Abstract
Purpose:
Protein kinases are known to play a prominent role in oncogenic progression across multiple cancer subtypes, yet their role in prostate cancer progression remains underexplored. The purpose of this study was to identify kinases that drive prostate cancer progression.
Experimental design:
To discover kinases that drive prostate cancer progression, we investigated the association between gene expression of all known kinases and long-term clinical outcomes in tumor samples from 545 patients with high-risk disease. We evaluated the impact of genetic and pharmacologic inhibition of the most significant kinase associated with metastatic progression in vitro and in vivo.
Results:
DNA-dependent protein kinase (DNAPK) was identified as the most significant kinase associated with metastatic progression in high-risk prostate cancer. Inhibition of DNAPK suppressed the growth of both AR-dependent and AR-independent prostate cancer cells. Gene set enrichment analysis nominated Wnt as the top pathway associated with DNAPK. We found that DNAPK interacts with the Wnt transcription factor LEF1 and is critical for LEF1-mediated transcription.
Conclusions:
Our data show that DNAPK drives prostate cancer progression through transcriptional regulation of Wnt signaling and is an attractive therapeutic target in aggressive prostate cancer.
Keywords: castration-resistant prostate cancer, DNA-dependent protein kinase, Wnt signaling pathway, prognostic biomarker, LEF1
Introduction
Androgen deprivation therapy is the standard of care for patients with advanced prostate cancer. However, responses are not durable and tumors invariably progress to castration-resistant prostate cancer (CRPC). CRPC has limited treatment options and ultimately undergoes metastatic progression to become a lethal disease. Although the development of next-generation anti-androgen therapy, chemotherapy, and immunotherapy have improved overall survival for metastatic CRPC (mCRPC) patients (1-6), the mortality rates of patients with mCRPC remain high. Thus, there is a clear need to identify novel therapeutic targets to prevent progression to mCRPC and to improve outcomes.
Kinases play a vital role in driving oncogenic pathways and have been the target for therapeutic intervention across multiple cancer subtypes (7). However, the role of kinases in aggressive prostate cancer remains poorly understood. Moreover, there are currently no kinase inhibitors that are known to increase survival of patients with prostate cancer (8). The discrepancy between the utility of kinase inhibitors in multiple cancer subtypes and the lack of effective kinase inhibitors in prostate cancer strongly suggests the need for a more systematic exploration of kinases in prostate cancer.
To identify novel driver kinases involved in prostate cancer progression, we performed an unbiased analysis evaluating the association between expression of all known kinases and clinical outcomes using prostate cancer samples from a large cohort of patients with high-risk disease treated with prostatectomy. We identified DNA-dependent protein kinase (DNAPK) as the most significantly associated kinase with metastatic progression in localized high-risk disease and validated the prognostic value of high levels of DNAPK expression across multiple datasets. DNAPK, a nuclear serine/threonine protein kinase, is well known for its role in DNA repair via the non-homologous end joining (NHEJ) pathway (9). However, numerous studies have demonstrated the importance of DNAPK in a variety of other processes, including the modulation of chromatin structure and transcription through its interaction with a variety of receptors, polymerases, and transcription factors (10-14). We have previously found that DNAPK transcriptionally regulates multiple pathways involved in prostate cancer progression (15); however, the dominant pathways mediated by DNAPK signaling have not been elucidated.
To interrogate the dominant mechanism by which DNAPK drives tumor progression, we utilized Gene Set Enrichment Analysis (GSEA) on gene expression data from both clinical prostate cancer specimens and prostate cancer cell lines to nominate Wnt signaling as a key DNAPK-regulated pathway. We demonstrate that genetic or pharmacological inhibition of DNAPK suppresses Wnt3A-induced migration and invasion in prostate cancer cells. We also show that DNAPK interacts with the Wnt transcription factor LEF1 and that inhibition of DNAPK reduces LEF1-mediated transcription. Using multiple DNAPK inhibitors, we demonstrate that inhibition of DNAPK reduces growth of prostate cancer xenografts in vivo as well as proliferation of human prostate cancer explants ex vivo. Together, our data show that DNAPK is a viable prognostic biomarker that drives disease progression through transcriptional regulation of Wnt signaling and support the use of DNAPK inhibitors as a therapeutic strategy in aggressive prostate cancer.
Materials and Methods
Human studies
Patient demographics are described in Table S1. Data collection at each institution (Mayo Clinic and Johns Hopkins Medical Institute) was approved by the corresponding institutional review board (IRB) and patient consent was waived by the IRBs. The studies were conducted in accordance with the U.S. common rule.
Cell lines
LNCaP, C4–2B, PC-3, VCaP, DU-145 cells (all male in gender) were purchased from the American Type Culture Collection (ATCC), grown in respective ATCC recommended culture media, and maintained at 37°C in 5% CO2. All cell lines were regularly tested for mycoplasma contamination and tested negative. For androgen deprivation studies, LNCaP cells were grown in RPMI media without phenol red supplemented with 5% charcoal-stripped serum (Thermo Fisher Scientific). LNCaP-AR-enza-res cells were generated by treating LNCaP-AR parental cells with increasing doses of enzalutamide (starting from 1 μM to 50 μM) until resistance emerged.
Clinical cohorts and microarray analysis
Radical prostatectomy samples were obtained from the Mayo Clinic (MC, N = 545) and Johns Hopkins Medical Institute (JHMI, N = 355). Patient demographics are included in Table S1. MC was a nested case-control study of metastatic progression (16), and JHMI was a cohort study of observation until metastatic progression (17). Stratification of patients into luminal and basal subtypes was determined as previously described (18). Samples were processed and gene expression was analyzed with the Affymetrix Human Exon 1.0 ST array in a CLIA-certified facility as described previously (16,17). All human kinases (19) were evaluated for differential expression associated with metastatic progression by Fisher’s exact test on high and low expression clusters determined by k-medoids clustering (Figure 1A) (19,20) using R software. Kaplan-Meier curves for freedom from metastasis (Figure 1B-C), prostate cancer specific survival, and overall survival were generated with high expression defined as the 80th percentile in each cohort as described previously (15) and P-values were calculated using the log-rank test. The association of DNAPK with metastasis was also examined independently in oncomine (www.oncomine.org) in prostate cancer versus metastatic samples. One cohort by Chandran et al. (21) was excluded because the DNAPK probe information was internally discordant. All statistical analyses were performed in R. A p < 0.05 was considered statistically significant.
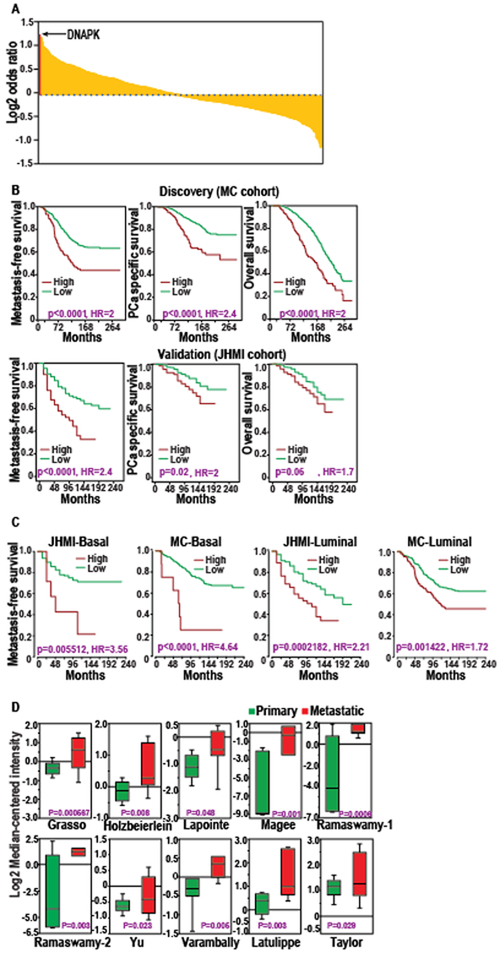
Figure 1. DNAPK is the most significant kinase associated with metastatic progression in prostate cancer.
