Significance
Central tolerance can generate holes in the CD4 T cell repertoire or divert cells into the Foxp3+ T regulatory (Treg) cell lineage. Little is known with regard to how these diametrically different cell fate decisions of autoreactive cells shape the size and composition of polyclonal cohorts of antigen-specific T cells. Here we generated inventories of very rare autoreactive T cells in the naive versus tolerant polyclonal repertoire and identified T cell receptors (TCRs) that were lost, whereas others mediated Treg cell differentiation. The antigen responsiveness of these TCRs revealed a TCR hierarchy that not only separates deleted from diverted TCRs but also generates a Treg cell compartment with high antigen reactivity.
Keywords: T cell tolerance, clonal deletion, clonal diversion, regulatory T cell, MHC class II tetramer
Abstract
Deletion or Treg cell differentiation are alternative fates of autoreactive MHCII-restricted thymocytes. How these different modes of tolerance determine the size and composition of polyclonal cohorts of autoreactive T cells with shared specificity is poorly understood. We addressed how tolerance to a naturally expressed autoantigen of the central nervous system shapes the CD4 T cell repertoire. Specific cells in the tolerant peripheral repertoire either were Foxp3+ or displayed anergy hallmarks and, surprisingly, were at least as frequent as in the nontolerant repertoire. Despite this apparent lack of deletional tolerance, repertoire inventories uncovered that some T cell receptors (TCRs) were lost from the CD4 T cell pool, whereas others mediated Treg cell differentiation. The antigen responsiveness of these TCRs supported an affinity model of central tolerance. Importantly, the contribution of different diverter TCRs to the nascent thymic Treg cell population reflected their antigen reactivity rather than their frequency among precursors. This reveals a multilayered TCR hierarchy in CD4 T cell tolerance that separates deleted and diverted TCRs and assures that the Treg cell compartment is filled with cells of maximal permissive antigen reactivity.
T cell receptor (TCR) stimulation of thymocytes can result in their deletion but can also specify the diversion of MHCII-restricted T cells into the Foxp3+ regulatory T (Treg) cell lineage (1). The determinants that specify these alternative cell fates are incompletely understood, and little is known about how clonal deletion and clonal diversion shape the composition of polyclonal cohorts of autoreactive cells of shared antigen specificity.
TCR/model antigen transgenic systems showed that Treg cell differentiation, similar to clonal deletion, can be instructed by TCR agonists (2, 3), and this was later confirmed for polyclonal T cells and natural self-antigens (4). In some models, low antigen levels favored Treg cell generation, whereas high antigen doses favored deletion, suggesting that Treg cell differentiation occurs within an avidity window between positive selection and deletion (5–8). Other observations, however, are difficult to reconcile with a reductionist, purely signal strength-based model. For instance, concomitant deletion is seen in essentially all TCR transgenic models of neoantigen-driven Treg cell diversion (2, 3, 7, 9), and it remains possible that Treg cell differentiation entails stochastic/selective elements (10).
Global TCR repertoire analyses supported the idea that Treg differentiation is instructed by self-antigen recognition, although some investigators came to opposing conclusions (11–13). Repertoire analyses also indicated that Aire-dependent expression of tissue-restricted antigens (TRAs) in thymic epithelial cells (TECs), a phenomenon that had previously been associated with deletional tolerance (14–17), also shapes the Treg cell repertoire (18–20).
MHCII tetramers were employed to trace minute cohorts of CD4 T cells when their cognate antigens were transgenically expressed to emulate a ubiquitous or TRA-like expression pattern (21, 22). Widespread antigen expression was associated with deletion, whereas a more restricted TRA-like expression ensued in the emergence of Foxp3+ Treg cells. While these findings suggested that the mechanism of tolerance is somehow dictated by the antigen expression pattern, it remains to be seen whether this is a generally applicable rule (23).
TCR transgenics, global repertoire sequencing, and MHC tetramers each entail distinct limitations. Monoclonal systems with unphysiologically high frequencies of antigen-specific cells may not faithfully recapitulate T cell fate decisions in polyclonal settings (24, 25). Large-scale repertoire analyses are limited in their potential to relate cell fate to antigen specificity. MHC tetramers may fail to reveal holes in the tolerant repertoire, as the loss of certain TCRs might be numerically compensated by expansion of cells carrying others TCRs. Moreover, although the emergence of Foxp3+ cells among Tetramer (Tet)+ cells indicates that Treg cell induction occurs, it remains unclear whether this selectively applies to cells bearing distinct TCRs.
