Abstract.
Head and neck squamous cell carcinoma (SCC) is primarily managed by surgical cancer resection. Recurrence rates after surgery can be as high as 55%, if residual cancer is present. Hyperspectral imaging (HSI) is evaluated for detection of SCC in ex-vivo surgical specimens. Several machine learning methods are investigated, including convolutional neural networks (CNNs) and a spectral–spatial classification framework based on support vector machines. Quantitative results demonstrate that additional data preprocessing and unsupervised segmentation can improve CNN results to achieve optimal performance. The methods are trained in two paradigms, with and without specular glare. Classifying regions that include specular glare degrade the overall results, but the combination of the CNN probability maps and unsupervised segmentation using a majority voting method produces an area under the curve value of 0.81 [0.80, 0.83]. As the wavelengths of light used in HSI can penetrate different depths into biological tissue, cancer margins may change with depth and create uncertainty in the ground truth. Through serial histological sectioning, the variance in the cancer margin with depth is investigated and paired with qualitative classification heat maps using the methods proposed for the testing group of SCC patients. The results determined that the validity of the top section alone as the ground truth may be limited to 1 to 2 mm. The study of specular glare and margin variation provided better understanding of the potential of HSI for the use in the operating room.
Keywords: hyperspectral imaging, head and neck cancer, squamous cell carcinoma, convolutional neural networks, histology, cancer margin
1. Introduction
Head and neck (H&N) cancer is the sixth most-common cancer worldwide, with majority of cancers of the upper aerodigestive tract, comprising the oral and nasal cavities, pharynx, and larynx, being squamous cell carcinoma (SCC).1 Approximately two-thirds of the SCC patients present with advanced disease, either stage III or IV.2 Surgical resection is the primary management of SCC of the aerodigestive tract, potentially with concurrent chemoradiation therapy depending on the extent of the disease.3 The local recurrence rates for SCC cases depend on the successful removal of the cancer. For surgeries with negative cancer margins, the local recurrence rate is 12% to 18%, compared to local recurrence rates of up to 30% to 55%, if positive cancer margins are determined.4–6 Moreover, positive cancer margins have a greatly reduced disease-free survival, with estimates ranging from 7% to 52%, compared to disease-free survival rates of 39% to 73% for negative margins.7,8 Disease recurrence greatly affects the likelihood for additional surgeries, reduced quality of life, complications from surgery, and increased mortality rates.9
Hyperspectral imaging (HSI) is a noncontact optical imaging modality capable of acquiring a single image of potentially hundreds of discrete wavelengths. Preliminary research demonstrates that HSI has the potential for providing diagnostic information for various diseases.10 Preliminary studies from our group show that HSI combined with machine learning (ML) may yield diagnostic information with potential applications for surgical use in head and neck cancers.11–14 Fabelo et al.15–17 have demonstrated the need and utility of HSI for in-vivo brain cancer detection and developed a visualization system that could lead to near-real-time guidance by using a machine learning-based classification algorithm. Moreover, the use of deep learning techniques to process the same dataset was studied, demonstrating that they outperform the traditional machine learning methods.17 Many applications of HSI for gastrointestinal procedures have been explored, including anatomical organ identification, anastomosis and ischemia classification, and cancer detection.18 Recently, the implementation of HSI for guided colorectal surgery as a replacement for tactile information lost in laparoscopic surgeries has been explored in an ex-vivo study, and Baltussen et al.19 demonstrated that HSI could distinguish between normal healthy colon wall and colorectal adenocarcinoma. Much work has been explored in head and neck HSI as well. Salmivuori et al.20 detailed the assistance of HSI in ill-defined, cutaneous basal cell carcinomas of the head and neck, and the authors contend that HSI offers clinical utility in more accurately assessing margins. Farah et al.21 demonstrated that narrow-band imaging (NBI) at green (400 to 430 nm) and blue (525 to 555 nm) visible light could reveal the extent of oral SCC under white light. Nonencoding portions of RNA called micro-RNA function to regulate the expression of DNA.22 Moreover, Farah et al.23,24 obtained micro-RNA and mRNA expression levels from both the primary tumor core and the near-tumor normal tissues; it was determined that NBI spatially correlated with tumor-like expression levels in detecting the abnormality in near-tumor normal tissues.
Machine learning methods, including support vector machines (SVMs) and convolutional neural networks (CNNs), the latter being an implementation of artificial intelligence, have demonstrated near human-level ability for image classification tasks.25,26 These experiments have been conducted on gross-level HSI acquired of ex-vivo tissue specimens from patients undergoing surgical cancer resection. However, HSI uses wavelengths of light that can penetrate different depths into the biological tissue, so it is possible that the superficial cancer margin may change with depth and create uncertainty in ground truth, which is used to obtain evaluation metrics for these experiments.
Selected previous works from our group performing head and neck squamous cell carcinoma (HNSCC) detection in ex-vivo samples from the upper aerodigestive tract used only single-class tissue specimens (purely tumor or normal tissues) to demonstrate classification potential with HIS.27,28 Lu et al.14 incorporated tumor-involved cancer margin specimens from HNSCC, but these methods were implemented on selected regions of interest (ROIs) of normal and tumor near the margin. Therefore, the area under the curve (AUC) reported for HNSCC detection thus far from our group ranges from 0.8 to 0.95, but these results may be limited to ROIs or single-class tissues. Similarly, Manni et al.29 used ROIs from tumor-margin specimens of seven patients with tongue SCC and obtained an AUC of 0.92. Weijtmans et al.30 developed a deep learning architecture that separately extracts spectral and spatial features from HSI, a dual-streamed approach, also validated on seven patients with tongue SCC; the proposed model performed better with both feature streams (AUC of 0.90) compared to individual stream. A few works have investigated SCC detection at the actual cancer margin; Halicek et al.31 performed SCC detection at the cancer margin, upon which the proposed method in this paper expands, but this previous work was limited by ignoring regions of specular glare. Trajanovski et al.32 performed semantic segmentation with deep learning of entire ex-vivo tissue specimens with excellent validation performance (AUC of 0.93) on gross specimens of the cancer margin from 14 tongue SCC samples.
This study aims to investigate the ability of HSI to detect SCC in surgical specimens from the upper aerodigestive tract using several distinct machine learning pipelines. In addition, another objective of this work is to investigate the limiting factors of HSI-based SCC detection, including specular glare, noise and blur, and uncertainty in the ground truth due to changes in superficial cancer margin with depth, all of which must be thoroughly explored to understand the potential of HSI in the operating room. A preliminary version of this work was presented at the 2019 SPIE Medical Imaging Conference.31 The contributions of this journal paper include a proposed algorithm combining deep learning and unsupervised machine learning; additionally, five testing patients have been serially sectioned to reveal changes in the cancer margin. This work expands upon previous cross-validation experiments with CNN-only methods on our H&N dataset to include multiple machine learning pipelines involving CNNs and other state-of-the-art methods. The proposed methods are tested on five HSI of five SCC patients, and the accuracy of the corresponding ground truths from these five tissues are discussed in detail along with potential directions for incorporating the outcomes of this work to improve future studies with HSI for cancer detection.
