Significance
Heart regeneration is restricted to the first week after birth in mice, and the underlying molecular mechanisms remain poorly understood. We compared the transcriptomes and epigenomes of the regenerative and nonregenerative mouse hearts over a 7-d time period after myocardial infarction. We show that injury of the regenerative heart triggers a unique immune response involving a macrophage-secreted factor, CCL24, that promotes regeneration. We also describe a developmental gene program that is active in the regenerative heart, within which we found an RNA-binding protein, IGF2BP3, that enhances regeneration. Our study reveals a detailed blueprint of the transcriptional basis of heart regeneration and represents a resource for the identification of genes that may facilitate cardiac repair in response to injury.
Keywords: transcriptome profiling, epigenome profiling, immune response, myocardial infarction, cardiogenic gene program
Abstract
The adult mammalian heart has limited capacity for regeneration following injury, whereas the neonatal heart can readily regenerate within a short period after birth. To uncover the molecular mechanisms underlying neonatal heart regeneration, we compared the transcriptomes and epigenomes of regenerative and nonregenerative mouse hearts over a 7-d time period following myocardial infarction injury. By integrating gene expression profiles with histone marks associated with active or repressed chromatin, we identified transcriptional programs underlying neonatal heart regeneration, and the blockade to regeneration in later life. Our results reveal a unique immune response in regenerative hearts and a retained embryonic cardiogenic gene program that is active during neonatal heart regeneration. Among the unique immune factors and embryonic genes associated with cardiac regeneration, we identified Ccl24, which encodes a cytokine, and Igf2bp3, which encodes an RNA-binding protein, as previously unrecognized regulators of cardiomyocyte proliferation. Our data provide insights into the molecular basis of neonatal heart regeneration and identify genes that can be modulated to promote heart regeneration.
Heart disease is the leading cause of death worldwide (1). In response to a myocardial infarction (MI), the adult human heart can lose up to a billion cardiomyocytes (CMs) that cannot be replenished due to the inability of adult CMs to proliferate. Loss of CMs leads to diminished cardiac contractility, scar formation, and heart failure with potentially fatal complications (2). In contrast, the neonatal mouse heart can efficiently regenerate following apical resection as well as MI, but this capacity is lost as early as postnatal day 7 (P7) (3, 4). Deciphering the molecular underpinnings of neonatal heart regeneration and the blockade to regeneration in later life may provide new strategies for heart repair.
Neonatal heart regeneration is orchestrated by multiple cell types, including cardiac resident cells and immune cells that infiltrate the heart after injury (5). Macrophages promote heart regeneration via a paracrine effect (6, 7), and neural innervation has been shown to be required for neonatal heart regeneration (8). Although these studies underscore the importance of multiple cell types for neonatal heart regeneration, this process ultimately relies on the activation of CM proliferation following injury. It has been shown that a significant proportion of CMs in the neonatal heart remain proliferative but exit the cell cycle during the first week after birth (9, 10). The timing of CM cell cycle withdrawal is concurrent with the loss of regenerative capacity of the postnatal heart. Whether the ability for CMs to proliferate after injury relies on their retained proliferation potential or injury-induced environmental cues unique to regenerative hearts, or both, remains poorly understood.
Here, we examine the transcriptome and active chromatin landscapes in regenerative and nonregenerative mouse hearts, and their responses to cardiac injury during an initial 7-d time course. Our analyses identified genes and biological processes that are uniquely activated in injured regenerative hearts, among which the immune response is dramatically distinct between regenerative and nonregenerative hearts. From the secreted factors expressed in regenerative neonatal hearts, we found that C-C motif chemokine ligand 24 (CCL24) protein can promote CM proliferation. Furthermore, we showed that the embryonic cardiac gene program is retained during the neonatal regenerative time window, and identified insulin-like growth factor 2 messenger RNA-binding protein 3 (Igf2bp3), which encodes an RNA-binding protein, as a developmental regulator of postnatal CM proliferation. Our data suggest that neonatal heart regeneration is associated with specific injury responses and expression of cardiac developmental genes. The molecular decoding of cardiac regeneration provides numerous inroads whereby this process could be therapeutically manipulated to enhance cardiac function in response to injury.
Results
Transcriptomic Changes during Neonatal Heart Regeneration.
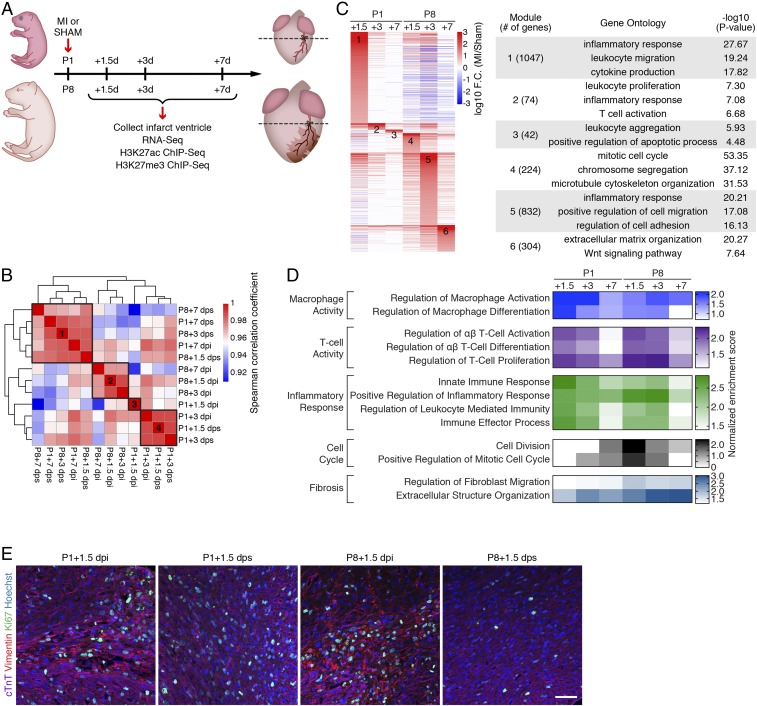
To investigate the mechanistic basis of neonatal heart regeneration, we profiled changes in gene expression spanning the first 7 d post-MI injury in regenerative and nonregenerative mouse hearts at P1 and P8, respectively. P1 and P8 hearts were subjected to permanent ligation of the left anterior descending (LAD) artery or sham surgery; ventricles were collected at 1.5, 3, and 7 d postinjury (dpi) or postsham (dps); and gene expression was analyzed by RNA sequencing (RNA-Seq) (Fig. 1A). Trichrome staining revealed massive loss of CMs in both P1 and P8 hearts at 1.5 dpi (SI Appendix, Fig. S1A). However, P1 hearts showed progressive regeneration and recovery, with minimal fibrosis at 7 dpi. In contrast, P8 hearts exhibited extensive fibrosis, wall thinning, and ventricular dilation at 3 and 7 dpi (SI Appendix, Fig. S1A). For RNA-Seq analysis, heart tissue was harvested below the LAD ligation plane. Analysis of gene expression in hearts subjected to MI or sham surgery identified differentially expressed genes (DEGs) that showed either higher expression (MI-up genes) or lower expression (MI-down genes) in MI samples compared with age-matched sham control samples (SI Appendix, Fig. S1B). Examination by Venn diagram of MI-up genes in P1 and P8 hearts showed that the majority of the MI-up genes were unique to a specific time point. Moreover, of all of the 2,523 MI-up genes in P1 and P8 hearts, 621 genes were shared, 874 genes were unique to the P1 hearts, and 1,028 genes were unique to the P8 hearts (SI Appendix, Fig. S1C).
Fig. 1.
Transcriptomic analysis reveals distinct and stage-specific responses in regenerative and nonregenerative hearts. (A) Schematic illustration of experimental design and time points for sample collection. (B) Spearman correlation of RNA-Seq datasets showing the transcriptomic similarity among samples. Four distinct clusters were identified. (C, Left) Heatmap showing log10 (fold change [F.C.]) of genes induced by MI. Fold change was calculated by comparing the expression of each gene in MI samples over sham samples for each time point. MI-induced genes from each time point were merged and sorted into 6 groups based on the time point that has the highest fold change. (C, Right) Selected top enriched GO terms for MI-up genes from clusters 1 to 6. (D) Heatmap showing normalized enrichment scores of selected biological pathways that were significantly enriched from the GSEA. (E) Immunostaining of cTnT (purple), vimentin (red), and Ki67 (green) proteins on transverse sections at the level of suture from P1+1.5 dpi, P1+1.5 dps, P8+1.5 dpi, and P8+1.5 dps hearts. Nuclei were counterstained with Hoechst (blue). (Scale bar, 50 μm.)