Here we addressed how clonal deletion and Treg induction impact not only the size but also the TCR composition of polyclonal CD4 T cells recognizing the natural CNS autoantigen myelin proteolipid protein (PLP). BL/6 mice are largely resistant to PLP-induced experimental autoimmune encephalomyelitis (EAE), suggesting a robust state of antigen-specific tolerance. In line with this, immunization of Plp1KO mice, to which PLP is a foreign antigen, elicits a CD4 T cell response against 2 epitopes spanning amino acids 11 to 19 and 240 to 248, whereas Plp1WT mice are functionally tolerant to these epitopes (26, 27). We assessed how far this unresponsiveness of the censored repertoire reflected the deletion, Treg cell diversion, or anergy of PLP-specific CD4 T cells and whether these tolerance modes selectively applied to cells carrying distinct TCRs.
Results
PLP-specific CD4 T Cells Are Not Reduced in the Tolerant Repertoire but Contain Foxp3+ Treg Cells.
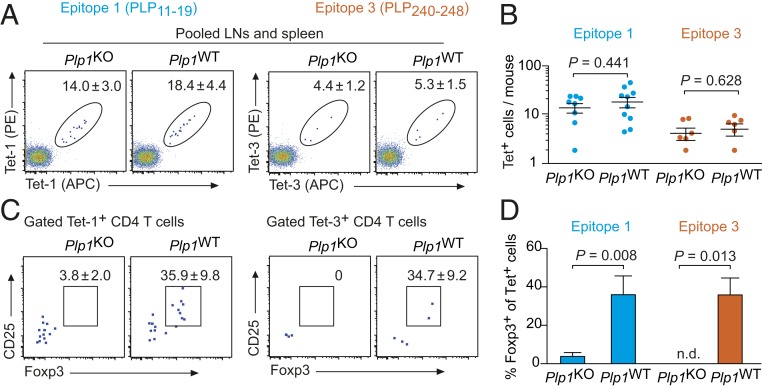
Using a combination of tetramer staining and magnetic enrichment (SI Appendix, Fig. S1) (28), we enumerated PLP11–19 (Tet-1) or PLP240–248 (Tet-3) specific CD4 T cells in pooled spleen and lymph node cells of Plp1KO and Plp1WT mice. The uncensored peripheral repertoire of Plp1KO mice contained 14.0 ± 3.0 Tet-1+ CD4 T cells and 4.4 ± 1.2 Tet-3+ cells (Fig. 1 A and B). In Plp1WT mice, PLP11–19– or PLP240–248–specific cells were not diminished; surprisingly, there was even a tendency toward elevated numbers of Tet+ cells in the tolerant repertoire, although these differences did not reach significance (Fig. 1 A and B). Whereas Foxp3+ cells were barely detectable among tetramer-positive T cells of Plp1KO mice, 30 to 40% of cells specific for either epitope were Foxp3+ in Plp1WT mice (Fig. 1 C and D).
Fig. 1.
PLP-specific peripheral CD4 T cells in Plp1KO or Plp1WT mice. (A) Analysis of pooled spleen and lymph node cells after enrichment of PLP11–19– or PLP240–248−specific CD4 T cells. Dot plots are gated on CD4+CD8−Dump− cells (SI Appendix, Fig. S1). The calculated mean number ± SEM of tetramer-positive cells/mouse is indicated (n ≥ 6 each). (B) Number of PLP11–19– or PLP240–248–specific CD4 T cells/mouse. Each data point represents an individual mouse. (C) Foxp3GFP and CD25 expression in peripheral tetramer-positive cells. Numbers indicate the mean frequency ± SEM of Foxp3+CD25+ cells (n ≥ 6 each). (D) Summary of data in C.
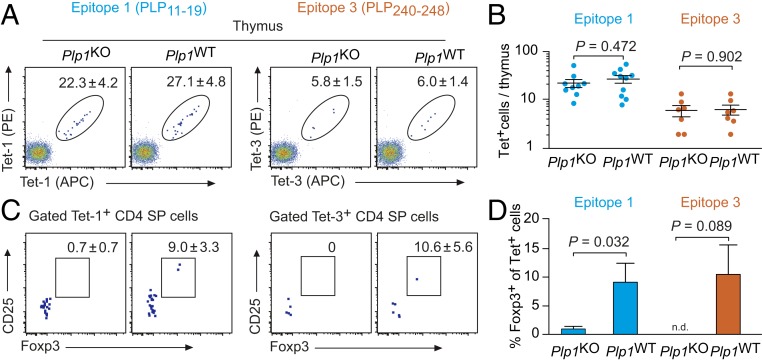
Also in the thymus, the number of Tet-1– or Tet-3–positive cells did not significantly differ between Plp1KO or Plp1WT mice (Fig. 2 A and B). The Tet-1+ or Tet-3+ CD4 SP population of Plp1KO mice was essentially devoid of Foxp3+ cells. By contrast, in Plp1WT mice, around 10% of CD4 SP cells specific for epitope 1 or 3 expressed Foxp3 (Fig. 2 C and D).
Fig. 2.
