). This last observation underlines the importance of paired metagenomic evaluation to distinguish infection- and non–infection-related host injury.
We next evaluated all samples from patients that were screened for UTI via conventional urine culture (no-UTI group and UTI group, n = 12 each). These samples were collected between days 8 and 55 posttransplant, a period in which we observed significant host injury in absence of infection (Fig. 3A), and were evaluated for hematuria by microscopy, a clinical marker of injury. We found that both bladder and leukocyte-derived cfDNA were elevated in samples from patients diagnosed with bacterial UTI and hematuria (red blood cell [RBC] counts per high power field [HPF] greater than 4) compared to patients diagnosed with UTI in the absence of hematuria (Fig. 3D; receiver operator characteristic analysis, area under the curve [AUC] = 0.91 for bladder cfDNA). We further found correlations between RBC counts and the concentrations of bladder cfDNA in urine (all samples for which RBC per HPF was measured, n = 24; Spearman’s rho = 0.43, P = 3.5 × 10−2). Together, these observations demonstrate the utility of our assay to assess the severity of injury due to bacterial UTI. The performance of the concentration of bladder cfDNA in distinguishing bacterial UTI and absence of UTI with and without hematuria was modest (AUC = 0.64 for bladder cfDNA; Fig. 3D), which is likely explained by the significant noninfection related injury in this patient population in the sample time window.
Last, we asked whether the concentration of kidney-derived cfDNA correlated with serum creatinine, a clinical marker of kidney function. Creatinine is a waste product of muscle metabolism and elevated serum creatinine levels are an indication of poor kidney function. We found good agreement between kidney-specific cfDNA in urine and serum creatinine (all samples for which serum creatinine was measured, n = 50; Spearman’s rho = 0.51, P = 1.5 × 10−4). Together, the data presented in Fig. 3 provide strong support for the use of WGBS of cfDNA to quantify host injury due to viral and bacterial UTI.
The pattern of degradation of cfDNA in plasma has previously been shown to depend on their origin and pathology. For example, tumor-derived cfDNA was found to be significantly shorter than cfDNA from normal tissue (35, 36). Here, we sized urinary cfDNA using paired-end read mapping (Materials and Methods). Bisulfite treatment is known to lead to DNA degradation (37), and this was corroborated by our cfDNA sizing assay. We found that bisulfite-treated cfDNA is on average 10.1 bp shorter than untreated cfDNA (n = 38 matched samples). We furthermore found that the mean fragment length of host-derived cfDNA was negatively correlated with the white blood cell proportion (Spearman’s rho = −0.45; P = 1.1 × 10−3; n = 51) and was shorter for samples from patients with bacterial infection than from patients without bacterial UTI (P = 2.4 × 10−2; mean length of 93 and 101 bp, respectively; Fig. 3F and SI Appendix, Fig. S3). This observation is in line with previous reports that leukocytes create a degradative environment for DNA (38, 39). The fragment size profile of cfDNA may offer an additional metric by which patients with different infectious pathologies can be stratified.
WGBS of cfDNA Identifies Clinically Relevant Pathogens.
Bisulfite conversion of DNA followed by PCR converts unmethylated cytosines into thymines. A corollary of bisulfite treatment is a decrease in cytosine content, and a reduction in overall read complexity. To determine whether WGBS can be used to identify specific uropathogens despite the reduction in sequence complexity inherent to bisulfite conversion, we compared pathogen abundances measured after shotgun sequencing of bisulfite-treated and untreated cfDNA (matched samples, n = 38). We determined the relative representation of bacteria and viruses in these datasets, using approaches previously described (2, 3, 40) (Materials and Methods). We computed the representation of microbial genomes relative to human genome copies and expressed this quantity as relative genome equivalents (RGE) (Materials and Methods). Fig. 4A shows a close quantitative agreement between the species abundance measured for bisulfite-treated and untreated cfDNA, confirming that it is possible to broadly identify microbial cfDNA via shotgun sequencing of bisulfite-treated cfDNA (Fig. 4A; Spearman’s rho = 0.72, P < 2.2 × 10−16).
Fig. 4.
WGBS of microbial cfDNA for clinical pathogen identification. (A) Scatterplot of relative genomic abundance of bacteria (green) and viruses (purple) measured by WGBS and conventional cfDNA sequencing. Spearman’s rho = 0.72; P < 2.2 × 10−16. Each data point represents the genomic abundance of a single microbe in matched bisulfite and untreated urinary cfDNA. (B) Violin plots of BKV sequence abundance in all samples. (C) Relative genomic abundance of microbes identified through urine culture. (D) Heat map of tissue proportions in samples where a microbe was detected through WGBS. Rows are hierarchically clustered according to tissue composition.
