Significance
Neutrophil-mediated inflammation is often detrimental to the host; thus, new methods to mitigate inflammation while preserving immunity are needed. We performed a genetic screen in zebrafish with neutrophil-specific overexpression of individual microRNAs, and from there, we identified miR-199, which can mitigate neutrophil migration and relieve systemic inflammation without generating an overall immunocompromised state. The primary mechanism of action of miR-199 is through direct suppression of cyclin-dependent kinase 2 (cdk2), a canonical cell cycle regulator, in the context of terminally differentiated neutrophils. In addition, MIR-199 overexpression and CDK2 inhibition dampened human neutrophil migration. Together, our work suggests MIR-199 and CDK2 as new targets for treating inflammatory ailments and introduces an avenue of the function for CDK2 outside the cell cycle regulation.
Keywords: microRNA, cyclin-dependent kinase 2, chemotaxis, zebrafish, innate immunity
Abstract
Neutrophil migration is essential for inflammatory responses to kill pathogens; however, excessive neutrophilic inflammation also leads to tissue injury and adverse effects. To discover novel therapeutic targets that modulate neutrophil migration, we performed a neutrophil-specific microRNA (miRNA) overexpression screen in zebrafish and identified 8 miRNAs as potent suppressors of neutrophil migration. Among those, miR-199 decreases neutrophil chemotaxis in zebrafish and human neutrophil-like cells. Intriguingly, in terminally differentiated neutrophils, miR-199 alters the cell cycle-related pathways and directly suppresses cyclin-dependent kinase 2 (Cdk2), whose known activity is restricted to cell cycle progression and cell differentiation. Inhibiting Cdk2, but not DNA replication, disrupts cell polarity and chemotaxis of zebrafish neutrophils without inducing cell death. Human neutrophil-like cells deficient in CDK2 fail to polarize and display altered signaling downstream of the formyl peptide receptor. Chemotaxis of primary human neutrophils is also reduced upon CDK2 inhibition. Furthermore, miR-199 overexpression or CDK2 inhibition significantly improves the outcome of lethal systemic inflammation challenges in zebrafish. Our results therefore reveal previously unknown functions of miR-199 and CDK2 in regulating neutrophil migration and provide directions in alleviating systemic inflammation.
Neutrophils are the first cells recruited to an immune stimulus stemming from infection or sterile injuries via a mixture of chemoattractant cues (1). In addition to eliminating pathogens, neutrophils coordinate inflammation by activating and producing inflammatory signals in the tissue and, in some cases, cause adverse tissue damage. Overamplified or chronic neutrophil recruitment directly leads to autoimmune diseases rheumatic arthritis, diabetes, neurodegenerative diseases, and cancer (2). Dampening neutrophil recruitment is thus a strategy to intervene in neutrophil-orchestrated chronic inflammation (3). Despite intensive research over the past several decades, clinical studies targeting neutrophil migration have been largely unsuccessful, possibly due to the prominent redundancy of adhesion receptors and chemokines (4, 5). Additional challenges also lie in the balance of dampening detrimental inflammation while preserving immunity (6). Furthermore, neutrophil migration is governed by spatially and temporally complex dynamic signaling networks (7), adding yet another layer of complexity in studying them. Immune cells use both mesenchymal migration (adhesive migration on substrates) and amoeboid migration (actin protrusion driven with weak substrate interaction) to infiltrate tissue using distinct signaling molecules (8, 9). Thus, extensive research is required to understand the machinery regulating neutrophil recruitment and the interactions between signals and chemotactic cues, which will lead to the development of exogenous inhibitors for clinical use.
MicroRNAs (miRNAs) are short (20 to 22 nucleotides), conserved, noncoding RNAs that are epigenetic regulators of the transcriptome. By binding primarily to the 3′ untranslated regions (3′UTRs) of their target transcripts and recruiting the RNA-induced silencing complex, miRNAs down-regulate gene expression (10). miRNAs are generally fine-tuners that suppress gene expression at modest levels, and also master regulators that target the expression of a network of genes (11). Recently, miRNAs and anti-miRNAs are used in clinical trials to treat cancer and infection (12). In addition, they are used as screening tools to identify the underlying mechanisms of diseases and cell behavior (13, 14). In immune cells, an overexpression screen of miRNA was performed to identify miRNAs that regulate B cell tolerance (15). In neutrophils, the functions of the highly expressed miRNAs miR-223 (16) and miR-142 (17) have been well characterized. However, other miRNAs and their targets as regulators of neutrophil recruitment remain largely unknown. The absence of this knowledge potentially leads to missed opportunities in harnessing miRNAs and their targets in restraining neutrophilic inflammation.
The zebrafish is a suitable model organism to study neutrophil biology. It has an evolutionarily conserved innate immune system, including phagocytes and their signaling molecules, including miRNAs (17, 18). The transparent property of zebrafish embryos enables noninvasive imaging of neutrophil behavior under physiological conditions (19, 20). Additionally, high fecundity and established genetic tools make the zebrafish an ideal platform for genetic screens and drug discovery (21).
We have previously established a system to express individual miRNAs in zebrafish neutrophils to assess their function in vivo (22). Here, we performed a miRNA overexpression screen and identified miR-199 as a regulator of neutrophil chemotaxis. Through miR-199 target analysis, we identified the canonical cell cycle-related cyclin-dependent kinase 2 (cdk2) gene as a previously unrecognized regulator of neutrophil migration. Overexpression of miR-199 or inhibition of Cdk2 in neutrophils improved survival upon infection and sterile inflammatory challenges. Our results were further validated in human neutrophil-like cells and primary human neutrophils, supporting the evolutionary conservation of miR-199 and Cdk2 in vertebrae neutrophil biology. These discoveries expand our current understanding of neutrophil migration and suggest a novel strategy to manage neutrophilic inflammation.
Results
Identification and Characterization of miRNAs That Regulate Neutrophil Recruitment.
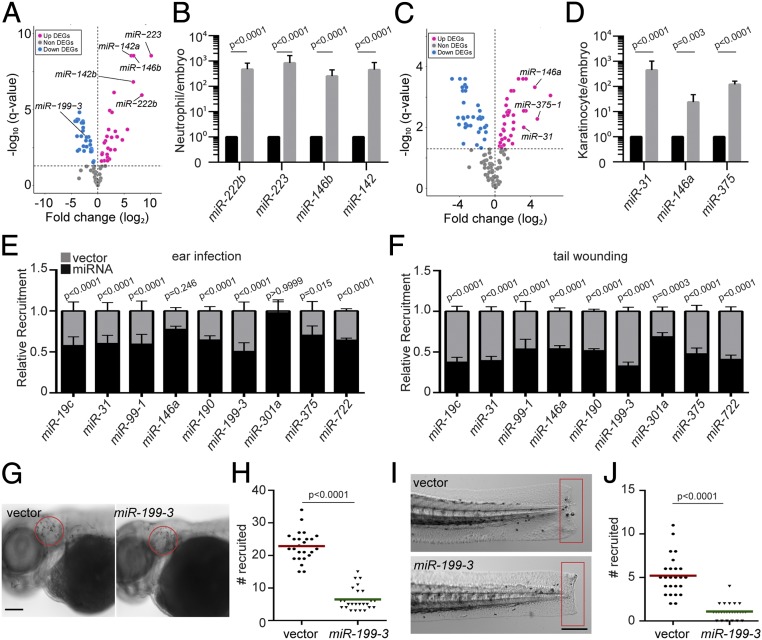
As a first step to understand miRNA-mediated gene regulation in zebrafish neutrophils, we sequenced miRNAs in zebrafish neutrophils sorted from 3-d-old embryos. Consistent with previous reports in humans and mice, miR-223 and miR-142 were among the miRNAs highly expressed in zebrafish neutrophils (Fig. 1 A and B and Dataset S1). As a control, we sequenced miRNAs in the immotile epical keratinocytes and indeed detected a distinct profile (Fig. 1 C and D). We next performed a functional genetic screen to identify a set of miRNAs that, when overexpressed, moderate neutrophil migration (SI Appendix, Fig. S1). Forty-one miRNAs with lower expression levels compared with the whole embryo or keratinocytes were selected. Individual miRNA candidates were cloned and transiently expressed specifically in zebrafish neutrophils, together with a fluorescent reporter gene. The miRNAs whose overexpression resulted in a >50% reduction in neutrophil recruitment to infection sites were selected (Dataset S2) for further validation. We generated transgenic zebrafish lines stably expressing each of the 8 miRNAs that passed the initial screen. To rule out potential artifacts associated with the transgene integration sites, more than 2 independent founders were obtained for each line. In 7 of the 8 lines, we found a significant decrease in neutrophil recruitment to ear infection sites, regardless of the choice of the founder (Fig. 1E). Similarly, neutrophil recruitment was also significantly reduced in all 8 lines in a tailfin amputation inflammation model (Fig. 1F and Dataset S3), demonstrating that the miRNA phenotypic screen was successful in identifying miRNAs that could modulate neutrophil migration. A miRNA characterized in our previous work, miR-722, served as a positive control (22). Among the 8 newly identified miRNAs, miR-146 is a known suppressor of immune cell activation (23), whereas the other 7 miRNAs have not been characterized for their roles in neutrophil migration. The overexpression of miR-199-3 resulted in the most robust inhibition in both models (Fig. 1 E–J), and therefore was chosen for further characterization.