Number and phenotype of PLP-specific CD4 SP thymocytes in Plp1KO or Plp1WT mice. (A) Flow cytometry of thymocytes after enrichment of PLP11–19– or PLP240–248–specific cells, gated on CD4+CD8−Dump− cells. The calculated mean number ± SEM of tetramer-positive cells/thymus is indicated (n ≥ 7 each). (B) Summary of data in A. Each data point represents an individual thymus. (C) Foxp3GFP and CD25 expression in tetramer-positive CD4 SP cells. Numbers indicate the mean frequency ± SEM of Foxp3+CD25+ cells (n ≥ 7 each). (D) Summary of data in C.
Treg Cells and Anergy in a Repertoire of Reduced Complexity.
Our findings were consistent with the idea that tolerance of PLP11–19– and PLP240–248–specific CD4 T cells predominantly operated through Treg cell induction, which was in line with previous observations with TRA-like expressed model antigens (21, 22). However, it remained possible that clonal deletion also shaped the composition of PLP-specific cells, yet was numerically masked, for instance, by compensatory expansion of other PLP reactive cells. Distinguishing these possibilities requires a comprehensive comparison of TCRs of Tet+ cells in Plp1KO versus Plp1WT mice. However, the sheer number of TCR α- and β-chain combinations that recognize a given antigen limits the feasibility of such an approach in fully polyclonal repertoires. Several hundred naive cells specific for a foreign antigen each expressed a different TCR (29, 30). It is likely that the same applies to PLP-specific CD4 T cells. Together with the paucity of Tet+ cells, this poses a significant hurdle to a conclusive comparison of antigen-specific TCRs in the absence or presence of a given autoantigen.
To circumvent these inherent limitations of fully polyclonal TCR repertoires, we introduced a transgenic TCR β chain derived from a PLP11–19–specific TCR (27). The rationale was to reduce the repertoire complexity to the diversity of TCRα rearrangements and impose a bias toward higher numbers of PLP11–19–specific cells. The genotype of these compound transgenic mice (TCRβ-PLP1TG::Tcra+/−::Foxp3GFP) will in the following be referred to as “Fixed-β.”
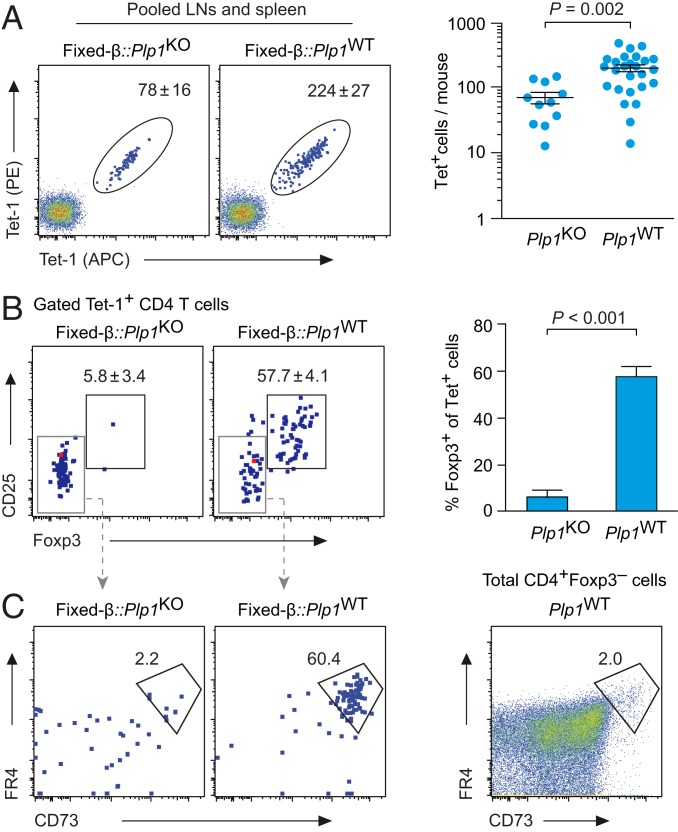
As expected, essentially all T cells in Fixed-β mice expressed the transgenic TCR Vβ6 chain (SI Appendix, Fig. S2). The peripheral repertoire of Fixed-β mice on Plp1KO background contained on average 78 ± 16 Tet-1+ CD4 T cells, which is about 5-fold more than in the corresponding fully polyclonal repertoire (Fig. 3A, compare Fig. 1A). Thus, the fixed β chain indeed imposed a certain repertoire bias, but the resulting frequency of PLP11–19–specific CD4 T cells was not unphysiologically high (31–33).
Fig. 3.
