Significance
Anterograde axonal transport supplies organelles and protein complexes throughout axonal processes to support neuronal morphology and function. It has been observed that reduced anterograde axonal transport is associated with neuronal diseases. In contrast, here we show that particular disease-associated mutations in KIF1A, an anterograde axonal motor for synaptic vesicle precursors, induce hyperactivation of KIF1A motor activity and increased axonal transport of synaptic vesicle precursors. Our results advance the existing knowledge of the regulation of motor proteins in axonal transport and provide insight into the cell biology of motor neuron diseases.
Keywords: axonal transport, hereditary spastic paraplegia, kinesin, KIF1A, UNC-104
Abstract
KIF1A is a kinesin family motor involved in the axonal transport of synaptic vesicle precursors (SVPs) along microtubules (MTs). In humans, more than 10 point mutations in KIF1A are associated with the motor neuron disease hereditary spastic paraplegia (SPG). However, not all of these mutations appear to inhibit the motility of the KIF1A motor, and thus a cogent molecular explanation for how KIF1A mutations lead to neuropathy is not available. In this study, we established in vitro motility assays with purified full-length human KIF1A and found that KIF1A mutations associated with the hereditary SPG lead to hyperactivation of KIF1A motility. Introduction of the corresponding mutations into the Caenorhabditis elegans KIF1A homolog unc-104 revealed abnormal accumulation of SVPs at the tips of axons and increased anterograde axonal transport of SVPs. Our data reveal that hyperactivation of kinesin motor activity, rather than its loss of function, is a cause of motor neuron disease in humans.
Axonal transport along microtubules (MTs) is fundamental for the development and maintenance of neuronal cells. Kinesin superfamily proteins (KIFs) are a large family of MT-dependent molecular motors, some of which are integral in driving anterograde axonal transport (1, 2). The kinesin-3 family has been shown to transport a wide variety of cargoes in axons (3–6). The constituents of synaptic vesicles (SVs) are synthesized in the cell body and transported to synapses by vesicle carriers called synaptic vesicle precursors (SVPs) by the kinesin-3 family motors KIF1A and KIF1Bβ (5, 6). Genetic and cell biological studies in Caenorhabditis elegans have revealed the basic molecular mechanism of SVP axonal transport. UNC-104, a founding member of the kinesin-3 family and a C. elegans ortholog of KIF1A and KIF1Bβ, was originally discovered in C. elegans through genetic screening (7, 8). In loss-of-function unc-104 mutants, SVPs are not properly transported to synapses, leading to abnormal accumulation of SVs in cell bodies and dendrites (8).
Several kinesin motors are known to be regulated through autoinhibitory mechanisms (9), and the axonal transport of SVPs is known to be controlled through autoinhibition of UNC-104/KIF1A motor activity (10, 11). We previously obtained gain-of-function unc-104 mutants via suppressor screening of arl-8 mutants (11). These gain-of-function mutations were mapped to the motor domain, coiled-coil 1 (CC1) domain, and coiled-coil 2 (CC2) domain (10, 12) and appear to lead to constitutive activation of UNC-104 motor activity (11). In mutant C. elegans that have gain-of-function unc-104 mutations, the axonal transport of SVPs is significantly increased. The mutated amino acid residues that lead to hyperactivation of UNC-104 are well conserved in mammalian KIF1A, and analogous mutations disrupt the autoinhibition of mammalian KIF1A expressed in COS-7 cells (11).
Hereditary spastic paraplegia (SPG) is a human motor neuron disease associated with mutations in more than 60 genes (13). Familial mutations in KIF1A have been identified as causes of SPG (14). In addition to familial SPG, de novo mutations in KIF1A cause SPG associated with intellectual disabilities (15). Most KIF1A mutations causing these neuropathies are located within the conserved motor domain. Because the motor domains of KIFs convert the energy of ATP hydrolysis into directional motility along the MT (2), it is thought that disease-associated mutations in the motor domain most likely disrupt the motile mechanism of KIF1A and consequently anterograde axonal transport of SVPs (13, 14, 16). Here we show that, in contrast to this concept, some disease-associated mutations in KIF1A actually lead to hyperactivation of motor activity, resulting in overactive anterograde transport of SVPs. Our results highlight how proper cellular control over the motile activity of MT-based motors is essential for neuronal homeostasis in humans.
Results
Not All Disease-Associated Mutations in KIF1A Are Loss of Function.
We noticed that the gain-of-function V6I mutation in unc-104, identified in our previous C. elegans genetic screen, is equivalent to the dominant SPG-associated mutation, V8M, in human KIF1A (11, 17) (Fig. 1A and SI Appendix, Fig. S1 A and B). We thus reasoned that not all disease-associated mutations in human KIF1A result in a simple loss of motor activity. To explore this possibility further, we first performed unc-104 complementation experiments using human KIF1A cDNA (18) (Fig. 1 B–D). A loss-of-function allele of unc-104, unc-104(e1265), displayed strong defects in worm movement on agar plates (Fig. 1 C and D). When human KIF1A was expressed under the unc-104 promoter, the motility in unc-104(e1265) worms was recovered to the wild-type (WT) level (Fig. 1 C and D).