Fig. 1.
Identification of miRNAs that suppress neutrophil recruitment in vivo. (A) DEGs of miRNAs in zebrafish neutrophils compared with whole embryo. (B) Quantification of selected miRs that were differentially expressed in neutrophils. (C) DEGs of miRNAs in zebrafish epical keratinocytes compared with whole embryo. (D) Quantification of selected miRs that were differentially expressed in keratinocytes. The assay was done with 3 biological repeats, each containing cells sorted from at least 100 larvae per repeat. The result is presented as mean ± SD, using the Holm–Sidak test. (E) Neutrophil recruitment to the infected ear in transgenic lines with neutrophil-specific overexpression of individual miRNAs. Results were normalized to the number of neutrophils recruited in the vector-expressing control lines in each individual experiment (set as a factor of 1). (F) Neutrophil recruitment to tailfin transection sites in transgenic lines as described in C. Representative images (G) and quantification (H) of neutrophils recruited to the infected ear in the vector or miR-199-3–overexpressing zebrafish line are shown. (Scale bar, 100 μm.) Representative images (I) and quantification (J) of neutrophils recruited to tailfin transection sites in the vector or miR-199-3–overexpressing zebrafish line are shown. (Scale bar, 200 μm.) In G–J, the assays were done with at least 2 individual founders with 3 biological repeats, each containing 25 fish per group. The result is presented as mean ± SD, using the Kruskal–Wallis test.
miR-199-3 Overexpression Inhibits Neutrophil Motility and Chemotaxis.
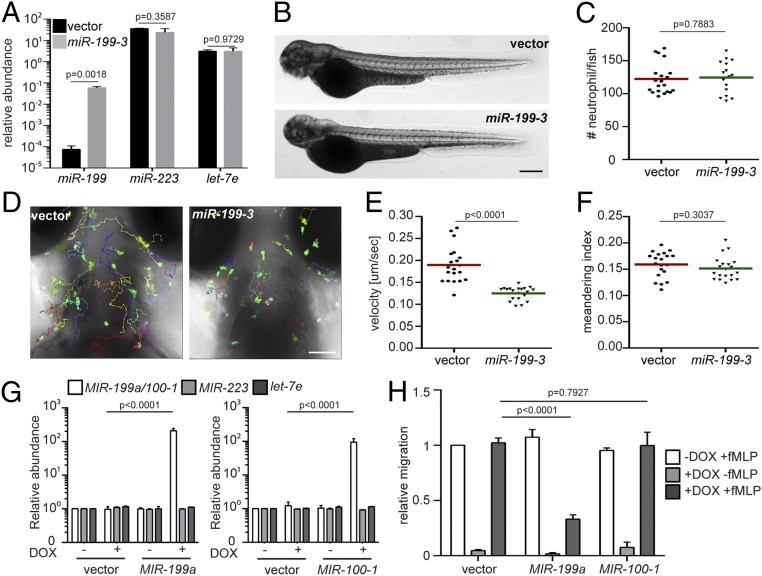
To characterize the zebrafish lines Tg(lyzC: miR-199-3-dendra2)pu19 and Tg(lyzC:vector-dendra2)pu7, henceforth referred to as the miR-199 and vector lines, we first isolated neutrophils from these lines and performed miRNA qRT-PCR to confirm the miR-199 overexpression in the miR-199 line (Fig. 2A). The levels of 2 abundant neutrophil miRNAs, miR-223 and let-7e, were not affected, demonstrating that the overexpression did not disrupt physiological miRNA levels. Total neutrophil numbers in embryos were comparable between the 2 lines, suggesting that the observed decreased neutrophil recruitment was not an artifact of a reduced number of neutrophils (Fig. 2 B and C). In addition to the defective recruitment to inflammatory sites, the velocity of neutrophil spontaneous motility in the head mesenchyme was significantly decreased in the miR-199 line (Fig. 2 D and E and Movie S1), whereas neutrophil directionality was not affected (Fig. 2F). The sequences of the mature zebrafish and human miR-199-5p isoforms are identical (Fig. 3D). We therefore sought to determine whether MIR-199 inhibits human neutrophil migration. We used MIR-100, whose overexpression did not result in any phenotype in the initial screen, as a negative control. We expressed human MIR-199a, MIR-100, or the vector alone under the control of a Tet-responsive element in HL-60 cells, a myeloid leukemia cell line that can be differentiated into neutrophil-like cells, as our model (24, 25) (SI Appendix, Fig. S2A). MIR-100 and MIR-199 were individually induced in the respective lines without affecting cell differentiation; cell death; or the expression of 2 other miRNAs, MIR-223 and LET-7 (Fig. 2G and SI Appendix, Fig. S2 B–D). The differentiated HL-60 (dHL-60) cells were then allowed to migrate toward N-formylmethionyl-leucyl-phenylalanine (fMLP) in transwells. Only the cells overexpressing MIR-199, but not the vector control or cells overexpressing MIR-100, displayed significant defects in chemotaxis (Fig. 2H). Together, our results show that overexpression of MIR-199 inhibits neutrophil migration in both zebrafish and human cell models.
Fig. 2.
miR-199-3–overexpression reduces neutrophil migration in zebrafish and humans. (A) Quantification of miR-199, miR-223, and let-7e levels in neutrophils sorted from the vector or miR-199-3 zebrafish lines. Neutrophils were isolated from 2 adult kidney marrows from 2 different founders. Results are normalized to u6 expression levels and presented as mean ± SD, using the Holm–Sidak test. Representative images (B) and quantification (C) of total neutrophils in the vector or miR-199-3 zebrafish lines are shown. (Scale bar, 500 μm.) The assays were done with 3 individual founders with 3 biological repeats, each containing 20 fish per group. The result from one representative experiment is shown as mean ± SD, using the Mann–Whitney test. Representative images (D), velocity (E), and meandering index (F) of neutrophil motility in vector or miR-199-3 zebrafish lines are shown. (Scale bar, 100 μm.) Three embryos each from 3 different founders were imaged, and quantification of neutrophils in one representative video is shown. The Kruskal–Wallis test was used. (G) Quantification of MIR-100/199, MIR-233, and LET-7E in dHL-60 cell lines with/without induced expression of the vector control or MIR-100/199. The results from 3 independent experiments are normalized to U6 expression levels and presented as mean ± SD, using the Holm–Sidak test. DOX, doxycycline. (H) Transwell migration of dHL-60 cells with/without induced expression of the vector control, MIR-199, or MIR-100 toward fMLP. Results are presented as mean ± SD from 3 independent experiments and normalized to the vector −DOX + fMLP, using the Kruskal–Wallis test.
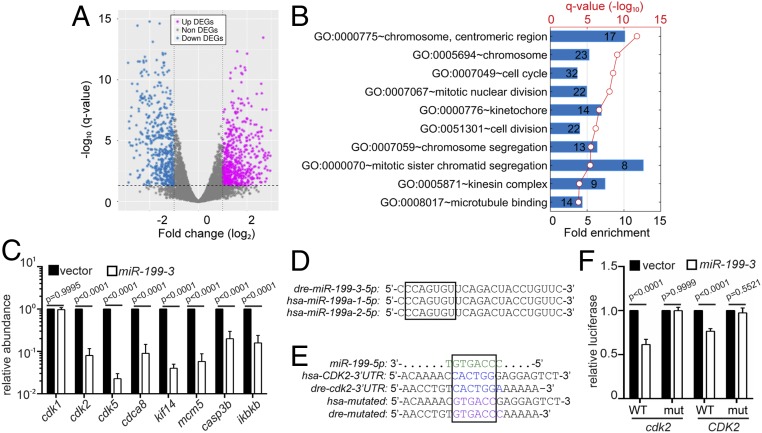
Fig. 3.
miR-199-3 overexpression suppresses the expression of cdk2 in neutrophils. (A) Volcano blot of DEGs with significant expression changes in miR-199–expressing neutrophils. Down-regulated DEGs are shown in cyan, and up-regulated DEGs are shown in magenta. (B) Pathway analysis of genes down-regulated by miR-199 in neutrophils. (C) Quantification of the transcript levels of down-regulated genes in neutrophils sorted from the vector or miR-199-3 zebrafish line (at least 100 embryos per group per repeat). Results from 3 independent experiments are normalized to rpl32 and presented as mean ± SD, using the Holm–Sidak test. (D) Alignment of mature human and zebrafish miR-199-5p sequences. (E) Alignment of miR-199-5p and predicted miR-199 binding sites in human and zebrafish CDK2 3′UTRs. (F) Suppression of Renilla luciferase expression via binding to both zebrafish and human CDK2 3′UTRs by miR-199. Results of Renilla luciferase activity normalized with firefly luciferase activity from 3 independent experiments and presented as mean ± SD, using the Kruskal–Wallis test. mut, mutation.
miR-199 Suppresses the Expression of Cell Cycle-Related Genes, Including cdk2.