Number and phenotype of PLP11–19–specific peripheral CD4 T cells in Plp1KO or Plp1WT mice expressing a fixed TCRβ chain. (A) Flow cytometry of pooled spleen and lymph node cells after enrichment of PLP11–19–specific CD4 T cells, gated on CD4+CD8−Dump− cells. The calculated mean number ± SEM of tetramer-positive cells/mouse is indicated (n ≥ 11 each). The graph on the right shows a summary. Each data point represents an individual mouse. (B) Foxp3GFP and CD25 expression in tetramer-positive cells from pooled spleen and lymph node cells. Numbers indicate the mean frequency ± SEM of Foxp3+CD25+ cells (n ≥ 11 each). Data are summarized on the right. (C) FR4 and CD73 expression on gated Foxp3− PLP11–19–specific CD4 T cells from pooled spleen and lymph node cells. The dot plot on the right shows FR4 and CD73 expression in total Foxp3− peripheral CD4 T cells for comparison (n ≥ 6 each).
In line with our findings in the fully polyclonal repertoire, Fixed-β::Plp1WT mice had higher numbers of peripheral Tet-1+ CD4 T cells than their PLP-deficient counterparts (Fig. 3A), and more than 50% of Tet-1+ cells were Foxp3+. Among cells from Plp1WT mice that did not express Foxp3, the majority were FR4+CD73+, a characteristic of anergic CD4 T cells (Fig. 3C) (34, 35), whereas essentially all Tet-1+Foxp3− cells in Plp1KO mice displayed a naive FR4−CD73− phenotype (Fig. 3 B and C). Moreover, most Tet-1+Foxp3− cells in Plp1WT mice were CD44hi and had lower levels of CD5 as compared to Tet-1+Foxp3− cells from Plp1KO mice, corroborating the notion that they were antigen-experienced and intrinsically tolerant rather than naive and ignorant (SI Appendix, Fig. S3).
Evidence for Clonal Deletion and Anergy Induction in the Thymus.
The CD4 SP compartment of Fixed-β::Plp1KO mice contained around 750 Tet-1+ cells, essentially all of which were Foxp3− (Fig. 4 A and B). In Plp1WT mice, Tet-1+ CD4 SP cells were significantly reduced, and a substantial fraction of cells were Foxp3+ (Fig. 4 A and B). Around 30% of the thymic Treg cell compartment consists of recirculating cells from the periphery (36). These lack CCR7 expression (37). More than 80% of Foxp3+ Tet-1+ cells in Plp1WT mice were CCR7+, indicating that they were recently induced Treg cells (Fig. 4C).
Fig. 4.
Number and phenotype of PLP11–19–specific CD4 SP thymocytes in Plp1KO or Plp1WT mice expressing a fixed TCRβ chain. (A) Flow cytometry of thymocytes after enrichment of PLP11–19–specific cells, gated on Dump−CD8−CD4+ cells. The calculated mean number ± SEM of tetramer-positive cells/thymus is indicated (n ≥ 12 each). The graph on the right shows a summary. Each data point represents an individual mouse. (B) Foxp3GFP and CD25 in Tet-1+ CD4 SP cells. Mean ± SEM of n ≥ 12 each. (C) CCR7 expression on gated Foxp3+CD25+ Tet+ CD4 SP thymocytes. (D) CD69 and MHC class I (H-2Kb) on Tet-1+ CD4 SP thymocytes from Fixed-β::Plp1KO or Fixed-β::Plp1WT mice. Colors refer to gating as in B (representative of n ≥ 6 each). (E) FR4 and CD73 on mature (CD69−MHCI+) Tet-1+ CD4 SP thymocytes from Fixed-β::Plp1KO or Fixed-β::Plp1WT mice, gated as in D (representative of n ≥ 5 each).
CD4 SP cells can be subdivided into sequential maturation stages according to expression of CD69 and MHCI (CD69+MHCI− → CD69+MHCI+ → CD69−MHCI+) (38). Tet-1+ cells were significantly reduced within the most mature CD69−MHCI+ subset of Plp1WT mice (2.7 ± 1.0% vs. 11.3 ± 2.1%; P = 0.004) (Fig. 4D). Together with the lower total number of Tet+ cells in Plp1WT thymi, this suggested that concomitant to the diversion of some cells into the Treg cell lineage, other cells were deleted at a relatively late stage of CD4 SP maturation. Moreover, the remaining mature Tet+ cells in Plp1WT thymi contained a substantial proportion of cells that were FR4+CD73+, consistent with anergy being a third antigen-instructed cell fate option of PLP-specific thymocytes.
Clonal Composition of Tet+ Cells in the Presence or Absence of PLP Expression.