A, the log2 odds ratio of having a metastatic event with expression of every kinase is shown. Each kinase is represented by a yellow bar. The enrichment of DNAPK is shown with the red bar. B, Kaplan-Meier curves demonstrate the clinical outcomes (metastasis-free survival, prostate cancer-specific survival, and overall survival) associated with DNAPK expression in discovery (MC cohort, N = 545) and validation (JHMI cohort, N = 355) cohorts. C, Kaplan-Meier curves demonstrate distant metastasis-free survival associated with DNAPK expression in MC and JHMI cohorts stratified into luminal and basal subtypes. D, Box plots show Oncomine analyses of DNAPK expression in primary (green) vs metastatic (red) prostate cancer. P-values for Kaplan-Meier survival curves were calculated using log-rank test.
DNAPK knockdown microarray
RNA was extracted using TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol and hybridized to Affymetrix Human Gene ST 2.1 microarrays. Expression data were normalized using the robust multi-array average (RMA) (22) and log2 transformed. Expression profiles were analyzed using standard GSEA comparing the controls to the DNAPK knockdown samples (23), the KEGG and Biocarta pathway gene sets, and using the GSEA Pre-Ranked algorithm on Pearson’s correlations between each gene and DNAPK in the clinical samples. The association of DNAPK with metastasis was also examined independently in Oncomine (www.oncomine.org) (24) in prostate cancer versus metastatic samples.
siRNA treatment and cell proliferation assays
ON-TARGETplus siRNA against DNAPK, LEF1, CTNNB1 and non-targeting control (siNTC) from Dharmacon were used at 100 nmol/L. Cells were transfected in 6-well plates at a density of 50,000 cells per well using Oligofectamine (Invitrogen), according to the manufacturer’s protocol. Transfection was repeated 24 hours later; the cells were grown for an additional 48 hours and re-plated at a density of 5,000–10,000 cells per well in 24-well plates. For proliferation assays with NU7441, cells were treated with NU7441 (1 μM) or DMSO for 24 hours before plating. Cells were then harvested and plated at the previously mentioned density in the presence of NU7441 or DMSO. Cells were counted in duplicates at indicated time points using a Beckman Coulter cell counter and growth curves were plotted as the mean ± S.D.
Quantitative real time-PCR assay
RNA was isolated from cell or tissue lysates by the RNeasy Micro Kit (Qiagen) and cDNA was synthesized from 1 μg RNA using SuperScript III (Invitrogen) and Random Primers (Invitrogen), per the manufacturer’s protocol. qRT-PCR was carried out on the StepOne Real-Time PCR system (Applied Biosystems) using gene-specific primers designed with Primer-BLAST (Supplementary Table S4) and synthesized by IDT Technologies. Validation of siRNA-mediated knockdown was carried out using Fast SYBR Green Master Mix (Thermo Fisher Scientific), per the manufacturer’s protocol. The Wnt gene list was adopted from the Wnt pathway array (SA Biosciences). GAPDH was used as an internal reference. qRT-PCR data were analyzed using the relative quantification method and plotted as the mean fold-change ± S.D. compared with the non-targeting siRNA (i.e. Relative Quantity or RQ) in duplicates.
Immunoblot analysis
Cell lysates were separated on 4% to 12% gradient SDS polyacrylamide gels (Thermo Fisher Scientific) and transferred to polyvinylidene difluoride membranes (GE Healthcare) by wet transfer. Antibodies to phospho-DNAPK (Abcam #18192), KU70 (BD Biosciences #611892), c-Myc (Cell Signaling Technology #5605S), GAPDH (Cell Signaling Technology # 2118L), total DNAPK (Ab-4, NeoMarkers), active β-catenin (Millipore # 05–665), and LEF1 (Millipore #17–604) were used at 1:1,000 dilutions. Membranes were blocked for one hour in 5% BSA or 5% low fat milk dissolved in 0.05% Tween-20/TBS (TBST) and then incubated with primary antibodies overnight at 4°C . Membranes were washed three times in TBST and incubated with HRP-linked secondary antibodies diluted 1:10,000 in TBST for one hour at room temperature. Membranes were then washed three times in TBST and developed using ECL Prime (GE Healthcare) detection reagent according to the manufacturer’s protocol.
Immunoprecipitation studies
For immuno-precipitation studies, nuclear pellets were prepared from cells using NE-PER kit (Thermo Fisher Scientific) according to manufacturer’s protocol. The nuclear pellets were directly lysed in immunoprecipitation buffer (IP buffer) containing 200 mM Tris pH 7.5, 1% Triton X-100, and 300 mM NaCl supplemented with Halt Protease/phosphatase inhibitor cocktail, (Thermo Fisher Scientific). Cell lysates were cleared with MagnaBind Beads (Thermo Fisher Scientific) for 2 h, and were incubated with antibodies against DNAPK or LEF1 antibodies overnight at 4°C . The following day, MagnaBind Beads were added to the lysates and incubated for 2 h at 4°C for the bead-antibody crosslinking. Beads were then washed three times with IP buffer and re-suspended in 2X loading buffer (BioRad). Immunoblot analysis for detection of DNAPK, KU70, and LEF1 were performed as described above.
TOPFLASH Reporter assay
TOPFLASH TCF/LEF1 reporter was a kind gift from Dr. Chandan Kumar-Sinha (University of Michigan). The reporter construct was co-transfected with Renilla luciferase (1:50 ratio) into PC-3 cells using FuGene HD transfection reagent (Promega). After 24 h, cells were treated with 1 μM NU7441, or were induced with 200 nM Wnt3A for 3 h, or were treated with a combination of NU7441 and Wnt3A. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Cat.#E1910) according to the manufacturer’s protocol. Levels of TOPFLASH firefly luciferase were normalized to Renilla luciferase and graphs were plotted as the mean ± S.D. in duplicates. Readings were recorded on GloMax 96 microplate luminometer (Promega).
Chromatin immuno-precipitation assay (ChIP)
ChIP assays were performed using HighCell CHIP kit (Diagenode #C01010061) following the manufacturer’s protocol. Briefly, PC-3 or C4–2B cells were treated with either DMSO or with 1 μM NU7441 for 24 hours. Cells were trypsinized and washed with PBS and were resuspended in 500 ul PBS. Cells were fixed by adding 13.5 μl of 36.5% formaldehyde for 8 minutes and the fixing was terminated by adding 57 μl, 1.25 M glycine. Cells were spun, washed thrice with PBS and resuspended in 1 ml of ice cold buffer L1. After 10 mins, cells were spun down, and the pellets were resuspended in 1 ml ice cold buffer L2. Cells were collected by centrifugation after 5 minutes and pellets were resuspended in 200 μl shearing buffer supplemented with protease inhibitors. Cells were sonicated in BioRuptor (Diagenode) for 4 runs of 10 cycles (30 seconds on and 30 seconds off). Lysates were resuspended in a total of 1 ml ChIP buffer C1 and were centrifuged. Cleared lysates were collected and 9.5 μl of lysate was used as input while 950 μl lysate was used for ChIP. ChIP was performed using protein G beads coated with 5 μg DNAPK cocktail antibody (Ab-4, Neomarkers) or IgG (Santa Cruz # SC-2025 ) overnight. The next day, beads were collected, washed three times with ChIP buffer C1 and once with wash buffer W1. The input samples and washed beads were incubated at 55°C (15 minutes) then 100°C (15 mins) in complete DIB buffer, supplemented with 1 μl proteinase K and were incubated. Beads were spun and supernatants containing DNA were collected to perform qRT-PCR. qRT-PCR data were analyzed using the relative quantification method after normalizing with respective input controls and plotted as the mean fold-change ± S.D. compared with the IgG (i.e. Relative Quantity or RQ). For the ChIP experiments in the backdrop of LEF1 knockdown, cells were treated with either siNTC or with siLEF1 and incubated for 48 hours. ChIP assays were performed as described above.