Fig. 1.
Complementation of an unc-104 mutant worm by human KIF1A cDNA. (A) Disease-associated mutations analyzed in this study. Mutations shown above and below in the KIF1A schematic indicate de novo and familial mutations, respectively. ID, patients with intellectual disability; no ID, patients without intellectual disability; SPG, patients with SPG. AD and AR indicate autosomal dominant and autosomal recessive mutations, respectively. (See also SI Appendix, Fig. S1.) (B and C) The phenotypes of unc-104(e1265) (B) and unc-104(e1265) expressing human KIF1A cDNA with the unc-104 promoter (C). (Scale bars: 1 mm.) (D) Bar graph showing worm movements. unc-104(e1265) mutant worms without KIF1A expression (control) exhibit severe movement defects compared with the WT N2 strain. Ectopic expression of WT KIF1A, KIF1A(V8M), KIF1A(A255V), and KIF1A(R350G) using the unc-104 promoter in the unc-104(e1265) background results in WT movement. Expression of other KIF1A with disease mutations (S58L, T99M, G199R, E253K, and R307Q) do not fully rescue unc-104(e1265) mutant worms. Asterisks indicate that the velocity is statistically not different from that of WT, indicating complete rescue (Tukey’s multiple comparison test). n = 60 worms for each genotype.
Next, we expressed human KIF1A with different disease-associated mutations in unc-104(e1265) mutant worms. Expression of KIF1A(S58L), KIF1A(T99M), KIF1A(G199R), KIF1A(E253K), and KIF1A(R307Q) could not fully rescue the unc-104 mutant phenotype (Fig. 1D). While we believe that these mutations are weak or strong loss of function, it is still possible that they change KIF1A expression, rather than motor activities, in worms. Further studies are needed to determine the exact nature of these mutations. In contrast, expression of KIF1A(V8M) completely rescued the unc-104 mutant (Fig. 1D), consistent with our initial hypothesis that KIF1A(V8M) may be a gain-of-function mutant. Interestingly, expression of KIF1A(A255V) and KIF1A(R350G) also completely rescued the unc-104(e1265) allele (Fig. 1D).
KIF1A Mutations Associated with SPG Hyperactivate the KIF1A Motor.
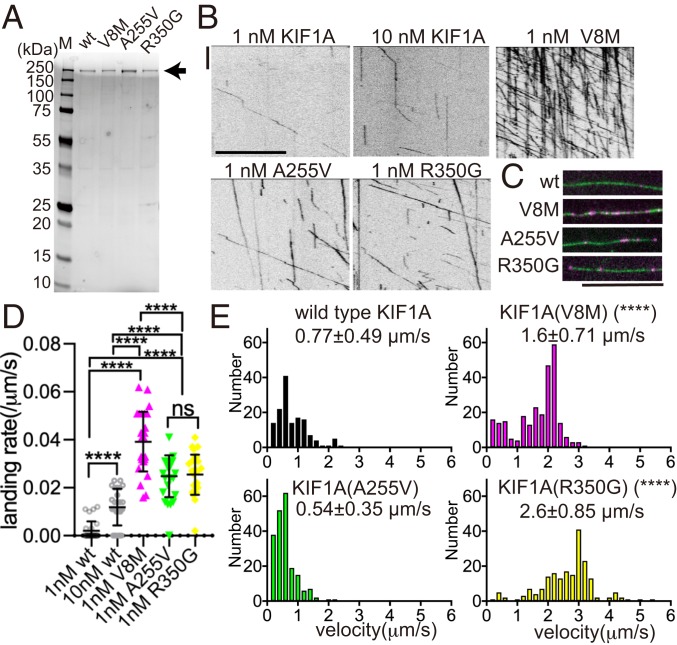
Behavioral rescue by KIF1A overexpression cannot tell whether these mutations are gain of function, very weak loss of function, or not loss of function. To discriminate among these possibilities, we next performed in vitro single molecule assays (Fig. 2). Previous studies have shown that CC1 and CC2 bind to the neck coiled-coil domain and motor domain of KIF1A and inhibit its binding to MTs (10–12). The landing rate—the number of molecules that bind to MTs normalized by time and MT length—is related to the ability of the motor domains to have productive interactions with the MT (10). Thus, if the autoinhibition of KIF1A is disrupted by disease-associated mutations, the landing rate of mutant motors on MTs should increase in vitro.
Fig. 2.
