Significance
Rice resistance against blast, a devastating fungal disease, is typically mediated by nucleotide-binding site leucine-rich repeat (NLR) proteins. Most previous studies focused on individual NLR genes, but single R genes typically confer no durable resistance owing to their narrow resistance spectrum. In this study, we sequenced the genome of Tetep, a widely used resistance donor, to decipher the molecular basis of its broad-spectrum and durable blast resistance. Large-scale cloning and functional analysis of annotated NLRs uncovered a large number of functional NLR genes and interactive NLR networks in the genome. Moreover, pedigree tracing of elite cultivars indicated the more NLRs inherited from Tetep the better resistance of the cultivar. Various datasets were provided for facilitating breeding for new resistant cultivars.
Keywords: Oryza sativa, NBS-LRR gene, blast resistance, R-gene cloning, NLR network
Abstract
Tetep is a rice cultivar known for broad-spectrum resistance to blast, a devastating fungal disease. The molecular basis for its broad-spectrum resistance is still poorly understood. Is it because Tetep has many more NLR genes than other cultivars? Or does Tetep possess multiple major NLR genes that can individually confer broad-spectrum resistance to blast? Moreover, are there many interacting NLR pairs in the Tetep genome? We sequenced its genome, obtained a high-quality assembly, and annotated 455 nucleotide-binding site leucine-rich repeat (NLR) genes. We cloned and tested 219 NLR genes as transgenes in 2 susceptible cultivars using 5 to 12 diversified pathogen strains; in many cases, fewer than 12 strains were successfully cultured for testing. Ninety cloned NLRs showed resistance to 1 or more pathogen strains and each strain was recognized by multiple NLRs. However, few NLRs showed resistance to >6 strains, so multiple NLRs are apparently required for Tetep’s broad-spectrum resistance to blast. This was further supported by the pedigree analyses, which suggested a correlation between resistance and the number of Tetep-derived NLRs. In developing a method to identify NLR pairs each of which functions as a unit, we found that >20% of the NLRs in the Tetep and 3 other rice genomes are paired. Finally, we designed an extensive set of molecular markers for rapidly introducing clustered and paired NLRs in the Tetep genome for breeding new resistant cultivars. This study increased our understanding of the genetic basis of broad-spectrum blast resistance in rice.
Tetep is a rice cultivar known for its broad-spectrum (i.e., conferring resistance to multiple and diverse blast pathogens) and durable (i.e., long lasting) resistance to blast, a devastating rice disease caused by the fungus Magnaporthe oryzae (1, 2). Currently Tetep is known to be susceptible to very few blast pathogens (1, 3). Resistance to blast is usually mediated by disease resistance genes (R genes), most of which are NLR (nucleotide-binding site leucine-rich repeat) genes (2), which either directly or indirectly recognize pathogen effector proteins and initiate a defense response (4). To date, over 100 blast R genes have been identified (5), but this number is likely an underestimate as some R genes with no strong effect might not have been identified. Although it is possible that instead of NLRs some physical/biochemical factors are responsible for the Tetep resistance, the many famous R genes that have already been cloned from Tetep (6) indicate that this rice variety is a useful target for resistance study. Although a number of NLR-type resistance genes have been cloned from Tetep or other cultivars, few of them conferred broad-spectrum resistance (7). Indeed, none of the cultivars derived from breeding programs using Tetep as the resistance donor could maintain durable resistance. This observation implies that many NLR genes are involved in the defense network of Tetep. So, does Tetep show broad and durable resistance to blast because it has many more NLR genes in its genome compared to other cultivars? Or are there multiple major NLR genes in Tetep that can individually confer broad-spectrum resistance to blast?
Another interesting question is whether NLR genes frequently interact with each other. In particular, how many of the NLR genes in the Tetep genome form interacting pairs? In an NLR pair, 1 gene (known as the helper) activates the defense signaling upon sensing the presence of a blast pathogen, while the other (known as the sensor) recognizes pathogen effectors and acts as an inhibitor on the helper to prevent autoimmunity in the absence of the pathogen (8). As many known NLR pairs confer strong blast resistance (8, 9), it is interesting to find if the Tetep genome carries many NLR pairs.
To answer these questions, we first obtained a high-quality assembly of the Tetep genome using PacBio RS II and Illumina sequencing. Second, we identified the NLR genes in the Tetep genome and compared them to those in Nipponbare, MH63 and R498, 3 well-annotated rice cultivars, to see whether there is an excess of NLR genes in Tetep (10). Third, we conducted a large-scale cloning, transformation, and functional study of Tetep NLRs in 2 susceptible rice cultivars to identify Tetep NLR genes that are potentially blast resistant. Cloning NLR genes is extremely labor intensive due to their long gene length and high numbers of polymorphisms among cultivars (11). We overcame this by using the newly built Tetep genome and extensively tested cloning vectors. Fourth, by combining pedigree tracing, identification of paired NLRs and functional evaluation by CRISPR knockout, we tried to decipher the NLR defense networks in Tetep. We also developed an efficient approach to identify NLR pairs in a genome. Through these studies, we sought to reveal the genetic basis of Tetep’s broad and durable resistance to blast and to provide an NLR gene repository. Finally, we designed and tested an extensive set of molecular breeding markers that will facilitate the breeding of new blast-resistant cultivars.
Results
Sequencing, Assembly, and Annotation of the Tetep Genome.
In order to assemble the Tetep genome, we generated ∼7.8 million PacBio RS II subreads with a total of 66G base pairs (SI Appendix, Table S1), equivalent to ∼150× genome coverage. The initial assembly of the genome by Canu (12) was ∼400 Mb in size, consisting of 1,119 contigs, with a N50 of ∼800 kb (SI Appendix, Fig. S1 and Table S2). With the Minghui63 (MH63) genome (13) as a reference (SI Appendix, SI Materials and Methods), 760 contigs, covering ≥80% of the sum of all contig lengths, were successfully ordered into 12 chromosome pseudomolecules. The assembled genome contained ∼98.0% of the 248 core eukaryotic genes (14), which is comparable to those in the MH63 (13) genome (95.2%) and the R498 (15) genome (99.2%) (SI Appendix, Table S3), indicating a high completeness of the assembled genome. We predicted 37,054 protein-coding genes by collecting evidence from homologous proteins, expressed sequence tags (ESTs), and RNA-seq data (SI Appendix, Table S1) using MAKER-P (16) (SI Appendix, SI Materials and Methods). This number is similar to those annotated in MH63 (37,324) (13) and R498 (37,549) (15) (SI Appendix, Table S4).