Hierarchical clustering of all of the samples based on their transcriptomic similarities segregated them into 4 clusters (Fig. 1B and SI Appendix, Fig. S1D). The P1+1.5 dpi sample formed its own cluster (cluster 3), reflecting the unique MI response in P1 hearts. Interestingly, the P1+3 dpi sample clustered with P1+1.5 dps and P1+3 dps controls (cluster 4), whereas the P1+7 dpi sample clustered with its corresponding sham control and all sham samples from P8 (cluster 1). This suggests that by 3 and 7 d after MI, the regenerative hearts are already similar to the uninjured hearts of the same age with respect to gene expression patterns. P8 hearts post-MI formed a distinct cluster (cluster 2). The uninjured P1 and P8 hearts clustered into different groups (cluster 4 and cluster 1, respectively), indicating transcriptional changes reflective of postnatal development of the heart (Fig. 1B). These findings indicated that the key transcriptional regulation of regeneration occurs in the initial response following cardiac injury, and that the P1 regenerative hearts rapidly revert to a gene expression pattern similar to that of uninjured control samples.
Distinct Injury-Responsive Pathways Associated with Neonatal Heart Regeneration.
To explore the injury-responsive genes unique to the P1 heart, we clustered the MI-up genes based on the time point when they showed highest expression relative to age-matched sham controls. This clustering analysis identified 6 modules of genes that were preferentially up-regulated at specific time points following injury of P1 and P8 hearts (Fig. 1C). For the P1 time points, gene ontology (GO) analysis identified immune processes, such as the inflammatory response and leukocyte activation, as the top GO terms. This is consistent with published reports that the innate immune response, mediated by macrophages, is essential for neonatal heart regeneration (6, 11). In contrast, the top enriched GO terms for the P8 time points included cell cycle process at +1.5 dpi, cell migration and adhesion at +3 dpi, and extracellular matrix organization at +7 dpi, reflecting active fibrotic remodeling of the injured P8 hearts (Fig. 1C). Clustering of the MI-down genes also revealed time point-specific responses (SI Appendix, Fig. S1E). MI-down genes at P1+1.5 dpi were associated with metabolic processes, including fatty acid metabolism and tricarboxylic acid (TCA) cycle. In contrast, MI-down genes at P8 time points were related to ion transport and muscle contraction, reflecting the compromised cardiac function of injured, nonregenerative hearts (SI Appendix, Fig. S1E).
We used gene set enrichment analysis (GSEA) to identify dynamically regulated biological processes in P1 and P8 hearts after injury (12) (Fig. 1D). Immune response pathways were highly enriched for both P1 and P8 time points. Intriguingly, the enrichment scores of the immune response pathways peaked in P1 hearts at 1.5 dpi, whereas in P8 hearts, the peak enrichment was observed at 3 dpi. In addition, P8 hearts showed strong enrichment of fibrotic remodeling processes at 3 and 7 dpi. Counterintuitively, cell cycle processes were induced in P8 hearts at 1.5 dpi, but were not induced in regenerative P1 hearts following injury. This may be attributed to the existing high levels of cell cycle-related gene expression at the regenerative stage when not all CMs withdraw from the cell cycle (9, 13). Indeed, our RNA-Seq data suggest that cell cycle-related genes are highly expressed at P1+1.5 and P1+3, regardless of MI or sham, and become down-regulated starting at P8 (P1+7 dps) when the regeneration capacity is lost (SI Appendix, Fig. S1F). In addition, immunohistochemistry of P1 and P8 hearts at 1.5 d after MI showed augmented cell cycle activation in fibroblasts of P8 hearts, whereas cell cycle activity was primarily detected in the CMs of P1 hearts (Fig. 1E). These data indicate that the endogenous cell cycle gene program is active in P1 CMs during heart regeneration. In contrast, the transient MI-induced cell cycle gene expression observed in the P8 hearts reflects fibroblast activation that contributes to cardiac fibrosis, as previously reported (14). Together, these findings reveal distinct injury-response processes and transcriptional dynamics in regenerative hearts.
Active Chromatin Regions Associated with Neonatal Heart Regeneration.
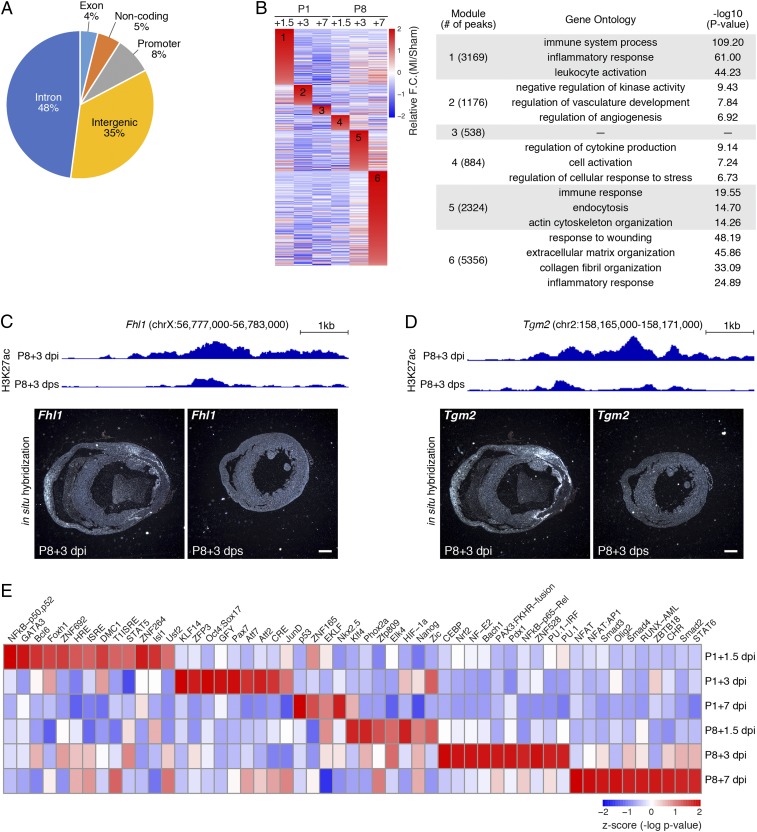
To understand the regulatory mechanisms underlying the distinct transcriptomic changes in response to injury, active chromatin regions of P1 and P8 hearts were profiled by chromatin immunoprecipitation followed by sequencing (ChIP-Seq) for histone H3 lysine-27 acetylation (H3K27ac), which marks active enhancers and promoters (Fig. 1A). H3K27ac signals were detected at the genomic loci of cardiac marker genes, such as Myh6, Tnnt2, Nppb, and Nppa, at similar levels in replicate samples, validating data reproducibility (SI Appendix, Fig. S2 A–C). Comparative analysis of the H3K27ac peaks between MI and sham samples of P1 and P8 hearts revealed genomic regions that showed increased (MI-gained) or decreased (MI-lost) H3K27ac signals at each time point following injury. The genomic distribution of the 13,447 MI-gained peaks showed that the majority were intronic (48%) or intergenic (35%), with only 8% being mapped to promoters (Fig. 2A). Similar to the trend observed for the DEGs (SI Appendix, Fig. S1B), the numbers of MI-gained and MI-lost peaks decreased significantly across the P1 time points following injury (SI Appendix, Fig. S2D). However, the number of peaks continuously increased in P8 hearts over time following injury (SI Appendix, Fig. S2D).
Fig. 2.
Injury-induced active chromatin regions revealed by H3K27ac ChIP-Seq. (A) Pie chart showing the genomic distribution of MI-gained H3K27ac peaks. (B, Left) Heatmap showing all MI-gained H3K27ac peaks, grouped and rearranged by the time point when they have the highest fold change compared with the sham control. (B, Right) Top GO terms associated with each group of peaks and the corresponding -log10 P value. (C, Upper) H3K27ac ChIP-Seq tracks showing the intronic peak activity in the Fhl1 gene locus at P8+3 dpi and P8+3 dps. (C, Lower) In situ hybridization images showing Fhl1 mRNA expression in transverse sections from P8+3 dpi and P8+3 dps hearts at the suture level. (Scale bar, 500 μm.) (D, Upper) H3K27ac ChIP-Seq tracks showing the Tgm2 enhancer peak activity at P8+3 dpi and P8+3 dps. (D, Lower) In situ hybridization images showing Tgm2 mRNA expression in transverse sections from P8+3 dpi and P8+3 dps hearts at the suture level. (Scale bar, 500 μm.) (E) Heatmap showing TF binding motifs preferentially enriched in MI-gained enhancers at each time point. For P8+3 dpi and P8+7 dpi, the top 10 enriched TF binding motifs are shown.