Single molecule motility assay in solution. (A) Coomassie blue-stained gel showing the purity of recombinant, full-length human KIF1A proteins. The arrow denotes the full-length protein (230 kDa). (B) Representative kymographs showing the motility of purified KIF1A::mScarlet along MTs in vitro. (Scale bars: vertical, 5 s; horizontal, 10 μm.) (C) Representative images showing KIF1A::mScarlet particles (magenta) on MTs (green). Wt, WT KIF1A; V8M, KIF1A(V8M); A255V, KIF1A(A255V); R350G, KIF1A(R350G). (Scale bar: 10 μm.) Note the strong increase in the number of mutant KIF1A molecules bound to MTs compared with the WT motors. (D) Landing rates of purified WT and mutant KIF1A motors: 0.002 ± 0.004/μm/s 1 nM KIF1A, 0.012 ± 0.007/μm/s for 10 nM KIF1A, 0.039 ± 0.012/μm/s for 1 nM KIF1A(V8M), 0.024 ± 0.008/μm/s for 1 nM KIF1A(A255V), and 0.025 ± 0.008/μm/s for 1 nM KIF1A(R350G). Lines show mean ± SD values, and each dot represents 1 counted molecule. n = 27 MTs from at least 3 trials per condition. ****Adjusted P < 0.0001 compared with WT KIF1A, Kruskal–Wallis 1-way ANOVA on ranks and Dunn’s multiple comparisons test. (E) Histograms showing the velocity of KIF1A mutants: 0.77 ± 0.48 μm/s for WT KIF1A, 1.6 ± 0.70 μm/s for KIF1A(V8M), 0.54 ± 0.35 μm/s for KIF1A(A255V), and 2.6 ± 0.85 μm/s for KIF1A(R350G), mean ± SD. n = 143, 260, 203, and 196 molecules, respectively, in >20 trials. ****Adjusted P < 0.0001 compared with WT KIF1A, 1-way ANOVA followed by Dunnett’s multiple comparison test. (See also SI Appendix, Fig. S2.)
Full-length human KIF1A, KIF1A(V8M), KIF1A(A255V), and KIF1A(R350G) fused to a C-terminal mScarlet-strepII tag were expressed in sf9 cells using baculovirus and purified (Fig. 2A). Purified motors were diluted to approximately 1 nM, and their behavior on MTs was observed using total internal reflection fluorescence microscopy. We observed processive movement of purified KIF1A on MTs, but with a relatively low landing rate (0.002 ± 0.004/μm/s at 1 nM) compared with previous data obtained by the analysis of purified tail-truncated and constitutively dimerized constructs of KIF1A (10, 19) (Fig. 2 B and C and SI Appendix, Fig. S2), consistent with the motor existing predominantly in an autoinhibited and possibly monomeric state (10, 12, 20). In contrast, full-length KIF1A(V8M), KIF1A(A255V), and KIF1A(R350G) showed significantly elevated landing rates compared with the WT motor (Fig. 2 B–D). KIF1A(V8M) had the highest landing rate (∼20-fold higher compared with WT), while KIF1A(A255V) and KIF1A(R350G) were activated to a lesser extent (∼10-fold less compared with WT) (Fig. 2D). While the velocity of KIF1A(A255V) was comparable to that of WT KIF1A, the velocity of KIF1A(V8M) and KIF1A(R350G) was ∼2- and 3-fold faster, respectively, than that of WT KIF1A (Fig. 2E), suggesting that either these mutations relieve the autoinhibitory mechanisms to various degrees or confer a gain-of-function increase in velocity. These data, together with the results from the rescue assays (Fig. 1), indicate that V8M, A255V, and R350G are gain-of-function rather than loss-of-function KIF1A mutations.
Establishment of SPG Model Worms.
To show that the autosomal-dominant SPG (ADSPG) mutation (V6M, corresponding to human V8M) and the autosomal-recessive SPG (ARSPG) mutation (A252V, corresponding to human A255V) activates C. elegans UNC-104, as is the case for human KIF1A, the localization of UNC-104::GFP with these mutations was observed in live C. elegans (Fig. 3 A–C and SI Appendix, Fig. S3). UNC-104::GFP specifically expressed in mechanosensory neurons was diffusely localized to neuronal cell bodies, proximal axons, and distal axons (Fig. 3B). In contrast, both UNC-104(V6M)::GFP and UNC-104(A252V)::GFP strongly accumulated in the distal part of anterior lateral MT (ALM) neurons in vivo (Fig. 3 A–C and SI Appendix, Fig. S3). Only a very dim fluorescent signal was detected in the proximal region of UNC-104(V6M)::GFP and UNC-104(A252V)::GFP neurons (Fig. 3B and SI Appendix, Fig. S3). This distal accumulation of mutant motors also has been observed in constitutively active KIF1A and UNC-104 in mammalian and worm cells (10, 11).
Fig. 3.