As we were especially interested in NLR genes, we added an ab initio prediction using Fgenesh (17) to capture more NLR genes. Through a combination of hidden Markov model (HMM)- and motif-based strategies (18, 19) (SI Appendix, SI Materials and Methods), 455 NLRs were identified in the assembled Tetep genome, which is similar to the numbers of NLR genes found in the Nipponbare (473) and MH63 (455) genomes, but more than the number (409, not statistically significant) found in the R498 genome (15) using the same prediction pipeline (SI Appendix, Table S4). Moreover, the 93 Tetep NLRs sequenced by the Sanger technique (averaged length over 3.7 kb, SI Appendix, SI Materials and Methods) could all be found in the annotated Tetep NLRs with the criteria of >99% sequence identity and >99% coverage.
Differences in NLR Genes among 4 Rice Genomes.
To evaluate the variation in NLR genes among different rice genomes, we searched for all possible homologs of Tetep NLR genes in the Nipponbare, MH63, and R498 genomes using OrthoFinder (20), and calculated their nucleotide diversities (SI Appendix, Table S5). We found that ∼20 to 27% of the annotated 455 NLR genes in Tetep lack a clear homolog in the Nipponbare, MH63, or R498 genome (SI Appendix, Table S5). Even if we only considered the nearest NLR gene orthologous pairs, we found on average 7- to 10-fold higher nucleotide diversity between the 2 genes of an NLR orthologous pair than the genomic average between Tetep and the other 3 genomes (SI Appendix, Table S5). At least 17 Tetep NLRs seem to be absent (defined as NBS domain protein sequence identity and coverage <50%) in Nipponbare; this number increases to 31 in MH63 and 19 in R498, possibly partly due to fewer annotated NLRs in MH63 and R498 (SI Appendix, Table S4). These observations suggest large differences in NLRs among the 4 rice cultivars, consistent with the high level of diversity found in NLRs (21, 22).
The differences in NLRs between Tetep and Nipponbare also include gene structure changes. When we mapped the Illumina reads of Tetep to the Nipponbare genome, 140 of the 455 NLR genes were predicted to harbor at least 1 disruptive change, such as a premature stop codon or a frameshift mutation in the coding frame. This result is consistent with the previous finding of frequent NLR pseudogenes in population analysis (23, 24). However, when the prediction was based on our assembled Tetep genome, those putative pseudogenes were actually not pseudogenes but spliced isoforms in the Tetep genome; the isoforms were supported by RNA-seq data.
Many Tetep NLRs Confer Blast Resistance to Susceptible Cultivars.
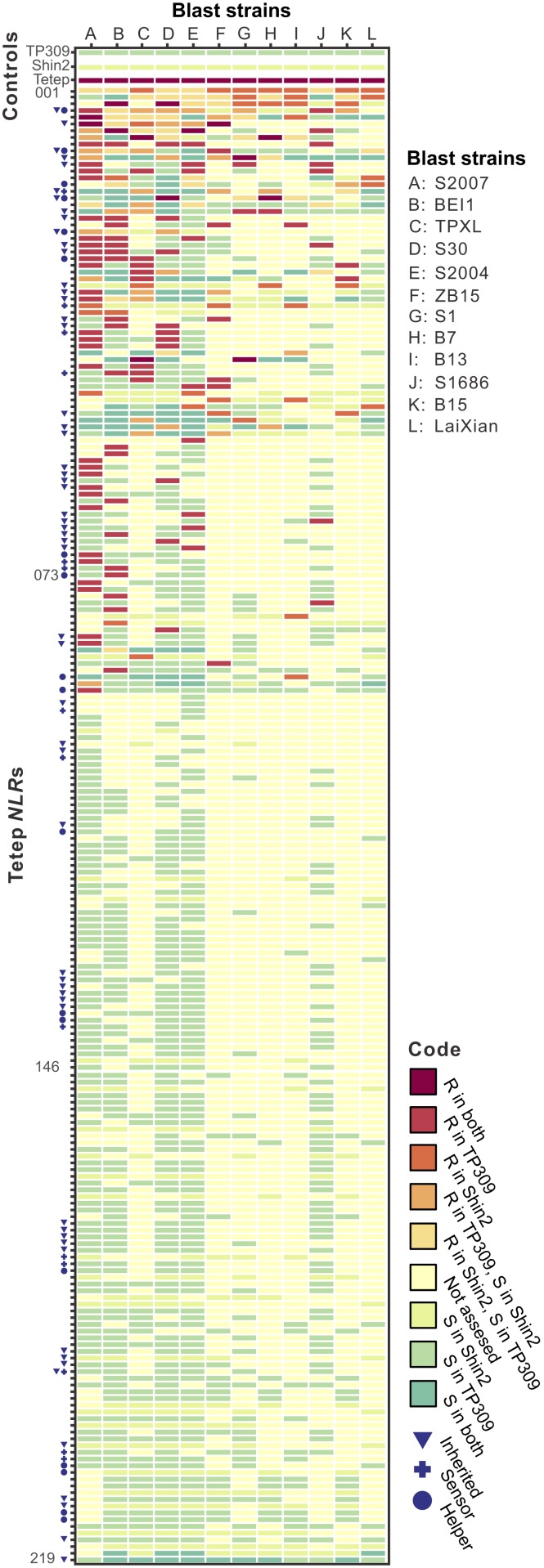
In order to understand the genetic basis of the durable and broad-spectrum blast resistance of Tetep, we did a genome-wide study of NLR genes in Tetep using gene cloning, transformation, and functional tests. We were able to clone 219 (∼50%) NLRs with their native promoters and terminators and transform them into at least 1 of 2 susceptible rice cultivars, Oryza sativa ssp. japonica cv. TP309 and O. sativa ssp. japonica cv. Shin2 (Fig. 1 and SI Appendix, Table S6). The transformed rice lines were tested for resistance using 5 to 12 independent strains of M. oryzae (SI Appendix, Fig. S2) as described previously (25, 26). Each test was repeated at least 3 times.
Fig. 1.
Resistance map of Tetep NLR genes. In a whole genome random survey, 219 NLRs (numbered 001 through 219) in the Tetep genome were cloned and tested for resistance against 12 highly diversified blast pathogen strains (from S2007 to LaiXian). Two susceptible rice cultivars, japonica cv. TP309 and Shin2, were used as transformation recipient cultivars. Each tested NLR is colored according to its resistance status, i.e., resistant (R) or susceptible (S), to the given blast strain in either cultivar. NLRs inherited in at least 1 of 5 Tetep-derived cultivars are marked by blue triangles. Paired NLRs are marked by a blue circle (a helper) or a cross (a sensor). A full list of the 219 NLR genes is given in SI Appendix, Table S6.