To identify MI-gained H3K27ac peaks unique to the regenerative heart, we clustered the peaks based on the time point when they showed the highest signal relative to the sham controls. This clustering analysis identified 6 modules of MI-gained H3K27ac peaks that were specific to each time point following injury for P1 and P8 hearts (Fig. 2B). Analysis of genomic regions in each group using genomic region enrichment of annotations tool revealed biological processes regulated by their associated genes (15). General immune responses, including inflammation and leukocyte activation, were highly enriched at P1+1.5 dpi (module 1), P8+3 dpi (module 5), and P8+7 dpi (module 6). This is consistent with the immune response dynamics observed in the GSEA (Fig. 1D). In addition, peaks in the P8+7 dpi group (module 6) were associated with fibrotic remodeling processes and represent the most pronounced injury response, as evidenced by the total number of peaks (Fig. 2B). Notably, vascular developmental processes were preferentially enriched at P1+3 dpi (module 2), highlighting the unique chromatin regions that respond to injury of the neonatal heart (Fig. 2B). In summary, these findings show that the epigenomic H3K27ac chromatin landscape responds differently to injury in regenerative and nonregenerative hearts, and highlight the unique dynamics of active chromatin landscape in regenerative hearts.
To determine whether the gene expression changes were attributable to changes in active chromatin, we examined whether there was a positive correlation between MI-gained H3K27ac peaks and the expression of their nearest genes (SI Appendix, Fig. S2E). Overall, genes associated with MI-gained H3K27ac peaks showed substantially higher expression levels in the MI samples compared with sham controls in RNA-Seq. Similarly, genes associated with MI-lost H3K27ac peaks showed markedly lower expression in the MI samples compared with sham controls. The positive correlation between the H3K27ac peaks and gene expression suggests transcriptional regulation at the epigenomic level. We found the GO terms derived from MI-up gene modules (Fig. 1C) and H3K27ac peak modules (Fig. 2B) to be mostly inclusive, with some discrepancy. This disparity could be caused by (1) overrepresentation of various active chromatin-associated genes due to enhancer redundancy, (2) delayed transcriptional response after enhancer activation, or (3) inaccurate mapping of target genes whose enhancers act at a long distance through chromatin looping. To validate that the MI-gained chromatin regions are associated with MI-up genes after injury, we selected 2 MI-gained H3K27ac peaks and examined the expression of their nearest genes using in situ hybridization. The first peak is located in the intronic region of the Fhl1 gene. Following MI of P1 and P8 hearts, this genomic region was activated at multiple time points, with the highest fold change at P8+3 dpi (Fig. 2C). Fhl1 was also an MI-up gene at this time point, as revealed by our RNA-Seq data. In situ hybridization of mouse heart sections at the same time point showed substantial Fhl1 up-regulation in regions surrounding the infarct, compared with the sham controls (Fig. 2C). The second candidate region is located ∼20 kb upstream of the Tgm2 gene. H3K27ac deposition on this region and Tgm2 gene expression were both significantly induced at P8+3 dpi, compared with P8+3 dps (Fig. 2D). In situ hybridization showed intense Tgm2 expression in the border zone as well as the right ventricular wall after MI (Fig. 2D). Together, the transcriptome and active chromatin profiles revealed various biological processes that are temporally activated after injury during heart regeneration and cardiac remodeling.
We further performed motif analysis to identify enhancer-associated transcription factors (TFs) controlling gene expression during heart regeneration. We first excluded H3K27ac peaks in promoter regions, and surveyed for TF-binding motifs that were preferentially enriched in MI-gained enhancers (referred to as MI-gained motifs) at each time point. This revealed TF-binding motifs that were significantly enriched in MI-gained enhancers at 1 or more time points examined, which were clustered based on the time point with highest enrichment to identify TF motifs that are unique to regenerative P1 hearts compared with nonregenerative P8 hearts (Fig. 2E). We found motifs such as nuclear factor κB (NFκB), Foxh1, and Stat5 to be preferentially enriched at P1+1.5 dpi; these motifs may play important roles during neonatal heart regeneration (16). Interestingly, we found motifs for class I NFκB (p50, p52) to be specifically enriched in P1+1.5 dpi hearts, whereas class II NFκB (p65, Rel) motifs were preferentially enriched in P8+3 dpi hearts (17, 18). This suggests that different classes of NFκB and their target genes may have opposing effects on heart regeneration. Consistent with the motif enrichment in the MI-gained enhancers, genes encoding some of the enriched TFs, such as Nfkb2 (p52), Gata3, Bcl6, Zfp692, Cebpa, Nfe2, Rel, Spi1 (PU.1), Nfatc3, Jun (AP1), Runx1, and multiple members of the IRF family (Irf1, Irf5, Irf7, and Irf9) were also up-regulated after MI (log2 fold change > 0.5) (SI Appendix, Fig. S2F). Although activation of the TF-directed gene network can be achieved by posttranslational modifications of TFs, such as phosphorylation and acetylation (19, 20), our data suggest that the transcriptional activation of these TFs can also contribute to the changes in the active chromatin landscape following injury.
Different Immune Responses in Regenerative and Nonregenerative Hearts.
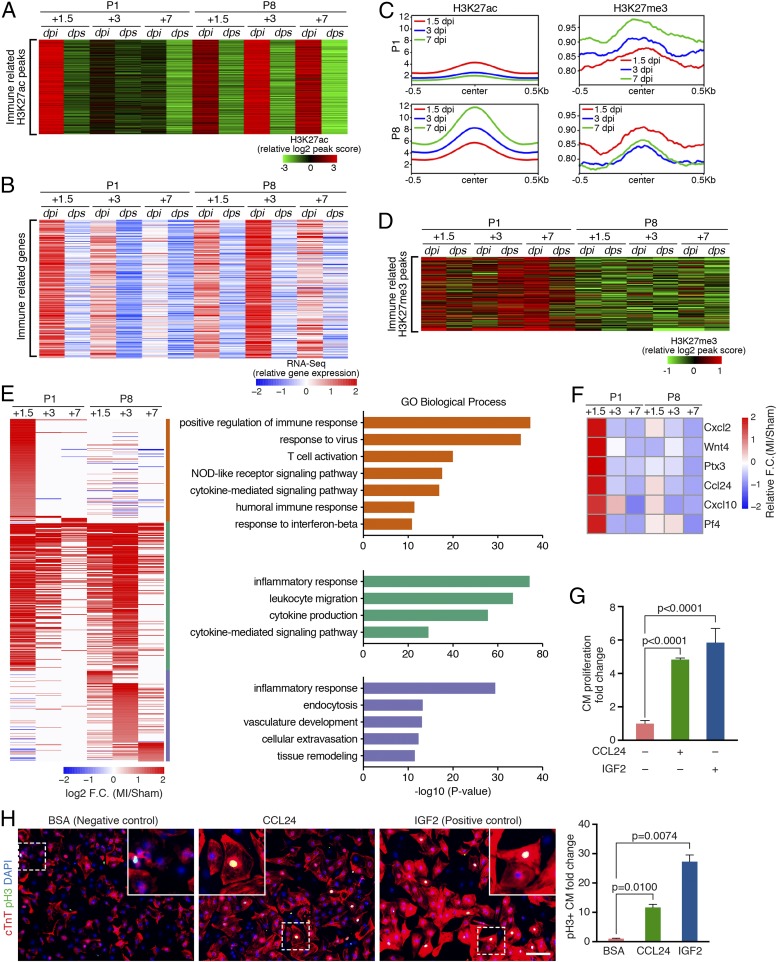
We identified a cluster of H3K27ac peaks that had markedly elevated signals in P1 hearts at 1.5 dpi and in P8 hearts at 1.5, 3, and 7 dpi (Fig. 3A). The top GO terms associated with this cluster are immune system process, the inflammatory response, and leukocyte activation. Furthermore, expression of their nearest genes positively correlated with the H3K27ac peak intensity and followed a similar trend (Fig. 3B). Thus, both ChIP-Seq and RNA-Seq revealed an acute and transient immune response in P1 hearts, and a slower but sustained response in P8 hearts (Fig. 3 A and B).