Boyden chamber-based invasion and migration assays
Boyden Transwell chambers for invasion and migration were purchased from Corning Inc. Cells were transfected with siRNAs as described above or treated with DMSO, 1 μM NU7441 (Selleckchem), 1 μM CC-115 (Celgene), 1 μM IWP-2 (Selleckchem), a combination of IWP-2 and CC-115 (1 μM each), or 10 μM enzalutamide (Selleckchem) for 24 h. Cells were then trypsinized and re-suspended in serum-free media at a density of 30,000 for PC-3 cells and at a density of 200,000 (for invasion, or 100,000 for migration) for LNCaP-AR and C4–2B cells, in the upper chamber. The lower chamber was supplied with media containing 10% FBS, supplemented with DMSO, 1 μM NU7441, 1 μM CC-115, or 200 ng/ml Wnt3A (R&D systems). For the experiments with IWP-2, lower chambers were supplemented with media containing 10% FBS and IWP-2 and/or CC-115 (1 μM each). Cells were incubated for 48 h and were then fixed using 4% paraformaldehyde for 30 min at room temperature. Wells were washed with PBS, and membranes were stained with 0.25% (wt/vol) crystal violet (Sigma-Aldrich) made in 25% (vol/vol) methanol (Fisher Scientific) for 20 mins and then washed (with PBS). For quantification, crystal violet trapped by the cells was dissolved in 10% acetic acid and absorbance was measured at 560 nm using a Spectramax 384 plus plate reader. Representative images were taken on an Olympus IX17 using DP Controller 3.1.1.267 software. Graphs were plotted as the mean ± S.D. and P values were calculated by two-way ANOVA using GraphPad Prism software.
In vivo animal studies
Four to six-week-old male CB17-SCID mice were provided by our lab’s in-house breeding colony and housed under pathogen-free conditions approved by the American Association for Accreditation of Laboratory Animal Care in accordance with current regulations and standards of the U.S. Department of Agriculture and Department of Health and Human Services. Animal experiments were approved by the University of Michigan’s University Committee on Use and Care of Animals (UCUCA) and carried out in accordance with established guidelines. For xenograft studies involving treatment with NU7441, pre-castrated mice were anesthetized with approximately 2% isoflurane and LNCaP-AR cells suspended in 100 μL of a 1:1 mixture of 1X PBS and Matrigel (BD Biosciences) were subcutaneously injected bilaterally into their flanks. When the tumor size reached 24 mm3, mice were randomized into control and treatment groups (n = 6 for the NU7441 arm and n = 5 for the vehicle arm). The DNAPK inhibitor NU7441 (Selleck Chemicals) was administered once daily via IP injection at 25 mg/kg body weight five times per week for four weeks. For the CC-115 and enzalutamide xenograft studies, LNCaP-AR cells were injected subcutaneously into the flanks of uncastrated mice as described above. Once tumors reached an average size of 80 mm3, mice were randomized and divided into treatment groups of vehicle control, CC-115 (2 mg/kg), enzalutamide (10 mg/kg), and a combination of CC-115 and enzalutamide. CC-115 and / or enzalutamide were administered once daily via oral gavage five times per week for six weeks. Tumor growth was monitored 2–3 times per week. After four (NU7441) or six (CC-115, enzalutamide) weeks of treatment, mice were weighed and euthanized and the tumors harvested. Tumor caliper measurements were converted into tumor volumes using the formula ½[length × width2] mm3 and plotted as mean tumor volume ± SEM using GraphPad Prism software.
Immunohistochemistry
Human prostate ex vivo explant cultures were conducted as previously described (25). Briefly, fresh primary tumor tissue was obtained from a clinical pathologist immediately following radical prostatectomy at Thomas Jefferson University Hospital in accordance with Institutional Review Board standards and in compliance with federal regulations governing research on deidentified specimens and/or clinical data (45 CFR 46.102(f)). The specimens were processed under a laminar flow hood using sterile technique and transported in culture media (IMEM, 5% FBS, 0.01 mg/mL insulin (Invitrogen #12585–014), 30 μmol/L hydrocortisone (Sigma #H-0888), and penicillin/streptomycin) on ice. Tissue was subdivided into approximately 1 mm3 pieces and placed in a 24-well plate on presoaked dental sponges (Novartis #96002) (2–3 pieces per sponge) placed into 0.5 mL culture media in presence or absence of drug (1 μM NU7441). Plates were placed in an incubator at 37°C and 5% CO2. Media were replaced every 48 hours with appropriate treatment, and explants were harvested on day 6. Tissue was either formalin fixed for immunohistochemical (IHC) analysis or placed in 1 mL RNAlater (Ambion #AM7020) (kept at 4°C for 24 hours, then stored at −80°C until processing) for RNA analysis. RNA extraction was performed using Trizol (Life Technologies #15596018) per manufacturer’s instructions. IHC samples were stained using Ki67 antibody (Cell signaling Technologies #9027) and Ki67 positive cells were counted . Graphs were prepared using GraphPad Prism software.
Data Availability
Microarray data is available at NCBI GEO accession GSE46691, GSE79956, GSE79957 and GSE116895.
Results
DNAPK is the most significantly associated kinase with metastatic progression in prostate cancer
To identify the kinases that potentially drive progression of aggressive prostate cancer, we first evaluated the association of expression of all known kinases with metastatic progression, using prostate cancer samples from a cohort of patients treated with prostatectomy at the Mayo Clinic. Patient demographics of this cohort are described in Supplementary Table S1. Notable features of this cohort include the long clinical follow-up (median of 13.4 years), large sample size (N = 545), and prevalence of high-risk characteristics as defined by National Comprehensive Cancer Network criteria (www.nccn.org) such as extracapsular extension (50%) and seminal vesicle invasion (32%). Additionally, and consistent with these high-risk features, 39% of the patients in this cohort experienced metastatic progression. In this discovery cohort, we ranked all kinases (39) by the relative enrichment for metastatic progression in cases with high versus low expression of each kinase, with expression cut-offs defined by an unbiased clustering approach as previously described (20). This analysis demonstrated that DNAPK was the most significant kinase that enriched for metastatic progression (OR = 2.19, Log2 OR 1.134, p < 0.0001, Fig. 1A; Supplementary Table S2). We next used Kaplan-Meier analyses to assess the prognostic value of elevated tumor DNAPK expression in this cohort using the same cutoff as described previously (15), and found that high expression of DNAPK was significantly associated with not only metastatic progression (HR = 2.0 [1.5–2.7], p < 0.0001), but also decreased rates of prostate cancer-specific survival (HR = 2.4 [1.7–3.5], p < 0.0001) and overall survival (HR = 2.0 [1.5–2.6], p < 0.0001) (Fig. 1B). We validated these findings in an independent cohort of patients from Johns Hopkins Medical Institute (JHMI) (Supplementary Table S1), where high DNAPK expression again was associated with increased metastatic progression (HR = 2.4 [1.7–3.6], p < 0.0001) and decreased prostate cancer-specific survival (HR = 2 [1.1–3.5], p = 0.02), with borderline significance for decreased overall survival (HR = 1.7 [0.97–2.8], p = 0.06) (Fig. 1B). Because we have previously found that DNAPK transcriptionally activates the androgen receptor (AR) to potentiate AR function, we next stratified our patient cohorts into luminal (more AR-driven) and basal (less AR-driven) subtypes (18,26). As expected, within the luminal subtype of prostate cancer, characterized by elevated AR expression and signaling activity, increased DNAPK expression was associated with poor metastasis-free survival. Unexpectedly, we found that increased DNAPK expression is also a strong prognostic indicator in the basal subtype of prostate cancer, which is thought to be more AR-independent (Fig. 1C). After identifying the prognostic value of DNAPK in localized disease and the luminal and basal subtypes of prostate cancer, we examined the expression of DNAPK in metastatic prostate cancer using Oncomine analysis (www.oncomine.org). DNAPK was significantly overexpressed in metastatic versus primary prostate cancer samples in 10 of the 12 cohorts that we examined (Fig. 1D). These results show that DNAPK is strongly prognostic in localized high-risk prostate cancer, the luminal and basal subtypes of prostate cancer, and is associated with development of metastatic disease.