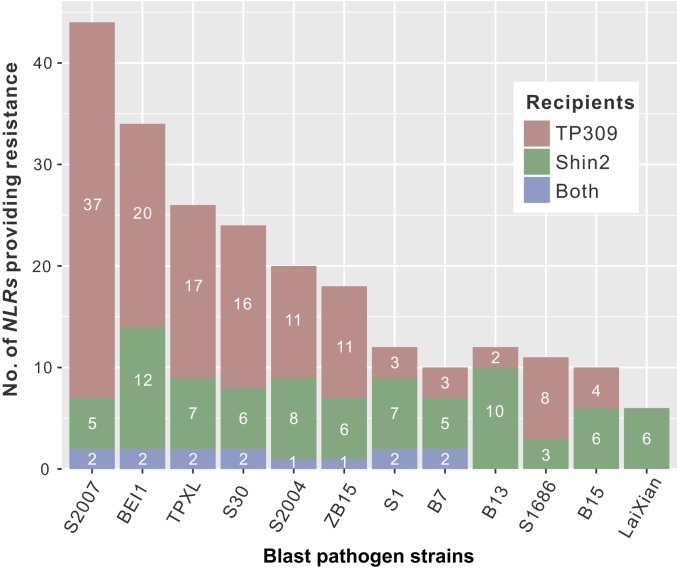
In total, at least 90 cloned NLRs (∼41% of the 219 cloned NLRs) conferred resistance to 1 or more of the 12 blast pathogen strains tested. (In many cases, fewer than 12 pathogen strains were tested due to experimental difficulties such as hard-to-control sporulation efficiency of a fungal strain under testing and unsatisfactory controls in some tests. See SI Appendix, SI Materials and Methods) For each pathogen strain, an average of 19 (ranging from 6 to 44) transgenic NLRs conferred resistance to a normally susceptible rice cultivar (Fig. 2), suggesting high overlapping resistance in the Tetep genome. This high overlapping resistance might contribute to the durable blast resistance in Tetep.
Fig. 2.
Numbers of tested Tetep NLRs that conferred resistance to 1 or more blast pathogen strains in 2 susceptible recipient cultivars TP309 and Shin2. The Tetep NLRs were cloned and transformed into TP309 and Shin2 separately and tested using 12 diversified blast pathogen strains. Each pathogen strain could be recognized by multiple NLRs. The number of recognized NLRs to each strain in TP309 or/and Shin2 is shown in the bar charts.
Interestingly, different NLRs had different resistance spectra. Among the 219 NLR genes cloned, 38 NLRs could recognize only 1 pathogen strain, while 19, 14, 8, and 11 could recognize 2, 3, 4, and ≥5 different blast strains, respectively (Fig. 1 and SI Appendix, Table S6). We used 7 of the 11 NLRs that conferred resistance to ≥5 blast strains to test 9 additional pathogen strains, increasing the maximum number of pathogen strains that could be tested to 21. We found that 3 NLR genes (chr12.fgenesh1062, chr06.fgenesh1195, and chr06.fgenesh377) could recognize 18, 14, and 11 of the 21 pathogen strains tested (SI Appendix, Table S7). However, no cloned NLR gene could recognize all tested pathogen strains. Therefore, to build a rice genome conferring broad-spectrum blast resistance, a combination of multiple NLR genes with different resistance spectra may be necessary.
Tetep NLRs Persisting in Modern Cultivars.
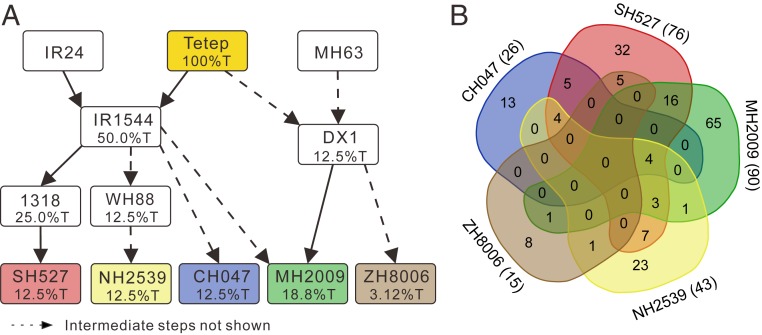
Tetep has been extensively used as a blast resistance donor in breeding programs. Which Tetep NLR genes were actually selected in these breeding programs? To answer this question, 5 elite cultivars, Chenghui047 (CH047), Mianhui2009 (MH2009), Neihui2539 (NH2539), Shuhui527 (SH527), and Zhonghui8006 (ZH8006), all of which used Tetep as the blast resistance donor, and their cross parents and intermediate lines were sequenced. From the pedigree in Fig. 3A, we inferred that 15 to 90 NLRs have been passed from Tetep to its descendants (Fig. 3B). Two cultivars, MH2009 and SH527, with better blast resistance, carry 90 and 76 Tetep NLR genes, respectively, while ZH8006, which is least resistant to blast, carries only 15 NLR genes from Tetep, suggesting that cultivars with more Tetep NLRs have broader and more durable resistance (http://www.ricedata.cn/variety/). There are 47 Tetep NLRs that have been passed to more than 1 cultivar; however, no Tetep NLR was shared by all 5 cultivars and only 4 Tetep NLRs were found in CH047, SH527, ZH8006, and MH2009 (Fig. 3B). These observations suggest that the resistance spectrum of a descendant cultivar is largely determined by its subset of the Tetep NLRs. This observation is consistent with the above view that multiple NLR genes are required to produce a broad-spectrum and durable resistant line.
Fig. 3.
Inheritance of NLR genes in breeding programs. (A) Pedigree of rice cultivars derived from Tetep. The percentage in each box shows the probability that a given locus was inherited from Tetep (T). The dashed lines indicate that some intermediate steps are not shown in this pedigree. (B) Inherited NLRs in 5 Tetep descendants. The Venn diagram shows the inheritance of Tetep NLRs in 5 modern cultivars derived from breeding programs that used Tetep as the resistant donor. The number of inherited NLRs in a cultivar is given in parentheses following the cultivar name.
Of all of the 188 inherited NLR genes, 56 were tested for blast resistance in the above genome-wide survey (Fig. 1 and SI Appendix, Table S6). Interestingly, ∼57% (32 genes) of the 56 genes were found to confer resistance to at least 1 blast pathogen strain, which was significantly higher than that observed in the genome survey (41%) (219 randomly sampled NLR genes, χ2 with Yates correction = 4.2, degrees of freedom [d.f.] = 1, P = 0.040). Moreover, nearly 80% of the inherited NLR genes were also found to be physically near each other (<300 kb); that is, they formed NLR gene clusters. These NLR gene-enriched clusters can be readily employed in resistance breeding, greatly facilitating breeding programs (see Discussion).