Conservation of disease-associate residues and establishment of disease models. (A) Residues mutated in AD and AR hereditary SPG are conserved in human KIF1A and worm UNC-104. (See also SI Appendix, Fig. S1B.) (B and C) Localization of UNC-104::GFP (UNC-104), UNC-104(V6M)::GFP (V6M), and UNC-104(A252V)::GFP (A252V) in the ALM neuron in vivo. (B) Representative images of proximal and distal regions of the ALM neuron. CB, cell body. (Scale bars: 10 μm.) (C) Mean fluorescent intensity in the cell body and distal axon was measured, and the intensity of cell body:intensity of distal axon ratio was calculated in UNC-104::GFP (wt), UNC-104(V6M)::GFP(V6M), and UNC-104(A252V)::GFP (A252V) expressed in the ALM neuron. n = 20 cells from 20 transgenic animals. Data are mean ± SD. ****Adjusted P < 0.001 compared with WT control, 1-way ANOVA followed by Dunnett’s multiple comparison test. Whole-cell images are shown in SI Appendix, Fig. S3. (D) Body bending assay. The number of body thrashings in M9 buffer in a 1-min observation period were counted in WT N2 (N2), unc-104(ADSPGV6M) (V6M), and unc-104(ARSPGA252V) (A252V) young adult worms (4 d after hatching) and old adult worms (10 d after hatching). Each dot represents an animal. Data are mean ± SD. **Adjusted P < 0.01, ****adjusted P < 0.0001, 1-way ANOVA followed by Tukey’s multiple comparison test. n = 50 worms for each genotype. (E) 4-d-old WT, unc-104(ADSPGV6M), and unc-104(ARSPGA252V) worms were transferred to agarose plates containing 1 mM aldicarb, and worm viability was monitored and plotted. A representative result of 3 independent assays is shown. (See also SI Appendix, Fig. S4.)
We established SPG model worms by introducing the corresponding human mutations into worm UNC-104 using genome editing (SI Appendix, Fig. S4 A and B). We refer to the edited unc-104(V6M) and unc-104(A252V) strains as unc-104(ADSPGV6M) and unc-104(ARSPGA252V), respectively. First, worm movement was analyzed by counting the number of body bends in M9 buffer. We observed that young mutant worms (4 d old) did not show overt motility defects, but in older worms (10 d old), body movement was reduced by both disease mutations (Fig. 3D).
Second, to test for presynaptic defects, we performed an aldicarb resistance assay (21). Aldicarb is an acetylcholine esterase inhibitor, and exposure of worms to aldicarb causes an accumulation of acetylcholine in the synaptic cleft, resulting in muscle excitation and paralysis. Mutants with presynaptic defects are more resistant to aldicarb, because defects in synaptic transmission cause reduced SV exocytosis and slower accumulation of acetylcholine in the synaptic cleft. We consistently observed that both unc-104(ADSPGV6M) and unc-104(ARSPGA252V) worms were more resistant to aldicarb compared with WT worms (Fig. 3E and SI Appendix, Fig. S4 C and D), indicating that human disease-associated mutations in unc-104 cause presynaptic defects.
Abnormal Accumulation of SVs in the Distal Axon in SPG Model Worms.
ALM neurons have a long axon with a single branch that extends to the nerve ring. Synapses are formed along this axon branch that can be visualized by the SV marker SNB-1::GFP expressed under the mec-7 promoter (22) (Fig. 4A). No SV accumulation was seen in the tip of the primary neurite in WT worms (Fig. 4 A and B). In contrast, adult worms harboring the ADSPG and ARSPG mutations often displayed abnormal accumulation of SVs at the tip of the primary neurite (Fig. 4 C–G, arrows). This phenotype was similar to (albeit less severe than) that of loss-of-function dynein mutants (Fig. 4 F and G). Moreover, the phenotype was age-dependent. At the larval 4 (L4) stage, no aberrant accumulation of SVs was observed in unc-104(ADSPGV6M) or unc-104(ARSPGA252V) worms (SI Appendix, Fig. S5), suggesting that the phenotype in adult worms results from a slow accumulation over time.
Fig. 4.