Genetic and pharmacologic inhibition of DNAPK suppresses aggressive cancer phenotypes in vitro
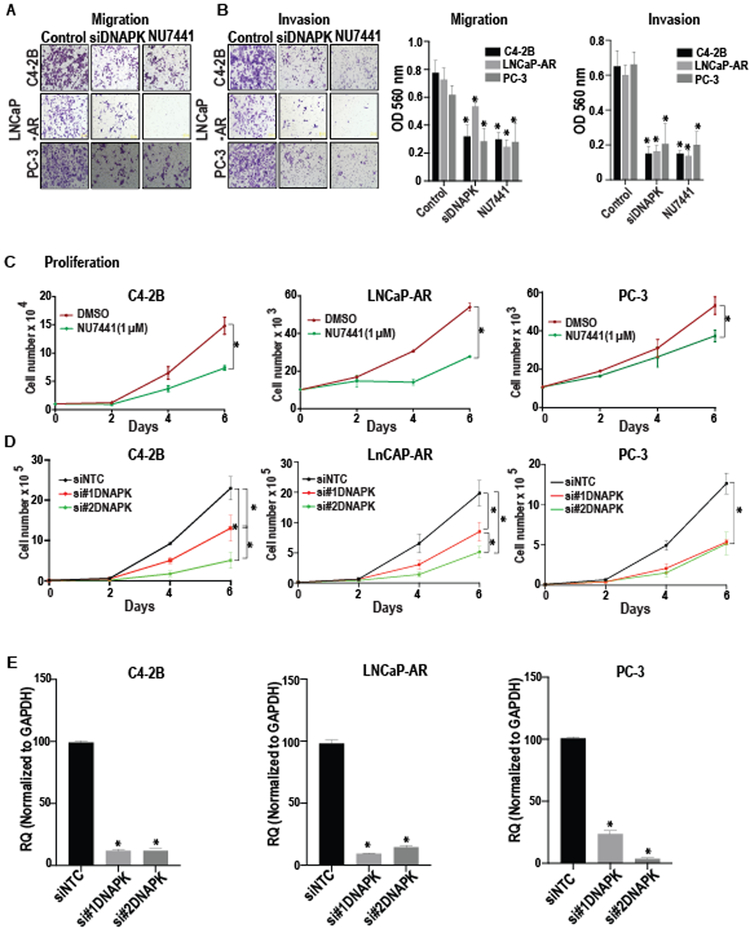
Given the prognostic value of DNAPK expression in localized disease and the luminal and basal subtypes of prostate cancer, we next assessed the functional significance of DNAPK in both AR-dependent and AR-independent prostate cancer cell line models. Genetic (via siRNA-mediated knockdown) or pharmacological inhibition (via the small molecule inhibitor NU7441) of DNAPK drastically diminished migration, invasion, and proliferation of both AR-positive cells (LNCaP-AR and C4–2B), as well as AR-negative PC-3 cells (Fig. 2A-D). Efficient knockdown of DNAPK was achieved in these experiments (Fig. 2E). Given our previous findings that DNAPK is a required co-factor for AR (12), our results from the AR-positive LNCaP-AR and C4–2B cells were expected; however, the reduction in aggressive cancer phenotypes in AR-negative PC-3 cells indicate that DNAPK has functions beyond regulating AR activity that may contribute to prostate cancer progression.
Figure 2. DNAPK inhibition suppresses aggressive cancer phenotypes in vitro.
(A and B), Representative images (magnification 20X) show the effects of genetic (via siRNA-mediate knockdown) or chemical inhibition (via NU7441, 1 μM) of DNAPK on migration and invasion of C4-2B, LNCaP-AR, and PC-3 cells, assessed by Boyden chamber-based assays. Bar graphs show the mean ± S.D. of invasion and migration where treatment groups are compared to their respective control cell lines (N=3 in duplicates). (C and D), The growth curves display the effect of chemical (via NU7441, 1 μM) or genetic (via siRNA-mediated knockdown) inhibition of DNAPK on proliferation of C4-2B, LNCaP-AR, and PC-3 cells. *p < 0.05, compared to their respective controls. (N=3 in duplicates) (E) Bar graphs show the genetic inhibition of DNAPK (via siRNA-mediated knockdown) in indicated cell lines, compared to a scrambled control and respective control cell lines. *p < 0.05. P-values were calculated using a Students t-test (2C, 2E) or two-way ANOVA (Figure 2A-B, 2D) in Graph Pad Prism software. All data are represented as mean ± S.D. NTC = non-targeting control siRNA.
The Wnt pathway is associated with DNAPK in prostate cancer samples
Recently, we investigated transcriptional networks that are modulated by DNAPK using cell line models (15) in the context of AR. Guided by our observation that DNAPK is a prognostic indicator in the luminal and basal subtypes of prostate cancer and that DNAPK inhibition significantly reduces aggressive cancer phenotypes in both AR-dependent and AR-independent cells, we extended these studies by investigating DNAPK associated pathways in clinical samples as well as pre-clinical models. We performed guilt-by-association analyses to identify the role of DNAPK in modulating pathways involved in promoting metastasis. We measured changes in gene expression, due to siRNA-mediated DNAPK knockdown in AR-dependent (VCaP, C4–2B) and AR-independent (PC-3, DU145) cell lines, using microarray analyses and ran standard GSEA analyses on these data. We also correlated each gene with DNAPK expression in clinical samples from the Mayo Clinic cohort and ran GSEA pre-ranked using the same gene sets. A scatterplot of the GSEA normalized enrichment scores (NES) for each pathway gene set was then generated with in vitro NES values on the y-axis, and in vivo NES values on the x-axis (Fig. 3A). The Wnt pathway had the highest average NES (knockdown NES = 1.48, MC NES = 2.47, average = 1.97, Fig. 3A; Supplementary Table S3). Furthermore, we found that expression of DNAPK and β-catenin (CTNNB1), a critical Wnt effector, was tightly correlated in clinical samples (Pearson’s correlation coefficient = 0.7, Supplementary fig. S1A). These data nominate Wnt pathway as highly significant pathway associated with DNAPK, supporting a role of DNAPK in mediating the regulation of the Wnt signaling pathway.
Figure 3. DNAPK regulates Wnt signaling pathway.