Fig. 3.
Different immune responses in regenerative and nonregenerative hearts. (A) Heatmap showing the peak scores of the H3K27ac peaks associated with immune response-related GO terms across all of the samples. (B) Heatmap showing expression levels of the nearest gene of each peak in A across all samples. (C) Metagene plots showing the normalized average signal (y axis) of H3K27ac (Left) and H3K27me3 (Right) at the immune-related peaks, identified in A, in P1 and P8 MI hearts at each time point following injury. An increase of the H3K27me3 signal was observed in the P1 regenerative heart at 1.5, 3, and 7 dpi, whereas decreased H3K27me3 signal was observed in the P8 nonregenerative heart after injury. The average signal of H3K27ac and H3K27me3 in the sham hearts at each time point is shown in SI Appendix, Fig. S3A. (D) Heatmap showing H3K27me3 signal at a cluster of MI-gained H3K27me3 peaks enriched for immune response-related GO terms. (E, Left) Heatmap showing log2 (fold change [F.C.]) of immune-related MI-up genes, which were further clustered into P1-enriched (orange bar, 156 genes), common (green bar, 221 genes), and P8-enriched (blue bar, 135 genes) clusters. (E, Right) Top enriched GO terms for P1-enriched, common, or P8-enriched immune-related gene clusters. (F) Heatmap showing z-score–transformed fold change of selected immune-related secreted factors at different time points post-MI. (G) CCL24 recombinant protein promotes CM proliferation. NRVMs were treated with 200 ng/mL bovine serum albumin (BSA) (negative control), 5 ng/mL CCL24, or 20 ng/mL IGF2 (positive control) recombinant proteins and pulse-labeled with EdU. The percentage of cTnT+/EdU+ double-positive cells was counted from immunofluorescent staining images (SI Appendix, Fig. S3E) to calculate the fold change of CM proliferation (n = 2 for each group). (H, Left) Immunofluorescent staining of cTnT (red) and pH3 (green) proteins on NRVMs treated with 200 ng/mL BSA (negative control), 5 ng/mL CCL24, or 20 ng/mL IGF2 (positive control) recombinant proteins (n = 2 for each group). Nuclei were counterstained with DAPI (blue). (Scale bar, 100 μm.) (H, Right) Fold change of the pH3+/cTnT+ double-positive cell proportion in CCL24- or IGF2-treated groups compared with BSA control was calculated.
The strong inactivation of immune-related H3K27ac peaks, as seen in P1 hearts at 3 and 7 dpi (Fig. 3A), led us to test whether cis-regulatory elements occupied by these peaks might be repressed as a means of protecting the heart from immune-related damage. We therefore profiled genomic regions bound by the repressive histone mark H3K27me3 (as schematized in Fig. 1A) and plotted the average H3K27me3 signals on the identified immune regulatory regions (Fig. 3C). For samples at various time points post-MI of P1, we observed a gradual increase in the average H3K27me3 signals from 1.5 to 7 dpi, accompanied by a decrease in the average H3K27ac signals. In contrast, this trend was reversed in P8 hearts, in which H3K27me3 signals were decreased at 3 and 7 dpi concurrent with an increase in H3K27ac signals (Fig. 3C). Interestingly, in P1 hearts at 7 dpi, the average H3K27me3 signal at these immune response regions was higher following MI than in sham control hearts, coinciding with the substantial loss of H3K27ac signals on these immune response regions (SI Appendix, Fig. S3A). As a metagene control, we selected a group of genomic regions where the H3K27ac signals were unchanged after MI and confirmed no increase in H3K27me3 deposition (SI Appendix, Fig. S3B). This indicates that the accumulation of H3K27me3 on immune-related loci is specific to P1 hearts, instead of a global effect at the genomic level during postnatal heart development. Therefore, the increased H3K27me3 signals likely contribute to the inactivation of these immune-related cis-regulatory elements in P1 hearts. We also identified a group of MI-gained H3K27me3 peaks with significantly higher H3K27me3 signals in P1 hearts at all time points post-MI and sham (Fig. 3D). Notably, the top GO terms associated with this cluster were all related to the immune response (SI Appendix, Fig. S3C). Moreover, differential peak analysis of the MI-gained H3K27me3 peaks revealed that the immune-related GO terms were enriched only in the P1 hearts, but not in the P8 hearts (SI Appendix, Fig. S3D). Together, these findings suggest that the increased H3K27me3 repressive marks in the P1 hearts postinjury attenuate the immune response during neonatal heart regeneration.
To further characterize the differential immune response at various time points post-MI of P1 and P8 hearts, we identified 512 immune system process-related genes (under GO:0009987) that were either up-regulated in P1 hearts (156 genes), P8 hearts (135 genes), or both (221 genes) after MI from RNA-Seq (Fig. 3E). GO analyses of each group of these immune genes revealed that certain immune response pathways were preferentially enriched in P1 or P8 hearts (Fig. 3E). For example, response to virus, T cell activation, nucleotide-binding oligomerization domain (NOD)-like receptor signaling, and humoral immune response pathways were preferentially enriched for immune genes up-regulated only in P1 hearts, whereas cellular extravasation, vasculature development, endocytosis, and tissue remodeling pathways were preferentially enriched for P8-unique MI-up immune genes.
Immune cells regulate wound healing and tissue regeneration via paracrine effects (21). Therefore, we sought to identify secreted immune factors that might facilitate neonatal heart regeneration by promoting CM proliferation. Among all of the 1,163 genes preferentially induced in P1 hearts after MI (modules 1 to 3 in Fig. 1C), we found 47 immune-related genes that were annotated to be secreted in GO analysis. Among these genes, Osm, Csf1, and Enpp2 have been previously shown to promote CM dedifferentiation and/or survival after injury (22–24). We next selected 6 factors that were previously uncharacterized during heart regeneration, and tested their ability to promote proliferation of neonatal rat ventricular cardiomyocytes (NRVMs) in vitro (Fig. 3F). We treated NRVMs with adenoviruses overexpressing these factors or recombinant proteins, followed by 5-ethynyl-2′-deoxyuridine (EdU) pulse-chasing to assess DNA synthesis as an indication of cell proliferation. We found that recombinant CCL24 enhanced EdU incorporation into NRVMs in a dose-dependent manner (SI Appendix, Fig. S3E). We tested different concentrations of CCL24 (1 ng/mL, 2.5 ng/mL, 5 ng/mL, or 10 ng/mL) and found that 5 ng/mL CCL24 recombinant protein resulted in the greatest increase (∼5-fold) of EdU+/cTnT+ NRVMs compared with the bovine serum albumin (BSA)-treated control, which is comparable to the effects of IGF2, a known cardiac mitogen (25, 26) (Fig. 3G and SI Appendix, Fig. S3E). Moreover, this proproliferation effect of CCL24 is more substantial in CMs than in noncardiomyocytes (NCMs) (SI Appendix, Fig. S3E). CCL24, encoded by the Ccl24 gene, is specifically expressed in macrophages (27) (SI Appendix, Fig. S3F). In addition, immunostaining for mitosis marker phospho-H3 (pH3) revealed enhanced NRVM proliferation after recombinant CCL24 treatment (Fig. 3H), and immunostaining for cytokinesis marker Aurora B suggested a trend in increased cytokinesis (SI Appendix, Fig. S3G). Together, these data suggest that the injury-induced factor CCL24 secreted from macrophages in P1 hearts could function as a mitogen to promote CM proliferation during neonatal heart regeneration.
Retention of the Developmental Gene Program during Neonatal Heart Regeneration.