2. Methods
2.1. Experimental Design
In collaboration with the Otolaryngology Department and the Department of Pathology and Laboratory Medicine at Emory University Hospital Midtown, head and neck cancer patients undergoing surgical cancer resection were recruited for our HSI studies. Written and informed consent was obtained from all patients before acquiring surgical tissue specimens for inclusion in our study, which was used for research purposes only and deidentified by a clinical research coordinator. The experimental methods and protocols were approved by the Institutional Review Board at Emory University. In previous studies, we have evaluated the efficacy of using HSI for optical biopsy of head and neck tissues.11,33 Excised tissue samples of HNSCCs and normal tissues were collected from the upper aerodigestive tract sites, including tongue, larynx, pharynx, and mandible. Three tissue samples, with approximate size of , were collected from each patient: a sample of the primary tumor specimen, a normal tissue sample, and a sample at the tumor-involved cancer margin with adjacent normal tissue; these specimens were scanned with an HSI system.14,28
In this study, we selected 26 tissue specimens from 12 patients with moderate-to-poorly differentiated primary SCC of the upper aerodigestive tract for this analysis, including primary origin sites of the larynx, pharynx, tongue, floor of mouth, alveolar ridge, buccal mucosa, and maxillary sinus. The patients were divided into two groups, cross-validation and testing groups. The first seven patients and corresponding 21 tissues samples were collected early in the course of this project that met the criteria of having the ideal distribution of tumor, tumor–normal, and normal tissues and were used for evaluating quantitative results, expanding from previously published results with this group of patients.13 In addition, five tumor-involved margin tissue specimens from five patients acquired at the end of the data collection period were selected to comprise the testing group. These tissue specimens were selected because they had approximately equal amounts of cancer and normal tissues in each specimen and were verified by the collaborating pathologist as of interest to undergo serial histological sectioning through the depth of the tissue. These five patients’ tissues were classified using models trained from the first seven patient cross-validation group (following a leave-one-patient-out cross-validation method), and these five patients’ results are presented qualitatively to compare with the variability in the superficial cancer margin.
2.2. Hyperspectral Imaging
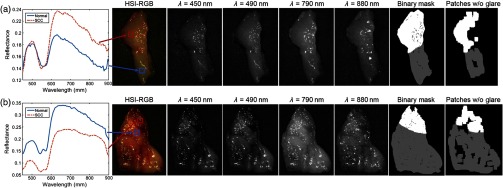
Hyperspectral images were acquired of ex-vivo surgical specimens using a previously described CRI Maestro imaging system (Perkin Elmer Inc., Waltham, Massachusetts), which captures images having and a spatial resolution of .11,28,34,35 Each hypercube contains 91 spectral bands, ranging from 450 to 900 nm with a 5-nm spectral sampling interval. The HS data were normalized by a standard white–dark calibration normalization, which involved subtracting the inherent dark current from the measured spectra and dividing it by a white reference spectra for all wavelengths sampled for all pixels.11,35 Figure 1 shows the RGBs, grayscale images at representative bands, and spectral signatures of the patients’ HSI data. The grayscale images at selected bands highlight the choice of the cutoffs due to noise at the ends of the broadband spectrum. The HSI was used to construct RGB composite images by implementing a Gaussian function centered on each color component of the spectrum: red (625 to 700 nm), green (520 to 560 nm), and blue (450 to 490 nm).
Fig. 1.
Two representative tissue specimens from different patients. Left: spectral signatures of SCC and normal ROIs are shown for both patients in (a) and (b). The HSI-RGB composite images and selected spectral bands are shown to highlight the noisy band cutoffs during preprocessing. Binary ground-truth masks including glare regions, generated by only removing patches centered on specular glare is shown; binary ground-truth mask excluding glare regions, generated by sufficient area to extract patches and avoiding specular glare (white: SCC, and normal: gray).
2.3. Histological Imaging
To obtain a ground-truth labeling for the tissue specimens imaged with HSI, tissues were inked after imaging to preserve orientation, fixed in formalin, paraffin embedded, sectioned from the top of the imaging surface, hematoxylin and eosin stained, and digitized using a Hamamatsu Photonics NanoZoomer at objective (specimen-level pixel size, ). The ex-vivo tissue sections were reviewed by a board-certified pathologist with expertise in H&N pathology and the cancer margins were annotated directly on the slide using digitized histology in Aperio ImageScope (Leica Biosystems Inc., Buffalo Grove, Illinois).
There exists the possibility for substantial deformation of the histological images during processing relative to the HSI setup. Previously, the registration challenges were explored by using a deformation-based image registration pipeline of the histological image with known registration landmarks, and an error of 0.5 mm for SCC tissues was calculated.36 For the cross-validation group patients in this work, the histological images were automatically registered using this deformable registration pipeline to produce the HSI ground truth. However, to calculate the error of histology ground truths for HSI in millimeters, a more accurate registration is needed than the automated approach. For the testing group patients since each had six histological slices, we manually registered each histological image using mutual landmarks to the HSI that were serially sectioned to reduce error.
2.4. Hyperspectral Imaging Binary Ground Truth
Specular glare is detected in each spectral band by fitting a gamma distribution to the pixel intensities of that band. Next, the top 1% of intensities are identified in this distribution, and any pixels with intensities of these bins are identified as specular glare. There were two types of specular glare observed: near-infrared (NIR) and visible, which usually were spatially independent. Visible glare occurs between 450 and 790 nm, and NIR glare occurs after 800 nm, which can be seen in the selected bands in Fig. 1. A pixel was identified as a specular glare pixel and removed if there was a glare identified using the distribution method in any spectral band from 490 to 790 nm. This range was selected because bands before and after this range were removed during preprocessing. NIR glare is observed beyond this range, so it is not relevant.
