Abstract
Purpose:
Radiation therapy (RT) has long been and remains the only treatment option for diffuse intrinsic pontine glioma (DIPG). However, all patients show evidence of disease progression within months of completing radiation therapy. No further clinical benefit has been achieved using alternative radiation strategies. Here we tested the hypothesis that histone demethylase inhibition by GSK-J4 enhances radiation-induced DNA damage, making it a potential radiosensitizer in the treatment of DIPG.
Experimental Design:
We evaluated the effects of GSK-J4 on genes associated with DNA double-strand break (DSB) repair in DIPG cells by RNA sequence, ATAC sequence, and quantitative real-time PCR. Radiation-induced DNA DSB repair was analyzed by immunocytochemistry of DSB markers γH2AX and 53BP1, DNA repair assay, and cell cycle distribution. Clonogenic survival assay was used to determine the effect of GSK-J4 on radiation response of DIPG cells. In vivo response to radiation monotherapy and combination therapy of RT and GSK-J4 were evaluated in patient-derived DIPG xenografts.
RESULTS:
GSK-J4 significantly reduced the expression of DNA DSB repair genes and DNA accessibility in DIPG cells. GSK-J4 sustained high levels of γH2AX and 53BP1 in irradiated DIPG cells, thereby inhibiting DNA DSB repair through homologous recombination pathway. GSK-J4 reduced clonogenic survival and enhanced radiation effect in DIPG cells. In vivo studies revealed increased survival of animals treated with combination therapy of RT and GSK-J4 in compared to either monotherapy.
Conclusions:
Together, these results highlight GSK-J4 as a potential radiosensitizer and provide a rationale for developing combination therapy with radiation in the treatment of DIPG.
Keywords: DIPG, Radiation, Histone demethylase, GSK-J4, DNA repair
INTRODUCTION
Diffuse midline glioma (DMG) with H3K27M mutation, resulting in substitution of lysine 27 with methionine (K27M) in the genes encoding histone H3 protein, is a newly defined entity that comprises all diffusely infiltrating astrocytic neoplasms affecting midline structures of the central nervous system (1). The pontine category of DMG, diffuse intrinsic pontine glioma (DIPG), mainly affects children of 4 to 7 years of age, presenting with cranial nerve deficits, balance issues and other brainstem signs and symptoms (1–3). DIPGs are among the most devastating childhood tumors with a median survival of 9 to 12 months from diagnosis. Investigations into curative therapies have been extremely challenging, and over 250 clinical trials involving different combinations of chemotherapeutic agents commonly used in glioma treatment have demonstrated no response in DIPG (3). As their predilection for the brainstem precludes surgical resection, fractionated focal radiation therapy to a total dose of 54–60 gray (Gy) over a six-week period remains the only treatment modality which can alleviate symptoms. This provides a “honeymoon period” of temporary symptomatic relief and a delay in tumor progression in about 70–80% of DIPG patients. No further clinical benefit has been achieved using alternative radiation strategies or combination therapies with radiation sensitizers (4,5). Unfortunately, no effective salvage therapy is available at the time of tumor progression, though re-irradiation may provide transient symptom improvement in some patients (6). Given these dire circumstances, the identification of efficacious therapeutic agents that enhance the anti-tumor effects of radiation is of high importance for improving treatment outcomes for this patient population.
DIPGs with H3K27M mutation (K27M DIPGs) show a global reduction in K27 methylation that is believed to modify cellular gene expression to promote tumor development (7–9). We have previously shown that JMJD3 demethylase inhibitor, GSK-J4, acts to restore K27 methylation in K27M DIPG cells, while demonstrating potent anti-tumor activity, in vitro and in vivo (10). In addition to its anti-tumor activity, GSK-J4 resulted in significant changes in K27M DIPG cell transcriptional profiles (10).
Current comparison of untreated vs. GSK-J4 treated expression profiles of K27M DIPG shows several significant decreases in transcripts from genes whose encoded proteins are known to be involved with DNA damage repair, including DNA double-strand break (DSB) repair. These results provide a possibility to test whether GSK-J4 inhibits DNA damage repair mediated by chromatin modification and enhances the radiation effect. We investigated the effect of GSK-J4 on radiation-induced DNA damage, DNA repair pathways, and chromatin accessibility in K27M DIPG cells, and used this information in pre-clinical testing. We used human K27M DIPG xenografts to study the effects of GSK-J4 on in vivo tumor growth in association with therapeutic combination of GSK-J4 and radiation. Together our data suggests that GSK-J4 is a potential radiosensitizer and provides a rationale for developing combination therapy with GSK-J4 and radiation in the treatment of K27M DIPG.