To understand the underlying molecular mechanism of how miR-199 regulates neutrophil migration, neutrophils from the miR-199 or vector lines were isolated and their transcripts subjected to RNA sequencing (RNAseq). Since miRNAs generally suppress the expression of target transcripts (26), we focused on the 512 genes significantly repressed by miR-199. The top 10 pathways associated with the down-regulated transcripts were related to cell cycle processes (Fig. 3 A and B and Datasets S4 and S5). This result was surprising because neutrophils are terminally differentiated, nonproliferative, and have exited the cell cycle (27). To validate the RNAseq results, we selected 5 genes (cdk2, cdk5, cdca8, kif14, and mcm5) and confirmed their reduced transcript levels in the miR-199–overexpressing neutrophils using qRT-PCR (Fig. 3C). The zebrafish ortholog of IKBKB (inhibitor of nuclear factor kappa B kinase subunit beta), a known target of human MIR-199 (28) and a critical component in the nuclear factor κB (NF-κB) pathway, was also down-regulated, supporting the conservation of miRNA-targeted interactions in zebrafish. The level of cdk1 messenger RNA (mRNA) was not altered. Among the down-regulated cell cycle genes, the 3′UTRs of both human and zebrafish cdk2 contain predicted binding sites for miR-199-5p, while the protein sequence of CDK2 is more than 90% identical between the 2 species, showing a high degree of conservation (Fig. 3D). In addition, among the 69 down-regulated genes in the top 10 pathways (Fig. 3B), 36 (52.1%) have interactions with cdk2, presenting a significant enrichment (P = 2.87 × 10−32) over the background of 4.1% (326 cdk2-interacting genes of 7,927 genes expressed in zebrafish neutrophils). On the contrary, cdk5, despite its down-regulation upon miR-199 overexpression, is not a classical cell cycle-related gene and does not have significant interactions with other down-regulated genes. To demonstrate the direct targeting of cdk2 mRNAs by miR-199-5p, we performed dual-luciferase reporter assays. The miR-199 significantly reduced the relative luciferase activity controlled by both the human and zebrafish cdk2 3′UTRs but did not show the repression of luciferase activity when the predicted binding sites were mutated (Fig. 3 E and F). These results indicate that miR-199 suppresses classical cell cycle pathways in neutrophils and could directly target cdk2 in both zebrafish and humans.
Pharmacological Inhibition of Cdk2 Decreases Neutrophil Motility and Chemotaxis.
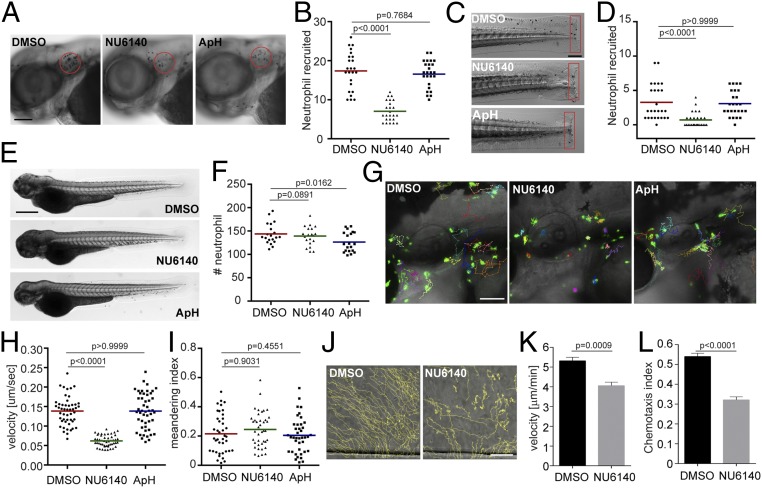
CDK2 is expressed and active in human neutrophils (29), but its function in neutrophils was previously unknown as CDK2 is traditionally thought to be involved in regulating the G1/S phase in the cell cycle (30), where neutrophils have exited the cell cycle and are terminally differentiated. To test whether cdk2 regulates neutrophil migration, we used NU6140 (31), a selective CDK2 inhibitor, with at least 10-fold selectivity over other CDKs. At a concentration of 100 μM, NU6140 did not affect zebrafish survival or neutrophil numbers (SI Appendix, Fig. S3). As a control for inhibiting cell cycle progression and DNA replication, we used a mixture of aphidicolin and hydroxyurea, as previously reported (32, 33), to uncouple the role of Cdk2 from DNA replication. Neutrophil recruitment to the infected ear (Fig. 4 A and B) or tailfin amputation sites (Fig. 4 C and D), as well as neutrophil motility (Fig. 4 G–I and Movie S2), was significantly reduced specifically by the Cdk2 inhibition, recapitulating the phenotypes caused by miR-199 overexpression. As a control, Cdk2 inhibition did not affect total neutrophil numbers in the fish (Fig. 4 E and F). To increase our confidence in the results obtained with pharmacological Cdk2 inhibition, 2 additional Cdk2 inhibitors (34) were used. A CDK2-selective inhibitor, CTV313, and a pan-CDK inhibitor, roscovitine, also blocked neutrophil motility and recruitment in inflammation models (SI Appendix, Fig. S4 and Movie S2). A previous report showed that high doses and prolonged treatment with roscovitine could lead to neutrophil apoptosis (29). To rule out the possibility that the reduced migration observed was due to cell apoptosis, we washed out the reversible inhibitors CTV313 and roscovitine after neutrophil motility was reduced. Neutrophil motility recovered after the removal of the inhibitors (SI Appendix, Fig. S5 and Movie S3), indicating that CDK2 inhibitor treatments do not result in terminal apoptosis. We then tested whether the neutrophils under investigation had indeed exited the cell cycle. No cell division was observed during wounding or infection (SI Appendix, Fig. S6 A and B and Movie S4), suggesting that neutrophil division is a rare event. Additionally, less than 3% of the neutrophils in adult kidney marrow were in the S or G2 phase of the cell cycle (SI Appendix, Fig. S6 C–E). Because cdk2 has not been previously implicated in regulating neutrophil migration, we thus determined if our findings could apply to humans. Primary human neutrophils were isolated and treated with NU6140. Inhibition of CDK2 significantly reduced chemotaxis toward interleukin-8, with a decreased velocity and chemotaxis index (Fig. 4 J–L and Movie S5). In summary, we report a previously unknown role for CDK2 in regulating neutrophil chemotaxis in a way that is separable from its function in DNA replication.
Fig. 4.
Inhibition of CDK2 reduces neutrophil motility and chemotaxis in zebrafish and humans. Representative images (A) and quantification (B) of neutrophils recruited to the infected ear in zebrafish larvae treated with CDK2 inhibitor (NU6140) or the DNA replication inhibitor aphidicolin + hydroxyurea (ApH) are shown. (Scale bar, 100 μm.) Representative images (C) and quantification (D) of neutrophils recruited to tailfin transection sites in zebrafish larvae treated with NU6140 or ApH are shown. (Scale bar, 200 μm.) Representative images (E) and quantification (F) of total neutrophil number in zebrafish larvae treated with NU6140 or ApH are shown. (Scale bar, 500 μm.) In A–F, assays were done with 3 individual founders with 3 biological repeats, each containing 20 (for motility) or 25 (for neutrophil recruitment) fish per group. The result from one representative experiment is shown as mean ± SD, using the Mann–Whitney test. Representative images (G), velocity (H), and meandering index (I) of neutrophil motility in zebrafish larvae treated with NU6140 or ApH are shown. (Scale bar, 100 μm.) Three embryos each from 3 different founders were imaged, and quantification of neutrophils in one representative video is shown, using the Kruskal–Wallis test. Representative tracks (J), mean velocity (K), and chemotaxis index (L) of primary human neutrophils treated with DMSO or NU6140 (50 μM) migrating toward interleukin-8 are shown. (Scale bar, 100 μm.) Results representative of 3 separate trials are shown. The result is presented as mean ± SEM, using a 2-way paired Welch’s t test.
Catalytic Activity of Cdk2 Is Required for Neutrophil Motility.