Binary masks are constructed from the histological images and used to construct the ground truth for HSI in two methods. The first method is to investigate only ideal quality pixels, which is constructed by avoiding HSI regions with a large amount of specular glare. For the ground-truth mask avoiding glare, only regions that contain sufficient area to extract spatial patches without any specular glare are included. The second method is to investigate the degrading effect of specular glare on classification accuracy. For the “ground-truth mask including glare” regions, the entire tissue area is included for patch-making, but the top 1% of glare pixels are identified by fitting a gamma distribution to pixel intensities, such that no patches are centered on a glare pixel. A binary mask of the specular glare pixels was generated, and patches were produced automatically using this mask in the regions around the specular glare. Figure 1 shows an example of the two masks generated from two different patients. Both masks will be used for evaluating quantitative testing results and are referred to as “ground-truth mask excluding glare” and ground-truth mask including glare, respectively. The specular glare pixels are represented by the black pixels within the first mask in the middle. The automatic patch-making process can be seen to avoid the regions of specular glare shown in the second mask on the right of Fig. 1.
2.5. Effect of Sectioning Depth on Cancer Margin Ground Truth
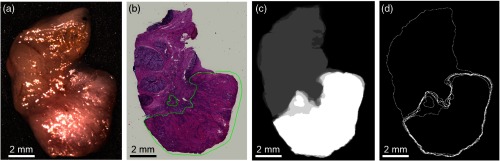
HSI uses wavelengths of light in the visible and NIR spectrum that can penetrate different depths into biological tissue. It is possible that the cancer margin may change with depth and create uncertainty in the classification results because of the variable penetration of the HSI broadband spectrum. To investigate this, additional histological sectioning was performed on five tumor-involved margin tissue specimens (see Fig. 2).
Fig. 2.
Representative tissue specimen that underwent serial histological sectioning to evaluate cancer margin variation with tissue depth. (a) HSI-RGB composite image; (b) first histological slice (the green outlined area is the pathologist annotation of the SCC region); (c) combined merged image of the six binary masks from the six histological images (cancer shown in white, normal in gray); (d) merged image of the SCC contours from the six histological images. The two rightmost images are used to depict the total variation of the cancer margin.
During histological sectioning, it is a standard procedure to discard the first sections until the first good-quality section is obtained that contains the entire perimeter of the tissue specimen. The five specimens used for additional histological sectioning were retroactively enrolled for serial sectioning, so it was unknown what initial depth of tissue was discarded to obtain the first good-quality slice. In discussion with the pathology laboratory technician, who performed the histological sectioning, and through observation of specimen sectioning, it was estimated that to were discarded on average during initial sectioning before obtaining the first high-quality, tissue-encompassing slide.
To produce serial histological sectioning further into the depth of the tissue specimens, the thickness of the remaining paraffin-embedded tissue was estimated. The microtome used for this study produced slices at , so the number of additional sections was documented and the distances of additional depth could be measured. From the sectioned surface, which corresponds to the top of the tissue that was optically imaged, five more sections were obtained, up to further into the depth of the tissue, by discarding nonincluded slices. The extent of the additional serial sections were obtained for 100 to beyond the first good-quality slice; the exact values for the five tissue specimens’ total additional sectioning depths were 100, 150, 200, and , depending on the unique tissue specimen. Therefore, the combined total estimate of additional sectioning is to into the tissue depth from the original HS image surface. Penetration depth depends on the tissue composition and wavelength of light. This sectioning depth was investigated because it represents the effective penetration depth for biological tissue in the shorter wavelengths of HSI used, which correspond to the most relevant features for HSI classification, such as hemoglobin between 550 and 600 nm.37–39
2.6. Machine Learning Techniques
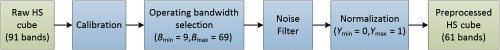
2.6.1. Data preprocessing
A further preprocessing chain was applied to the data mainly to reduce the noise in the spectral signatures (Fig. 3) and was compared with the standard white-reference calibration. The machine learning methods detailed below were tested with and without the following preprocessing steps. The proposed preprocessing chain is based on four steps: image calibration, operating bandwidth selection, noise filtering, and data normalization. In the first step, the previously described method to normalize the data using white reference and dark current is performed. Next, the spectral bands are truncated between 490 and 790 nm, so the final HS cube contains only 61 bands, which will be used for classification. Spectral bands outside this range were noisy because they were too close to the detectable limits of the HS camera. In the third step, a smoothing filter is applied to the data in order to reduce the spectral noise. A moving average with a span of 10 spectral bands was used as the smoothing filter, implemented as a sliding low-pass filter with coefficients equal to the reciprocal of the span. Finally, each pixel’s spectral signature is normalized between 0 and 1, using a function to map the minimum and maximum to these values.
Fig. 3.
Block diagram of the proposed preprocessing chain.
2.6.2. Convolutional neural networks
A deep CNN was used to detect SCC in the ex-vivo specimens of the upper aerodigestive tract, implemented using TensorFlow in Python on a NVIDIA Titan-XP GPU.13,25,26,40 In summary, an Inception-V1-style three-dimensional (3-D) CNN was designed and trained using leave-one-patient-out cross validation.26 The 3-D convolutional kernels that comprised the modified 3-D inception V1 modules were of sizes , , and , and in total, the CNN architecture contained two inception modules, three convolutional layers, and two fully connected layers using the 3-D patch input size of () for unprocessed HSI patches and for preprocessed HSI patches. The total number of HSI samples in the cross-validation group was 647,000 normal HSI pixels and 877,000 cancer HSI pixels, calculated from the binary masks excluding glare, from the 21 tissue specimens from 7 patients. Each HSI pixel served as the center for an image patch (). Dropout was employed to avoid overfitting in the CNN models.
In total, the CNN described was separately trained using four different scenarios using the information from two versions of the binary ground-truth masks and the two proposed processing methods, which produce different models: first, CNN trained without specular glare patches (trained separately both with and without added preprocessing of spectral data), and second, CNN trained with specular glare patches (trained separately both with and without added preprocessing of spectral data). The 95% confidence intervals (CIs) were calculated using a bootstrapping method by sampling 1000 pixels from each class with replacement from each patient and calculating the AUC of the receiver operator characteristic (ROC); the method was performed 1000 times for each patient and 2.5 and 97.5 percentiles were reported. The saved, trained models of the patients from the previous work were used to classify the HS images of the five testing group patients that underwent serial histological sectioning for surgical margin variation with depth evaluation. The probabilities of all models were averaged per patient to obtain qualitative probability heat maps, scaled from 0 to 1, where 0 represents normal class and 1 represents high probability that the tissue belongs to the cancer class.