Identifying NLR Pairs.
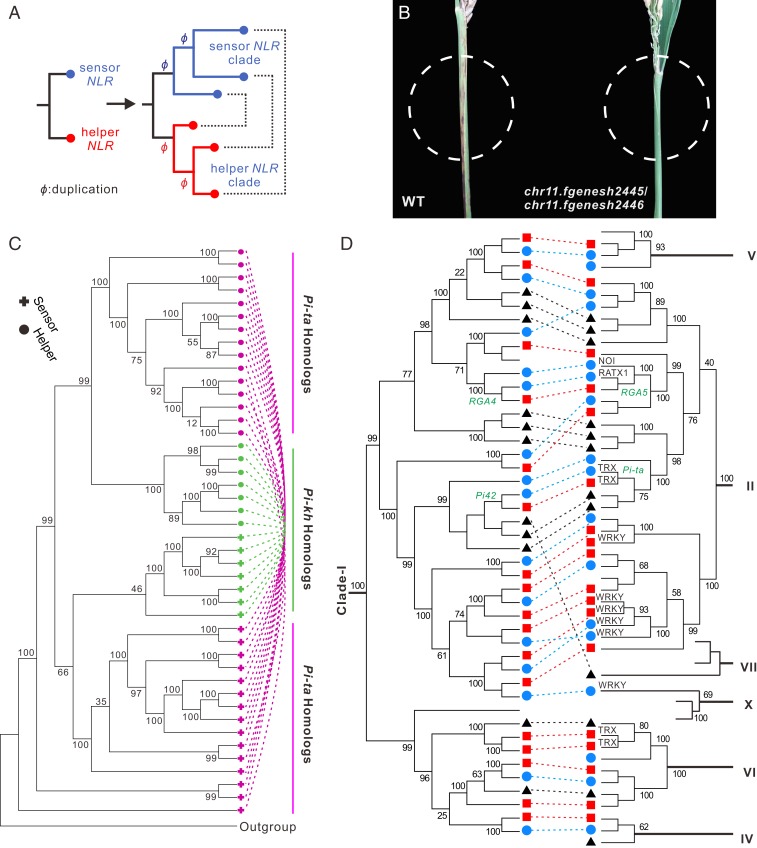
The findings of the RRS1/RPS4 pair in Arabidopsis (27) and the RGA4/RGA5 pair in rice (28) suggested 2 NLR genes could function together, known as a sensor/helper pair (8, 29). The sensor was often found to carry an extra domain, e.g., WRKY, RATX, NOI, and TRX, which could act as a decoy of pathogen effectors (30). However, it remains unclear as to 1) how often NLRs in a genome function in pairs, and 2) how such pairs have evolved. By searching for head-to-head NLR genes in the Tetep, Nipponbare, MH63, and R498 genomes (SI Appendix, SI Materials and Methods for details), we found 43 pairs in Tetep, 47 in Nipponbare, 33 in MH63, and 47 in R498, suggesting that ∼20% of NLRs are paired in a rice genome (Fig. 4 and SI Appendix, Fig. S3 and Table S8).
Fig. 4.
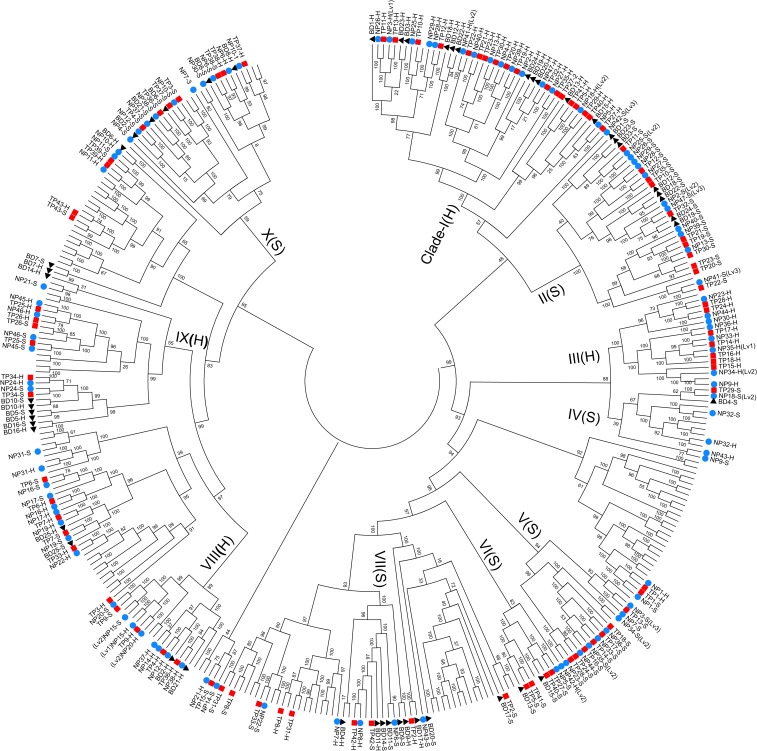
Features of paired NLRs. (A) A proposed scenario for the evolution of paired NLRs. Due to functional constraints, the sensor and the helper NLR were most likely duplicated simultaneously. This model is a simplified version of Fig. 5 in Wu et al. (8). (B) Functional tests of paired NLRs. (Left) TP309 wild type (WT); (Right) Transgenic plants with the Tetep NLR pair chr11.fgenesh2445/chr11.fgenesh2446. Both the WT and transgenic plants were tested using blast strains during the heading stage. The WT plants displayed severe neck blast symptoms, while the transgenic lines conferred strong resistance when mature (shown in dashed circles). (C) Paired NLRs identified in the Nipponbare genome that are Pi-ta or Pi-kh homologs. The 2 members of each pair are linked by a dashed line. These pairs appear approximately at mirrored positions to each other in the same phylogenetic tree. (D) A representative cladogram of the identified paired NLRs in B. distachyon (black triangle), Nipponbare (blue circle) and Tetep (red rectangle). Seven clades (clades I, II, IV, V, VI, VII, and X) are shown here. In nearly all cases, the 2 members of a pair (linked by a line in the figure) are clearly separated into different clades. Some unusual domains present in these NLRs are given on the branches. Four function-known NLRs—Pi-ta, Pi42, RGA4, and RGA5—are highlighted by green. Clade I consists of the helpers, while the remaining clades consist of sensors. A full phylogenetic tree of all paired NLRs and their homologs is given in Fig. 6, and a full list is given in SI Appendix, Table S8.