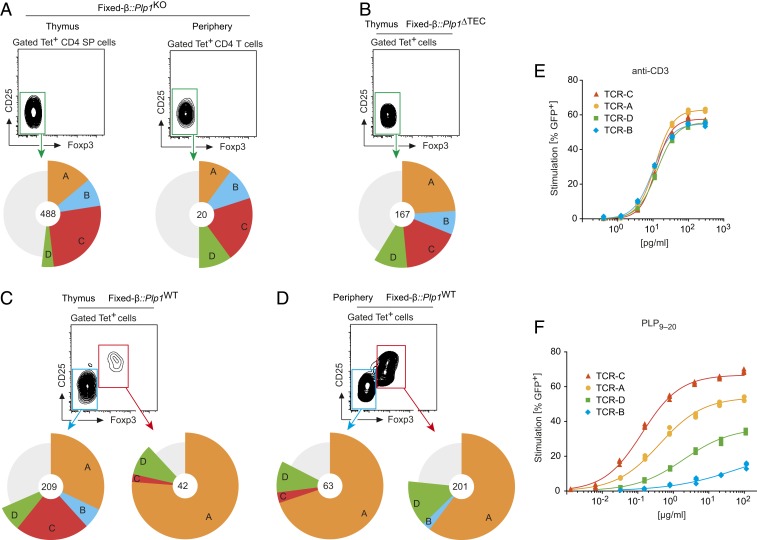
If deletion and diversion into the Treg cell lineage selectively applied to cells carrying different TCRs, some TCRs from the uncensored repertoire were expected to preferentially contribute to the Treg cell population in Plp1WT mice, whereas other TCRs would disappear from the repertoire. To investigate this, we generate an uncensored reference library of PLP11–19–specific TCRs from Tet+ thymocytes from Plp1KO mice by single-cell TCRα sequencing (39). These cells are neither exposed to tolerogenic forces nor subject to peripheral homeostatic influences of noncognate nature. We obtained TCRα sequences from 529 Tet-1+ CD4 SP cells, and of these, a total of 488 TCRα nucleotide sequences encoded for TCRα rearrangements that were each present at a frequency of at least 1% (Fig. 5A). Four public TCR entities, in the following referred to as TCR A, B, C, and D (SI Appendix, Fig. S4), were present at frequencies between 4 and 26% in the thymus and together accounted for roughly 50% of the Tet-1+ population in both the thymus and the periphery (Fig. 5A).
Fig. 5.
TCR inventories of PLP11–19–specific CD4 T cells from thymus or periphery of Plp1KO and Plp1WT mice expressing a fixed TCRβ chain. (A) Frequency of the 4 public TCRs A, B, C, and D in thymic or peripheral Tet-1+ T cells of Fixed-β::Plp1KO mice. The gray sector comprises all other TCR entities that were found at a frequency of >1%. Numbers in the center indicate the total number of cells that are represented in the respective pie chart. The graphs summarize data obtained from single cells from 111 mice analyzed in multiple pools. (B) Frequency of the TCRs A, B, C, and D in the thymic Tet-1+ CD4 SP cell population of Fixed-β::Plp1ΔTEC mice. Summary of data obtained from sorted single cells from 47 mice. (C) Frequency of the TCRs A, B, C, and D in Foxp3− or Foxp3+ Tet-1+ CD4 SP thymocytes from Fixed-β::Plp1WT mice. Summary of data obtained from sorted single cells from 114 mice. (D) Frequency of the TCRs in peripheral Foxp3− or Foxp3+ Tet-1+ CD4 T cells from Fixed-β::Plp1WT mice. Summary of cumulative data from sorted single cells from 132 mice. (E) Dose–response of CD4 T cell hybridoma cells expressing TCRs A, B, C, and D to stimulation with titrated amounts of anti-CD3 antibody. (F) Dose–response of CD4 T cell hybridoma cells expressing TCRs A, B, C, and D to stimulation with titrated amounts of cognate antigen. Data in E and F are representative of at least 3 experiments.
We next addressed the frequency of the TCRs A, B, C, and D in mice with a conditional deletion of Plp1 in thymic epithelial cells (Plp1ΔTEC). We previously showed that expression of PLP in TECs is necessary for central tolerance (26, 27). Indeed, the Tet-1+ CD4 SP cell population in Plp1ΔTEC mice was significantly larger as compared to Plp1WT mice and essentially devoid of Foxp3+ cells, confirming that both manifestations of central tolerance in Plp1WT thymi required PLP expression in TECs (SI Appendix, Fig. S5 A and B). The relative abundance of the TCRs A to D was similar to that in Plp1KO mice (Fig. 5B). Thus, the 4 public PLP-specific TCRs were present at stereotypic frequencies in 2 independently generated uncensored inventories.
In the thymus of Fixed-β::Plp1WT mice, the relative abundance of the TCRs A to D among Tet-1+ Foxp3− CD4 SP cells resembled their distribution in the uncensored reference inventories (Fig. 5C). By contrast, the TCR composition of the Tet-1+ Treg cell population was strikingly different. TCR A was strongly overrepresented, whereas TCRs B and C were absent or substantially underrepresented, respectively. Only TCR D was similarly abundant among both Foxp3+ and Foxp3− CD4 SP cells from Plp1WT mice (and as in the uncensored repertoire). This revealed a differential propensity of TCRs A, B, C, and D to divert CD4 SP cells into the Treg cell lineage in the presence of cognate antigen.