2.6.3. HELICoiD algorithms
The results of the CNN classification method and the generated probability maps were compared to the results obtained by a machine learning pipeline previously developed for intraoperative detection of brain cancer using HIS.15,41–44 In summary, a spatial–spectral classification algorithm, here referred to as HELICoiD [Fig. 4(a)], was implemented using a classification map obtained by an SVM classifier that is spatially homogenized by employing a combination of a one-band representation obtained from the first principal component analysis (PCA) decomposition through a K-nearest neighbors (KNN) filtering method [Fig. 4(a), part A]. After that, the result of the KNN filtering is merged with an unsupervised segmentation map generated by a hierarchical K-means (HKM) algorithm through a majority voting (MV) method.16 The result of this algorithm is a classification map that includes both the spatial and the spectral features of the HS images. In addition, for this application, the KNN filtering is applied again to the MV probabilities and the PCA one-band representation to homogenize the results [Fig. 4(a), part B].
Fig. 4.
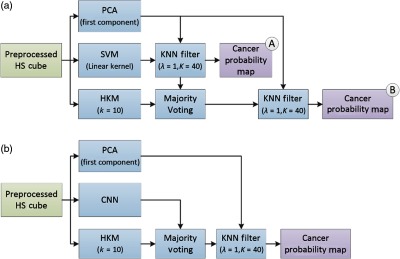
Block diagrams of the proposed classification frameworks. (a) HELICoiD algorithm with the additional KNN filter. (b) Pipeline of the mixed algorithm.
A component of the HELICoiD algorithm was isolated and referred to as spatial SVM, which uses both the spectral and the spatial components of HSI for machine learning through a combination of . This spectral–spatial implementation of SVM is performed with and without the additional preprocessing pipeline to be used as a surrogate for direct comparison to the CNN with and without preprocessing. Furthermore, a pipeline that combines the CNN architecture with the entire HELICoiD algorithm was proposed. In this case, the spatial–spectral stage of HELICoiD () is replaced by the CNN architecture trained with the preprocessed data [Fig. 4(b)]. Both HELICoiD and algorithms use the preprocessed HS data as input.
In summary, we present six algorithms for investigation of HSI machine learning: the CNN (with and without preprocessing), the spatial SVM (with and without preprocessing), the HELICoiD algorithm, and finally the algorithm. These six machine learning algorithms were tested first on the group of experiments using the binary ground-truth masks that exclude specular glare, and again the experiments were performed with the binary ground-truth masks that include specular glare. The six algorithms were compared using box plot distributions of the values and median AUC with 95% CIs, and, a paired, one-tailed -test on the classification results was used to determine statistical differences with a 0.05 threshold for significance. The quantitative classification results of seven patients are reported in Table 1, obtained by using leave-one-patient-out cross validation. In addition, like the CNN method, the saved, trained models from these patients were used to classify five tissue specimens that were imaged with HSI and underwent serial histological sectioning for cancer margin variability evaluation. The probabilities of all models were averaged per patient to obtain qualitative probability heat maps, scaled from 0 to 1, where 0 represents normal class and 1 represents high probability that the tissue belongs in the cancer class.
Table 1.
Results of interpatient cross validation of SCC versus normal, obtained using the leave-one-patient-out method. Average AUCs are reported with bootstrapped 95% CI.
| Ground truth | Classifier | Average AUC [95% CI] |
|---|---|---|
| Excluding glare | Spatial SVM | 0.71 [0.68, 0.74] |
| CNN | 0.86 [0.82, 0.89] | |
| Spatial SVM (preprocessed HSI) | 0.82 [0.80, 0.84] | |
| CNN (preprocessed HSI) | 0.84 [0.81, 0.86] | |
| HELICoiD (preprocessed HSI) | 0.82 [0.79, 0.84] | |
| CNN + HELICoiD (preprocessed HSI) | 0.82 [0.79, 0.85] | |
| Including glare | Spatial SVM | 0.69 [0.67, 0.71] |
| CNN | 0.73 [0.71, 0.76] | |
| Spatial SVM (preprocessed HSI) | 0.76 [0.74, 0.77] | |
| CNN (preprocessed HSI) | 0.78 [0.76, 0.81] | |
| HELICoiD (preprocessed HSI) | 0.79 [0.77, 0.81] | |
| CNN + HELICoiD (preprocessed HSI) | 0.81 [0.80, 0.83] |
3. Results
Quantitative results from the leave-one-patient-out cross validation, using both the ground-truth regions that include glare pixels and the subsampled masks that include only ideal quality regions that exclude glare, show that the CNN-based classifier group outperformed the SVM-based classifier group using the average AUC of the ROC, as shown in Table 1 and Fig. 5. The results are reported using sevenfold cross validation to validate on all seven patients.
Fig. 5.
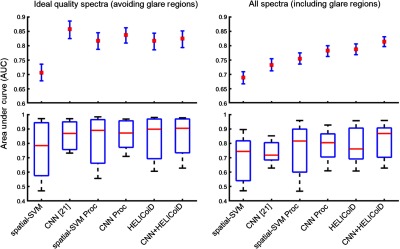
Results of interpatient cross validation of SCC versus normal, obtained using the leave-one-patient-out method. Top: average AUCs reported with 95% CI. Bottom: box plots with the range in black, 75th and 25th percentile in blue, and median in red.
When classification is performed on only ideal quality pixels (obtained from the subsampled mask, see Fig. 1), the results indicate that additional preprocessing of the spectral signature and addition of the HELICoiD method and KNN filtering do not significantly improve the results compared to only using the CNN [see Fig. 5(a)]. The average AUCs for the CNN groups are 0.86, 0.84, and 0.82 for the CNN, CNN with preprocessed input data, and method, respectively. The average AUCs for the SVM-based groups are 0.71, 0.82, and 0.82 for the spatial SVM without and with preprocessed input data, and HELICoiD method, respectively. The 95% CIs overlap for all groups except for spatial SVM, as shown in Table 1. In addition, all methods have a similar interquartile range and median distribution (see Fig. 5).
However, when the classification is performed over the entire HS image tissue area, which includes classification of specular glare pixels that contain more noise and variability, the CNN + HELICoiD method outperforms other methods tested with an average AUC of 0.81 for classification. This classification scheme, including specular glare pixels, represents a more realistic application of HSI. The CNN algorithm alone with or without preprocessing has an average AUC of 0.78 and 0.73, respectively, and both constitute a statistically significant decrease in performance compared the method ( and 0.04, respectively). The average AUC values range from 0.69 to 0.79 for the SVM-based groups, and the HELICoiD algorithm significantly outperforms the spatial SVM (). The additional preprocessing pipeline, as described in Fig. 3, only offers a statistically significant increase in performance for the spatial-SVM algorithm (), not the CNN () (see Table 1, for complete results with 95% CIs). In summary, from the cross-validation experiments, the best classification method was using the CNN as the input for the filtering method. For comparison, using the spectral–spatial for input to the HELICoiD algorithm instead of the CNN component yielded slightly lower results that were not statistically significant.