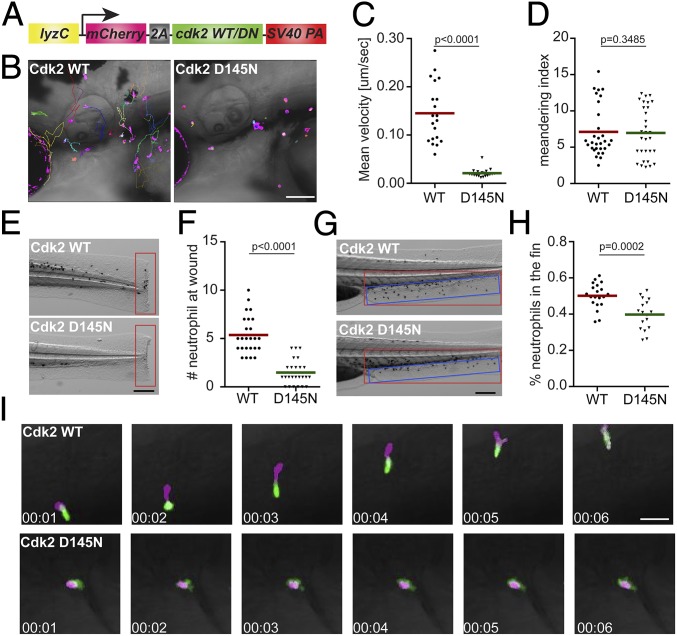
To further test our hypothesis that the kinase activity of Cdk2 is essential for neutrophil motility, we cloned the zebrafish cdk2 gene and tagged it with mCherry at the N terminus using a self-cleavable 2a peptide (Fig. 5A). Mutations at D145, a residue that chelates Mg2+ and facilitates adenosine 5′-triphosphate (ATP) binding to the catalytic cleft, would result in a catalytically dead and dominant-negative form of CDK2 (35). We constructed a zebrafish line overexpressing Cdk2 (D145N) in neutrophils, Tg(lyzC:mcherry-2a-cdk2-d145n)pu19, together with a control line, Tg(lyzC:mcherry-2a-cdk2)pu20, henceforth referred to as the Cdk2 D145N or wild-type (WT) line, respectively. Again, we observed a significant reduction of neutrophil motility (Fig. 5 B–D and Movie S6) and recruitment to the tail amputation site (Fig. 5 E and F) in the D145N line. To characterize neutrophil chemotaxis to a single chemoattractant, we bathed embryos in leukotriene B4 and quantified neutrophil emigration from the caudal hematopoietic tissue (CHT) to the caudal fin. The percentage of neutrophils migrating outside the CHT and the distance neutrophils traveled in the caudal fin were significantly reduced in the D145N line (Fig. 5 G and H, SI Appendix, Fig. S7 A and B, and Movie S7). In addition, neutrophil motility in the CHT was reduced (SI Appendix, Fig. S7 C and D and Movie S8). To visualize neutrophil polarization, the Cdk2 D145N or WT line was crossed with Tg(mpx:GFP-UtrCH), which labels stable actin in the trailing edge of migrating neutrophils (36). In Cdk2 D145N-expressing neutrophils, stable actin is enriched in transient cell protrusions that quickly retract, displaying disrupted cell polarity and actin dynamics (Fig. 5I and Movie S9). Hence, our results indicate that the catalytic activity of Cdk2 is required for neutrophil polarization and efficient chemotaxis.
Fig. 5.
Dominant-negative Cdk2 suppresses neutrophil motility and chemotaxis. (A) Schematic of the construct for neutrophil-specific expression of Cdk2 WT or D145N. Representative images (B), velocity (C), and meandering index (D) of neutrophil motility in Cdk2 WT or D145N zebrafish line are shown. (Scale bar, 100 μm.) Three embryos each from 3 different founders were imaged, and quantification of neutrophils in one representative video is shown, using the Kruskal–Wallis test. Representative images (E) and quantification (F) of neutrophil recruitment to the tailfin transection sites in Cdk2 WT or D145N zebrafish line are shown. (Scale bar, 200 μm.) Representative images (G) and quantification (H) of neutrophils migrated to the caudal fin (blue box) normalized to total neutrophils in the trunk (red box) in Cdk2 WT or D145N zebrafish line are shown. (Scale bar, 200 μm.) (I) Simultaneous imaging of the calponin-homology domain of utrophin (Utr-CH)-GFP distribution in neutrophils expressing either WT or D145N Cdk2. Data are representative of more than 3 separate time-lapse videos. (Scale bar, 50 μm.) In E–H, the assays were done with 3 individual founders with 3 biological repeats, each containing 20 (for motility) or 25 (for neutrophil recruitment) fish per group. The result is presented as mean ± SD, using the Mann–Whitney test.
To test whether cdk2 is the primary target of miR-199 that regulates cell migration, we restored Cdk2 expression in the miR-199–overexpressing zebrafish neutrophils. Cdk2 expression alone could partially rescue the migration defects of the neutrophils carrying miR-199, but not to the vector control level (SI Appendix, Fig. S8 and Movie S10), suggesting that other miR-199 targets possibly also regulate neutrophil migration. To further confirm the specific requirement of CDK2 activity in neutrophil migration, we incubated dHL-60 cells with increasing doses of inhibitors of CDK1, CDK1/5, CDK2, or CDK4/6. Indeed, only the CDK2 inhibitor reduced transwell migration in a dose-dependent manner (SI Appendix, Fig. S9). In addition, transient expression of dominant-negative Cdk2, but not dominant-negative Cdk5, reduced neutrophil migration in zebrafish (SI Appendix, Fig. S10 and Movie S11). Along the same line, knocking down CDK5 in dHL-60 cells did not affect transwell migration (SI Appendix, Fig. S11 C–H). These results suggest a specific requirement of CDK2 in regulating neutrophil migration.
CDK2 Regulates Polarization and Signaling in dHL-60 Cells.
To gain more insight into how CDK2 regulates neutrophil migration, 2 CDK2 knockdown dHL-60 cell lines were generated using 2 different short hairpin RNAs. The CDK2 protein levels in these 2 lines were significantly reduced, whereas the level of CDK1 or CDK5 remained relatively constant (SI Appendix, Fig. S11 A and B). Cells deficient in CDK2 were viable with comparable surface levels of CD11b, indicating that CDK2 knockdown did not affect cell differentiation (SI Appendix, Fig. S11 E–G). CDK2 knockdown cells exhibited hindered transwell migration toward fMLP (SI Appendix, Fig. S11H) and could not establish prominent actin-rich lamellipodia when stimulated with a uniform field of fMLP (SI Appendix, Fig. S12 A and B). Phospho (p)-myosin light chain 2, which labels the rear of the cell and drives tail retraction (37), was also mislocalized (SI Appendix, Fig. S12 A and C). After fMLP stimulation, the activation of Rac (indicated by the level of p-PAK [P21 (RAC1) activated kinase]) or the level of phosphatidylinositol (3–5)-trisphosphate (indicated by the level of p-AKT) (38) was also reduced in the CDK2 knockdown cells, whereas the p–extracellular signal-regulated kinase level (39) was not affected (SI Appendix, Fig. S12 D and E), indicating a specific requirement for CDK2 in activating the phosphatidylinositol 3-kinase (PI3K)/RAC axis. In addition, unlike CDK4 (40), CDK2 does not regulate neutrophil extracellular trap (NET) formation (SI Appendix, Fig. S13), reiterating that different CDKs have unique functions in neutrophils.
miR-199 Overexpression or Cdk2 Inhibition Ameliorates Systemic Inflammation.
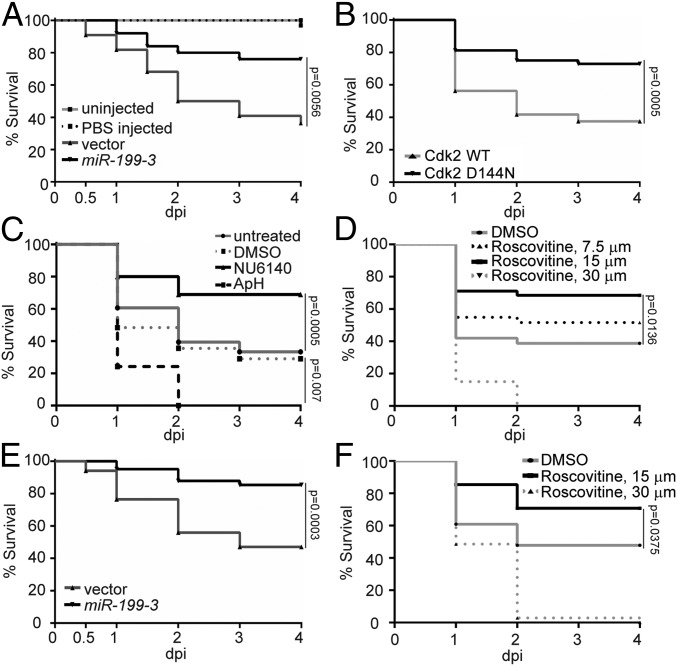
With the findings that miR-199 overexpression or Cdk2 abrogation hinders neutrophil chemotaxis, we sought to determine whether these manipulations could alleviate acute systemic inflammation caused by a lethal dose of Pseudomonas aeruginosa (PAK) in the circulation (41). Survival of the zebrafish embryos was significantly improved in the miR-199 line and the Cdk2 D145N line compared with their respective controls (Fig. 6 A and B). Consistently, NU6140 treatment improved the infection outcome, whereas DNA replication inhibitors were detrimental to the infected host (Fig. 6C). Roscovitine, the pan-CDK inhibitor (42) currently in phase II trial, also displayed a protective effect at 15 μM but was detrimental at 30 μM, possibly due to the inhibitory effect on cell proliferation and tissue repair as previously reported in cell culture and ex vivo bone marrow isolations (43) (Fig. 6D). To further assess the protective role of miR-199 overexpression or Cdk2 inhibition in another model, we utilized a previously described sterile inflammation model of endotoxemia (44). Neutrophil-specific miR-199 overexpression or roscovitine treatment again reduced host mortality (Fig. 6 E and F). These results indicate that miR-199 overexpression or Cdk2 inhibition can have a protective effect during acute systemic inflammation and infection, without generating an immune suppression status with defects in pathogen clearance.