To test whether an NLR gene pair functions as a unit, we randomly picked 25 NLR pairs to do single NLR gene knockout using the CRISPR/Cas9 system in a japonica cultivar Wuyungeng24 because transformation in Tetep often produces nonviable progeny. In 4 of these genes from 18 NLR pairs, we failed to design rational spacers or verification primers. In addition, among the same 18 NLR pairs, the transformation failed in 16 genes after ∼3 to 4 trials, and 12 other genes were found to have not mutated (SI Appendix, Table S9). The general low successful editing rate (∼29.6% vs. ∼67.5% for non-NLR genes surveyed, χ2 with Yates’ correction = 38.85, d.f. = 1, P = 4.57 × 10−10, SI Appendix, SI Materials and Methods and Table S10) may imply a high fitness reduction when 1 of the 2 genes in a pair was knocked out. Such resistance cost was also observed when individual NLRs were transformed into susceptible hosts (SI Appendix, SI Materials and Methods, Table S11, and Fig. S4). In the end, we successfully generated 8 helper NLR knockouts and 10 sensor NLR knockouts (SI Appendix, Table S9). Seven of the 8 helper genes are paired with 7 of the 10 sensor genes. As explained below, 9 of these 10 sensor knockouts showed deleterious phenotypic effects.
Of the 7 pairs for which both sensor and helper knockouts were available, 3 sensor knockouts became very weak as seedlings and died quickly (a lethal phenotype), 3 sensor knockouts had the phenotypes of dwarfing and infertility or early heading, and 1 sensor knockout had the phenotype of smaller spikes and a shorter stature. In contrast, their corresponding helper knockouts displayed much milder phenotypic effects, e.g., just a little shorter or even no phenotypic changes compared with its wild type (Fig. 5 and SI Appendix, Table S9). Interestingly, 3 of the 7 helper knockouts, Os07g29820, Os11g46070, and Os11g39230, were slightly susceptible to blast in natural conditions, suggesting that these NLR pairs can confer blast resistance. Taking together, these observations suggest that these sensor/helper pairs play an important role in response to rice blast, and that a helper may function with 1 or more sensors (8).
Fig. 5.
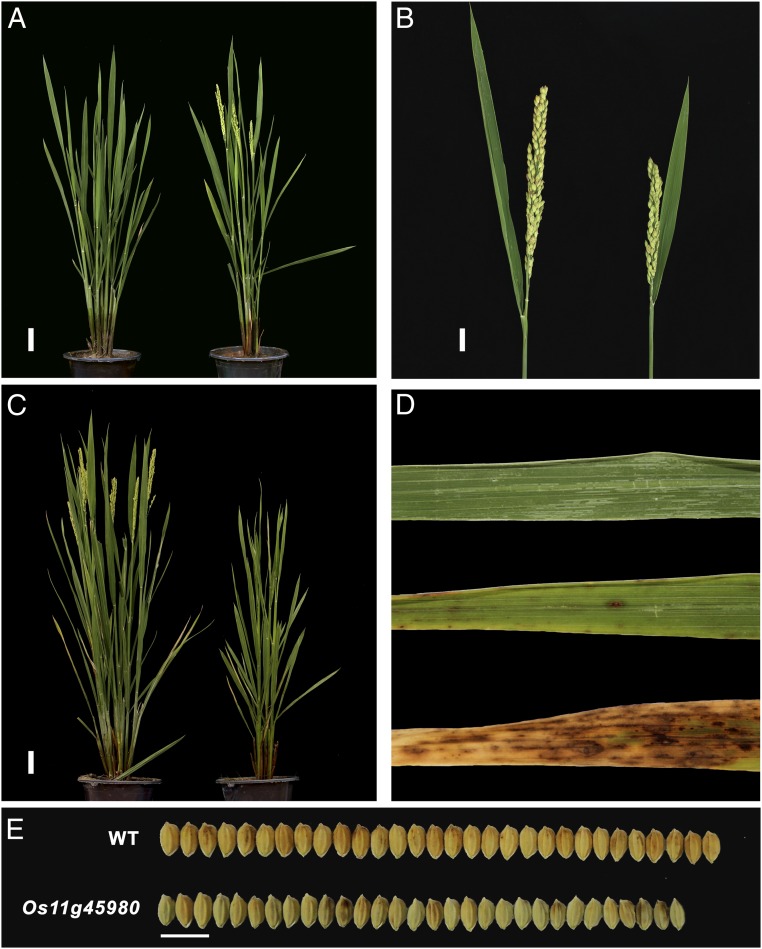
Phenotypes of CRISPR/Cas9 knockouts of paired-NLR sensor or helper genes. The sgRNA:Cas9 vector was transformed into O. sativa L. ssp. japonica cultivar Wuyungeng24 via Agrobacterium-mediated transformation. (A) Phenotypic analysis of the LOC_Os08g14810 mutant. (Left) Wild type; (Right) LOC_Os08g14810 (sensor) T1 mutant with the early heading phenotype; 90 d old. (B) Phenotypic analysis of the LOC_Os11g39290 mutant. (Left) Wild type; (Right) LOC_Os11g39290 (sensor) T1 mutant with a smaller spike; 125 d old. (C) Phenotypic analysis of the LOC_Os11g45980 mutant. (Left) Wild type; (Right) LOC_Os11g45980 (helper) T1 mutant with the dwarf and late heading phenotypes; 110 d old. (D) Phenotypic analysis of the LOC_Os10g22484 mutant. (Top) Wild type; (Middle) rice blast lesion; (Bottom) LOC_Os10g22484 (sensor) T1 mutant displayed a pseudolesion, mimics those caused by blast, no T2 plants were obtained for this mutant because T1 plants were infertile; 6 weeks old. (E) Phenotypic analysis of the LOC_Os11g45980 mutant. The LOC_Os11g45980 (helper) T1 mutant displayed a smaller seed size compared to the WT. These phenotypes were consistent between T1, T2, and T3 generations. (Scale bars, 5 cm [A and C], 2 cm [B], and 1 cm [E].)
In the remaining 3 sensor knockouts for which the paired helper member was found not mutated, 2, Os08g30634 and Os10g22484, were observed with “blast pseudolesions” (Fig. 5D). No hyphae were observed in these lesions under microscopic examination, indicating that those spots were most likely owing to programmed cell death (PCD) initiated by helper NLR proteins after losing their sensor suppressors. Furthermore, RT-qPCR experiments revealed no significant changes in the expression levels of paired NLR knockouts (SI Appendix, Fig. S5), suggesting that this process was likely initiated at the protein level. The effect of knocking out 1 member of the paired NLRs was more severe in some sensor mutants that eventually led to the death of the plants, including the mutants with the Pi-ta gene knocked out (SI Appendix, Table S9). Additional transgenic experiments which transformed 2 pairs each as a unit (1 with the chr11.fgenesh1896/1897 pair and another with the chr11.fgenesh2445/2446 pair) into a susceptible cultivar either conferred strong or enhanced resistance to rice blast strains (see Fig. 4B and SI Appendix, SI Materials and Methods for details), further confirming the resistance function of the paired NLRs.