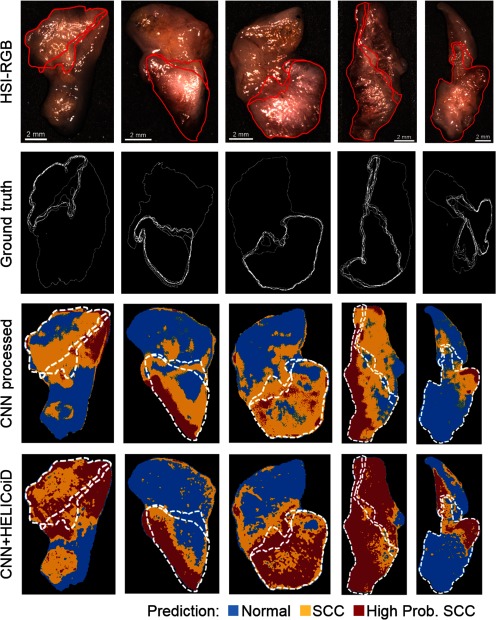
For generalization and application, HS images from five tissue specimens from five patients with SCC comprised the hold-out testing group and were classified using the saved models that were trained and cross validated using the seven patient cross-validation group. The testing patients were classified by all cross-validation models and averaged to obtain qualitative probability heat maps. Qualitative investigation of the five testing patients, classified with the CNN trained with preprocessed data alone and the filtering method is shown in Fig. 6. As shown in Figure 6, the technique performs better on the five testing patients, in agreement with the quantitative metrics from the cross-validation group. The SCC probability heat maps are shown with binary masks depicting the uncertainty and variation in the cancer margin with depth. Depending on the tissue, as demonstrated, the margin changes by about 1 mm depending on the sectioning depth. Therefore, our qualitative results can be interpreted within the range of uncertainty of the ground truth to provide more insight into the classification potential of machine learning methods using HSI for cancer detection.
Fig. 6.
Representative results of binary cancer classification of the five testing patients. From top to bottom: HSI-RGB composite; histological ground truth showing variation in cancer margins with cancer area outlined; heat maps for cancer probability for and techniques. The extremes in the superficial cancer margin are overlaid on to the heat maps.
4. Discussion
In this work, we presented and quantified the combination of two state-of-the-art machine learning-based classification methods for HSI of ex-vivo HNSCC surgical specimens. In summary, there are two methods for generating SCC prediction probability maps: the first uses a CNN, and the second uses a combination of . After generating the predicted cancer probabilities on a pixel level, the probability map is combined with an HKM unsupervised segmentation layer through an MV algorithm that determines the class belonging to the ROI. Therefore, two distinct methods are compared using leave-one-patient-out cross validation to obtain quantitative evaluation metrics. In addition, the methods are tested on a group of HSI obtained from five SCC patients who underwent serial histological sectioning to evaluate the variation in the cancer margin with penetration depth of the light wavelengths.
The quantitative results of this paper suggest that when working with ideal quality pixels, such as the spectral signatures generated from flat-surface-tissue surfaces with no glare, the CNN techniques and spatial–spectral machine learning algorithms will perform with no significant difference and no additional preprocessing will be necessary. The average AUCs for these methods using the preprocessed input data range from 0.82 to 0.86 with overlapping CIs. However, when the pixels classified contain noise, for example, due to sloping of the tissue edges or specular glare from completely reflected incident light, additional spectral smoothing and additional filtering of the classifier improve classification results of the CNN. The best performing method tested was with an average AUC of 0.81 [0.80, 0.83]. These tested methods outperform the traditional spectral–spatial machine learning methods employed in this study. Therefore, the techniques using both the CNN and for cancer probability maps performed best for the seven cross-validation patients, and so both were employed on the five patients testing set. One major limitation of the approach used in this paper was the small sample size, and therefore, the proposed ML models could be prone to overfitting and lack generalization to larger testing datasets. To investigate potential overfitting in this experiment, we analyzed the training and cross-validation accuracies for the CNN trained with preprocessed HSI data. The average training accuracy was 85%, and the average cross-validation accuracy with CI was 79% [78, 81] for excluding glare and 74% [73, 76] for including glare. This result indicates that our models did not suffer substantially from overfitting.
Excluding glare, the CNN alone without preprocessing performs best. It is hypothesized that CNN and deep learning methods should be expected to outperform traditional ML techniques because a substantially large dataset should allow learning of variance, tolerance of noise, and removal of the need for preprocessing. Also, the method performs only slightly better than the original HELICoiD; this could be produced because the CNN probability maps are out-weighted by the PCA and KNN filtering that are applied afterward. After the inclusion of specular glare, the results change relative to each method, with the CNN with preprocessing outperforming the CNN without preprocessing. This outcome could be the result of the classification problem becoming more challenging compared to excluding glare, so the dataset is now too small for the CNN to perform well without preprocessing. Expanding the training dataset with more patients with specular glare and large amounts of noise may allow original CNN methods to outperform other techniques.
To test the general application of the proposed methods, HSI from five testing group patients with SCC were classified using the models that were trained and cross validated using the seven-patient group. To qualitatively investigate these results, histological images of the five ex-vivo tissue specimens were obtained down to about 0.3 mm to determine how the superficial cancer margin may change with depth. As shown in Fig. 6, the technique performs best on the five-patient SCC testing group, which was the same result obtained from the sevenfold quantitative cross-validation experiment.
The five patients’ tumor-involved margin specimens were classified with the CNN trained with the fully preprocessed image patches that were extracted from the complete binary ground-truth mask including specular glare pixels. These probability maps are shown in Fig. 6, and the CNN map is also used as the input for the HELICoiD + KNN-filtering technique. In the top three rows of Fig. 6, there are regions of glare with tissue specimens that are classified incorrectly as normal with the CNN, but the correctly classifies these ROIs as SCC. It is also possible to observe that the method tends to overpredict SCC at regions of normal tissue near the cancer margin. We hypothesize that the technique outperforms the patch-based CNN alone because it can incorporate more local and regional spatial and spectral information to overcome the degrading effect of specular glare. Therefore, future work could involve the application of a full CNN for SCC detection on tissues with specular glare. This algorithm requires more data necessary for training as it produces labels that are end to end, a full pixel classification map for a full HSI input, and so it would require the entire HSI dataset acquired for this project.