A, Scatter plot represents the GSEA normalized enrichment scores (NES) for all pathways associated with DNAPK in both the in vitro knockdown experiment (using VCaP, C4-2B, PC-3, DU145 cells) as well as in the clinical samples from the Mayo Clinic. The Wnt pathway had the top average NES, with high scores in both the clinical and in vitro knockdown results, and is shown in red. Dots in black denote other pathways. B, the bar graph shows the expression of select Wnt pathway genes in LNCaP-AR and C4-2B (CRPC) cells compared to LNCaP (hormone-naive) cells. GAPDH was used as an internal reference (N=3 in duplicates). C, the growth curves show the effect of androgen depletion on proliferation of LNCaP cells, compared to LNCaP cells grown under normal serum (N.S.) conditions (N=3 in duplicates). D Bar graphs represent the expression of Wnt genes in LNCaP cells grown under androgen-depleted (charcoal-stripped serum) conditions, LNCaP cells grown under normal serum conditions, and after DNAPK knockdown in LNCaP cells grown under charcoal-stripped serum conditions, compared to LNCaP cells grown under charcoal-stripped conditions. (N=3, in duplicates). E, Cell growth curves demonstrate the effect of DNAPK knockdown on proliferation of LNCaP cells that developed ADT resistance (from C) (N=2 in duplicates). A subset of these cells was switched to normal serum conditions as a reference. *p < 0.05, **p < 0.001. P-values were calculated using students t-test (Figure 3B, D) or two-way ANOVA (Figure 3C, 3E). All data are represented as mean ± S.D. RQ = relative quantity.
siRNA-mediated knockdown of DNAPK suppresses ADT-induced Wnt signaling
To investigate the relationship between DNAPK and Wnt signaling in prostate cancer, we first evaluated the expression of Wnt pathway genes in prostate cancer across all stages of progression as represented by the following cell lines: 1) LNCaP, which are AR-positive, androgen-sensitive cells that represent hormone-sensitive disease; 2) LNCaP-AR and C4–2B, which are AR-positive, androgen-insensitive cells that represent CRPC; and 3) PC-3 cells, which are AR-negative, androgen-insensitive cells that represent androgen-independent disease. We found that increased Wnt pathway gene expression associated with models of more aggressive disease (LNCaP-AR, C4–2B, and PC-3) compared to the hormone-sensitive LNCaP cells (Fig. 3B; Fig. S1B), suggesting that progression on ADT could be mediated by Wnt signaling. Consistent with clinical observations that hormone-naïve cells eventually overcome ADT, we found that LNCaP cells continued to grow despite androgen depletion using charcoal-stripped serum, albeit at a much slower rate (Fig. 3C). Interestingly, growth under androgen-depleted conditions triggered the expression of multiple Wnt genes in LNCaP cells (Fig. 3D, gray bars), further supporting a role for Wnt-mediated disease progression in response to ADT.
Given our earlier observation that DNAPK modulates the Wnt pathway, we next asked if DNAPK is involved in ADT-induced Wnt signaling. siRNA-mediated knockdown of DNAPK in LNCaP cells grown under androgen-depleted conditions caused a marked decrease in expression of Wnt genes (Fig. 3D, red bars), suggesting that DNAPK is required for ADT-induced Wnt signaling. Furthermore, DNAPK knockdown (via siRNA) prevented the emergence of ADT-resistant LNCaP cells (Fig. 3E), demonstrating the importance of DNAPK-mediated signaling in hormone resistance. Taken together, these data suggest that DNAPK inhibition is a viable therapeutic strategy to prevent the emergence of ADT resistance in prostate cancer and likely acts through inhibiting Wnt signaling.
DNAPK inhibition reduces Wnt-induced invasion and migration in CRPC cells
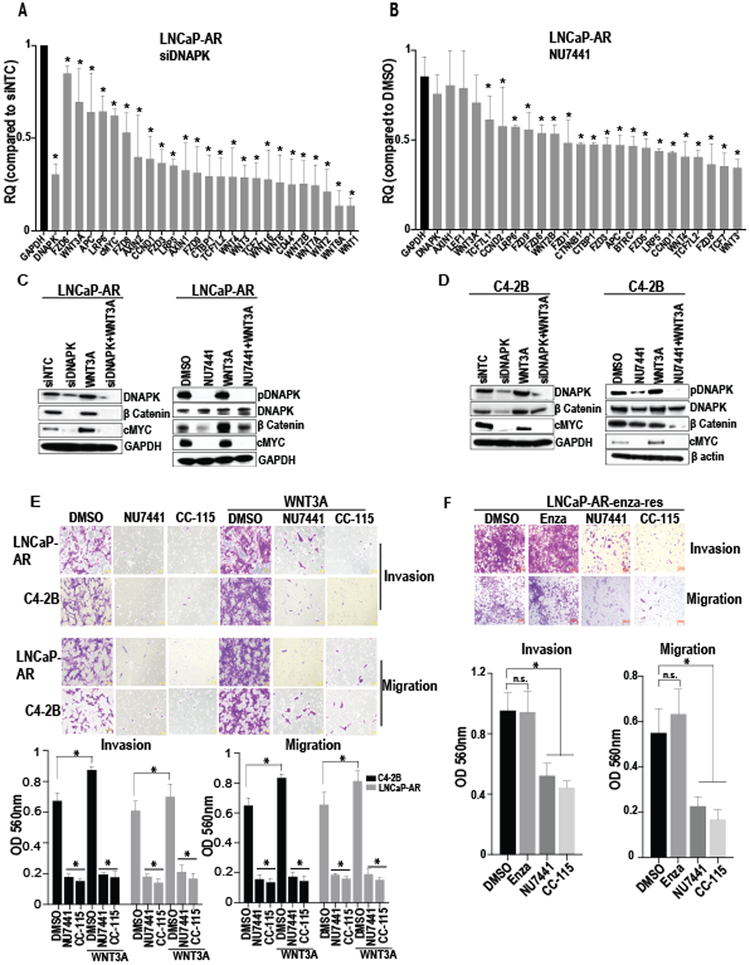
Since our results implicate DNAPK-mediated Wnt signaling as a contributor to ADT resistance, we next investigated the relationship between DNAPK and Wnt signaling in cell line models of frank castration-resistance. Genetic or pharmacological inhibition of DNAPK in CRPC cells (LNCaP-AR and C4–2B) using siRNAs targeting DNAPK or the DNAPK inhibitor NU7441 caused a marked reduction in mRNA levels of Wnt pathway genes (Fig. 4A-B; Supplementary fig. S2A-B), whereas mRNA levels of genes implicated in other pathways remained broadly unchanged (Supplementary fig. S2C). Genetic and pharmacological inhibition of DNAPK also reduced steady state levels of active (dephosphorylated) β-catenin and c-Myc protein (a Wnt target gene) (27) and suppressed Wnt3A-mediated induction of active β-catenin and c-Myc protein levels (Fig. 4C-D). Pharmacological inhibition using NU7441 or the DNAPK inhibitor CC-115 (28) (United States Patent No. 8,110,578), which is currently being tested in early clinical trials for solid tumors (, , ), reduced invasion and migration of CRPC cells and suppressed Wnt3A-mediated induction of invasion and migration of CRPC cells (Fig. 4E). Interestingly, a combination of CC-115 and IWP-2 (a laboratory-grade Wnt antagonist) reduces the migration of PCa cells similar to the extent of CC-115 alone (Supplementary fig. S3). Together, these data show that inhibition of DNAPK suppresses Wnt signaling and Wnt-mediated metastatic phenotypes in CRPC cells, suggesting that DNAPK may be a clinically relevant therapeutic target in the current absence of effective and tolerable clinical agents that target β-catenin or Wnt signaling.
Figure 4. DNAPK inhibition reduces Wnt signaling in CRPC cells.