We then checked whether the NLR pairs have been favored in breeding programs. By tracing the pedigree described above, at least 13 Tetep NLR pairs were found in those resistant lines. The tig00011732.fgenesh48/tig00011732.fgenesh49 pair was inherited in MH2009, while the Pi-ta/Pi42 (tig00012489.fgenesh13/tig00012489.fgenesh15) pair was found in both SH527 and MH2009. Another pair, the Pik-1/Pik-2 (chr11.fgenesh2455/chr11.fgenesh2457) pair, which has also been functionally characterized, was found in NH2539.
The above results suggest that these NLR pairs have been retained in the elite cultivars by strong selection and play an important role in response to the rapidly evolving pathogens.
Evolutionary and Functional Analyses of Sensor/Helper Pairs.
We traced the evolutionary trajectories of the paired NLRs through phylogenetic analysis using the coding sequences. Surprisingly, most NLR pairs were clustered within only a few clades; a clade is defined as a group of descendant branches with a bootstrap value ≧ 80%, SI Appendix, Fig. S3), suggesting that these pairs were derived from a few common ancestral pairs. We then reconstructed the phylogenetic tree using only the sequences of these NLR pairs and their homologs (Figs. 4 C and D and 6). The reconstructed tree contains several well-supported clades. For example, a subclade of the biggest clade, clade I, includes 13, 12, and 9 members from Nipponbare, Tetep, and Brachypodium distachyon, respectively. Interestingly, each of these 34 members only came from 1 member of the 34 different NLR pairs. In other words, each of the 34 NLR pairs strictly has only 1 member in this clade. Among them, 2 well-characterized genes, RGA4 from the RGA4/RGA5 pair and Pi42 from the Pi42/Pi-ta pair, were also found in this clade, and both of them are helpers (hereafter a clade mainly consisting of helper NLRs is defined as a helper clade). In contrast, both of their paired counterparts, including RGA5 and Pi-ta, which are sensors (hereafter a clade mainly consisting of sensor NLRs is defined as a sensor clade), were grouped into another clade (Figs. 4D and 6). In accordance with this evolutionary pattern, these pairs belong to 4 helper and 6 sensor clades (Figs. 4D and 6).
Fig. 6.
Phylogenetic tree of paired NLRs. The tree is constructed using all identified paired NLRs from B. distachyon (BD, marked by black triangles), Nipponbare (NP, marked by blue circles), and Tetep (TP, marked by red rectangles) genomes and their homologs (terminal branches without any marks). The large tree contains different subclades, numbered from I to X. Each member of a pair is presented as “-H” if it’s a helper or “-S” if it’s a sensor, e.g., “TP1-H” means the helper of the Tetep paired NLR TP1, while “TP1-S” means the sensor of TP1. Each subclade is marked as a sensor (S) or helper (H) clade based on those NLRs included in this clade. The phylogenetic tree was constructed using the ML (maximum likelihood) method with 1,000 bootstrap replicates. The bootstrap percentages are given above each branch. The phenotypic changes of CRISPR/Cas9 mutants were marked as level 1 (Lv1): slight or no phenotypic changes; level 2 (Lv2): moderate phenotypic changes including obvious dwarfing, pseudoblast lesions, low setting rates, etc.; and level 3 (Lv 3): lethal phenotypes.
To test whether an NLR pair truly functions as a unit, we considered 4 expectations. First, the effector-targeted decoy domains, WRKY, RATX, NOI, TRX, etc., each of which is frequently fused with a sensor NLR member (30), should only be found in sensor NLR clades but not in any helper clade. In fact, these unusual domains were exclusively present in sensor NLR clades (Fig. 4D and SI Appendix, Table S8).
Second, it has been found that paired NLR proteins generally form heterogeneous protein complexes (31), in which the sensor NLR is essential for suppressing the helper NLR’s autoactivation in the absence of the pathogen. Therefore, loss of a sensor NLR may lead to autoactivation of the helper NLR, which could be extremely deleterious to the host plant. In contrast, loss of a helper NLR should be less harmful. The phylogenetic tree is consistent with this expectation in that all members in the helper NLR clade have their paired counterparts in the sensor clades, but not vice versa. Some genes (i.e., those homologous genes of NLR pairs) in sensor clades had neither a potential helper counterpart in the helper NLR clade nor a tightly linked NLR with a head-to-head orientation in the genome.
Third, knocking out a sensor gene in an NLR pair should show a more severe effect than knocking out a helper gene. Indeed, in the 7 NLR pairs with successful sensor or helper knockouts, 5 of their T1 transgenic lines with the sensor member knocked out displayed various abnormal phenotypes, such as lethal, dwarfing or semidwarfing, infertile, lesion mimic, late or early heading, and so on (Fig. 5 and SI Appendix, Table S9). In contrast, knocking out the helper of an NLR pair only caused a slight phenotypic change (Fig. 6, paired Wilcoxon signed rank test for phenotypic change levels in 7 paired mutants, 1-tail, P = 0.0239).
Finally, since the sensor members are involved in direct-binding fast-evolving AVR effectors (28, 32), they are more likely to possess greater genetic diversity or divergence compared to helper members. This has been observed when comparing orthologous pairs between Nipponbare and Tetep. A significantly higher diversity (SI Appendix, Table S12, paired Wilcoxon signed rank test, 1-tail, P = 0.0478) has been detected in orthologous sensor members between Nipponbare and Tetep than that between orthologous helper members. This was also in agreement with their phylogenetic structure, where most helper members were clustered in a few well-supported clades, but the sensor members were distributed in various clades.
Genome-Wide Molecular Markers for Resistance Breeding.
The above findings indicate that introducing a large number of Tetep NLR genes into the genome of a recipient cultivar is an efficient means to establish a new cultivar with broad-spectrum resistance to blast. Therefore, we designed 1,909 pairs of PCR primers to amplify ∼430 of the 455 annotated Tetep NLRs (SI Appendix, SI Materials and Methods and Fig. S6 and Dataset S1). Three types of PCR primer pairs were designed to target single NLRs (type I), paired NLRs (type II), and clustered NLRs (type III) in the Tetep genome (SI Appendix, Fig. S6). We tested 320 PCR primer pairs that targeted 240 NLRs, including all 23 NLR clusters and 43 NLR pairs, which are the most desirable NLRs to include in a breeding program. We found a success rate of 88.1% (SI Appendix, SI Materials and Methods and Fig. S7 and Dataset S1). Importantly, all of the 23 NLR clusters and 43 NLR pairs could be verified using as few as 146 of the 320 tested PCR primer pairs (Dataset S1). Thus, with this set of 1,909 designed PCR markers, one may be able to introduce desired combinations of NLR genes, especially the paired and clustered NLR genes and those that have been functionally confirmed, into a recipient cultivar to breed a new resistant line.