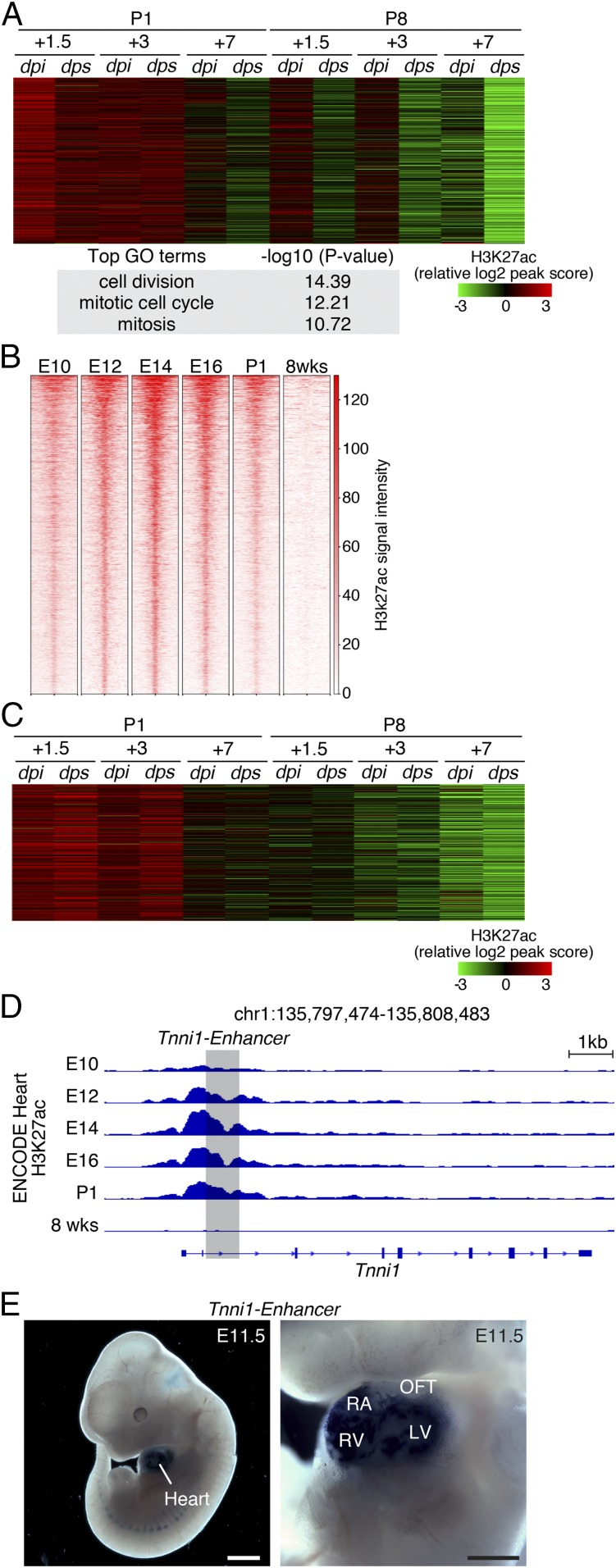
Comparison of transcriptomes of the P1+1.5 and P8+1.5 sham hearts revealed high transcriptional activity of cell cycle genes in P1+1.5 hearts (SI Appendix, Fig. S4 A and B). We also identified a cluster of H3K27ac peaks enriched for cell cycle GO terms, whose peak intensity mirrored the dynamics of cell cycle gene expression (Fig. 4A and SI Appendix, Fig. S1F). Together, by transcriptome and epigenome profiling, we observed that the active cell cycle process of the P1 heart is retained following injury, which infers that the neonatal heart may retain the potential to regenerate by leveraging the existing developmental gene program. We next sought to analyze whether a developmental gene program is also active in the P1 heart, contributing to the regenerative potential. We compared H3K27ac ChIP-Seq data from hearts at different embryonic stages (embryonic day 10.5 [E10.5] to P1) as well as 8 wk of age, generated by the Encyclopedia of DNA Elements (ENCODE) Consortium (28), and identified a group of H3K27ac peaks that were highly active in all embryonic stages and silenced in the adult heart (Fig. 4B). GO analysis confirmed their association with cardiac development processes (SI Appendix, Fig. S4C). We further plotted the activities of these cardiac development-associated peaks in our H3K27ac ChIP-Seq data from P1 and P8 MI/sham hearts, and found that they remained active during the early neonatal stage, precisely overlapping with the regenerative time window (Fig. 4C). Of note, the overall activity of these peaks and their target genes was not induced by MI (Fig. 4C and SI Appendix, Fig. S4D). One of these peaks is an intronic enhancer of the Tnni1 locus, which displayed strong H3K27ac signals in embryonic hearts but not 8-wk-old hearts (Fig. 4D). The strong H3K27ac signal in this region was retained in the P1 heart at 1.5 and 3 dpi, and is not induced by injury (SI Appendix, Fig. S4E). Additionally, transgenic mouse embryos carrying a lacZ reporter driven by this enhancer showed beta-galactosidase activity specific to the heart at E11.5, confirming this sequence as a bona fide cardiac developmental enhancer (Fig. 4E), which we have further characterized in a recent publication (29). Together, these data uncover an embryonic developmental gene program that is transiently retained in the neonatal heart.
Fig. 4.
Intrinsic developmental gene program is retained in regenerative hearts and is not induced by MI. (A) Heatmap showing the signal intensity of the H3K27ac peaks associated with cell cycle GO terms across all of the samples. (B) Analysis of ENCODE H3K27ac ChIP-Seq data from developing hearts and 8-wk-old hearts identified a developmental gene program composed of active chromatin regions that showed high H3K27ac signals at E10, E12, E14, E16, and P1. (C) H3K27ac signals suggesting the activity of the developmental gene program in hearts subjected to MI or sham surgery at P1 or P8. The developmental gene program activity was retained at the early neonatal stages (P1+1.5 and P1+3) and was not induced by injury. Plotted values are z-scores of normalized peak intensities. (D) ENCODE H3K27ac ChIP-Seq tracks at the Tnni1 gene locus. Tracks representing H3K27ac signals at different embryonic stages are indicated. The region in shadow was cloned and tested for enhancer activity in a transgenic model shown in E. (E) Transgenic enhancer assay showing Tnni1 enhancer-directed heart-specific lacZ expression in E11.5 mouse embryos. (Scale bars: Left, 1 mm; Right, 500 μm.) LV, left ventricle; OFT, outflow tract; RA, right atrium; RV, right ventricle.
Igf2bp3 Is Highly Expressed in the Regenerative Heart and Promotes CM Proliferation.
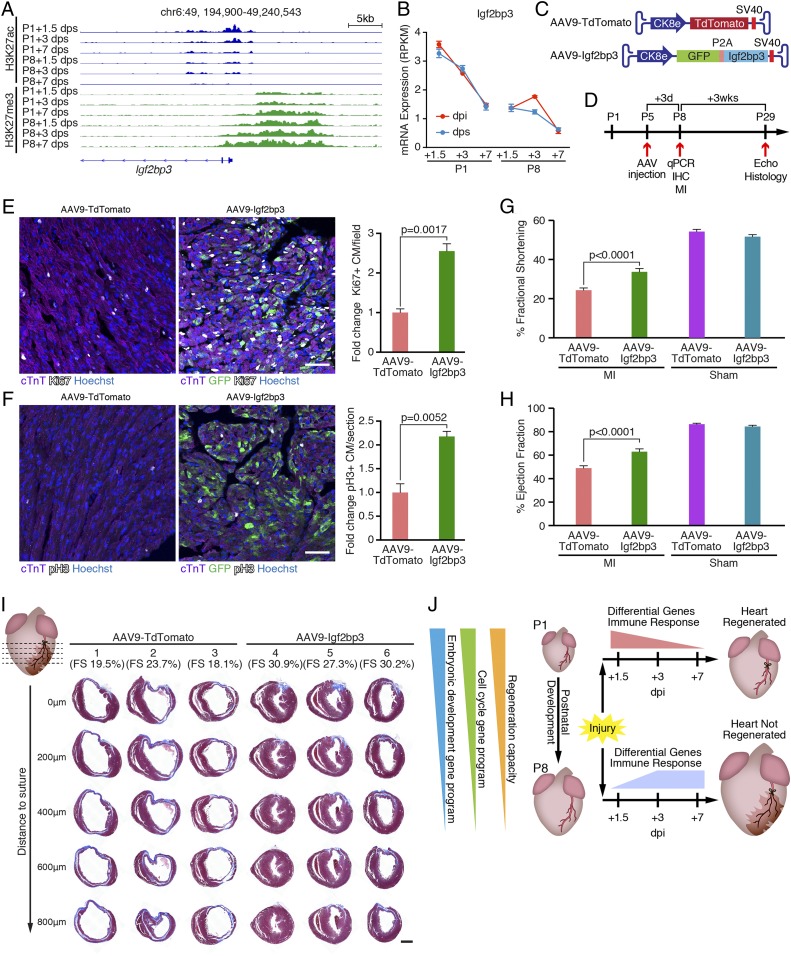
To explore the functional relevance of the identified developmental gene program in cardiac development and regeneration (Fig. 4B), we further overlapped its associated genes with the neonatal enriched genes identified comparing RNA-Seq data from P1+1.5 dps and P8+1.5 dps hearts (SI Appendix, Fig. S4A and Table S1.). One of these genes is Igf2bp3. As shown in Fig. 5A, H3K27ac signals decreased concurrent with increased H3K27me3 signals at the Igf2bp3 locus during postnatal development. Importantly, the expression of Igf2bp3 was sharply decreased from P1 to P8, and was not induced by MI, indicating that its expression is intrinsic to the P1 hearts (Fig. 5B). Igf2bp3 expression can be found in most cell types of the neonatal heart, including CMs, but it becomes undetectable in CMs from the adult stage (14) (SI Appendix, Fig. S5A). IGF2BP3 is a known regulator of IGF2 signaling that enhances IGF2 mRNA translation by stabilizing the 3′ untranslated region of IGF2 mRNA, and IGF2 signaling has been implicated in the regulation of embryonic heart development by promoting CM proliferation (25, 30). IGF2BP3 is also a known regulator of MYC, which is involved in cell cycle regulation (31–33). Therefore, we assessed whether IGF2BP3 overexpression promotes NRVM proliferation and compared it with overexpression of constitutively active YAP (YAP1-S112A), a known activator of CM proliferation (34, 35). Indeed, we found that overexpression of IGF2BP3 by adenoviral delivery in NRVMs resulted in a significant increase in EdU incorporation compared with overexpression of YFP as a negative control, as measured by immunostaining quantification and flow cytometry (SI Appendix, Fig. S5 B and C). Notably, this effect of IGF2BP3 is more substantial in CMs compared with NCMs (SI Appendix, Fig. S5C). In addition, IGF2BP3 overexpression significantly enhanced mitosis and cytokinesis as revealed by pH3 staining and Aurora B staining (SI Appendix, Fig. S5 D and E).