Synaptic phenotypes of SPG disease models. (A–G) The localization of SV marker SNB-1::GFP in the ALM neuron. A stably integrated marker, jsIs37, was used. (A) Schema showing the morphology of the ALM neuron. The large and small dotted boxes delineate the areas shown in B–D and E, respectively. (B–E) Representative images of the head region (B–D) and the tip of primary neurite (E) of WT and disease model worms. (Scale bars: 10 μm.) (F) Quantification of the phenotype in each animal. Young adult worms were scored. n = 40. (See also SI Appendix, Fig. S5.) (G) Quantification of the size of SNB-1::GFP puncta at the tip of axons in WT, unc-104(ADSPGV6M) heterozygote, unc-104(ADSPGV6M) homozygote, unc-104(ARSPGA252V) heterozygote, unc-104(ARSPGA252V) homozygote, dhc-1 homozygote, and dnc-1 homozygote. Only axonal tips showing the aberrant accumulation phenotype in F were compared. All data are plotted along with mean ± SD values. WT and unc-104(ARSPGA252V) heterozygote were not statistically analyzed because of no accumulation. ****Adjusted P < 0.0001. (H–J) Synaptic phenotype of DA9 neurons. Stably integrated marker wyIs85 was used. (H) Line scan images of DA9 neurons. Ten DA9 neurons from independent animals were scanned and aligned. (Scale bar: 5 μm.) Representative synaptic images in the worm body are shown in SI Appendix, Fig. S6 A–C. (I) Plots of intersynaptic distances. Each dot represents each intersynaptic distance. n = 60 intersynapses from 3 independent animals. Data are mean ± SD. **Adjusted P < 0.01, Tukey’s multiple comparison test. (J) Mean fluorescent intensity of each dorsal synapse. Data are mean ± SD. *Adjusted P < 0.05, Tukey’s multiple comparison test. n = 102, 102, and 87 synapses from 5 independent animals in WT, unc-104(ADSPGV6M), and unc-104(ARSPGA252V), respectively.
Synaptic Morphology Is Affected in Dorsal Synapses in SPG Model Worms.
Worm motor neurons in the dorsal nerve cord have en passant synapses. We visualized these synapses in the DA9 motor neuron using a DA9-dominant promoter, itr-1, and an SV marker, GFP::RAB-3 (wyIs85[Pitr-1::gfp::rab-3]) (11) in both unc-104(ADSPGV6M) and unc-104(ARSPGA252V) worms (Fig. 4 H–J and SI Appendix, Fig. S6 A–C). First, we found that the length between synapses is longer in these disease models (Fig. 4 H and I and SI Appendix, Fig. S6 A–C), similar to previously characterized gain-of-function mutants of unc-104 (11). Second, we noticed that the fluorescent intensity of GFP::RAB-3 in dorsal synapses is dimmer in unc-104(ADSPGV6M) and unc-104(ARSPGA252V) compared with WT (Fig. 4J). To confirm this phenotype, we analyzed the synapses in the dorsal nerve cord by electron microscopy (Fig. 5 A–C and SI Appendix, Fig. S6 D–F).
Fig. 5.
Electron microscopy analyses of dorsal synapses in WT and unc-104(ADSPGV6M) young adult worms (4 d after hatching). (A and B) Representative images of dorsal synapses in WT (A) and unc-104(ADSPGV6M) (B). AZ, active zone. (Scale bars: 100 nm.) (C) Bar graphs showing the number of SVs in each synaptic bouton reconstituted by serial sections. n = 15 synapses from 2 WT worms and 24 synapses from 3 unc-104(ADSPGV6M) worms reconstituted from 100 serial ultrathin sections. Data are mean ± SEM. *P < 0.05, Welch’s t test. (See also SI Appendix, Fig. S6 D–G).
To analyze the number of SVs, the length of active zones and the volume of synaptic boutons, 100 continuous ultrathin sections were observed. Consistent with the reduced GFP::RAB-3 signals in synapses (Fig. 4J), the number of SVs in each synapse was significantly reduced in unc-104(ADSPGV6M) compared with WT (Fig. 5 A–C); however, synaptic volume and length of the active zone in unc-104(ADSPGV6M) were not significantly affected (SI Appendix, Fig. S6 D–G). These data are consistent with the results of aldicarb assays and a model in which overactive UNC-104 motility leads to reduced SVs in en passant synapses in worm motor neurons.
Human Disease Mutations Lead to Aberrant Activation of SVP Transport.
In mutant worms that lack ARL-8, UNC-104 motor activity is not properly activated (11). As a result, SVs accumulate abnormally in the commissure and proximal asynaptic region of the DA9 axon due to reduced anterograde transport (23, 24) (Fig. 6 A–C). This phenotype is suppressed by gain-of-function mutations in unc-104, which disrupt the autoinhibition of UNC-104 (11).
Fig. 6.
SPG mutations suppress the arl-8 phenotype. (A) Schematic drawing of the DA9 neuron, showing the dorsal asynaptic region and the commissure that were observed and analyzed. The asterisk indicates the location where the commissure joins the dorsal nerve cord. (B–F) Representative images showing the localization of GFP::RAB-3 in WT (wt) (B), arl-8(wy271) (C), arl-8(wy271);unc-104(ADSPGV6M) (D), arl-8(wy271);unc-104(ARSPGA252V) (E), and arl-8(wy271);unc-104(ADSPGV6M)/+ (F). Integrated marker wyIs85 [Pitr-1::GFP::RAB-3] was used. Asterisks indicate the commissure bend shown in A. (Scale bars: 50 μm.) (G–I) Statistical analysis of mutant phenotypes. Shown are the length of the asynaptic region (G), the length of synaptic region in the dorsal axon (H), and the number of puncta misaccumulated at the commissure (I). Lines represent the median, and each dot represents 1 animal. Numbers above show actual median values. **Adjusted P < 0.01, ****adjusted P < 0.0001 compared with arl-8, Kruskal–Wallis 1-way ANOVA on ranks and Dunn’s multiple comparisons test. n = 30 animals for each genotype.