(A and B), Bar graphs show the expression of Wnt pathway genes after knockdown (via siRNA) or inhibition (via NU7441, 1 μM) of DNAPK in LNCaP-AR cells (N=2, in duplicates). (C and D), Immunoblot analyses show the effect of DNAPK knockdown (via siRNA) or inhibition (via NU7441 1 μM) on indicated proteins in LNCaP-AR and C4-2B cells. E, Representative images (magnification 20X) demonstrate the effect of DNAPK inhibition (via NU7441 or the clinical-grade DNAPK inhibitor CC-115, 1 μM) on invasion and migration of LNCaP-AR and C4-2B cells (N=3 in duplicates). Recombinant human WNT3A (200 ng/ml) was used to stimulate Wnt signaling (C-E). Bar graphs represent the quantification of invaded/migrated cells. F, Images (magnification 20X) display the effect of DNAPK inhibition (via NU7441 or CC-115) on invasion and migration of enzalutamide resistant LNCaP-AR (LNCaP-AR-enza-res) cells. Bar graphs show quantification of invaded/migrated cells. All data are represented as mean ± SEM *= p < 0.05 compared to respective controls and were calculated using two-way ANOVA (Figure 4A, 4B, 4E) or one way AVOVA (Figure 4F). Graphs and statistical analyses were done on GraphPad Prism software. RQ = relative quantity.
Second-generation ADTs including abiraterone and enzalutamide provide only modest survival benefits in CRPC (1-4). Based on our findings, we hypothesized that Wnt signaling could be an important mechanism of resistance to second-generation ADTs. Interestingly, we found that expression of Wnt pathway genes was broadly increased in enzalutamide-resistant LNCaP-AR cells (LNCaP-AR-enza-res; Supplementary fig. S4). Similar to our results in CRPC cells presented in Fig. 4E, DNAPK inhibition with NU7441 or CC-115 dramatically reduced invasion and migration of LNCaP-AR-enza-res cells (Fig. 4F). Thus, DNAPK inhibition is a potential therapeutic strategy not only in CRPC but also in enzalutamide-resistant CRPC, for which there are currently limited treatment options.
DNAPK transcriptionally regulates Wnt signaling through interaction with LEF1 to control metastatic phenotypes in CRPC cells
LEF1 is the primary transcription factor involved in canonical Wnt-mediated signaling (27). To investigate the molecular mechanisms underlying DNAPK-mediated regulation of Wnt signaling, we performed co-immunoprecipitation studies and found that DNAPK interacts with LEF1 in a DNA-independent manner (Fig. 5A). Using a TOPFLASH reporter assay that measures TCF/LEF1-mediated transcription, we also found that DNAPK inhibition with NU7441 significantly reduced basal and Wnt3A-induced TOPFLASH reporter activity in AR-independent PC-3 cells (Fig. 5B). Chromatin immuno-precipitation (ChIP) studies showed that DNAPK is enriched on the regulatory regions of Wnt genes and that this enrichment is drastically reduced after NU7441 treatment (Fig. 5C) but remains unaltered in the backdrop of LEF1 knockdown (Supplementary fig. S5). Taken together, these results suggest that DNAPK facilitates Wnt signaling through interactions with the transcription factor LEF1. Moreover, treatment with siRNAs targeting LEF1 or β-catenin (CTNNB1) phenocopies the reductions in migration observed with DNAPK siRNA treatment in LNCaP-AR, C4–2B, and PC-3 cells (Fig. 5D), suggesting that the Wnt pathway is a major downstream effector of DNAPK. Together, these findings suggest that DNAPK-transcriptionally regulates Wnt signaling to control metastatic phenotypes in CRPC cells.
Figure 5. DNAPK interacts with LEF1.
A, Immunoblot analyses of DNAPK immunoprecipitates probed with DNAPK, KU70 and LEF1 antibodies and LEF1 immunoprecipitates probed with DNAPK and LEF1 antibodies in LNCaP-AR cells, in the presence or absence of ethidium bromide is shown. B, Bar graph displays the TOPFLASH reporter activity in PC-3 cells (N=3). Luminescence is normalized to renilla luciferase. Data are represented as mean ± SEM. *= p < 0.05 compared to DMSO. ** =p<0.05 compared to WNT3A using one way ANOVA. C, Enrichment of DNAPK on regulatory regions (by Chromatin immuno-precipitation assay, using DNAPK antibody) of select Wnt genes is shown after the treatment with DMSO or NU7441. En=enhancer. Prom= promoter. n.s.= not significant. Data are represented as mean ± SEM.*p<0.05 compared to IgG control. **=p<0.05 compared to DMSO treatment. (N=3 in duplicates). P values were calculated by two-way ANOVA. D, Representative images (magnification 20X) show the effect of DNAPK, LEF1 or beta catenin (CTNNB1) knockdown on invasion and migration of LNCaP-AR, C4-2B and PC-3 cells (N=2 in duplicates). Bar graphs represent the mean ± S.D of migration where siRNA knockdowns are compared to NTC control of respective cell lines. *=p<0.05 using two way ANOVA. Graphs and statistical analyses were completed using GraphPad Prism software.
DNAPK inhibition is effective in both in vivo and ex vivo models of prostate cancer
To further study the potential clinical impact of DNAPK inhibition, we assessed its effect in a CRPC xenograft model. Treatment of palpable LNCaP-AR tumors in mice with the DNAPK inhibitor NU7441 caused a 44% reduction in tumor growth in the NU7441 treated group compared to the control group (Fig. 6A-B). This growth inhibition translated into a significant delay in the median tumor doubling time in the NU7441 arm (21.5 days for NU7441 vs. 7 days for vehicle, Fig. 6C), with minimal signs of toxicity as measured by body weight (Supplementary fig. S6A).
Figure 6. DNAPK inhibition reduces CRPC xenograft growth in vivo.
A, Growth curves demonstrate the growth of LNCaP-AR xenografts, treated daily with vehicle (black, n = 5) or 25 mg/kg NU7441 (red, n = 6). B, The bar graph shows the fold change in tumor volume at the end of the experiment. C, Kaplan-Meier curves display the tumor doubling time in vehicle or NU7441 (25 mg/kg body weight) treated xenografts. D, Growth curves show the effect of enzalutamide (10 mg/kg, n = 16), CC-115 (2 mg/kg, n = 14), enzalutamide + CC-115 (n = 14) treatment on LNCaP-AR xenografts compared to vehicle (n = 14) control. CC115 and enzalutamide were administered once daily via oral gavage 5 times per week for 6 weeks. The bar graph shows the percent increase in tumor volumes, compared to starting volume, at the end point of treatments. E, Kaplan-Meier curves demonstrating the tumor tripling time in vehicle, enzalutamide, CC-115, and enzalutamide + CC-115 treated xenografts. Tumors were measured at indicated time points using caliper measurements. F, The bar graph displays the effect on select Wnt target genes in human explants treated with NU7441 (n = 6) or vehicle control (n=6). G, Representative images showing the percent Ki67 nuclei (red arrows) that were scored in the explant tissue after treatment with either vehicle or NU7441 (n = 6). All data are represented as mean ± S.E.M. *= p < 0.05 compared to respective control, #= p < 0.05 enzalutamide compared to CC-115 + enzalutamide, ** = p < 0.01 CC-115 compared to Vehicle. P-values were calculated using the Student’s t-test for tumor growth studies and log-rank test for Kaplan-Meier survival curves.