Fig. 6.
Cdk2 inhibition and miR-199 overexpression improve zebrafish survival during systemic bacterial infection or sterile inflammation. (A) Survival of zebrafish larvae after infection of 1,000 CFU of PAK administered intravenously (i.v.) in the vector or miR-199 zebrafish line. Vector lines were left uninfected or injected with PBS as a control. (B) Survival of zebrafish larvae after infection of 1,000 CFU of PAK administered i.v. in the Cdk2 WT or D145N zebrafish line. (C) Survival of zebrafish larvae treated with vehicle, NU6140 (100 μM), or ApH (150 μM/20 mM) or left untreated after infection with 1,000 CFU of PAK administered i.v. (D) Survival of zebrafish larvae treated with indicated dilutions of roscovitine infected with 1,000 CFU of PAK administered i.v. (E) Survival of zebrafish larvae after injection of 25 ng of LPS administered i.v. in the vector or miR-199 zebrafish line. (F) Survival of zebrafish larvae treated with indicated dilutions of roscovitine injected with 25 ng of LPS i.v. In A–F, 1 representative experiment of 3 independent experiments (n ≥ 20 each group) is shown, using the Gehan–Breslow–Wilcoxon test.
Discussion
miRNAs play a pivotal role in diseases driven by neutrophilic inflammation (45–47), where both protective and detrimental effects are observed. Currently, there is no systemic screen to identify miRNAs that regulate neutrophil migration, possibly because neutrophils are terminally differentiated and not genetically tractable in cell culture. In the present study, we performed an in vivo functional screen in zebrafish and identified miR-199 as a critical regulator of neutrophil migration and chemotaxis. miR-199 hinders neutrophil motility by downregulating genes in the canonical cell cycle pathways, and directly targets cdk2. We further demonstrate that the catalytic activity of Cdk2 is critical for neutrophil migration and chemotaxis, whereas DNA replication and an active cell cycle are not essential (SI Appendix, Fig. S7E).
Although no prior studies have investigated the role of miR-199 in neutrophils, its role in suppressing inflammation and cell migration has been reported in cancer cells. In ovarian cancer cells, MIR-199 inhibits canonical NF-κB signaling by targeting IKBKB, thus preventing the production of proinflammatory cytokines (28). In our current work, the miR-199/ikbkb axis was also validated in neutrophils, further supporting the evolutionary conservation of miRNA–target interaction in vertebrates. Suppression of ikbkb may also contribute to the survival of zebrafish during systemic inflammatory challenge due to its central role in inflammation. From the perspective of cell migration and recruitment, miR-199 hinders cancer cell migration and invasion by downregulating β1 integrin levels (48), decreasing CD44 expression and hence interaction with Ezrin (49), or by directly targeting α3 integrin (50) in various cancer cells. Introduction of miR-199 alleviated invasiveness in vitro through targeting the Wnt or HIF-1α/VEGF pathway (51, 52). Here, we provide evidence that miR-199 is a suppressor of cell migration in leukocytes, expanding its role beyond cancer biology. The MIR-199a-5p levels were increased in the plasma of patients with neutrophilic asthma (53) and inflammatory bowel disease (54), suggesting that MIR-199 possibly plays physiological functions in inflammation. In addition, MIR-199b suppresses myeloid differentiation (55), which can also contribute to its role in suppressing the innate immunity.
The most surprising finding of our study is that miR-199 predominantly regulates the cell cycle-dependent kinase cdk2 in terminally differentiated neutrophils. A significant inverse relationship of MIR-199 and CDK2 was observed in samples of patients with breast cancer or head and neck cancer, indicating that MIR-199 possibly regulates CDK2 in human pathological conditions (SI Appendix, Fig. S14). CDK2 is a cell cycle-dependent kinase whose cofactors, cyclin E and cyclin A, are expressed during mitosis, activating its kinase activity and cell cycle progression (30). CDK2 phosphorylates transcriptional factors to drive cell cycle progression and cell differentiation, and to alter cell metabolism and DNA double-strand break repair (reviewed in ref. 56). Although the literature of CDK2 in the context of innate immunity and inflammation is scarce (as reviewed in ref. 57), CDK2 possibly regulates the secretion of inflammatory cytokines in macrophages. In addition, a single-nucleotide polymorphism of CDK2 is significantly enriched in asthma patients (58), supporting the possible function of CDK2 in regulating inflammation.
Here, we provide evidence that the kinase activity of CDK2 is required for neutrophil migration. Although the cytosolic CDK inhibitors have been implicated in regulating cell migration (59), there are no previous reports showing that CDK2 directly regulates cell migration. However, there is a limitation in our work that the substrates directly phosphorylated by CDK2 in neutrophils are not yet identified. CDK2 is a serine/threonine protein kinase, which uses the KLAD*FGLA kinase consensus domain to phosphorylate the “Ser/Thr-Pro-X-H/K/R” motif of target proteins, with additional prevalent noncanonical target motifs (60). Over 100 proteins are likely phosphorylated by CDK2-cyclinA in human embryonic kidney (HEK) cells (61), and the CDK2-cyclinE substrates are not well characterized. Extensive work will be required to characterize how CDK2 contributes to the signaling network required for neutrophil chemotaxis. CDK2 is possibly present in the neutrophil cytosol, and its expression is up-regulated upon lipopolysaccharide (LPS) stimulation (62), supporting the possibility that CDK2 ties in the signaling pathways downstream of the chemokine receptors. As a step closer to the potential mechanism, we have observed defective cell polarization in both zebrafish neutrophils and dHL-60 cells upon CDK2 abrogation. In addition, a specific defect in activating the PI3K/RAC2 axis is observed in CDK2-deficient dHL-60 cells, providing a direction for our future research.
Acute neutrophilic overinflammation is a pressing concern in many disease pathologies (1), and methods that simultaneously hinder neutrophil migration and inflammation while preserving immune integrity are highly desired. Our study presents an acute method to dampen neutrophil recruitment by inhibiting Cdk2, which reduces the lethality caused by acute systemic inflammation. Although reduced migration alone can contribute to reduced inflammation, alterations in other neutrophil functions, such as NETosis, phagocytosis, oxidative burst, degranulation, and cytokine release, may also contribute to increased survival. Previous studies have demonstrated roles for CDK4/6 in the formation of neutrophil extracellular traps (40), CDK7/9 in neutrophil apoptosis (62), and CDK5 in the secretion of neutrophil-specific granules (63). It remains to be determined whether other neutrophil functions in addition to migration are regulated by CDK2. The pan-CDK inhibitor roscovitine competes with ATP for the occupation of the kinase active site (64) and is currently in multiple phase II clinical trials to inhibit cystic fibrosis and multiple types of tumors (https://clinicaltrials.gov). Previous studies in mice have shown that treatment with roscovitine at 10 to 100 mg/kg for 3 to 7 d after inflammation induction resulted in decreased neutrophil infiltration into the inflamed tissue and better clinical outcome (29). This reduced neutrophil recruitment was attributed to CDK9 inhibition and neutrophil apoptosis (65), whereas a link to CDK2 was not shown. Our data add to the list of non–cell cycle-related functions of CDKs, where acute inhibition of CDK2 suppresses neutrophil migration without affecting their viability. In Cdk2 knockout mice, normal myeloid cell numbers during resting and stressed conditions were observed (66), and no developmental defects other than a smaller body size and sterility were noted (67). Thus, CDK2 inhibition is not expected to cause adverse side effects, and thus presents an attractive strategy for short-term and long-term control of inflammation, which may be better tolerated than a pan-CDK inhibitor, such as roscovitine. In summary, our study demonstrates previously unappreciated roles of miR-199 and CDK2 in regulating neutrophil migration and points to new directions in managing neutrophilic inflammation.