Fig. 5.
Igf2bp3 is highly expressed in regenerative hearts and promotes CM proliferation. (A) Decreasing H3K27ac and increasing H3K27me3 signals at the Igf2bp3 genomic locus in sham control hearts during postnatal development. chr, chromosome. (B) Igf2bp3 mRNA expression in hearts decreases from P1 to P8, and is not induced by MI (n = 3 for each condition). RPKM, reads per kilobase of transcript per million mapped reads. (C) Illustration of AAV9 vectors for overexpressing TdTomato (AAV9-TdTomato, control) and Igf2bp3 (AAV9-Igf2bp3). Ck8e, muscle creatine kinase promoter; P2A, porcine teschovirus-1 2A self-cleaving peptide coding sequence; SV40, simian virus 40 poly-A signal. (D) Strategy of AAV9 injection experiment. C57BL6 male mice were injected with AAV9-TdTomato or AAV9-Igf2bp3 at P5. At 3 d postinjection (P8), some mice (randomly selected) were euthanized for immunohistochemistry (IHC) analysis of proliferating CMs or qPCR analysis to examine Igf2bp3 overexpression. The remaining mice were subjected to MI surgery at P8, their cardiac function was examined by echocardiography (Echo) 3 wk post-MI (P29), and they were euthanized for histological examination. (E, Left) Immunostaining of cTnT (purple), Ki67 (white), and GFP (green) proteins on transverse sections from AAV9-TdTomato and AAV9-Igf2bp3 hearts at P8. Nuclei were counterstained with Hoechst (blue). (Scale bar, 50 μm.) (E, Right) Quantification of immunostaining images showing the fold change of Ki67+/cTnT+ double-positive CM percentage in AAV9-Igf2bp3 hearts compared with AAV9-TdTomato hearts (n = 3 for each group). (F, Left) Immunostaining of cTnT (purple), pH3 (white), and GFP (green) proteins on transverse sections from AAV9-TdTomato and AAV9-Igf2bp3 hearts at P8. Nuclei were counterstained with Hoechst (blue). (Scale bar, 50 μm.) (F, Right) Quantification of immunostaining images showing the fold change of pH3+/cTnT+ double-positive CM percentage in AAV9-Igf2bp3 hearts compared with AAV9-TdTomato hearts (n = 3 for each group). (G) Fractional shortening measurements of mice injected with AAV9-TdTomato (control) or AAV9-Igf2bp3 at P5 and analyzed at 3 wk after P8 MI, measured by echocardiography (n = 20 for AAV9-TdTomato MI group, n = 22 for AAV9-Igf2bp3 MI group, n = 2 for AAV9-TdTomato sham group, and n = 6 for AAV9-Igf2bp3 sham group). (H) Ejection fraction measurements of mice injected with AAV9-TdTomato (control) or AAV9-Igf2bp3 at P5 and analyzed at 3 wk after P8 MI, measured by echocardiography (n = 20 for AAV9-TdTomato MI group, n = 22 for AAV9-Igf2bp3 MI group, n = 2 for AAV9-TdTomato sham group, and n = 6 for AAV9-Igf2bp3 sham group). (I) Masson’s trichrome staining showing AAV9-Igf2bp3 promotes heart regeneration. P29 hearts injected with AAV9-TdTomato or AAV9-Igf2bp3 were transversely sectioned at the level of LAD ligation (0 μm), and at 200, 400, 600, and 800 μm below the suture. Three representative hearts are shown for each group, and their fractional shortening (FS) measurements are noted. (Scale bar, 500 μm.) (J) Model showing that both P1-unique injury responses (differential gene usage and distinct immune response dynamics) and the intrinsic properties of the regenerative heart (retained cell cycle activity and embryonic developmental gene program) define neonatal mouse heart regeneration.
We next determined whether overexpression of IGF2BP3 could promote cardiac regeneration in nonregenerative hearts. To achieve overexpression in postnatal CMs in vivo, we used adeno-associated virus 9 (AAV9) as a delivery system, which displays cardiac and skeletal muscle tropism (36). We generated an AAV9 vector that expresses IGF2BP3 or TdTomato (control) regulated by the muscle creatine kinase 8 (CK8e) promoter (37) (Fig. 5C). A single dose of 5E13 viral genome (vg)/kg of AAV9-Igf2bp3 was administered to C57BL/6 mice at P5 by intraperitoneal (i.p.) injection (Fig. 5D). This resulted in >40-fold overexpression of Igf2bp3 mRNA in the ventricles at 3 d postinjection compared with littermates injected with control AAV9, as measured by qRT-PCR (SI Appendix, Fig. S5F). Immunohistochemistry of heart sections revealed a significant increase in both Ki67+ and pH3+ CM numbers in hearts overexpressing IGF2BP3, validating that IGF2BP3 promotes CM proliferation in vivo (Fig. 5 E and F). To examine the potential proregenerative role of IGF2BP3 following cardiac injury, we performed MI on P8 mice that were preinjected with control AAV9-TdTomato or AAV9-Igf2bp3 at P5 (Fig. 5D). At 3 wk postinjury (P29), cardiac function, indicated by fractional shortening and ejection fraction, was significantly improved in mice overexpressing Igf2bp3 compared with controls (Fig. 5 G and H). Furthermore, trichrome staining revealed less fibrosis and reduced ventricular dilation in AAV9-Igf2bp3 hearts compared with control hearts (Fig. 5I).
Together, these data suggest that Igf2bp3, which is highly expressed in regenerative hearts and significantly down-regulated in nonregenerative hearts, contributes to CM proliferation and cardiac regeneration. As Igf2bp3 is part of the retained developmental gene program in neonatal hearts, these data also suggest that a permissive state in P1 hearts enables CM proliferation during regeneration by leveraging an embryonic developmental gene program.
Discussion
In this study, we generated a resource database of the transcriptome and epigenetic landscapes for 2 histone marks in both regenerative P1 hearts and nonregenerative P8 mouse hearts following MI injury across a 7-d time window. We found that regenerative P1 hearts undergo an acute and transient injury response, and retain embryonic developmental properties and cell cycle gene programs. In contrast, nonregenerative P8 hearts undergo a sustained response to MI and have lost the activity of the embryonic gene program. The differential injury response and retained developmental gene program highlight the unique neonatal properties that enable P1 hearts to regenerate (Fig. 5J).
Distinct Immune Responses between Regenerative and Nonregenerative Hearts.