We assessed the relative genomic abundance of cfDNA from bacterial and viral pathogens identified by conventional diagnostic assays. In 9 out of 9 BKVN samples and 6 out of 7 BKV+/N− samples, we identified high BK viral loads (RGE > 103; Fig. 4B). These values correlated strongly with matched plasma BKV copies as determined by quantitative PCR (Spearman’s rho = 0.81; P = 6.2 × 10−6). We next compared the relative genomic abundance of bacterial cfDNA for patients diagnosed with bacterial infection (12 samples matched with a positive clean-catch midstream urine culture, UTI group), to the relative genomic abundance measured for 12 negative clean-catch midstream urine cultures (no-UTI group). Of these positive urine cultures, 11 had a single identifiable bacterium (Escherichia coli, n = 4; Enterococcus faecalis, n = 3; Pseudomonas aeruginosa, n = 2; Enterococcus faecium, n = 1; Klebsiella oxytoca, n = 1), while a single sample presented 2 different bacterial species (E. faecalis and coagulase-negative Staphylococcus). We found agreement between the urinary cfDNA assay described here and conventional bacterial culture (detection accuracy, 100%; no information rate, 16.0%; P [accuracy > no information rate] < 2.2 × 10−16; Fig. 4C). In 1 sample associated with an E. coli-positive urine culture, we also detected E. faecalis and Staphylococcus, at low and high abundance, respectively (Fig. 4C). These organisms were detected at similar abundance in the same sample using conventional metagenomic cfDNA sequencing (RGE of 0.25 and 3.9 by conventional sequencing versus 0.17 and 2.5 by WGBS for E. faecalis and Staphylococcus, respectively), but not by conventional culture, which may be due to the limited sensitivity of conventional urine culture (2). Together, these results support the use of WGBS of cfDNA as an assay to screen for potential pathogens in clinical isolates.
Finally, we tested whether the cfDNA cell and tissue type composition depended on the presence or absence of viral and bacterial pathogens as determined by WGBS. We used unsupervised hierarchical clustering of the cfDNA cell and tissue type composition for all samples in which BK virus (RGE > 103) or a potential bacterial uropathogen was detected (RGE > 0.09, the lowest corresponding relative genomic abundance observed in the comparison to clinical metrics described above). This analysis, shown in Fig. 4D, summarizes the major layers of information that are made accessible with the cfDNA assay reported here, and shows that the cfDNA tissue and cell type composition is associated with the presence or absence of viral or bacterial uropathogens.
Discussion
We have described a metagenomic assay that simultaneously quantifies the abundance of a large array of viruses and bacteria in clinical samples, and the degree of host injury. This work is motivated by the need to integrate information about host–microbe interactions in clinical metagenomic assays in order to distinguish infection from infectious disease, and to assess the severity of disease. The assay reported here takes advantage of genome-wide profiling of CpG methylation marks comprised within cfDNA to quantify the contributions of different cell and tissue types to the mixture of cfDNA in the sample, and thereby the degree of host damage. Compared to conventional metagenomic cfDNA assays (2, 10), this assay requires a single additional experimental step, bisulfite treatment of the cfDNA isolate, which is inexpensive and can be completed within approximately 2 h. The library preparation requires DNA isolated from just 1 mL of urine and costs approximately $65. A minimum number of 60 million paired-end reads were collected per sample, which can be achieved for $100 to $300, depending on the sequencing platform used. We expect advances in DNA sequencing technology to further decrease the cost of this assay.
We tested the utility of this assay to monitor viral and bacterial infections of the urinary tract in a cohort of kidney transplant recipients. We found that the concentration of cfDNA derived from different cell and tissue types was a function of infection status in these patients. Patients diagnosed with BKVN had elevated kidney-specific cfDNA in urine compared to a normal control group. BKV reactivation without nephropathy was also characterized by elevated levels of kidney-specific DNA but not to the same degree as BKVN. Our findings suggest that there may be kidney damage occurring before the onset of nephropathy. Biopsies provide information only on the sampled region of the kidney and do not capture the inherent heterogeneity of BKV infection within the kidney allograft and disease progression. The assay described here may find use as a noninvasive alternative to conventional biopsy to screen for BKV-related kidney injury.