Materials and Methods
miRNA Sequencing.
Neutrophil and apical epithelial cells were sorted from transgenic zebrafish lines Tg(lyzC:GFP) and Tg(Krt4:GFP) at 3 d postfertilization (dpf) using fluorescence-activated cell sorting (FACS). Total RNA was exacted using an Ambion mirVana kit (without enrichment for small RNA). A library was prepared using a TruSeq Small RNA Preparation Kit (Illumina). DNA of 145 to 160 bp was excised from Invitrogen 6% Novex TBE Gel and sequenced using an Illumina HiSeq 2000 system with paired-end reads of 50 bp with a total of 2 to 3 million per sample. The miRNA sequencing was performed at the University of Wisconsin Collaborative Genomics Core. For analysis, sequencing reads were mapped to the zebrafish genome (GRCz11) using an RNAseq aligner from STAR (v2.5) (68), using previously described parameters (69) to evaluate expression levels of zebrafish mature miRNAs based on uniquely mapped reads with the parameters “-s 1 –Q 10.” Before trimmed mean of M values (TMM) normalization and differential expression (DE) analysis by edgeR (v3.20.8) (70), miRNAs whose count per million (cpm) was less than 10 were removed from all samples. Differentially expressed miRNAs were selected if their false discovery rate (FDR)-adjusted P values were less than 0.05.
RNASeq.
Kidney marrow was dissected from 2 adults from Tg(lyzC: miR-199-3-dendra2)pu19 or Tg(lyzC: vector-dendra2)pu7, and neutrophils were sorted using FACS. Total RNA was extracted using a RNeasy Plus Mini Kit (no. 74104; Qiagen). RNAseq was performed at the Center for Medical Genomics at Indiana University School of Medicine. Samples were polyA-enriched and sequenced with an Illumina HiSeq 4000 ultralow system with reads ranging from 37 to 44 million. The RNAseq aligner from STAR (v2.5) (71) was employed to map RNAseq reads to the reference genome, zebrafish (GRCz11), with the following parameter: “–outSAMmapqUnique 60.” Uniquely mapped sequencing reads were assigned to genes using featureCounts (from subread v1.5.1) (71) with the following parameter: “-p -Q 10.” The genes were filtered for further analysis if their read was less than 0.5 cpm in more than 3 samples. The TMM method was adopted for gene expression normalization across all samples, followed by DE analysis between different conditions using edgeR (v3.20.8). Differentially expressed genes (DEGs) were determined for the comparison if its FDR-adjusted P value was less than 0.05 and the amplitude of fold change (FC) was larger than linear 2-fold. The functional analysis was performed on DEGs of our interest with a cutoff of FDR < 0.05 to identify significantly overrepresented gene ontology and/or Kyoto Encyclopedia of Genes and Genomes pathways, using the Database for Annotation, Visualization, and Integrated Discovery (72).
Protein–Protein Interactions.
Genes/proteins interacting with cdk2 or cdk5 in Danio rerio were retrieved from the protein–protein interactions database, STRING v11 (https://string-db.org/) (73). The pairs were filtered if their confidence scores were less than 0.4. The significance value of enrichment was calculated based on the hypergeometric model.
qRT-PCR.
MicroRNAs were extracted using a mirVana miRNA purification kit (Thermo Fisher Scientific) and reverse-transcribed with a Universal cDNA Synthesis Kit II (Exiqon). qRT-PCR was performed with ExiLENT SYBR Green Master Mix (Exiqon) using predesigned primers (Exiqon) in a LightCycler 96 Real-Time PCR System (Roche Life Science). U6 was used as a loading control. Messenger RNAs were extracted using the RNeasy Plus Mini Kit (no. 74104; Qiagen). One-step RT-qPCR was performed using a SuperScript III Platinum SYBR Green kit (Invitrogen) in a LightCycler 96 Real-Time PCR System with the loading control rpl32. The relative levels of microRNAs and mRNA were calculated using the deltaCt method. The specificity of the primers was verified as a single peak in the melt-curves. The relative fold change with correction of the primer efficiencies was calculated following instructions provided by Real-time PCR Miner (http://ewindup.info/miner/data_submit.htm) (74). The following primers were used: dre-cdk1+: 5′-ttggggtcccagtaagagtcta-3′, dre-cdk1−: 5′-ggtgtggaatagcgtgaagc-3′, dre-cdk2+: 5′-agatggcagagaccttctcg-3′, dre-cdk2−: 5′-cggaaaaaccgatgaacaag-3′, dre-cdk5+: 5′-tgggaacgccaacagaag-3′, dre-cdk5−: 5′-agctggatacatcgggtacg-3′, dre-cdca8+: 5′-atggcaccgctgaagtctac-3′, dre-cdca8−: 5′-cgttgaaggagctttctttga-3′; dre-kif14+: 5′-aggaggctcgcttgaagg-3′; dre-kif14−: 5′-ctctttagccacctgaatgc-3′, dre-mcm5+: 5′-aagctattgcctgcctgct-3′, dre-mcm5−: 5′-ccctcttcgtgtcagaccat-3′, dre-casp3b+: 5′-caggatattacgcatggagga-3′, dre-casp3b−: 5′-tcgcacagcgaggagataa-3′, dre-ikbkb+, 5′-gaggagcgagaaacaactgc-3′, dre-ikbkb−: 5′-tggactgcgaactttactgc-3′, dre-rpl32+: 5′-tcagtctgaccgctatgtcaa-3′, and dre-rpl32−: 5′-tgcgcactctgttgtcaatac-3′.
Generation of Transgenic Zebrafish Lines.
Genomic DNA fragments (300 to 600 bp) flanking the respective miRNAs were PCR-amplified from zebrafish genomic DNA using specific primers and inserted into the BbsI site in the vector backbone as described (22). Briefly, the miRNA, along with the flanking sequences, was inserted into an intron cassette in a Tol2 backbone containing the lyzC promoter and SV40 polyA for neutrophil-specific expression. The zebrafish cdk2 gene was cloned from zebrafish mRNA using a SuperScript III RT kit (no. 18080044; Invitrogen), amplified with primers (zCdk2+: 5′-gaaaaccccggtcctatggagtcttttcagaaagtggag-3′ and zCdk2−: 5′-catggctgattatgattataggcgtaaaggaggcactgg-3′), and inserted into a Tol2-mcherry-2A backbone. Point mutation was generated using infusion (no. 638920; Takara) site-directed mutagenesis with zCdk2 DN+: 5′-actggctaactttggtttggccagagcgttc-3′ and zCdk2 DN−: 5′-ccaaagttagccagtttgatctcgccctg-3′. Microinjections of fish embryos were performed by injecting 1 nL of mixture containing 25 ng/μL plasmid and 35 ng/μL Tol2 transposase mRNA in an isotonic solution into the cytoplasm of embryos at the 1-cell stage. The stable lines were generated as previously described (36). At least 2 founders (F0) for each line were obtained. Experiments were performed using F2 larvae produced by F1 fish derived from multiple founders to minimize the artifacts associated with random insertion sites.
Dual-Luciferase Reporter Assay.
Both zebrafish and human CDK2 3′UTRs were amplified from zebrafish or HL-60 cell genomic DNA with a SuperScript III RT kit (no. 18080044; Invitrogen) and cloned into psiCHECK2 (Promega) at XhoI and NotI sites with the following primers: zCDK2+:5′-taggcgatcgctcgagaacgagatcaactttggcaag-3′, zCdk2−: 5′-ttgcggccagcggccgctgtaacaacataaaccaaatgtt-3, hCDK2+: 5′-taggcgatcgctcgagagccttcttgaagccccca-3′, and hCdk2−: 5′-ttgcggccagcggccgctataaaactaggcacatttttttaa-3′. Mutated 3′UTR constructs were generated using an In-Fusion HD Cloning Kit (Clontech) with the following primers: zCdk2 mut+: 5′-cctgtgtgacccaaaaaaacagctgtgctaatagt-3′, zCdk2 mut−: 5′-tttgggtcacacaggttgcaatggataatcttca-3′, and hCdk2 mut+: 5′-aaaacgtgaccgaggagtctattttaaagaattcgg-3′. DNA encoding miR-199 or vector was amplified from the construct used for expression in zebrafish and inserted into pcDNA3.1 at the HindIII/XbaI cloning sites using the following primers: pcDNA-199+: 5′-gtttaaacttaagcttgccaccatggatgaggaaatcgc-3′ and pcDNA-199−: 5′-aaacgggccctctagagaccggtacccccgggctgc-3′.
Live Imaging.