The innate immune response plays a critical role in initiating tissue regeneration in several animal models, including Xenopus, salamander, and zebrafish (21). We have also shown that the innate immune response is required for neonatal heart regeneration, as neonatal mice depleted of macrophages fail to regenerate and form fibrotic scars (6). An inverse relationship between maturation of the immune system and the ability to regenerate has been proposed (38, 39). However, the molecular differences between the immune responses in the regenerative stage versus the nonregenerative stage are largely unknown. By analyzing enhancer and transcriptome landscapes, we have uncovered key differences in the immune responses between regenerative P1 and nonregenerative P8 hearts in mice. The acute activation of immune genes evidenced by H3K27ac deposition and the rapid shutdown of these genes by H3K27me3 modification in P1 hearts represent key steps in initiating regeneration. In contrast, the slow and persistent activation of the immune response in P8 hearts may prevent the regenerative response. In addition, these acutely activated immune factors in the regenerative heart may serve an overall proregenerative role, and the absence of those factors may hinder regeneration in the P8 heart. We also identified the cytokine CCL24 as a previously unrecognized inducer of CM proliferation. Ccl24 is preferentially expressed in P1 macrophages compared with P14 macrophages (6). How CCL24 and its downstream signaling pathway activate CM proliferation remains to be defined. Furthermore, the injury-induced secreted factors may facilitate heart regeneration by regulating other cellular processes, such as endothelial cell (EC) migration or inhibiting myofibroblast activation, which remain to be further studied.
The distinct immune responses of P1 and P8 hearts may partially result from heterogeneity in immune cell populations. There are marked differences in the compositions of immune cell types between P1 and P14 hearts (6). The P1 heart harbors more (F4/80/CD11c/I-Ab)loLy-6Chi monocytes and (F4/80/CD11c/I-Ab)hiLy-6Clo macrophages than the P14 heart, whereas the P14 heart has more (F4/80/CD11c/I-Ab)loLy-6Clo monocytes. B lymphocytes and T cells have also been shown to infiltrate the heart after MI (40, 41). Besides immune cells, cardiac resident fibroblasts, ECs, and epicardial cells also contribute to the immune response and heart regeneration by secreting cytokines and responding to immune effectors (42–45). Thus, the dynamic transcriptome and epigenome responses in P1 and P8 hearts after MI are likely due to a combination of cellular compositional changes (e.g., infiltration of immune cells, cell death) and gene expression changes in resident cardiac cells. Further investigation of the cellular compositional changes in injured hearts is currently ongoing.
An Active Developmental Gene Program during Neonatal Regeneration.
Unlike the adult zebrafish heart in which CMs reengage developmental programs upon injury, neonatal mouse hearts do not deploy a global induction of the developmental program following injury (46, 47). Instead, our findings indicate that the P1 heart retains developmental and cell cycle gene activities, which allow P1 hearts to fully regenerate after injury. Conversely, the loss of these gene programs leads to the loss of regenerative capacity. This is supported by previous findings that an active cell cycle gene program was identified in neonatal CMs (both MI and sham), but not in adult CMs, indicating a developmentally permissive transcription state in neonatal hearts (14). Thus, reactivation of the developmental and cell cycle gene programs in adult CMs serves as a potential therapeutic approach to replenish lost CMs after ischemic injury. In this regard, overexpression of cell cycle regulators or inhibition of the Hippo pathway unlocks cell cycle arrest and induces CM proliferation (35, 48–50). Although global activation of the developmental gene program was not observed during neonatal heart regeneration, we cannot exclude the possibility that it can be activated in a small subpopulation of CMs that undergo dedifferentiation.
Regulation of CM Proliferation by IGF2BP3.
We identified Igf2bp3 as a regulator of CM proliferation with higher expression in regenerative hearts than nonregenerative hearts. Overexpression of Igf2bp3 leads to enhanced CM proliferation both in vitro and in vivo. Interestingly, the Igf2bp3 locus was shown to progressively accumulate DNA methylation during postnatal heart development, suggesting epigenetic regulation of Igf2bp3 expression at multiple levels (51). IGF2BP3 may promote CM proliferation, at least in part, by enhancing IGF2 and MYC signaling within the regeneration time window. IGF2BP3 has been shown to promote Igf2 mRNA translation in various cell lines (30, 52), and IGF2 signaling is involved in the CM proliferation during embryonic heart development (25). In zebrafish, Igf2 signaling is up-regulated and required during heart regeneration following apical resection (53). Recently, proteins of the IGF2BP family have been identified as readers of N6-methyladenosine modification on mRNA that enhance target mRNA stability and translation (32). The cell cycle regulator Myc is one of the known mRNA targets of IGF2BP3 (32). Thus, it is possible that IGF2BP3 regulates the expression of a broad spectrum of proregeneration factors (including IGF2 and MYC), which remains to be further investigated. As previously seen by modulating the Hippo pathway (35, 50), overexpression of Igf2bp3 in the adult heart may offer an approach to enhance the regenerative capacity of the heart following injury.
In conclusion, this work provides comprehensive transcriptomic and epigenomic insight into the molecular underpinnings of neonatal heart regeneration. Our findings highlight the unique neonatal features that are involved in the mechanistic basis of heart regeneration and uncover regulatory gene programs that might be manipulated to promote cardiac repair.
Materials and Methods
A full description of experimental materials and methods is provided in SI Appendix, SI Methods.
Experimental Animals.
Animal work described in this manuscript has been approved and conducted under the oversight of the University of Texas Southwestern Institutional Animal Care and Use Committee. Timed-pregnant ICR/CD-1 mice (Charles River Laboratories) were used to deliver pups for surgical procedures on P1 or P8. The day of birth is considered P1. B6C3F1 mice (Charles River Laboratories) were bred to collect embryos for transgenic enhancer assay. Neonatal Sprague–Dawley rats (Envigo) were used to isolate NRVMs. Mice were housed in a 12-h light/dark cycle in a temperature-controlled room in the Animal Research Center of the University of Texas Southwestern Medical Center, with free access to water and food. In each experiment, the age of the mouse is indicated in the text and in the figure. Sex was not determined for embryos or neonatal pups.
Cell Lines.
All cells were cultured at 37 °C with 5% CO2. NRVMs were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma, D5796)/199 medium (Gibco, 11150-059) (3:1) with 3% fetal bovine serum (FBS; Gemini Bio Products, 100-106) and 1% penicillin/streptomycin (Sigma, P0781). Adeno-X 293 cells (Clontech, 632271) were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin. Platinum E cells (Cell Biolabs, RV-101) were cultured in DMEM with 10% FBS, 1% penicillin/streptomycin, 1 μg/mL puromycin (Thermo Fisher Scientific, A1113803), and 10 μg/mL blasticidin (Thermo Fisher Scientific, A1113903).
Murine Model of MI and Sample Collection.
MI was performed as previously described, with modifications detailed in SI Appendix, SI Methods (3, 4, 54). Mice were euthanized at 1.5 d (36 h), 3 d, or 7 d postsurgery. Each heart was dissected in ice-cold phosphate-buffered saline, and a transverse cut on the ligation plane was made, separating it into an above-ligation plane part and a below-ligation plane (BLP) part. BLP parts were flash-frozen with liquid nitrogen and stored at −80 °C for RNA-Seq or ChIP-Seq. Each tissue collected was treated as a biological replicate, and no pooling of samples was performed throughout the study.
Generation and Injection of AAV9.
AAV9 vectors expressing TdTomato or Igf2bp3 driven by the muscle-specific CK8 promoter were cloned by replacing the Cas9 coding sequence of the AAV9-CK8-Cas9 vector with TdTomato or the GFP-P2A-Igf2bp3 coding sequence, and were packaged by the Harvard Medical School/Boston Children’s Hospital Viral Core, as previously described (37, 55). C57BL/6 mice were injected with AAV9 at P5 at a titer of 5E13 vg/kg by i.p. injection.
RNA-Seq.
For each time point and condition, BLP ventricles from 3 mouse hearts were used as biological triplicates for the RNA-Seq experiments. RNA extraction was performed as previously described, with modifications (56). Briefly, the collected samples were ground to powder in liquid nitrogen, resuspended in 1 mL of TRIzol per 50 mg of tissue, and homogenized using 20G1/2 needles. After chloroform extraction, the aqueous phase was mixed with an equal volume of 75% ethanol and further purified using a RNeasy Mini Kit (QIAGEN, 74104) according to the manufacturer’s protocol. An Agilent 2100 TapeStation was used for RNA quality analysis. Stranded mRNA-Seq libraries were generated using a KAPA mRNA HyperPrep Kit (Roche, KK8581) following manufacturer’s protocol. Sequencing was performed on an Illumina NextSeq 500 system using a 75-bp, high-output sequencing kit for single-end sequencing. Methods for RNA-Seq data analysis are described in detail in SI Appendix, SI Methods.