The TCR composition of peripheral Tet-1+ Treg cells in Fixed-β::Plp1WT was very similar to the nascent Tet-1+ Treg cell repertoire in the thymus (Fig. 5D, compare Fig. 5C). Again, TCR A was far more abundant than TCR D, and TCRs B and C were extremely rare or absent. Remarkably, the distribution of the TCRs A, B, C, and D among peripheral Foxp3− CD4 T cells (most of which displayed anergy hallmarks) was essentially a mirror image of that on Treg cells; that is, TCRs A and D were dominant, while TCRs B and C were largely lacking (Fig. 5D).
A Reactivity Hierarchy Among Deleter and Diverter TCRs.
TCR A dominated the Treg cell compartment of tolerant mice, indicating that it acted as an efficient diverter TCR. By contrast, TCR C, albeit being at least as abundant in the uncensored repertoire as TCR A, was largely absent from the tolerant peripheral repertoire, suggesting that it was a deleter TCR. To address whether these distinct cell fate-specifying properties correlated with TCR reactivity, we reexpressed the 4 public TCRs in CD4 T cell hybridoma cells carrying a GFP IL-2 reporter (40). All 4 hybridoma cell lines showed identical control responses to noncognate polyclonal stimulation with anti-CD3 antibody (Fig. 5E). Upon stimulation with titrated amounts of PLP peptide, the deleter TCR C displayed the highest functional avidity to cognate antigen stimulation, with an IC50 that was about 50-fold or 250-fold lower than that of the diverter TCR A or D, respectively. TCR B had the lowest responsiveness to antigen.
Discussion
Our findings corroborate the idea that CD4 T cell tolerance to TRAs may preferentially operate through Treg cell induction rather than deletion (21, 22). However, an inspection of the TCR composition of PLP-specific T cells indicated that the mere number of Tet+ cells in tolerant and nontolerant repertoires only poorly reflected whether and to what extent deletion shaped this autorective T cell population (41). Moreover, our findings suggest that anergy is a third antigen-instructed cell fate option of TRA-specific CD4 T cells.
In line with a central prediction of the affinity model of clonal deletion versus Treg cell induction (1, 10), the tip of PLP reactivity was absent from the tolerant repertoire. Although TCR C was still detectable among CD4 SP thymocytes from Plp1WT mice, we deem it likely that its absence from the peripheral T cell pool at least in part resulted from thymic deletion. We showed previously that central tolerance to PLP involves direct antigen presentation by mTECs (27). Typically, TRAs are expressed by a small fraction of mTECs. This imposes spatial and temporal imitations on the likelihood with which a given antigen is encountered by developing T cells. Hence, the TCR inventory of Foxp3− thymocytes may to a considerable degree be derived from cells that are upstream of antigen encounter and the ensuing cell fate decision. In line with loss of PLP-specific deleter TCRs such as TCR C occurring at a very late CD4 SP cell stage, there was a strong reduction of mature Tet+ CD4 SP cells in Plp1WT mice, whereas more immature CD4 SP stages were unaffected by the presence of cognate antigen. The paucity of mature thymic Tet+ cells in Plp1WT mice precluded testing the prediction that TCR C should be largely absent from the mature CD4 SP cell compartment.
The contribution of distinct diverter TCRs to the Treg cell population reflected their relative antigen responsiveness rather than their frequency in the uncensored thymic precursor pool. One possible explanation is that mature Treg cells of the highest relative reactivity may possess a competitive advantage in the periphery (42). However, we deem it equally if not more likely that the correlation between a TCR’s relative reactivity and its abundance among Treg cells reflects a differential efficacy of Treg lineage specification itself, as it is already evident in the nascent thymic Treg cell repertoire. Consistent with this idea, the extent of Treg cell differentiation upon injection of precursors with different OVA-specific TCRs into OVA expressing thymi directly correlated with their in vitro reactivity to antigen (43). In TCR transgenic models, cells with identical TCRs compete for a limiting niche during thymic Treg cell differentiation (24, 25). It will be interesting to address whether Treg cell precursors with higher-affinity TCRs may outcompete lower-affinity cells for a shared developmental niche or whether each clonal specificity occupies a private niche whose size increases with TCR reactivity. Continuous TCR stimulation fuels Treg cell fitness and function (44, 45). Hence, filling of the Treg cell repertoire with cells of the highest permissive TCR affinity may be crucial for optimal immune regulation.