Larvae at 3 dpf were settled on a glass-bottom dish, and imaging was performed at 28 °C. Time-lapse fluorescence images in the head mesenchyme were acquired with a laser-scanning confocal microscope (LSM710; Zeiss) with a Plan-Apochromat 20×/0.8 M27 objective. Neutrophil motility at the CHT was imaged using a Zeiss EC Plan-NEOFLUAR 10×/0.3 objective. For neutrophil nucleus and cytosol reporter line imaging, an LD C-Apochromat 40×/1.1 W Korr M27 objective was used. The green and red channels were acquired sequentially with 0.1 ∼ 0.3% power of the 488-nm laser and 0.5 ∼ 2% power of the 561-nm laser, respectively, with a 200-μm pinhole at a speed of 1.27 μs per pixel and averaged (line 2). The fluorescent stacks were flattened using the maximum intensity projection and overlaid with a single slice of the bright-field image. Neutrophil chemotaxis upon LTB4 treatment was captured with a Zeiss ZV16 dissection microscope at a magnification of 2× every 15 s for 30 min.
Inflammation Assays in Zebrafish.
Zebrafish wounding and infection were performed as described (22). Briefly, 3-dpf larvae were amputated posterior to the notochord or inoculated with PAK into the left otic vesicle or into the vasculature at 1,000 colony-forming units (CFU) per embryo. The larvae were fixed in 4% paraformaldehyde at 1 h postwounding or postinfection. Neutrophils were stained with Sudan Black, and the numbers at the indicated regions were quantified. For LTB4 recruitment assays, 3-dpf zebrafish larvae were treated with 30 nM LTB4 for 15 min and fixed. Neutrophils were stained with Sudan Black, and the numbers at the indicated regions were quantified.
Generation of Stable HL-60 Cell Lines.
HL-60 cells were obtained from the American Type Culture Collection (CCL-240) and cultured using Roswell Park Memorial Institute (RPMI)-1640 with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) supplemented with 10% fetal bovine serum (FBS) and sodium bicarbonate. The lentiviral backbone pLIX_403 was a gift from David Root, Broad Institute, Cambridge, MA (plasmid no. 41395; Addgene). The DNA sequence flanking MIR-199 was cloned from HL-60 cell genomic DNA and then cloned into a backbone containing dendra2. The miRNA and dendra2 reporter were then cloned into pLIX_403 vector using the NheI/AgeI sites with the following primer set: pLIX-mir+: 5′-tggagaattggctagcgccaccatggatgaggaaatcgc-3′ and pLIX-mir−: 5′-catacggataaccggttaccacacctggctgggc-3′. Stable HL-60 cell lines were generated as described (25, 75). Briefly, HEK-293 cells were transfected with pLIX_403 or pLKO.1, together with VSV-G and CMV8.2 using Lipofectamine 3000 (Thermo Fisher Scientific). The viral supernatant was harvested on day 3 and concentrated with Lenti-X concentrator (no. 631232; Clontech). HL-60 cells were infected with concentrated lentivirus in complete RPMI medium supplemented with 4 μg/mL polybrene (TR-1003-G; Sigma) at 2,500 × g for 1.5 h at 32 °C and then selected with 1 μg/mL puromycin (A1113803; Gibco) to generate stable lines.
Inhibitor Treatment for Zebrafish Larvae.
CDK2 inhibitors NU6140 (no. ALX-270-441; Enzo), CTV313 (no. 238803; Sigma), and roscovitine (no. S1153; Selleckchem) were dissolved in dimethyl sulfoxide (DMSO) to make a 100 mM stock and then further diluted in E3 to working concentrations (100 μM for recruitment and motility assays and indicated concentration for survival assays). For inhibiting DNA replication, 150 μM aphidicolin and 20 mM hydroxyurea (no. A0781; Sigma) in 1% DMSO were used as previously described (32). For neutrophil recruitment and random motility assays, larvae were pretreated with the inhibitor for 1 h before experimental procedures. For survival assays, larvae were pretreated and kept in the inhibitors from 1 h postinfection.
Transwell Migration Assay.
The dHL-60 transwell assays were performed as described (75). Briefly, 2 × 106 differentiated cells per milliliter were resuspended in Hanks’ balanced salt solution (HBSS) with 0.5% FBS and 20 mM Hepes. One hundred microliters was placed into a 6.5-mm-diameter, 3-μm pore size transwell inset (no. 3415; Corning) and allowed to migrate for 2 h at 37 °C toward 100 nM fMLP in 500 μL of HBSS in a 24-well plate. Loading controls were done by directly adding 100 μL of cells to 400 μL of HBSS. Cells that migrated to the lower chamber were released with 0.5 M ethylenediaminetetraacetic acid and counted using a BD LSRFortessa flow cytometer with an acquisition time of 30 s. The counts were normalized with the total numbers of cells added to each well, and the data were then gated for live cells and analyzed with Beckman Kaluza 2.1 software.
Primary Neutrophil Isolation and Chemotaxis Assay.
Primary human neutrophils were isolated with a Miltenyi MACSxpress Neutrophil Isolation Kit. Cells were stained for 10 min with calcein AM and washed with phosphate-buffered saline (PBS). A total of 107 cells per milliliter were incubated in RPMI + 0.1% human serum albumin (HSA) containing 50 μM NU6104 or DMSO at 37 °C/5% CO2 for 45 min. Microfluidic devices were fabricated as previously described (76). Microfluidic chambers were coated with 10 μg/mL fibrinogen for 1 h at 37 °C in PBS. Three microliters of neutrophil suspension (4 × 106 cells per milliliter) was added to the device, and 5 μm of interleukin-8 was added to the source port. The gradient was allowed to set up and equilibrate in a 37 °C humidified chamber for 15 min before imaging. Time-lapse imaging was performed using a 10× numerical aperture (NA) of 0.45, or 60× NA of 1.4 objective and motorized stage (Ludl Electronic Products) on an inverted microscope (Eclipse TE300), using a charge-coupled device camera (CoolSNAP ES2), and captured into MetaVue imaging software v6.2. Images were taken every 30 s for 30 to 45 min, with up to 8 devices being imaged simultaneously. Tracking and velocity quantification were performed as described (9).
Survival Assay.
Larvae at 3 dpf were injected with 1 nL of 25 ng/nL LPS or 1,000 CFU of PAK into the tail vein and incubated individually in 96-well plates. Survival was tracked for 4 d or when a group reached 100% mortality. Representative results of at least 3 independent experiments (n ≥ 20 larvae in each experiment) were shown.
Statistical Analysis.
Statistical analysis was carried out using PRISM 6 (GraphPad). The Mann–Whitney test (when comparing 2 groups), Kruskal–Wallis test (when comparing with a single group), and Gehan–Breslow–Wilcoxon test (for survival curve comparison) were used in this study as indicated in the figure legends. Individual P values are indicated in the figures, with no data points excluded from statistical analysis. One representative experiment of at least 3 independent repeats is shown. For qPCR, each gene was normalized to a reference gene and compared with the Holm–Sidak test (individual comparison with paired value), where each P value and degree of freedom was adjusted for respective multiple comparisons.
Supplementary Material
Acknowledgments
The work was supported by research funding from the NIH (Grant R35GM119787 to Q.D., Grant R35GM118027 to A.H., Grant R01HD073156 to D.M.U., and Grant P30CA023168 to the Purdue Center for Cancer Research) for shared resources. Bioinformatics analysis was conducted by the Collaborative Core for Cancer Bioinformatics shared by the Indiana University Simon Cancer Center (Grant P30CA082709) and the Purdue University Center for Cancer Research with support from the Walther Cancer Foundation. A.Y.H. is supported by the Purdue Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.D.L. is a guest editor invited by the Editorial Board.
Data deposition: The microRNA-sequencing and RNA-sequencing raw and processed data are publicly accessible in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE127174).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905221116/-/DCSupplemental.