To test whether ARSPG and ADSPG mutations lead to a similar gain of function, we generated arl-8; unc-104(ARSPGA252V) and arl-8; unc-104(ADSPGV6M) double-mutant worms (Fig. 6 D–I). Both unc-104(ARSPGA252V) and unc-104(ADSPGV6M) mutations suppressed the aberrant localization of SVs observed in arl-8 mutants (Fig. 6 D, E, and G–I). unc-104(ADSPGV6M) heterozygotes also suppressed the arl-8 mutant phenotype (Fig. 6 F–I), indicating the dominant nature of the V6M mutation. The arl-8; unc-104(ARSPGA252V)/+ phenotype was comparable to the arl-8 single mutant (Fig. 6 G–I), indicating that a heterozygous A252V mutation cannot completely suppress the mislocalization of SVs. These data indicate that both unc-104(ARSPGA252V) and unc-104(ADSPGV6M) mutations lead to activation of the UNC-104 motor even in the absence of the UNC-104 activator ARL-8, resulting in increased axonal transport of SVPs.
Increase in Anterograde Transport of SVPs in SPG Model Worms.
Finally, we analyzed the axonal transport of SVPs in SPG model worms by time-lapse microscopy of GFP:RAB-3 in the asynaptic ventral region of the DA9 neuron (24) (Fig. 7 and SI Appendix, Fig. S7). First, the velocity of the anterograde transport was faster in unc-104(ADSPGV6M) worms compared with WT worms, but the velocity of anterograde transport was comparable in unc-104(ARSPGA252V) and WT worms (Fig. 7 A and B). These data are consistent with results from single molecule analysis showing that the velocity of recombinant KIF1A(V8M) is faster than that of WT KIF1A, while the velocity of KIF1A(A255V) is similar to that of WT (Fig. 2).
Fig. 7.
Axonal transport is more active in SPG model worms. (A) Representative kymographs showing the axonal transport of SVPs in WT, unc-104(ADSPGV6M), and unc-104(ARSPGA252V) worms. wyIs251 that stably expresses GFP::RAB-3 under the mig-13 promoter was used. Drawings highlight the locations of processive transport events in the original kymograph. Horizontal and vertical lines represent 5 μm and 10 s, respectively. The left and right sides of the kymographs are the proximal and distal sides, respectively. (B) Histogram showing the velocity of anterogradely transported vesicles. Vesicles that moved >1 μm in 1 min are included. Data are mean ± SD. n = 145, 162 and 150 moving vesicles from more than 20 movies in WT, unc-104(ADSPGV6M), and unc-104(ARSPGA252V), respectively. unc-104(ADSPGV6M), but not unc-104(ARSPGA252V), is statistically faster than WT. P < 0.0001, Tukey’s multiple comparisons test. (C and D) The number of vesicles that were anterogradely (C) or retrogradely (D) moving were plotted and adjusted by axon length and time. Vesicles that moved >1 μm in 1 min are included. *Adjusted P < 0.05, **adjusted P < 0.01, ***adjusted P < 0.001. Kruskal–Wallis 1-way ANOVA on ranks and Dunn’s multiple comparisons test. n = 35 movies from 35 worms. Bars represent mean ± SD. (E and F) Quantification of the dissociation rate (E) and capture probability (F) in the ventral axon in WT, unc-104(ADSPGV6M), and unc-104(ARSPGA252V) worms. The kymographs show a representative dissociation event, capture event, and passing event; 2 μm (horizontal) and 6 s (vertical). Bars represent mean ± SD. *Adjusted P < 0.05, ***adjusted P < 0.001, Kruskal–Wallis 1-way ANOVA on ranks and Dunn’s multiple comparisons test. Actual adjusted P values are 0.0008 (WT vs. ADSPG) and 0.04 (WT vs. ARSPG). n = 42 vesicle pools from 3 movies from 3 worms.