Global TCR sequencing revealed some overlap between the TCR repertoires of Foxp3-positive and Foxp3-negative cells (12, 46, 47). It was concluded that a substantial number of autoreactive cells evade from tolerance induction in the thymus, yet are accompanied by Treg cells of identical specificity that prevent their unwanted activation (10). Reminiscent of this buddy hypothesis, the distribution of TCRs among Foxp3-negative Tet+ cells in the tolerant repertoire was essentially identical to that of Treg cells. Importantly, though, rather than having escaped from tolerance induction, these Foxp3-negative cells mostly displayed phenotypic hallmarks of anergy. Where and how the anergic phenotype of peripheral PLP-specific cells is imprinted remains to be established. The presence of a considerable fraction of FR4+CD73+ cells among mature Foxp3− CD4 SP cells in PLPWT thymi is consistent with a role of the thymus in anergy induction, and the strikingly similar TCR composition of Foxp3+ and anergic Tet+ cells raises the possibility that their developmental trajectories are interconnected (48). Downstream of the initial TCR stimulus, Treg cell precursors compete for nonantigenic factors such as IL-2 to complete their differentiation (1). It is tempting to speculate that at least some of the primed Treg precursors that lose this competition exit the thymus in an anergic state.
In sum, our findings reveal a multilayered TCR hierarchy that shapes the size, composition, and functional state of an autoreactive T cell repertoire. Optimizing the efficacy of the generation of TCR inventories of rare antigen-specific cells will pave the way toward deciphering interindividual variations in the composition of autoreactive T cell cohorts, more precisely understanding where and when deleter TCRs are lost from the repertoire, and unraveling whether and which TRA-specific Treg cells might be strategically positioned in the vicinity of their cognate autoantigen.
Materials and Methods
Animals.
Foxp3GFP reporter mice (DEREG) (49), Tcra−/− mice (50), and Plp1KO (51) mice have been described previously. Plp1ΔTEC mice (27) were obtained by crossing Foxn1-Cre mice (52) to mice carrying a conditional Plp1 allele (Plp1fl) (53). The transgenic fixed TCRβ allele was from the αβ TCR transgenic mouse strain TCR-PLP1 (27). For more details, see SI Appendix. Animal studies were approved by local authorities (Regierung von Oberbayern, Az 7-08 and 142-13).
Flow Cytometry.
Single-cell suspensions of spleen, lymph nodes, or thymus were surface stained according to standard procedures. For details on antibodies, flow cytometry and software, see SI Appendix.
Generation of I-Ab Tetramers.
MHCII tetramers were produced as described previously (33). For details, see SI Appendix.
Enrichment of Tet+ Cells.
Tet–labeled cells were enriched using anti-PE and anti-APC microbeads and magnetized columns (Miltenyi Biotech) as described previously (33). For details, see SI Appendix.
Single-Cell TCRα Sequencing.
TCRα-chain sequencing was performed with a protocol modified from ref. 39. For more details, see SI Appendix.
Reexpression and Functional Testing of TCRs.
TCRs were reconstituted by viral transduction of NFAT-GFP reporter hybridoma cells (40) stably expressing the fixed TCRβ chain. Hybridoma cells were stimulated with titrated amounts of peptide or anti-CD3 antibody, and GFP expression was measured by flow cytometry. For more details, see SI Appendix.
Statistical Analyses.
Statistical significances were calculated with Prism7 using the 2-tailed unpaired Student’s t test.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council (ERC-2016-ADG 742290–TOLERANCE FOOTPRINT to L.K.). M.H. and E.U. received support from the Deutsche Forschungsgemeinschaft (SFB 1054, projects A01 and IRTG). M.K.J. is supported by the US National Institutes of Health (P01 AI035296). D.H.B. received support from the Deutsche Forschungsgemeinschaft (SFB 1054, project B09). We thank L. Richter (FlowCyt Core Facility, BioMedical Center, Munich, Germany) for flow cytometry support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907615116/-/DCSupplemental.
References
- 1.Klein L., Robey E. A., Hsieh C. S., Central CD4+ T cell tolerance: Deletion versus regulatory T cell differentiation. Nat. Rev. Immunol. 19, 7–18 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Apostolou I., Sarukhan A., Klein L., von Boehmer H., Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 3, 756–763 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Jordan M. S., et al. , Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2, 301–306 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Kieback E., et al. , Thymus-derived regulatory T cells are positively selected on natural self-antigen through cognate interactions of high functional avidity. Immunity 44, 1114–1126 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Feuerer M., et al. , Enhanced thymic selection of FoxP3+ regulatory T cells in the NOD mouse model of autoimmune diabetes. Proc. Natl. Acad. Sci. U.S.A. 104, 18181–18186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinterberger M., et al. , Autonomous role of medullary thymic epithelial cells in central CD4(+) T cell tolerance. Nat. Immunol. 11, 512–519 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Picca C. C., et al. , Thymocyte deletion can bias Treg formation toward low-abundance self-peptide. Eur. J. Immunol. 39, 3301–3306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simons D. M., et al. , How specificity for self-peptides shapes the development and function of regulatory T cells. J. Leukoc. Biol. 88, 1099–1107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein L., Khazaie K., von Boehmer H., In vivo dynamics of antigen-specific regulatory T cells not predicted from behavior in vitro. Proc. Natl. Acad. Sci. U.S.A. 100, 8886–8891 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh C. S., Lee H. M., Lio C. W., Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 12, 157–167 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Hsieh C. S., et al. , Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity 21, 267–277 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Hsieh C. S., Zheng Y., Liang Y., Fontenot J. D., Rudensky A. Y., An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 7, 401–410 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Pacholczyk R., et al. , Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity 27, 493–504 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson M. S., et al. , The cellular mechanism of Aire control of T cell tolerance. Immunity 23, 227–239 