References
- 1.Sadik C. D., Kim N. D., Luster A. D., Neutrophils cascading their way to inflammation. Trends Immunol. 32, 452–460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan C., Neutrophils and immunity: Challenges and opportunities. Nat. Rev. Immunol. 6, 173–182 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Soehnlein O., Steffens S., Hidalgo A., Weber C., Neutrophils as protagonists and targets in chronic inflammation. Nat. Rev. Immunol. 17, 248–261 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Remick D. G., et al. , CXC chemokine redundancy ensures local neutrophil recruitment during acute inflammation. Am. J. Pathol. 159, 1149–1157 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams M. R., Azcutia V., Newton G., Alcaide P., Luscinskas F. W., Emerging mechanisms of neutrophil recruitment across endothelium. Trends Immunol. 32, 461–469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sônego F., et al. , Paradoxical roles of the neutrophil in sepsis: Protective and deleterious. Front. Immunol. 7, 155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mócsai A., Walzog B., Lowell C. A., Intracellular signalling during neutrophil recruitment. Cardiovasc. Res. 107, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lämmermann T., et al. , Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Yamahashi Y., et al. , Integrin associated proteins differentially regulate neutrophil polarity and directed migration in 2D and 3D. Biomed. Microdevices 17, 100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman R. C., Farh K. K. H., Burge C. B., Bartel D. P., Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurol T., Zhou W., Deng Q., MicroRNAs in neutrophils: Potential next generation therapeutics for inflammatory ailments. Immunol. Rev. 273, 29–47 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Rupaimoole R., Slack F. J., MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Olarerin-George A. O., Anton L., Hwang Y. C., Elovitz M. A., Hogenesch J. B., A functional genomics screen for microRNA regulators of NF-kappaB signaling. BMC Biol. 11, 19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K., Vinayagam A., Perrimon N., A rapid genome-wide microRNA screen identifies miR-14 as a modulator of Hedgehog signaling. Cell Rep. 7, 2066–2077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Martin A., et al. , The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat. Immunol. 17, 433–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnnidis J. B., et al. , Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Fan H. B., et al. , miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood 124, 1320–1330 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Zhou W., et al. , MicroRNA-223 suppresses the canonical NF-κB pathway in basal keratinocytes to dampen neutrophilic inflammation. Cell Rep. 22, 1810–1823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan C., Kim C. H., Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 25, 341–350 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Lieschke G. J., Currie P. D., Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Takaki K., Cosma C. L., Troll M. A., Ramakrishnan L., An in vivo platform for rapid high-throughput antitubercular drug discovery. Cell Rep. 2, 175–184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu A. Y., et al. , Overexpression of microRNA-722 fine-tunes neutrophilic inflammation by inhibiting Rac2 in zebrafish. Dis. Model. Mech. 10, 1323–1332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D., NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob C., et al. , DMSO-treated HL60 cells: A model of neutrophil-like cells mainly expressing PDE4B subtype. Int. Immunopharmacol. 2, 1647–1656 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Hsu A. Y., et al. , Inducible overexpression of zebrafish microRNA-722 suppresses chemotaxis of human neutrophil like cells. Mol. Immunol. 112, 206–214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L., Hannon G. J., MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Lahoz-Beneytez J., et al. , Human neutrophil kinetics: Modeling of stable isotope labeling data supports short blood neutrophil half-lives. Blood 127, 3431–3438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R., et al. , Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene 27, 4712–4723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi A. G., et al. , Cyclin-dependent kinase inhibitors enhance the resolution of inflammation by promoting inflammatory cell apoptosis. Nat. Med. 12, 1056–1064 (2006). Correction in: Nat. Med. 12, 1434 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Malumbres M., Barbacid M., Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Pennati M., et al. , Potentiation of paclitaxel-induced apoptosis by the novel cyclin-dependent kinase inhibitor NU6140: A possible role for survivin down-regulation. Mol. Cancer Ther. 4, 1328–1337 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Zhang L., Kendrick C., Jülich D., Holley S. A., Cell cycle progression is required for zebrafish somite morphogenesis but not segmentation clock function. Development 135, 2065–2070 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuhara S., et al. , Visualizing the cell-cycle progression of endothelial cells in zebrafish. Dev. Biol. 393, 10–23 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharjee R. N., Banks G. C., Trotter K. W., Lee H. L., Archer T. K., Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol. Cell. Biol. 21, 5417–5425 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Heuvel S., Harlow E., Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262, 2050–2054 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Deng Q., Yoo S. K., Cavnar P. J., Green J. M., Huttenlocher A., Dual roles for Rac2 in neutrophil motility and active retention in zebrafish hematopoietic tissue. Dev. Cell 21, 735–745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J., et al. , Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 114, 201–214 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Graziano B. R., et al. , A module for Rac temporal signal integration revealed with optogenetics. J. Cell Biol. 216, 2515–2531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang E. R., Liu S., Wu L. F., Altschuler S. J., Cobb M. H., Chemoattractant concentration-dependent tuning of ERK signaling dynamics in migrating neutrophils. Sci. Signal. 9, ra122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amulic B., et al. , Cell-cycle proteins control production of neutrophil extracellular traps. Dev. Cell 43, 449–462.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Brannon M. K., et al. , Pseudomonas aeruginosa type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell. Microbiol. 11, 755–768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bach S., et al. , Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 280, 31208–31219 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Cui C., Wang Y., Wang Y., Zhao M., Peng S., Exploring the relationship between the inhibition selectivity and the apoptosis of roscovitine-treated cancer cells. J. Anal. Methods Chem. 2013, 389–390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu A. Y., et al. , Development and characterization of an endotoxemia model in zebra fish. Front. Immunol. 9, 607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao M., et al. , Mechanisms of impaired neutrophil migration by microRNAs in myelodysplastic syndromes. J. Immunol. 198, 1887–1899 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Surmiak M., Hubalewska-Mazgaj M., Wawrzycka-Adamczyk K., Musiał J., Sanak M., Neutrophil MiRNA-128-3p is decreased during active phase of granulo-matosis with polyangiitis. Curr. Genomics 16, 359–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arroyo A. B., et al. , MiR-146a regulates neutrophil extracellular trap formation that predicts adverse cardiovascular events in patients with atrial fibrillation. Arterioscler. Thromb. Vasc. Biol. 38, 892–902 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Li W., et al. , miR-199a-5p regulates β1 integrin through Ets-1 to suppress invasion in breast cancer. Cancer Sci. 107, 916–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S. H., et al. , MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int. J. Clin. Exp. Pathol. 7, 7131–7141 (2014). [PMC free article] [PubMed] [Google Scholar]
- 50.Koshizuka K., et al. , Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 108, 1681–1692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song J., et al. , MiR-199a regulates cell proliferation and survival by targeting FZD7. PLoS One 9, e110074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye H., et al. , A critical role of mir-199a in the cell biological behaviors of colorectal cancer. Diagn. Pathol. 10, 65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y., et al. , Plasma miR-199a-5p is increased in neutrophilic phenotype asthma patients and negatively correlated with pulmonary function. PLoS One 13, e0193502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X. M., Zhang H. J., miRNAs as new molecular insights into inflammatory bowel disease: Crucial regulators in autoimmunity and inflammation. World J. Gastroenterol. 22, 2206–2218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Favreau A. J., McGlauflin R. E., Duarte C. W., Sathyanarayana P., miR-199b, a novel tumor suppressor miRNA in acute myeloid leukemia with prognostic implications. Exp. Hematol. Oncol. 5, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hydbring P., Malumbres M., Sicinski P., Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat. Rev. Mol. Cell Biol. 17, 280–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laphanuwat P., Jirawatnotai S., Immunomodulatory roles of cell cycle regulators. Front. Cell Dev. Biol. 7, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirota T., et al. , Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat. Genet. 43, 893–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gui P., et al. , Rho/ROCK pathway inhibition by the CDK inhibitor p27(kip1) participates in the onset of macrophage 3D-mesenchymal migration. J. Cell Sci. 127, 4009–4023 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Higashi H., et al. , Differences in substrate specificity between Cdk2-cyclin A and Cdk2-cyclin E in vitro. Biochem. Biophys. Res. Commun. 216, 520–525 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Chi Y., et al. , Identification of CDK2 substrates in human cell lysates. Genome Biol. 9, R149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leitch A. E., et al. , Cyclin-dependent kinases 7 and 9 specifically regulate neutrophil transcription and their inhibition drives apoptosis to promote resolution of inflammation. Cell Death Differ. 19, 1950–1961 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosales J. L., Ernst J. D., Hallows J., Lee K. Y., GTP-dependent secretion from neutrophils is regulated by Cdk5. J. Biol. Chem. 279, 53932–53936 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Otyepka M., Bártová I., Kríz Z., Koca J., Different mechanisms of CDK5 and CDK2 activation as revealed by CDK5/p25 and CDK2/cyclin A dynamics. J. Biol. Chem. 281, 7271–7281 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Hoodless L. J., et al. , Genetic and pharmacological inhibition of CDK9 drives neutrophil apoptosis to resolve inflammation in zebrafish in vivo. Sci. Rep. 5, 36980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jayapal S. R., et al. , Hematopoiesis specific loss of Cdk2 and Cdk4 results in increased erythrocyte size and delayed platelet recovery following stress. Haematologica 100, 431–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis P., Cdk2 knockout mice are viable. Curr. Biol. 13, 1775–1785 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simopoulos A. P., Diagnostic implications of cleft palate in the newborn. J. Am. Med. Womens Assoc. 21, 921–923 (1966). [PubMed] [Google Scholar]
- 70.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Huang da W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao S., Fernald R. D., Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 12, 1045–1062 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cavnar P. J., Berthier E., Beebe D. J., Huttenlocher A., Hax1 regulates neutrophil adhesion and motility through RhoA. J. Cell Biol. 193, 465–473 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berthier E., et al. , Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog. 9, e1003289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.