We tested this idea using an experimental cross between Tetep and Jingeng698 by introducing a resistance block from Tetep containing the NLR pair, tig00011732.fgenesh48/tig00011732.fgenesh49 (Os05g40160/Os05g40150), into Jingeng698 (SI Appendix, Fig. S8). Jingeng698 is a japonica cultivar with excellent yielding traits (SI Appendix, Fig. S8C), but is susceptible to blast disease. To overcome the indica–japonica hybrid sterility, the experiment crosses begin from 3 repeat backcrossings with Jingeng698. The progeny in BC3F5 generation (3 backcrossings followed by 5 selfing generations) displayed enhanced blast resistance compared with Jingeng698 when growing in the field (SI Appendix, Fig. S8D).
Discussion
In this study we assembled the complete genome of Tetep and identified 455 NLR genes. From an analysis of these genes, we made the following observations:
First, we found no excess of NLRs in the Tetep genome compared to those in Nipponbare, MH63, and R498, challenging the notion of “more NLRs, more resistance (10).” However, we did find extensive divergence of NLRs between genomes, showing a much higher diversity in NLRs than the genomic average (e.g., ∼5.96% in NLRs vs. 0.84% for the genomic average divergence between Tetep and Nipponbare), consistent with their high variations among rice populations (33). As mentioned above, if we map Tetep Illumina reads to the Nipponbare genome, we would misclassify nearly a third of the NLRs as pseudogenes due to numerous frameshifts and premature stop codons found in those regions. Thus, the NLR genes in the Tetep genome are quite different from those in the Nipponbare genome and tend to confer stronger resistance to blast than those in the Nipponbare genome.
Second, a genome-wide resistance survey of NLRs gave us some clues about how they contribute to Tetep’s defense system. We found that at least 90 of the 219 cloned NLRs showed resistance to 1 or more of the 12 pathogen strains tested. This high abundance of functional NLR genes in the Tetep genome is likely the genetic basis for the broad-spectrum resistance of Tetep. However, as none of the 219 cloned NLR genes showed resistance to all of the 12 pathogen strains, multiple NLR genes are apparently required for broad-spectrum resistance, as also suggested by recent studies (5, 34, 35). On the other hand, each of the 12 pathogen strains was resisted on average by 19 cloned NLR genes. This resistance redundancy (overlapping resistance) might be the genetic basis for the durable resistance of Tetep. These inferences are supported by the results from pedigree tracing of NLRs in widely used cultivars derived from breeding programs that used Tetep as the resistant donor. The pedigree analysis revealed that different lines preserved distinct sets of NLR genes from Tetep and that the resistance strength and breadth of a line showed a positive correlation with the number of NLRs it inherited from Tetep. Moreover, the frequently observed NLR interactions in plants (36) would indicate that redundant networks are a general mechanism contributing to a robust immune system.
Third, the genome-wide resistance survey and pedigree tracing revealed that a large portion of effective NLRs are present in clusters, as observed previously (18, 33, 37). By targeting NLR clusters, multiple NLRs could be introduced as a single functional unit without disrupting any putative interactions within them. Thus, the clustering may facilitate breeding for new resistant strains. In order to breed a cultivar with durable and broad-spectrum resistance, one may design a combination of NLR clusters from the Tetep genome. The combination can be selected through widely used pedigrees. The experimental cross between Tetep and Jingeng698 is an example that made use of those clusters for resistance breeding. Through rapid selection for clusters derived from the Tetep genome (SI Appendix, Fig. S8) using those carefully designed molecular markers (SI Appendix, Fig. S6 and Dataset S1), we were able to generate an improved resistance line without causing obvious harm to their yield or other agronomic traits.
Fourth, our study reinforced the notion that a long-lasting broad-spectrum resistance cannot be achieved without multiple NLRs. The observation of varied spectra in resistance of different genes provided evidence for a simple pyramiding of NLRs. However, a cultivar’s genetic background affects the performance of NLRs as many of the transgenic NLRs only confer resistance to 1 susceptible line but not the other. In our 219 cloned NLR genes, 33 were transformed into both TP309 and Shin2, yielding 187 bilateral testing results across all 12 tested blast strains (Fig. 1 and SI Appendix, Table S6). Only in 14 bilateral tests (involving 8 NLR genes) did an NLR gene confer blast resistance in both cultivars (Figs. 1 and 2 and SI Appendix, Table S6). In contrast, about 77 bilateral tests (involving 27 NLR genes) displayed resistance only in 1 of the 2 cultivars tested (Fig. 1 and SI Appendix, Table S6). Among the remaining 96 bilateral tests (also involving 27 genes), the tested gene was susceptible to blast in both TP309 and Shin2 (Figs. 1 and 2 and SI Appendix, Table S6). We also noticed that, for an NLR gene, the phenotypic changes could differ between a transformant and a CRISPR mutant. A possible explanation for this difference is that, when transforming an NLR gene into a recipient cultivar, its phenotypical impact could be alleviated by the recipient cultivar’s native NLR networks. In contrast, the phenotypical impact can more readily manifest in a knockout mutant, possible due to a direct interruption of the original NLR network. These observations reinforce the importance of NLR networks over a single NLR gene.
Finally, an NLR pair is an important form of interaction between NLRs. A genome scan of 13 domesticated and wild rice varieties revealed significantly enriched head-to-head configuration in adjacent NLRs (33). Another observation is that head-to-head NLR pairs frequently carry decoy domains (33, 38), consistent with characteristics of known NLR pairs. These observations suggest that NLR pairs are not rare in a rice genome. However, a more detailed functional assessment is still required, especially when considering the pair as a single functional unit. Our study made full use of these known characteristics to develop a method to identify paired NLRs and confirm their functions with knockout experiments. We detected 43 NLR pairs in Tetep and 47 pairs in Nipponbare using a phylogenetic-guided strategy, greatly increasing the number of known NLR pairs. This strategy can be readily adopted to other cultivars or species. An enlarged NLR pair repository can facilitate both the functional and evolutionary study of NLRs.
Materials and Methods
Plant Materials, Sequencing, and De Novo Assembly.