An additional aim of this investigation was to determine the variation of the superficial cancer margin through the depth of the tissue. From the five testing patient tissue specimens, it can also be seen that the superficial cancer margin of the ex-vivo tissue specimens can vary from the extreme near and far margins in the range of 1 to 2 mm. These results allow interpretation of the cancer prediction probability maps with observed variation in the ground truth. However, additional possible uncertainty may exist in the histological ground truth. For example, if the angle of the sectioning plane is skewed from the tissue plane, then the ground truth could be warped, which could lead to errors that cannot be corrected by deformable registration. In a previous work, we explored a pipeline of affine and deformable demons-based registration for alignment of the histological ground truth to gross-level images of specimens, and it was determined via needle-bored control points that target registration error was .36 The combination of these two main sources of uncertainty in the histological ground truth for the ex-vivo tissue specimens allows for error propagation of up to 2.5 mm in the binary mask. This would greatly affect the results of the pixel-wise evaluation metrics such as accuracy, sensitivity, and specificity.
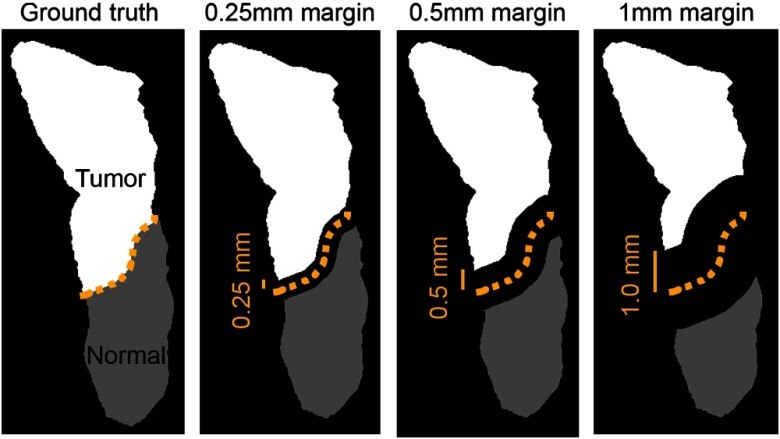
Future work includes development and implementation of a new performance evaluation method to handle this margin uncertainty, for instance, evaluating primary tumor clearance at several millimeter increments from the ideal tumor margin. Currently, the standard for surgeons’ opinions on margin adequacy is 5 mm for HNSCC, with margins between 1 and 5 mm defined as “close margins.”45 However, some studies suggest that outcomes may be similar for margin of 2.2 to 5 mm.46 It is evident that the question of margin adequacy is still being determined, and surgeons would be interested in the performance of optical imaging methods at different margin distances. Therefore, the proposed error metric should extend to about 2 to 3 mm. Figure 7 demonstrates how the cancer margin could be systematically eroded and be used to determine accuracies at several distances from the cancer margin to better provide surgeons and physicians with a method for interpreting results for HSI studies, given the conclusions of this paper on the uncertainty of the cancer margin. The outcomes of this work suggest the use of this error metric for future studies.
Fig. 7.
Proposed evaluation metric demonstrates millimetric, systematic cancer-margin erosion that could provide performance updates at several distances from the cancer margin to provide physicians with an easily interpretable method for understanding the results of HSI studies.
Another avenue of future work involves the rethinking of the definition of normal tissues in the tissue specimens of the tumor-involved margin. In oral SCC, it has been studied that normal tissue directly adjacent to the primary SCC is molecularly distinct from normal tissues farther from the SCC, and additional resection to this extended margin may lead to increased disease-free survival and reduced local recurrence.23,24 Moreover, this result was obtained by investigation that NBI at 400 to 430 nm and 525 to 555 nm reveals changes in normal tissue that correlate to significantly different levels of micro-RNA epigenetic regulation compared to primary tumor and normal tissue that are not visible under white light alone.21,23,24 Therefore, it may be possible to extract micro-RNA expression levels to determine a molecular ground truth for certain tissues employed in this study, as Liu and Xu47 have demonstrated that micro-RNAs can be reliably obtained from formalin-fixed, paraffin-embedded tissue samples.
5. Conclusion
This study investigated the effects of specular glare, noise, blurring, and tissue-edge sloping artifacts on HSI-based cancer detection. It explored the potential of HSI and machine learning for the detection of head and neck cancer. According to the experimental results, the CNN seems to be more robust against the environmental conditions of the acquired images and provides better classification results even without data preprocessing. Future work includes experimenting with deep learning methods that can incorporate more contextual information of the tissue, such as a full CNN. In addition, another objective was to evaluate the general efficacy on example test cases with uncertainty in the ground truth as the superficial cancer margin varies with penetration depth. This was tested by serially sectioning the tissue samples in the testing group to reveal the variation of the cancer margin through the depth of the tissue. This determined that the validity of the top section alone as the ground truth may be limited to 1 to 2 mm, suggesting that an alternative approach for obtaining performance metrics should be developed. All of the above factors are necessary to explore and understand the potential of HSI in the operating room. The proposed deep learning and machine learning methods employed to study these objectives require sufficiently large patient datasets for training, validation, and testing. Therefore, the preliminary results of this study encourage the inclusion of more data and further exploration into the ability of HSI for cancer detection.
Acknowledgments
This research was supported in part by the U.S. National Institutes of Health (NIH) Grants Nos. R21CA176684, R01CA156775, R01CA204254, and R01HL140325. H.F., S.O., and G.M.C. were supported in part by the Canary Islands Government through the ACIISI (Canarian Agency for Research, Innovation and the Information Society), ITHACA project under Grant Agreement ProID2017010164, the Spanish Government and European Union (FEDER funds) as part of support program in the context of Distributed HW/SW Platform for Intelligent Processing of Heterogeneous Sensor Data in Large Open Areas Surveillance Applications (PLATINO) project, under Contract No. TEC2017-86722-C4-1-R. This work has been also supported in part by the European Commission through the FP7 FET (Future Emerging Technologies) Open Programme ICT-2011.9.2, European Project HELICoiD under Grant Agreement No. 618080. In addition, this work has been supported in part by the 2016 PhD Training Program for Research Staff of the University of Las Palmas de Gran Canaria. Finally, this work was completed while Samuel Ortega was beneficiary of a predoctoral grant given by the “Agencia Canaria de Investigacion, Innovacion y Sociedad de la Información” (ACIISI) of the “Conserjería de Economía, Industria, Comercio y Conocimiento” of the “Gobierno de Canarias,” which is part-financed by the European Social Fund (FSE) [POC 2014-2020, Eje 3 Tema Prioritario 74 (85%)]. The authors would like to thank the surgical pathology team at Emory University Hospital Midtown for their help in collecting fresh tissue specimens. The authors would also like to thank the members of Quantitative BioImaging Laboratory at the University of Texas at Dallas and UT Southwestern Medical Center.