The frequency of anterograde transport was increased in both AD and AR model worms (Fig. 7C). In contrast, the frequency of retrograde transport of SVPs was reduced in unc-104(ADSPGV6M) and unc-104(ARSPGA252V) strains (Fig. 7D). Previous studies have shown that the amount of axonal transport is determined by the dissociation rate and capture rate at vesicle pools along the axon (24). In axonal transport, vesicles are occasionally dissociated from vesicle pools along the axon. The number of events was counted and analyzed (Fig. 7E); however, the dissociation rate was not significantly affected in unc-104(ADSPGV6M) and unc-104(ARSPGA252V) strains. When moving vesicles reach stationary vesicle pools along the axon, they are either captured or pass through the stationary pool. We assessed the probability of capture events in our data (Fig. 7F) and found that a decreased capture rate in both disease model strains, suggesting overactive axonal transport in the mutant axons. Taken together, our data indicate that mutant UNC-104 motor activity is hyperactive, resulting in increased anterograde transport of SVPs in disease model axons.
Discussion
Mutations in the motor domain of KIF1A are associated with numerous neuronal diseases, including both pure and complicated SPG as well as intellectual disabilities (15–17). However, why different mutations cause different types of diseases remains unclear. Previous studies, along with the data presented here, suggest that KIF1A mutations that cause complicated SPG with intellectual disabilities are loss-of-function mutations (16, 25) (Fig. 1). In contrast, KIF1A(V8M), KIF1A(A255V), and KIF1A(R350G) cause overactivation of KIF1A motor activity in vitro and are associated with pure or complicated SPG without intellectual disabilities, suggesting that gain-of-function mutations cause milder neuropathies. Consistent with human symptoms, the neuronal phenotype of unc-104(ADSPGV6M) and unc-104(ARSPGA252V) shown here in worms is weaker than that of loss-of-function unc-104 mutants. Although the loss-of-function allele of unc-104 shows very strong motility defects (Fig. 1D), unc-104(ADSPGV6M) and unc-104(ARSPGA252V) worms can still move on agar plates (Fig. 3D), while these mutants have behavioral and morphological defects (Figs. 3 D and E, 4, and 5).
In vitro experiments suggest that an autoinhibitory mechanism that regulates MT binding (10) is disrupted by these mutations. The landing rate of KIF1A(V8M) was much higher than the rates of KIF1A(A255V) and KIF1A(R350G) in vitro (Fig. 2 A–D). This may explain the AD nature of KIF1A(V8M) and the AR nature of KIF1A(A255V) and KIF1A(R350G). Because the activity of KIF1A(V8M) is very high, even heterozygous mutation induces neuropathy. Moreover, the velocities of KIF1A(V8M) and KIF1A(R350G) were faster than the velocity of WT KIF1A (Fig. 2E). Consistent with this, anterograde transport was faster in unc-104(ADSPGV6M) worms compared with WT worms. The molecular mechanism for these velocity changes remains unclear, but the mutations could conceivably alter the motor enzymatic rate or lead to varying degrees of release of the autoinhibition mechanisms.
Interestingly, a previous study did not detect defects in the motor activity of KIF1A(A255V) (16). This result is reasonable, because the study used a deletion mutant of KIF1A that lacked the entire tail domain and thus any autoregulatory elements. Moreover, in MT-gliding assays, motors are attached to a glass surface, which can force motors into their active conformations (26). Here we analyzed full-length KIF1A by single-molecule motility assays in solution, which made it possible to detect defects in KIF1A autoregulation. (A further discussion on KIF1A motility is provided in SI Appendix, Supplementary Note).
Previous studies have found disease-associated mutations in KIF21A and dynein-binding protein BICD2 to be gain of function rather than loss of function (27–29). KIF21A is a member of the kinesin-4 family, which regulates MT polymerization. Gain-of-function mutations in KIF21A cause congenital fibrosis of extraocular muscle type 1 (CFEOM1). Disease-associated mutations hyperactivate KIF21A and inhibit MT polymerization in neurons (27, 28). BICD2 activates cytoplasmic dynein to preform retrograde axonal transport (30, 31). Mutations in BICD2 are associated with the motor neuron disease, spinal muscular atrophy, and mutant BICD2 proteins hyperactivate the motility of dynein (29). Thus, overly active MT-based transport in either direction leads to neurodegeneration, highlighting the importance of a proper balance of motor activity for neuronal health.
Materials and Methods
Detailed descriptions of all materials and methods used in this study are provided in SI Appendix, Materials and Methods.
Worm Genetics and Transformation.
The strains and vectors used in this study are listed in SI Appendix, Tables S1 and S2. To visualize UNC-104::GFP in ALM neurons (Fig. 3), antibiotic selection with Nourseothricin was performed as described previously (32). Disease model worms were established by genome editing as described previously (33).
Light Microscopy.
Worms were analyzed by light microscopy as described previously (24). Steady-state imaging was obtained using an inverted Carl Zeiss Axio Observer Z1 microscope equipped with a 40×/1.4 objective and an LSM710 confocal scanning unit (Fig. 5) or LSM 800 confocal scanning unit (Figs. 3 and 4 and SI Appendix, Figs. S3 and S6). Time-lapse imaging was provided by an inverted Carl Zeiss Axio Observer Z1 microscope equipped with a Plan-Apochromat 100×/1.4 objective and a Hamamatsu ORCA flash v3 sCMOS camera. Movies were taken at 5 fps for 1 min and analyzed using Fiji software as described previously (34).