The genomic DNA was extracted from young Tetep leaves using the cetyltrimethyl ammonium bromide (CTAB) method (39). For PacBio sequencing, 5 libraries (20-kb templates) were built and sequenced on a PacBio RS II platform, yielding 50 single molecule real-time sequencing cells. For Illumina sequencing, 2 libraries were sequenced on Hiseq 2000 and Hiseq 4000 platforms, respectively (SI Appendix, Table S1). Two Tetep RNA samples were sequenced on the Illumina Hiseq 4000 platform, each with ∼120 M clean reads (SI Appendix, Table S1). A Tetep genome draft was built using PacBio RSII subreads (SI Appendix, Fig. S1) and anchored to 12 chromosomes according to the MH63 genome (13). Detailed procedures are provided in SI Appendix, SI Materials and Methods. Related pipelines for assembly and downstream analyses as well as accompanying scripts have been deposited in the Github page https://github.com/wl13/Tetep_Genome.
NLR Genes Analyses.
Protein-coding genes were identified using both MAKER-P pipeline (16) and Fgenesh (17). The NLR gene was predicted using hmmscan (40) and NLR-parser (41). Phylogenetic trees were constructed using FastTree (42). The candidate NLR pairs were identified through searching for NLR genes near each other in the Tetep, Nipponbare, MH63, R498, and B. distachyon genomes, and confirmed through phylogenetic analyses. The detailed procedures are described in SI Appendix, SI Materials and Methods.
Cloning, Transformation, and Testing of NLR Genes.
Each clone was transformed into 2 blast-susceptible cultivars, TP309 and Shin2. The 12 pathogen strains used in this study were chosen in consideration of their geographic origin, sequence diversity, and sporulation efficiency (SI Appendix, Fig. S2). For each gene, 8 to 10 independent transformants were selected. The detailed procedures of each steps are described in SI Appendix, SI Materials and Methods.
Generation of Knockout Mutants of Paired NLRs.
Unique single-guide RNA (sgRNA) spacer sequences were designed to target the NLR. Each sgRNA was then incorporated into the Cas9 vector and transformed into the japonica cultivar Wuyungeng24. Details about the knockout steps, efficiency, as well as off-target evaluation are provided in SI Appendix, SI Materials and Methods.
Design of a PCR Marker Set for Verifying Tetep-Derived NLRs.
The PCR primer pairs were designed to amplify nearly every NLR gene in the Tetep genome (Datasets S1–S3). For each single or paired NLR, 1 or 2 primer pairs were designed (SI Appendix, Fig. S6). For a NLR cluster, usually 3 or 4 pairs of primers were designed to target the first and last genes plus 1 to target a middle region (SI Appendix, Fig. S6A). Detailed information and evaluation of the marker set are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Major Special Project on New Varieties Cultivation for Transgenic Organisms (No. 2016ZX08009001-003), National Natural Science Foundation of China (31671322, 31601041, 31571267, 31870204, and 31570368), and Jiangsu Collaborative Innovation Center for Modern Crop Production.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Whole Genome sequencing data were deposited at DDBJ/ENA/GenBank under the accession QQAJ00000000. The raw sequencing data generated for this study have been submitted to the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject/) under accession nos. SRP168526 and SRP051581. All processed data for NLR analyses have been submitted to the Figshare database (https://doi.org/10.6084/m9.figshare.7775810.v1). Phylogenetic trees can be viewed at https://itol.embl.de/shared/wanglong_nju.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910229116/-/DCSupplemental.
References
- 1.Ou S. H., Pathogen variability and host resistance in rice blast disease. Annu. Rev. Phytopathol. 18, 167–187 (1980). [Google Scholar]
- 2.Skamnioti P., Gurr S. J., Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Ou S. H., Jennings P. R., Progress in the development of disease-resistant rice. Annu. Rev. Phytopathol. 7, 383–410 (1969). [Google Scholar]
- 4.Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Zhao H., et al. , The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9, 2039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh A., et al. , Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012, pls029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., et al. , Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol. Plant Pathol. 11, 419–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C.-H., et al. , NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 114, 8113–8118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baggs E., Dagdas G., Krasileva K. V., NLR diversity, helpers and integrated domains: Making sense of the NLR IDentity. Curr. Opin. Plant Biol. 38, 59–67 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Green G. J., Campbell A. B., Wheat cultivars resistant to Puccinia graminis tritici in western Canada: Their development, performance, and economic value. Can. J. Plant Pathol. 1, 3–11 (1979). [Google Scholar]
- 11.Ding J., et al. , Highly asymmetric rice genomes. BMC Genomics 8, 154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koren S., et al. , Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., et al. , Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc. Natl. Acad. Sci. U.S.A. 113, E5163–E5171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra G., Bradnam K., Korf I., CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Du H., et al. , Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 8, 15324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell M. S., et al. , MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salamov A. A., Solovyev V. V., Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyers B. C., Kozik A., Griego A., Kuang H., Michelmore R. W., Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yue J.-X., Meyers B. C., Chen J.-Q., Tian D., Yang S., Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 193, 1049–1063 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Emms D. M., Kelly S., OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers B. C., et al. , Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Yang S., et al. , Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol. Biol. 62, 181–193 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Clark R. M., et al. , Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317, 338–342 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Wang L., et al. , Genome-wide survey of pseudogenes in 80 fully re-sequenced Arabidopsis thaliana accessions. PLoS One 7, e51769 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., et al. , Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 110, 18572–18577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., et al. , A genome-wide survey reveals abundant rice blast R genes in resistant cultivars. Plant J. 84, 20–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narusaka M., et al. , RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Cesari S., et al. , The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C.-H., Krasileva K. V., Banfield M. J., Terauchi R., Kamoun S., The “sensor domains” of plant NLR proteins: More than decoys? Front. Plant Sci. 6, 134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura M. T., Monteiro F., Dangl J. L., Treasure your exceptions: Unusual domains in immune receptors reveal host virulence targets. Cell 161, 957–960 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Williams S. J., et al. , Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Césari S., et al. , The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein J. C., et al. , Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 1618 (2018). Erratum in: Nat. Genet.50, 285–296 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Xie Z., et al. , A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos. Trans. R Soc. B Biol. Sci. 374, 20180308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., et al. , Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against Magnaporthe oryzae in rice (Oryza sativa L.). Rice (N. Y.) 12, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wróblewski T., et al. , Genome-wide functional analyses of plant coiled-coil NLR-type pathogen receptors reveal essential roles of their N-terminal domain in oligomerization, networking, and immunity. PLoS Biol. 16, e2005821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michelmore R. W., Meyers B. C., Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Bailey P. C., et al. , Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray M. G., Thompson W. F., Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn R. D., et al. , The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steuernagel B., Jupe F., Witek K., Jones J. D. G., Wulff B. B. H., NLR-parser: Rapid annotation of plant NLR complements. Bioinformatics 31, 1665–1667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price M. N., Dehal P. S., Arkin A. P., FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.