Biographies
Martin Halicek is a PhD candidate in biomedical engineering at the Georgia Institute of Technology and Emory University. His PhD thesis research is supervised by Dr. Baowei Fei, who is a faculty member of the University of Texas at Dallas and Emory University. His research interests are medical imaging, biomedical optics, and machine learning. He is also a sixth-year MD/PhD student from the Medical College of Georgia at Augusta University.
Himar Fabelo received his PhD in telecommunication technologies from the University of Las Palmas de Gran Canaria (ULPGC), Spain, in 2019. Since 2015, he has conducted his research activity in the Institute for Applied Microelectronics at the ULPGC in the field of electronics and bioengineering. His research interest area includes the use of machine learning and deep learning techniques applied to hyperspectral images for cancer analysis.
Samuel Ortega is a PhD candidate in telecommunication technologies from the University of Las Palmas de Gran Canaria (ULPGC), Spain. Since 2015, he has conducted his research activity in the Institute for Applied Microelectronics at the ULPGC in the field of electronics and bioengineering. His current research interests are in the use of machine learning algorithms in medical applications using hyperspectral images.
Gustavo Marrero Callico (1970) is a telecommunications engineer (1995) and has a PhD European doctorate (2003). In 2000–2001 he stayed at the Philips Research Laboratories in Eindhoven (NL). He is currently an associate professor at the ULPGC. He has more than 170 publications in national and international journals, conferences and book chapters. He has participated in 18 research projects funded by the European Commission, governments and industries. He is a senior editor of the IEEE Transactions on Consumer Electronics (2009) and an associate editor of the IEEE Access (2016). He was the coordinator of the European project HELICoiD under FP7.
Baowei Fei is a professor and Cecil H. and Ida Green Chair in systems biology science in the Department of Bioengineering at the Erik Jonsson School of Engineering and Computer Science, University of Texas, Dallas. He is also a professor in the Department of Radiology at the University of Texas Southwestern Medical Center. Before he was recruited in Dallas, he was an associate professor with tenure at Emory University where he was the leader of the Precision Imaging: Quantitative, Molecular and Image-guided Technologies Program in the Department of Radiology and Imaging Sciences. He was also a primary faculty member in the Joint Department of Biomedical Engineering at Emory University and Georgia Institute of Technology. He was recognized as a distinguished investigator by the Academy for Radiology and Biomedical Imaging Research. He was also recognized as a distinguished cancer scholar by the Georgia Cancer Coalition and the Governor of Georgia. He serves as conference chair for the international conference of SPIE Medical Imaging—Image-Guided Procedures, Robotics Interventions, and Modeling from 2017 to 2020. He is a fellow of the International Society for Optics and Photonics (SPIE) and a fellow of the American Institute for Medical and Biological Engineering (AIMBE).
Biographies of the other authors are not available.
Disclosures
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose.
References
- 1.Joseph L. J., et al. , “Racial disparities in squamous cell carcinoma of the oral tongue among women: a SEER data analysis,” Oral Oncol. 51(6), 586–592 (2015). 10.1016/j.oraloncology.2015.03.010 [DOI] [PubMed] [Google Scholar]
- 2.Yao M., et al. , “Current surgical treatment of squamous cell carcinoma of the head and neck,” Oral Oncol. 43(3), 213–223 (2007). 10.1016/j.oraloncology.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 3.Kim B. Y., et al. , “Prognostic factors for recurrence of locally advanced differentiated thyroid cancer,” J. Surg. Oncol. 116(7), 877–883 (2017). 10.1002/jso.24740 [DOI] [PubMed] [Google Scholar]
- 4.Chen T. Y., Emrich L. J., Driscoll D. L., “The clinical significance of pathological findings in surgically resected margins of the primary tumor in head and neck carcinoma,” Int. J. Radiat. Oncol. Biol. Phys. 13(6), 833–837 (1987). 10.1016/0360-3016(87)90095-2 [DOI] [PubMed] [Google Scholar]
- 5.Loree T. R., Strong E. W., “Significance of positive margins in oral cavity squamous carcinoma,” Am. J. Surg. 160(4), 410–414 (1990). 10.1016/S0002-9610(05)80555-0 [DOI] [PubMed] [Google Scholar]
- 6.Sutton D. N., et al. , “The prognostic implications of the surgical margin in oral squamous cell carcinoma,” Int. J. Oral Maxillofac. Surg. 32(1), 30–34 (2003). 10.1054/ijom.2002.0313 [DOI] [PubMed] [Google Scholar]
- 7.Amaral T. M. P., et al. , “Predictive factors of occult metastasis and prognosis of clinical stages I and II squamous cell carcinoma of the tongue and floor of the mouth,” Oral Oncol. 40(8), 780–786 (2004). 10.1016/j.oraloncology.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 8.Garzino-Demo P., et al. , “Clinicopathological parameters and outcome of 245 patients operated for oral squamous cell carcinoma,” J. Cranio-Maxillo-Facial Surg. 34(6), 344–350 (2006). 10.1016/j.jcms.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 9.Rathod S., et al. , “A systematic review of quality of life in head and neck cancer treated with surgery with or without adjuvant treatment,” Oral Oncol. 51(10), 888–900 (2015). 10.1016/j.oraloncology.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 10.Lu G., Fei B., “Medical hyperspectral imaging: a review,” J. Biomed. Opt. 19(1), 010901 (2014). 10.1117/1.JBO.19.1.010901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fei B., et al. , “Label-free hyperspectral imaging and quantification methods for surgical margin assessment of tissue specimens of cancer patients,” in 39th Annu. Int. Conf. IEEE Eng. Med. and Biol. Soc., pp. 4041–4045 (2017). 10.1109/EMBC.2017.8037743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halicek M., et al. , “Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks,” Proc. SPIE 10469, 104690X (2018). 10.1117/12.2289023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halicek M., et al. , “Tumor margin classification of head and neck cancer using hyperspectral imaging and convolutional neural networks,” Proc. SPIE 10576, 1057605 (2018). 10.1117/12.2293167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G., et al. , “Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging,” Clin. Cancer Res. 23(18), 5426–5436 (2017). 10.1158/1078-0432.CCR-17-0906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabelo H., et al. , “An intraoperative visualization system using hyperspectral imaging to aid in brain tumor delineation,” Sensors 18(2), 430 (2018). 