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Anderson M. S., et al. , Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Derbinski J., Schulte A., Kyewski B., Klein L., Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2, 1032–1039 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Liston A., Lesage S., Wilson J., Peltonen L., Goodnow C. C., Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4, 350–354 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Malchow S., et al. , Aire enforces immune tolerance by directing autoreactive T cells into the regulatory T cell lineage. Immunity 44, 1102–1113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malchow S., et al. , Aire-dependent thymic development of tumor-associated regulatory T cells. Science 339, 1219–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry J. S. A., et al. , Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41, 414–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legoux F. P., et al. , CD4+ T cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory T cells rather than deletion. Immunity 43, 896–908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra D., et al. , Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol. 17, 187–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taniguchi R. T., et al. , Detection of an autoreactive T-cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)-mediated selection. Proc. Natl. Acad. Sci. U.S.A. 109, 7847–7852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bautista J. L., et al. , Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat. Immunol. 10, 610–617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung M. W., Shen S., Lafaille J. J., TCR-dependent differentiation of thymic Foxp3+ cells is limited to small clonal sizes. J. Exp. Med. 206, 2121–2130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein L., Klugmann M., Nave K. A., Tuohy V. K., Kyewski B., Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 6, 56–61 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Wang L., et al. , Epitope-specific tolerance modes differentially specify susceptibility to proteolipid protein-induced experimental autoimmune encephalomyelitis. Front. Immunol. 8, 1511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon J. J., et al. , Tracking epitope-specific T cells. Nat. Protoc. 4, 565–581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon J. J., et al. , Quantitative impact of thymic selection on Foxp3+ and Foxp3- subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14602–14607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tubo N. J., et al. , Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu H. H., Moon J. J., Kruse A. C., Pepper M., Jenkins M. K., Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J. Immunol. 185, 4705–4713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins M. K., Chu H. H., McLachlan J. B., Moon J. J., On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu. Rev. Immunol. 28, 275–294 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Moon J. J., et al. , Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalekar L. A., et al. , CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol. 17, 304–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez R. J., et al. , Arthritogenic self-reactive CD4+ T cells acquire an FR4hiCD73hi anergic state in the presence of Foxp3+ regulatory T cells. J. Immunol. 188, 170–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiault N., et al. , Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors. Nat. Immunol. 16, 628–634 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Cowan J. E., McCarthy N. I., Anderson G., CCR7 controls thymus recirculation, but not production and emigration, of Foxp3(+) T cells. Cell Rep. 14, 1041–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing Y., Wang X., Jameson S. C., Hogquist K. A., Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17, 565–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dössinger G., et al. , MHC multimer-guided and cell culture-independent isolation of functional T cell receptors from single cells facilitates TCR identification for immunotherapy. PLoS One 8, e61384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aschenbrenner K., et al. , Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8, 351–358 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Yu W., et al. , Clonal deletion prunes but does not eliminate self-specific αβ CD8(+) T lymphocytes. Immunity 42, 929–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lathrop S. K., Santacruz N. A., Pham D., Luo J., Hsieh C. S., Antigen-specific peripheral shaping of the natural regulatory T cell population. J. Exp. Med. 205, 3105–3117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee H. M., Bautista J. L., Scott-Browne J., Mohan J. F., Hsieh C. S., A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity 37, 475–486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine A. G., Arvey A., Jin W., Rudensky A. Y., Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vahl J. C., et al. , Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity 41, 722–736 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Pacholczyk R., Ignatowicz H., Kraj P., Ignatowicz L., Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity 25, 249–259 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Wong J., et al. , Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J. Immunol. 178, 7032–7041 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Kalekar L. A., Mueller D. L., Relationship between CD4 regulatory T cells and anergy in vivo. J. Immunol. 198, 2527–2533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahl K., et al. , Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 204, 57–63 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mombaerts P., et al. , Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature 360, 225–231 (1992). [DOI] [PubMed] [Google Scholar]
- 51.Klugmann M., et al. , Assembly of CNS myelin in the absence of proteolipid protein. Neuron 18, 59–70 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Rossi S. W., et al. , Keratinocyte growth factor (KGF) enhances postnatal T-cell development via enhancements in proliferation and function of thymic epithelial cells. Blood 109, 3803–3811 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lüders K. A., Patzig J., Simons M., Nave K. A., Werner H. B., Genetic dissection of oligodendroglial and neuronal Plp1 function in a novel mouse model of spastic paraplegia type 2. Glia 65, 1762–1776 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.