In addition, patients with bacterial UTIs show higher neutrophil contributions, suggesting immune activation and recruitment of neutrophils to the urinary tract, as well as elevated amounts of bladder cfDNA, indicating tissue injury. A common question in infectious diseases is how to interpret positive urine cultures. Outside of specific indications such as pregnancy, urological procedures, and being within 3 mo of transplant (41, 42), a positive urine culture is currently treated with antibiotics only in symptomatic individuals. By quantifying the release of cfDNA from different cell types and tissues, the assay reported here can provide clinicians with additional information to guide treatment decisions.
A quantitative measurement of the tissues of origin of cfDNA constitutes a generalizable noninvasive approach to identify injury to vascularized tissues and participation of the immune system and can find wide application in diagnostic medicine. Several studies have reported technologies to trace the tissue origin of cfDNA in a wide range of settings, targeting different epigenetic marks comprised within cfDNA, including the footprints of nucleosomes and transcription factors, cytosine methylation and hydroxymethylation (6–8, 43, 44). Here, we have applied this technology to the monitoring of host tissue damage due to infection.
Host-based molecular signatures have previously been considered as classifiers of infectious complications. In respiratory infection, host transcriptional profiling of the peripheral blood has been shown to provide a means to characterize the host response to viral and bacterial infection, and to discriminate between infectious and noninfectious states (45). Host-response molecular signatures thereby offer a diagnostic approach orthogonal to approaches that focus on viral and bacterial pathogens. Recently, Langelier et al. (46) described a diagnostic approach that combines transcriptional profiling and metagenomic sequencing of tracheal aspirate to classify lower respiratory tract infection. Here, we show that a cfDNA assay can be used to screen for infectious agents and to quantify the degree of host damage. The strength of this assay lies in its simplicity of implementation, its noninvasiveness, and its ability to directly interrogate host damage, the metric that is most relevant to classify infectious complications in the framework proposed by Casadevall and Pirofski (5).
In summary, we propose that WGBS of cfDNA can be used as a metagenomic sequencing assay to provide in-depth understanding of both the metagenome as well as the host injury response to infection. This assay is generalizable to multiple disease states and has the potential to distinguish colonization from infectious disease in a clinical setting.
Materials and Methods
Fifty-one urine samples were collected from 36 kidney transplant recipients treated at NewYork-Presbyterian Hospital–Weill Cornell Medical Center. The study was approved by the Weill Cornell Medicine Institutional Review Board (protocols 9402002786, 710009490, and 1207012730). All patients provided written informed consent. Clinical tests, urine collection, cfDNA extraction, sequencing protocols, and analysis workflows are described in detail in SI Appendix, SI Materials and Methods.
Data Availability.
The sequencing data that support the findings of this study are available in the database of Genotypes and Phenotypes (dbGaP) (accession number phs001564.v1.p1).
Code Availability.
Custom scripts are available at https://github.com/alexpcheng/bisulfite_cfDNA.
Supplementary Material
Acknowledgments
We thank Peter Schweitzer and colleagues at the Cornell Genomics Facility for help with sequencing assays. This work was supported by NIH Grant 1DP2AI138242 (to I.D.V.), NIH Grant 1R21AI133331 (to I.D.V. and J.R.L.), NIH Grant R37AI051652 (to M.S.), a National Sciences and Engineering Research Council of Canada fellowship (401236174) (to A.P.C.), and National Science Foundation Graduate Research Fellowship Program Grant DGE-1144153 (to P.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.T. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the database of Genotypes and Phenotypes (dbGaP) (accession no. phs001564). Scripts are available at https://github.com/alexpcheng/bisulfite_cfDNA.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906320116/-/DCSupplemental.
References
- 1.Weber G., Shendure J., Tanenbaum D. M., Church G. M., Meyerson M., Identification of foreign gene sequences by transcript filtering against the human genome. Nat. Genet. 30, 141–142 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Burnham P., et al. , Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 9, 2412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vlaminck I., et al. , Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155, 1178–1187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vlaminck I., et al. , Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. U.S.A. 112, 13336–13341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A., Pirofski L. A., Host-pathogen interactions: Basic concepts of microbial commensalism, colonization, infection, and disease. Infect. Immun. 68, 6511–6518 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun K., et al. , Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann-Werman R., et al. , Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E1826–E1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann-Werman R., et al. , Specific detection of cell-free DNA derived from intestinal epithelial cells using methylation patterns. bioRxiv:10.1101/409219 (5 September 2018).
- 9.Wilson M. R., et al. , Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 370, 2408–2417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blauwkamp T. A., et al. , Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4, 663–674 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Global Observatory on Donation and Transplantation , Activity data report—GODT (2016). http://www.transplant-observatory.org/download/2016-activity-data-report/. Accessed 12 February 2019.