Further investigation utilizing the DNAPK inhibitor CC-115 (28), currently in early phase clinical trials for solid tumors, in a CRPC xenograft model using the VCaP prostate cancer cell line, demonstrated that CC-115 causes a dose-dependent reduction in tumor growth. Maximal effects were observed at a non-toxic dose of 2 mg/kg (Supplementary fig. S6B-C). Because enzalutamide represents a standard treatment approach for metastatic CRPC, we also assessed the impact of CC-115 in combination with enzalutamide in a CRPC (LNCaP-AR) xenograft model. This combination caused a significant delay in tumor growth compared to enzalutamide monotherapy. Interestingly, CC-115 monotherapy resulted in a greater delay in tumor growth compared to enzalutamide alone (Fig. 6D). The tumor doubling time (Supplementary fig. S6D) and the tumor tripling times (Fig. 6E) in the groups receiving both CC-115 plus enzalutamide or CC-115 alone were the most delayed. The median tumor tripling times in the CC-115 plus enzalutamide or CC-115 alone treatment groups were 38 and 29 days, respectively, compared to 23 days for enzalutamide monotherapy and 15 days for vehicle (Fig. 6E). Additionally, DNAPK inhibition via CC-115 also resulted in significant regression of AR-independent PC-3 tumors (Supplementary fig. S6E), supporting the potential for treatment strategies based on DNAPK inhibition in AR-independent disease. Consistent with our xenograft studies, human prostate cancer explants treated with the DNAPK inhibitor NU7441 demonstrated a marked reduction in expression of Wnt target genes (Fig. 6F) and a significant decrease in Ki67 positive staining, a marker of proliferative index (Fig. 6G). Taken together, our results highlight the promise of DNAPK inhibition for the treatment of aggressive prostate cancer.
Discussion
Current clinical prognostic paradigms, using PSA level, Gleason score, tumor stage, and pathological features provide guidance in the treatment of PCa patients but are imperfect (29). The genomics era is revolutionizing decision making in the clinic by providing an unprecedented view into tumor biology and the promise of personalized medicine. Unbiased microarray expression studies have been used to identify numerous prognostic and/or predictive individual-gene biomarkers. More recently, multi-gene signatures have been developed that have performed strongly in several clinical contexts and are now available commercially, including Decipher and Prolaris (16,30). While these tools provide prognostic information, they have no direct association with therapeutic strategies. Currently, androgen-directed therapies remain a standard of care for patients with advanced prostate cancer but are not durable, with inevitable development of resistance to enzalutamide and other second-generation AR-targeting agents [reviewed in (31)]. Thus, novel therapeutic strategies that target AR as well as other driver pathways are desperately needed. Our findings provide strong pre-clinical evidence that DNAPK inhibition may be one such effective strategy.
Here, utilizing large discovery and validation cohorts, we identify a highly targetable kinase, DNAPK, as a top prognostic gene for aggressive CRPC. Our pre-clinical data show that DNAPK inhibition inhibits aggressive cancer phenotypes including proliferation, migration, and invasion. We further demonstrate that Wnt signaling is activated very early after initiation of ADT and that DNAPK inhibition blocks Wnt signaling to prevent the emergence of ADT resistance. These data provide compelling support for DNAPK inhibition as a promising therapeutic strategy during ADT initiation. We also show that DNAPK is a strong prognostic indicator not only for AR-enriched luminal PCa but for the AR-independent basal subtype of PCa as well and show that DNAPK inhibition suppresses aggressive cancer phenotypes in AR-independent cells in vitro and in vivo. Thus, DNAPK inhibition has strong potential for overcoming therapy resistance in AR-independent, advanced prostate cancer. The impact of these findings is amplified by the availability of the clinical-grade DNAPK inhibitor, CC-115, which is currently being assessed in patients with metastatic CRPC (clinicaltrials.gov identifier ). Moreover, based on our finding that DNAPK inhibition interrupts Wnt signaling at a very distal step, we strongly suspect that DNAPK inhibition will block aberrant Wnt signaling arising from genetic alterations upstream in the pathway making it effective against a wide variety of driver mutations.
Overcoming therapeutic resistance is dependent on identifying compensatory pathways prostate cancer cells use to evade current therapies. In this regard, our finding that Wnt signaling is activated upon ADT or enzalutamide treatment is consistent with a growing body of literature that Wnt signaling is a mechanism by which prostate cancer cells develop resistance to ADT (32-34). Wnt pathway genes have been shown to be upregulated in locally advanced and metastatic prostate cancer patients in the months after first receiving androgen-deprivation therapy (ADT) (35). Additionally, alterations in the Wnt signaling pathway, including genetic aberrations (36) and elevated expression (37), are virtually absent in treatment naïve prostate cancer, whereas they are prevalent in advanced disease (35). Furthermore, the Wnt pathway has a known tumor-promoting role in advanced prostate cancer (34,38) and, interestingly, the androgen receptor itself was reported to be a Wnt target gene (39). The relationship between AR and Wnt signaling remains complex with conflicting interpretations [reviewed in (33)], nevertheless, the observations listed above all support or are consistent with the hypothesis that activation of Wnt signaling is a key mechanism of resistance to AR directed therapies. Despite decades of effort, and numerous targets and compounds, pharmacological inhibitors of the Wnt pathway have not progressed in clinical trials due to toxicity. While we are actively working to characterize the DNAPK-Wnt axis, we hypothesize, based on our current data, that the inhibition of DNAPK activity interferes with its binding to active transcription sites of Wnt genes and suppresses LEF1-mediated Wnt signaling. Thus, the ability of DNAPK inhibition to block Wnt signaling represents a long-awaited opportunity to effectively inhibit the Wnt pathway and potentially AR transcription (12), co-targeting two dominant pathways in castration-resistant prostate cancer.
Advanced prostate cancer is further complicated by a growing recognition that ADT might drive transdifferentiation of prostate cancer cells toward a neuroendocrine phenotype [reviewed in (40)]. This very fast-growing and aggressive stage of disease is androgen/AR-independent, and is nearly always lethal due to a lack of effective treatment options. Thus, our observations that DNAPK is strongly prognostic in the basal subtype of prostate cancer and that DNAPK inhibition efficiently reduces the aggressive cancer phenotypes of PC-3 (AR-independent) cells in pre-clinical studies provide the rationale for DNAPK inhibition as a therapeutic strategy for AR-independent prostate cancer.
In addition to prostate cancer, DNAPK inhibition potentially has significant clinical relevance across several disease sites. In a recent study by the Cancer Genome Atlas Network characterizing the molecular landscape of colon and rectal cancer (CRC), aberrations in Wnt signaling pathways were found in approximately 93% of CRC tumors (41). Wnt signaling also appears to be a driver in gastric cancer, one of the leading causes of cancer-related deaths world-wide. Upregulation and genetic or epigenetic alterations of many Wnt genes are known to be associated with poor prognosis and resistance to therapies for this disease (42). Similarly, Wnt signaling is known to be an oncogenic driver in cancers of the lung, breast, liver, and ovary (43).
In summary, by systematically exploring the landscape of kinases in lethal prostate cancer, we uncovered the strong prognostic importance of DNAPK and demonstrated a novel role of DNAPK in regulating Wnt signaling in anti-androgen resistant and AR-independent models of prostate cancer. Moreover, a clinical-grade DNAPK inhibitor, currently being tested in humans, reduces aggressive cancer phenotypes and resistance. Thus, DNAPK is a unique and exciting “therapeutically targetable prognostic biomarker” in lethal prostate cancer with rapid translational potential.
Supplementary Material
Translational relevance.
Kinases are associated with oncogenic events in multiple cancer subtypes, but have not been systematically explored in prostate cancer. In an unbiased analysis, we identified DNA-dependent protein kinase (DNAPK) as the most significantly associated kinase with metastatic progression in prostate cancer. We found that inhibition of DNAPK suppresses growth of both androgen receptor (AR)-positive and AR-negative prostate cancer models. Our results suggest that DNAPK interacts with the Wnt transcription factor LEF1 and promotes disease aggressiveness through Wnt signaling. Together, these findings suggest that DNAPK represents an attractive therapeutic target in prostate cancer, and highlight the potential of DNAPK inhibition as a strategy for treating Wnt-addicted cancers. Our findings have led to the initiation of the first trial of DNAPK inhibition for patients with metastatic prostate cancer.