Supplementary Material
Acknowledgments
We thank Dr. Asako Sugimoto (Tohoku University) for helpful discussions and Jeremy Allen, PhD (Edanz Group; https://www.edanzediting.com) for editing a draft of this manuscript. This work is supported by Japan Society for the Promotion of Science (JSPS) Kakenhi Grants 17H05057 (to T.O.), 17KK0139 (to S.N.), 17H05010 (to S.N.), and 16H06536 (to S.N., S.A., and K.H.). This work is also supported by NIH Grant R35 GM124889 (to R.J.M.), an Osamu Hayaishi Memorial Scholarship for Study Abroad (to K.C.), a JSPS Overseas Research Fellowship (to K.C.), the Takeda Science Foundation (T.O.), the Uehara Memorial Foundation (T.O.), and the Naito Foundation (S.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905690116/-/DCSupplemental.
References
- 1.Holzbaur E. L., Scherer S. S., Microtubules, axonal transport, and neuropathy. N. Engl. J. Med. 365, 2330–2332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirokawa N., Noda Y., Tanaka Y., Niwa S., Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Lo K. Y., Kuzmin A., Unger S. M., Petersen J. D., Silverman M. A., KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci. Lett. 491, 168–173 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Stucchi R., et al. , Regulation of KIF1A-driven dense core vesicle transport: Ca2+/CaM controls DCV binding and Liprin-α/TANC2 recruits DCVs to postsynaptic sites. Cell Rep. 24, 685–700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada Y., Yamazaki H., Sekine-Aizawa Y., Hirokawa N., The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81, 769–780 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Niwa S., Tanaka Y., Hirokawa N., KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat. Cell Biol. 10, 1269–1279 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Otsuka A. J., et al. , The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron 6, 113–122 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Hall D. H., Hedgecock E. M., Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65, 837–847 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Verhey K. J., Hammond J. W., Traffic control: Regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10, 765–777 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Hammond J. W., et al. , Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7, e72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa S., et al. , Autoinhibition of a neuronal kinesin UNC-104/KIF1A regulates the size and density of synapses. Cell Rep. 16, 2129–2141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Bassam J., et al. , Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J. Cell Biol. 163, 743–753 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutry M., Morais S., Stevanin G., Update on the genetics of spastic paraplegias. Curr. Neurol. Neurosci. Rep. 19, 18 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Klebe S., et al. , KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: Distinct phenotypes according to the nature of the mutations. Eur. J. Hum. Genet. 20, 645–649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohba C., et al. , De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J. Hum. Genet. 60, 739–742 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Esmaeeli Nieh S., et al. , De novo mutations in KIF1A cause progressive encephalopathy and brain atrophy. Ann. Clin. Transl. Neurol. 2, 623–635 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iqbal Z., et al. , Targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. PLoS One 12, e0174667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klopfenstein D. R., Vale R. D., The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell 15, 3729–3739 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monroy B. Y., et al. , Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 9, 1487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomishige M., Klopfenstein D. R., Vale R. D., Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297, 2263–2267 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Oh K. H., Kim H., Aldicarb-induced paralysis assay to determine defects in synaptic transmission in Caenorhabditis elegans. Bio Protoc. 7, e2400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koushika S. P., et al. , Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J. Neurosci. 24, 3907–3916 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klassen M. P., et al. , An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron 66, 710–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y. E., Huo L., Maeder C. I., Feng W., Shen K., The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron 78, 994–1011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guedes-Dias P., et al. , Kinesin-3 responds to local microtubule dynamics to target synaptic cargo delivery to the presynapse. Curr. Biol. 29, 268–282.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vale R. D., Reese T. S., Sheetz M. P., Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42, 39–50 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng L., et al. , Human CFEOM1 mutations attenuate KIF21A autoinhibition and cause oculomotor axon stalling. Neuron 82, 334–349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Vaart B., et al. , CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell 27, 145–160 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Huynh W., Vale R. D., Disease-associated mutations in human BICD2 hyperactivate motility of dynein-dynactin. J. Cell Biol. 216, 3051–3060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlager M. A., Hoang H. T., Urnavicius L., Bullock S. L., Carter A. P., In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenney R. J., Huynh W., Tanenbaum M. E., Bhabha G., Vale R. D., Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obinata H., Sugimoto A., Niwa S., Streptothricin acetyl transferase 2 (Sat2): A dominant selection marker for Caenorhabditis elegans genome editing. PLoS One 13, e0197128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arribere J. A., et al. , Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa S., Immobilization of Caenorhabditis elegans to analyze intracellular transport in neurons. J. Vis. Exp., e56690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.