ChIP-Seq.
For each time point and condition, BLP ventricles from 2 mouse hearts were used as biological duplicates for the ChIP-Seq experiments. ChIP was performed as previously described, with modifications detailed in SI Appendix, SI Methods (57, 58). ChIP-Seq libraries were generated using a NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, E7645L) according to the manufacturer’s protocol. Final ChIP-Seq libraries were pooled and sequenced on an Illumina NextSeq 500 system using a 75-bp, high-output sequencing kit for single-end sequencing. Methods for ChIP-Seq data analysis are described in detail in SI Appendix, SI Methods.
Statistics and Data Availability.
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc.) using a 2-tailed unpaired t test, with P < 0.05 considered significant unless otherwise indicated. All data are displayed as mean ± SEM unless otherwise indicated. All data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE123868.
Supplementary Material
Acknowledgments
We dedicate this work to the memory of Wenduo Ye, an exceptional colleague and caring friend who generated data for this study. We thank Jose Cabrera for graphics, John McAnally for performing microinjection for the transgenic enhancer assay, Jian Xu and Xin Liu from the Sequencing Core Facility (Children’s Research Institute at University of Texas Southwestern Medical Center) for performing the Illumina sequencing, and the Molecular Histopathology Core (University of Texas Southwestern Medical Center) under the direction of James Richardson for help with histology. We thank Hisayuki Hashimoto, Catherine Makarewich, and other members of the E.N.O. laboratory for helpful discussions. We are grateful for the support, advice, and assistance from Beibei Chen, Min S. Kim, and Venkat Malladi from the Bioinformatics Core Facility (University of Texas Southwestern Medical Center). We thank the ENCODE Consortium and the ENCODE production laboratories for generating the datasets used in this study. This work was supported by grants from the NIH (AR-067294, HL-130253, HL-138426, and HD-087351), the Fondation Leducq Transatlantic Networks of Excellence in Cardiovascular Research, and the Robert A. Welch Foundation (Grant 1-0025 to E.N.O.). Z.W. was supported by a predoctoral fellowship from the American Heart Association and the Harry S. Moss Heart Trust (19PRE34380436).
Footnotes
Conflict of interest statement: E.N.O. and P.R.R. both receive funding from a Leducq Network grant, but they are not direct collaborators.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE123868).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905824116/-/DCSupplemental.
References
- 1.Roth G. A., et al. , Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 70, 1–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohn J. N., Ferrari R., Sharpe N., Cardiac remodeling–Concepts and clinical implications: A consensus paper from an international forum on cardiac remodeling. Behalf of an International Forum on Cardiac Remodeling. J. Am. Coll. Cardiol. 35, 569–582 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Porrello E. R., et al. , Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porrello E. R., et al. , Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. U.S.A. 110, 187–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cahill T. J., Choudhury R. P., Riley P. R., Heart regeneration and repair after myocardial infarction: Translational opportunities for novel therapeutics. Nat. Rev. Drug Discov. 16, 699–717 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Aurora A. B., et al. , Macrophages are required for neonatal heart regeneration. J. Clin. Invest. 124, 1382–1392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leor J., Palevski D., Amit U., Konfino T., Macrophages and regeneration: Lessons from the heart. Semin. Cell Dev. Biol. 58, 26–33 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Mahmoud A. I., et al. , Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev. Cell 34, 387–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F., Wang X., Capasso J. M., Gerdes A. M., Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 28, 1737–1746 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Walsh S., Pontén A., Fleischmann B. K., Jovinge S., Cardiomyocyte cell cycle control and growth estimation in vivo–An analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 86, 365–373 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Han C., et al. , Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. 25, 1137–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alkass K., et al. , No evidence for cardiomyocyte number expansion in preadolescent mice. Cell 163, 1026–1036 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Quaife-Ryan G. A., et al. , Multicellular transcriptional analysis of mammalian heart regeneration. Circulation 136, 1123–1139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean C. Y., et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karra R., Knecht A. K., Kikuchi K., Poss K. D., Myocardial NF-κB activation is essential for zebrafish heart regeneration. Proc. Natl. Acad. Sci. U.S.A. 112, 13255–13260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escoubet-Lozach L., et al. , Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet. 7, e1002401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins N. D., Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Filtz T. M., Vogel W. K., Leid M., Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 35, 76–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benayoun B. A., Veitia R. A., A post-translational modification code for transcription factors: Sorting through a sea of signals. Trends Cell Biol. 19, 189–197 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Aurora A. B., Olson E. N., Immune modulation of stem cells and regeneration. Cell Stem Cell 15, 14–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai Y.-T., et al. , ENPP2 protects cardiomyocytes from erastin-induced ferroptosis. Biochem. Biophys. Res. Commun. 499, 44–51 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Okazaki T., et al. , Macrophage colony-stimulating factor improves cardiac function after ischemic injury by inducing vascular endothelial growth factor production and survival of cardiomyocytes. Am. J. Pathol. 171, 1093–1103 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., et al. , Oncostatin M-induced cardiomyocyte dedifferentiation regulates the progression of diabetic cardiomyopathy through B-Raf/Mek/Erk signaling pathway. Acta Biochim. Biophys. Sin. (Shanghai) 48, 257–265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P., et al. , IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 138, 1795–1805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDevitt T. C., Laflamme M. A., Murry C. E., Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J. Mol. Cell. Cardiol. 39, 865–873 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skelly D. A., et al. , Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 22, 600–610 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Consortium E. P.; ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto H., et al. , Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell 25, 69–86.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suvasini R., et al. , Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J. Biol. Chem. 286, 25882–25890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bretones G., Delgado M. D., León J., Myc and cell cycle control. Biochim. Biophys. Acta 1849, 506–516 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Huang H., et al. , Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palanichamy J. K., et al. , RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Invest. 126, 1495–1511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishioka N., et al. , The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Xin M., et al. , Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. U.S.A. 110, 13839–13844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zincarelli C., Soltys S., Rengo G., Rabinowitz J. E., Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Martari M., et al. , Partial rescue of growth failure in growth hormone (GH)-deficient mice by a single injection of a double-stranded adeno-associated viral vector expressing the GH gene driven by a muscle-specific regulatory cassette. Hum. Gene Ther. 20, 759–766 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uygur A., Lee R. T., Mechanisms of cardiac regeneration. Dev. Cell 36, 362–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eming S. A., Hammerschmidt M., Krieg T., Roers A., Interrelation of immunity and tissue repair or regeneration. Semin. Cell Dev. Biol. 20, 517–527 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Yan X., et al. , Temporal dynamics of cardiac immune cell accumulation following acute myocardial infarction. J. Mol. Cell. Cardiol. 62, 24–35 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Zouggari Y., et al. , B lymphocytes trigger monocyte mobilization and impair heart function after acute myocardial infarction. Nat. Med. 19, 1273–1280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawaguchi M., et al. , Inflammasome activation of cardiac fibroblasts is essential for myocardial ischemia/reperfusion injury. Circulation 123, 594–604 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Zhu M., et al. , FoxO4 promotes early inflammatory response upon myocardial infarction via endothelial Arg1. Circ. Res. 117, 967–977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smart N., et al. , Thymosin β4 induces adult epicardial progenitor mobilization and neovascularization. Nature 445, 177–182 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Vieira J. M., et al. , The cardiac lymphatic system stimulates resolution of inflammation following myocardial infarction. J. Clin. Invest. 128, 3402–3412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta V., et al. , An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 23, 1221–1227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jopling C., et al. , Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohamed T. M. A., et al. , Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104–116.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Z., et al. , Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 115, 354–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leach J. P., et al. , Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sim C. B., et al. , Dynamic changes in the cardiac methylome during postnatal development. FASEB J. 29, 1329–1343 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Liao B., Hu Y., Herrick D. J., Brewer G., The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J. Biol. Chem. 280, 18517–18524 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Huang Y., et al. , Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS One 8, e67266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahmoud A. I., Porrello E. R., Kimura W., Olson E. N., Sadek H. A., Surgical models for cardiac regeneration in neonatal mice. Nat. Protoc. 9, 305–311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amoasii L., et al. , Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 9, eaan8081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu N., et al. , A twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat. Cell Biol. 19, 202–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J., et al. , Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36, 9–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X., et al. , In situ capture of chromatin interactions by biotinylated dCas9. Cell 170, 1028–1043.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.