Acknowledgements
The authors gratefully thank Dr. Rohit Malik (University of Michigan), Elai Davicioni (GenomeDx Biosciences), and members of the Feng and Knudsen laboratories for their input. We thank Steven Kronenberg for help with the figures. This work was supported by a grant to V. Kothari and S.G. Zhao from the PCF, to F.Y. Feng, K.E. Knudsen and S.A. Tomlins from the PCF/Evans Foundation, K.E. Knudsen from PA CURE and NCI (CA159945, CA176401), F.Y. Feng and V. Kothari from the DOD PCa Idea Development Award (W81XWH1810599).
Footnotes
Potential conflicts of interest:
S.G. Zhao has ownership interests (including patents) in Celgene and GenomeDx Biosciences. L. Chang is an employee of and has ownership interests (including patents) at PFS Genomics. S.A. Tomlins is an employee of and has ownership interests (including patents) at Strata Oncology, is a consultant/ advisory board member for Janssen, Astellas, Sanofi, Almac Diagnostics and AbbVie, reports receiving commercial research grants from GenomeDX and Astellas/Medivation, and is a co-inventor on a patent on ETS gene fusion which has been patented by the University of Michigan and whose diagnostic field of use has been licensed to Hologic/Gen-Probe, Inc.,which has sublicensed rights to Roche/Ventana Medical Systems. K. Hege has ownership interests (including patents) in and reports receiving commercial research support from Celgene. E.H. Filvaroff has ownership interests (including patents) in Celgene. J. Karnes has ownership interests (including patents) in Genome Dx. D. Rathkopf is a consultant/advisory board member for Janssen, Tracon, and AstraZeneca. D.E. Spratt is a consultant/advisory board member for Janssen and Blue Earth Diagnositics. A. Ashworth has ownership interests in Syncona, Tango, ProLynx, Gladiator and Bluestar Genomics, is a consultant/advisory board member for Tango, Pfizer, SPARC, Bluestar Genomics, Gladiator, and Genentech, reports receiving commercial research grants from Astra Zeneca and SPARC, and is a co-inventor/ holds patents on the use of PARP inhibitors held jointly with Astra Zeneca from which he has benefited financially (and may do so in the future) through the ICR Rewards to Inventors Scheme. G.V. Raj is an employee of and has ownership interests (including patents) at EtiraRx, reports receiving speakers bureau honoraria from Astellas, Pfizer and Bayer, and reports receiving commercial research grants from Bayer. K.E. Knudsen is a consultant/advisory board member for and reports receiving commercial research support from Celgene. F.Y. Feng is a co-founder of and has ownership interests (including patents) at PFS Genomics, is a consultant/advisory board member for Bayer, Blue Earth Diagnostics, Celgene, Clovis, Janssen, EMD Serono, Sanofi, Dendreon, Ferring, Astellas, and Reflexion, and reports receiving commercial research grants from Zenith and Amgen. No potential conflicts of interest were disclosed by the other authors.
References
- 1.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364(21):1995–2005 doi 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan CJ, Smith MR, Fizazi K, Saad F, Mulders PF, Sternberg CN, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol 2015;16(2):152–60 doi 10.1016/S1470-2045(14)71205-7. [DOI] [PubMed] [Google Scholar]
- 3.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367(13):1187–97 doi 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 4.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371(5):424–33 doi 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med 2010;363(5):411–22 doi 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 6.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004;351(15):1502–12 doi 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009;9(1):28–39 doi 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rask-Andersen M, Zhang J, Fabbro D, Schioth HB. Advances in kinase targeting: current clinical use and clinical trials. Trends Pharmacol Sci 2014;35(11):604–20 doi 10.1016/j.tips.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010;79:181–211 doi 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A 1992;89(24):11920–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Molecular cell 2013;51(2):185–99 doi 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov 2013;3(11):1254–71 doi 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson SP, MacDonald JJ, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell 1990;63(1):155–65. [DOI] [PubMed] [Google Scholar]
- 14.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol 2012;19(3):276–82 doi 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, et al. DNA-PKcs-Mediated Transcriptional Regulation Drives Prostate Cancer Progression and Metastasis. Cancer Cell 2015;28(1):97–113 doi 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 2013;8(6):e66855 doi 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, Marchionni L, et al. Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur Urol 2015. doi 10.1016/j.eururo.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 18.Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol 2017. doi 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kothari V, Wei I, Shankar S, Kalyana-Sundaram S, Wang L, Ma LW, et al. Outlier kinase expression by RNA sequencing as targets for precision therapy. Cancer Discov 2013;3(3):280–93 doi 10.1158/2159-8290.CD-12-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol 2014;15(13):1469–80 doi 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007;7:64 doi 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4(2):249–64 doi 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9(2):166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol 2013;10(8):483–7 doi 10.1038/nrurol.2013.126. [DOI] [PubMed] [Google Scholar]
- 26.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer 2001;1(1):34–45 doi 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 27.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008;8(5):387–98 doi 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 28.Thijssen R, Ter Burg J, Garrick B, van Bochove GG, Brown JR, Fernandes SM, et al. Dual TORK/DNA-PK inhibition blocks critical signaling pathways in chronic lymphocytic leukemia. Blood 2016;128(4):574–83 doi 10.1182/blood-2016-02-700328. [DOI] [PubMed] [Google Scholar]
- 29.Lu-Yao GL, Albertsen PC, Moore DF, Shih W, Lin Y, DiPaola RS, et al. Outcomes of localized prostate cancer following conservative management. JAMA 2009;302(11):1202–9 doi 10.1001/jama.2009.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, Reid JE, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer 2012;106(6):1095–9 doi 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claessens F, Helsen C, Prekovic S, Van den Broeck T, Spans L, Van Poppel H, et al. Emerging mechanisms of enzalutamide resistance in prostate cancer. Nat Rev Urol 2014;11(12):712–6 doi 10.1038/nrurol.2014.243. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Placencio V, Iturregui JM, Uwamariya C, Sharif-Afshar AR, Koyama T, et al. Prostate tumor progression is mediated by a paracrine TGF-beta/Wnt3a signaling axis. Oncogene 2008;27(56):7118–30 doi 10.1038/onc.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama NN, Shao S, Hoang BH, Mercola D, Zi X. Wnt signaling in castration-resistant prostate cancer: implications for therapy. Am J Clin Exp Urol 2014;2(1):27–44. [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med 2012;18(9):1359–68 doi 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajan P, Sudbery IM, Villasevil ME, Mui E, Fleming J, Davis M, et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol 2014;66(1):32–9 doi 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–28 doi 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Wang L, Zhang M, Melamed J, Liu X, Reiter R, et al. LEF1 in androgen-independent prostate cancer: regulation of androgen receptor expression, prostate cancer growth, and invasion. Cancer Res 2009;69(8):3332–8 doi 10.1158/0008-5472.CAN-08-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, Wang Y, DeGraff DJ, Wills ML, Matusik RJ. Wnt/beta-catenin activation promotes prostate tumor progression in a mouse model. Oncogene 2011;30(16):1868–79 doi 10.1038/onc.2010.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Chen MW, Terry S, Vacherot F, Bemis DL, Capodice J, et al. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene 2006;25(24):3436–44 doi 10.1038/sj.onc.1209366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terry S, Beltran H. The many faces of neuroendocrine differentiation in prostate cancer progression. Front Oncol 2014;4:60 doi 10.3389/fonc.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiurillo MA. Role of the Wnt/beta-catenin pathway in gastric cancer: An in-depth literature review. World J Exp Med 2015;5(2):84–102 doi 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013;13(1):11–26 doi 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data is available at NCBI GEO accession GSE46691, GSE79956, GSE79957 and GSE116895.