10.3390/s18020430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabelo H., et al. , “Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations,” PLoS One 13(3), e0193721 (2018). 10.1371/journal.pone.0193721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabelo H., et al. , “Deep learning-based framework for in vivo identification of glioblastoma tumor using hyperspectral images of human brain,” Sensors 19(4), 920 (2019). 10.3390/s19040920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega S., et al. , “Use of hyperspectral/multispectral imaging in gastroenterology. Shedding some different light into the dark,” J. Clin. Med. 8(1), 36 (2019). 10.3390/jcm8010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baltussen E. J. M., et al. , “Hyperspectral imaging for tissue classification, a way toward smart laparoscopic colorectal surgery,” J. Biomed. Opt. 24(1), 016002 (2019). 10.1117/1.JBO.24.1.016002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmivuori M., et al. , “Hyperspectral imaging system in the delineation of ill-defined basal cell carcinomas: a pilot study,” J. Eur. Acad. Dermatol. Venereol. 33(1), 71–78 (2019). 10.1111/jdv.2019.33.issue-1 [DOI] [PubMed] [Google Scholar]
- 21.Farah C. S., et al. , “Improved surgical margin definition by narrow band imaging for resection of oral squamous cell carcinoma: a prospective gene expression profiling study,” Head Neck 38(6), 832–839 (2016). 10.1002/hed.23989 [DOI] [PubMed] [Google Scholar]
- 22.Leigh-Ann M., Paul R. M., “MicroRNA: biogenesis, function and role in cancer,” Curr. Genomics 11(7), 537–561 (2010). 10.2174/138920210793175895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farah C. S., “Narrow band imaging-guided resection of oral cavity cancer decreases local recurrence and increases survival,” Oral Dis. 24(1–2), 89–97 (2018). 10.1111/odi.12745 [DOI] [PubMed] [Google Scholar]
- 24.Farah C. S., Fox S. A., Dalley A. J., “Integrated miRNA-mRNA spatial signature for oral squamous cell carcinoma: a prospective profiling study of narrow band imaging guided resection,” Sci. Rep. 8(1), 823 (2018). 10.1038/s41598-018-19341-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Proc. 25th Int. Conf. Neural Inf. Process. Syst., Vol. 1, pp. 1097–1105 (2012). [Google Scholar]
- 26.Szegedy C., et al. , “Going deeper with convolutions,” in IEEE Conf. Comput. Vision and Pattern Recognit. (2014). 10.1109/CVPR.2015.7298594 [DOI] [Google Scholar]
- 27.Halicek M., et al. , “Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks,” J. Biomed. Opt. 24(3), 036007 (2019). 10.1117/1.JBO.24.3.036007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halicek M., et al. , “Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging,” J. Biomed. Opt. 22(6), 060503 (2017). 10.1117/1.JBO.22.6.060503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manni F., et al. , “Automated tumor assessment of squamous cell carcinoma on tongue cancer patients with hyperspectral imaging,” Proc. SPIE 10951, 109512K (2019). 10.1117/12.2512238 [DOI] [Google Scholar]
- 30.Weijtmans P. J. C., et al. , “A dual stream network for tumor detection in hyperspectral images,” in IEEE 16th Int. Symp. Biomed. Imaging (2019). 10.1109/ISBI.2019.8759566 [DOI] [Google Scholar]
- 31.Halicek M., et al. , “Cancer margin evaluation using machine learning in hyperspectral images of head and neck cancer,” Proc. SPIE 10951, 109511A (2019). 10.1117/12.2512985 [DOI] [Google Scholar]
- 32.Trajanovski S., et al. , “Tumor semantic segmentation in hyperspectral images using deep learning,” in 2nd Int. Conf. Med. Imaging with Deep Learn., London, England: (2019). [Google Scholar]
- 33.Fei B., et al. , “Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients,” J. Biomed. Opt. 22(8), 086009 (2017). 10.1117/1.JBO.22.8.086009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu G., et al. , “Hyperspectral imaging of neoplastic progression in a mouse model of oral carcinogenesis,” Proc. SPIE 9788, 978812 (2016). 10.1117/12.2216553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu G., et al. , “Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery,” J. Biomed. Opt. 20(12), 126012 (2015). 10.1117/1.JBO.20.12.126012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halicek M., et al. , “Deformable registration of histological cancer margins to gross hyperspectral images using demons,” Proc. SPIE 10581, 105810N (2018). 10.1117/12.2293165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sordillo L. A., et al. , “Deep optical imaging of tissue using the second and third near-infrared spectral windows,” J. Biomed. Opt. 19(5), 056004 (2014). 10.1117/1.JBO.19.5.056004 [DOI] [PubMed] [Google Scholar]
- 38.Ash C., et al. , “Effect of wavelength and beam width on penetration in light-tissue interaction using computational methods,” Lasers Med. Sci. 32(8), 1909–1918 (2017). 10.1007/s10103-017-2317-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., et al. , “Penetration depth of photons in biological tissues from hyperspectral imaging in shortwave infrared in transmission and reflection geometries,” J. Biomed. Opt. 21(12), 126006 (2016). 10.1117/1.JBO.21.12.126006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abadi M., et al. , “TensorFlow: large-scale machine learning on heterogeneous systems,” 2015, https://www.tensorflow.org.
- 41.Florimbi G., et al. , “Accelerating the K-nearest neighbors filtering algorithm to optimize the real-time classification of human brain tumor in hyperspectral images,” Sensors 18(7), 2314 (2018). 10.3390/s18072314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravi D., et al. , “Manifold embedding and semantic segmentation for intraoperative guidance with hyperspectral brain imaging,” IEEE Trans. Med. Imaging 36(9), 1845–1857 (2017). 10.1109/TMI.42 [DOI] [PubMed] [Google Scholar]
- 43.Lazcano R., et al. , “Porting a PCA-based hyperspectral image dimensionality reduction algorithm for brain cancer detection on a manycore architecture,” J. Syst. Archit. 77, 101–111 (2017). 10.1016/j.sysarc.2017.05.001 [DOI] [Google Scholar]
- 44.Madroñal D., et al. , “SVM-based real-time hyperspectral image classifier on a manycore architecture,” J. Syst. Archit. 80, 30–40 (2017). 10.1016/j.sysarc.2017.08.002 [DOI] [Google Scholar]
- 45.Baddour H. M., Jr., Magliocca K. R., Chen A. Y., “The importance of margins in head and neck cancer,” J. Surg. Oncol. 113(3), 248–255 (2016). 10.1002/jso.v113.3 [DOI] [PubMed] [Google Scholar]
- 46.Zanoni D., et al. , “A proposal to redefine close surgical margins in squamous cell carcinoma of the oral tongue,” JAMA Otolaryngol. Head Neck Surg. 143(6), 555–560 (2017). 10.1001/jamaoto.2016.4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu A., Xu X., “MicroRNA isolation from formalin-fixed, paraffin-embedded tissues,” Methods Mol. Biol. 724, 259–267 (2011). 10.1007/978-1-61779-055-3 [DOI] [PMC free article] [PubMed] [Google Scholar]