- 12.Hirsch H. H., et al. , Polyomavirus-associated nephropathy in renal transplantation: Interdisciplinary analyses and recommendations. Transplantation 79, 1277–1286 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Drachenberg C. B., et al. , Polyomavirus BK versus JC replication and nephropathy in renal transplant recipients: A prospective evaluation. Transplantation 84, 323–330 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Chuang P., Parikh C. R., Langone A., Urinary tract infections after renal transplantation: A retrospective review at two US transplant centers. Clin. Transplant. 19, 230–235 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Cheng A. P., bisulfite_cfDNA. Github. https://github.com/alexpcheng/bisulfite_cfDNA. Deposited 15 April 2019.
- 16.Cheng A. P., Burnham P., Lee J. R., Suthanthiran M., Dadhania D., De Vlaminck I., Structure and diversity of urinary cell-free DNA informative of host-pathogen interactions in human urinary tract infectionof genotypes and phenotypes. https://www.ncbi.nlm.nih.gov/bioproject/445347. Deposted 7 August 2019.
- 17.Burnham P., et al. , Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci. Rep. 6, 27859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gansauge M.-T., Meyer M., Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Bujold D., et al. , The International Human Epigenome Consortium data portal. Cell Syst. 3, 496–499.e2 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Fernández J. M., et al. ; BLUEPRINT Consortium , The BLUEPRINT data analysis portal. Cell Syst. 3, 491–495.e5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albrecht F., List M., Bock C., Lengauer T., DeepBlue epigenomic data server: Programmatic data retrieval and analysis of epigenome region sets. Nucleic Acids Res. 44, W581–W586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein B. E., et al. , The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium , The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Jühling F., et al. , Metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 26, 256–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McInnes L., Healy J., UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 (6 December 2018).
- 26.Lokk K., et al. , DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 15, r54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng T. H. T., et al. , Genomewide bisulfite sequencing reveals the origin and time-dependent fragmentation of urinary cfDNA. Clin. Biochem. 50, 496–501 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Guo S., et al. , Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 49, 635–642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen B. S., Eyring K., De S., Yang I. V., Schwartz D. A., Fast and accurate alignment of long bisulfite-seq reads. arXiv:1401.1129 (13 May 2014).
- 30.He Y., Ecker J. R., Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 16, 55–77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Vlaminck I., et al. , Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 6, 241ra77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck J., et al. , Donor-derived cell-free DNA is a novel universal biomarker for allograft rejection in solid organ transplantation. Transplant. Proc. 47, 2400–2403 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Lo Y. M. D., et al. , Presence of fetal DNA in maternal plasma and serum. Lancet 350, 485–487 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Koh W., et al. , Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl. Acad. Sci. U.S.A. 111, 7361–7366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underhill H. R., et al. , Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouliere F., et al. , Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K., Okamoto A., Degradation of DNA by bisulfite treatment. Bioorg. Med. Chem. Lett. 17, 1912–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Nakazawa D., et al. , The responses of macrophages in interaction with neutrophils that undergo NETosis. J. Autoimmun. 67, 19–28 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Lamers M. C., De Groot E. R., Roos D., Phagocytosis and degradation of DNA-anti-DNA complexes by human phagocytes. I. Assay conditions, quantitative aspects and differences between human blood monocytes and neutrophils. Eur. J. Immunol. 11, 757–764 (1981). [DOI] [PubMed] [Google Scholar]
- 40.Xia L. C., Cram J. A., Chen T., Fuhrman J. A., Sun F., Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS One 6, e27992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolle L. E., et al. ; Infectious Diseases Society of America; American Society of Nephrology; American Geriatric Society , Infectious Diseases Society of America guidelines for the diagnosis and treatment of asymptomatic bacteriuria in adults. Clin. Infect. Dis. 40, 643–654 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Parasuraman R., Julian K.; AST Infectious Diseases Community of Practice , Urinary tract infections in solid organ transplantation. Am. J. Transplant. 13 (suppl. 4), 327–336 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Snyder M. W., Kircher M., Hill A. J., Daza R. M., Shendure J., Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song C.-X., et al. , 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 27, 1231–1242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsalik E. L., et al. , Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 8, 322ra11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langelier C., et al. , Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. U.S.A. 115, E12353–E12362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data that support the findings of this study are available in the database of Genotypes and Phenotypes (dbGaP) (accession number phs001564.v1.p1).