MATERIALS AND METHODS
Cell sources and propagation
Primary pediatric human glioma cell lines SF8628 (K27M DIPG) and SF9427 [H3 wild-type glioblastoma (GBM)] were obtained from the University of California, San Francisco (UCSF) medical center, and in accord with an institutionally approved protocol. Establishment of SF8628 and SF9427 cell cultures from surgical specimens, and tumor cell modification for expression of firefly luciferase for in vivo bioluminescence imaging, have been described (10–13). DIPG-007 (K27M DIPG) cell line was kindly provided by Dr. Angel Montero Carcaboso (Hospital Sant Joan de Déu, Barcelona, Spain). Human astrocytes expressing wild-type (Astro WT) or K27M H3F3A transgene (Astro KM) have been previously described (7, 10). GBM43 cell lines were established and propagated as subcutaneous xenografts as previously described (10, 12). The SF8628 and human astrocyte cells were propagated as monolayers in complete medium consisting of Dulbecco’s Modified Eagle’s medium (DMEM, 11965092) supplemented with 10% fetal bovine serum (FBS, A31604–02) and non-essential amino acids (11140–050) from ThermoFisher. SF9427 and DIPG-007 cell lines were grown adherently in tumor stem medium (TSM) base with 5% FBS. TSM base was prepared using the following: neurobasal-A medium (10888–022), DMEM/F-12 medium (11330–032), HEPES buffer (15630–080), sodium pyruvate (11360–070), MEM non-essential amino acids (11140–050), GlutaMAX-I supplement (35050–061), antibiotic-antimycotic (15240–096), B-27 supplement minus vitamin A (12587–010) from ThermoFisher, EGF and FGF (Shenandoah Biotech, 100–26 and 100–146), PDGF-A and PDGF-B (Shenandoah Biotech, 100–16 and 100–18), and 0.2% heparin (STEMCELL Technologies, 07980). Short tandem repeat (STR), using the Powerplex16HS System (Promega DC2101), were obtained to confirm the identity of the cell lines. All cells were cultured in an incubator at 37°C in a humidified atmosphere containing 95% O2 and 5% CO2 and were mycoplasma-free at the time of testing with a Mycoplasma Detection Kit (InvivoGen).
RNA sequencing and analysis
RNA sequencing was performed using NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs NEB E7490) and NEBNext® Ultra RNA Library Prep Kit for Illumina® (New England Biolabs E7530) according to the manufacturer’s instructions. Briefly, first strand cDNA was obtained from DNase 1-treated RNA samples from SF8628 and DIPG-007 cells treated with dimethyl sulfoxide (DMSO) (Sigma D2650) for 0 h (untreated) and 48 h or GSK-J4 (6 μM, R&D systems 4594) for 6, 24, 48, and 72 h (two replicates per time point). Second strand cDNA synthesis was performed followed by purification using 1.8 X Agencourt AMPure XP Beads (Beckman Coulter A63881). The cDNA library was end prepped, followed by immediate adaptor ligation and ligation reaction purification. Following PCR enrichment and reaction purification, the library quality was assessed on a Bioanalyzer® (Agilent High Sensitivity Chip). We used STAR 2.4 to align the RNA-sequencing samples to the reference genome (hg19) and counted the number of reads mapping to each gene in the ensemble GRCh37 gene model. Differential expression between the different groups was performed through the use of DESeq2 (14). Hierarchical clustering of the resulting matrix was done in R using heatmap2.
Quantitative real-time polymerase chain reaction (qPCR)
Six-well tissue culture plates were seeded with 10,000 cells. The cells were allowed to adhere for 16 hours and then treated with DMSO or 6 μM of GSK-J4 for 72 hours, followed by radiation at 6 Gy. Radiation was delivered by gamma irradiator. Cells were collected at 6 hours following radiation. Total RNA was extracted using the RNAeasy Mini kit (Qiagen 74104), and 2 μg of RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad 1708890). Real-time detection and quantification of cDNAs were performed with the iCycler instrument (Bio-Rad 1861096). qPCR was conducted using SYBR@GreenER qPCR Supermixes (Bio-Rad 1725270). Fluorescence data was collected at annealing stages and real-time analysis was performed with iCyclerTM iQ Optical System Software V3.0a. Gene expressions were normalized with that of GAPDH endogenous control gene. All experiments were repeated at least three times. For data analysis, the fold change in the target gene relative to the GAPDH was determined by: fold change=2–Δ(ΔCt) where ΔCt=Cttarget–CtGAPDH and Δ(ΔCt)=ΔCttreatment–ΔCtcontrol. Forward (F) and reverse (R) primers used for qPCR were 5’–GGCGTGAACCTCACCAGTAT–3’ (F) and 5’–TTCTCCTGGTTTGGTGCTTC–3’ (R) for PCNA, 5’–CAGCCCTACAGCAAGGACTC–3’ (F) and 5’–GCTGTGACTGGGGATGTCTT–3’ (R) for XRCC1, 5’–GGTGCAGAGCTACGAGAAGG–3’ (F) and 5’–ATGAAGAGTCCCGGATGTTG–3’ (R) for POLD1, 5’–CTCGTGCCTTTTGAGTCTCC–3’ (F) and 5’–GTCCCCAGCTGATGACAAAT–3’ (R) for FANCA, and 5’–TGCACCACCAACTGCTTAGC–3’ (F) and 5’–GGCATGGACTGTGGTCATGAG–3’ (R) for GAPDH. All primers were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/).
ATAC-sequence library preparation and analysis
We submitted 80,000 cells for ATAC-sequence according to the manufacturer’s instructions (Active Motif). On average, 35 million paired-end 42 base pair (bp) sequencing reads per sample were generated by Illumina sequencing (using NextSeq 500). The paired-end reads were mapped to the reference genome (hg19) using the bowtie2 algorithm. The overall alignment rate was over 99%. We used MACS1.4 to call the binding peaks (15). Normalized sample count matrix was used as input for DEseq2 (14) to call differential binding peaks between GSK-J4 (6 μM) and DMSO treated samples. We assigned the peaks according to the nearest genes using R-ChIPseeker package and separated the peaks into two broad categories: namely TSS-associated peaks including peaks within promoter or 5’UTR, and enhancer-associated peaks including peaks within distal, downstream, 3’UTR region. TSS-/Enhancer peaks with at least 10 reads support were used for GSEA analysis (16). GSEA analysis was applied in weighted mode against the gene sets collection in MSigDB (v5.1). Gene sets with over 5,000 genes or less than 10 genes were excluded for further analysis. Each gene set was permuted 1,000 times to calculate p value and FDR values.
Immunoblotting
Cells were cultured as described above and treated with 6 μM of GSK-J4 for 72 hours. Control cells and treated cells were irradiated with 6 Gy and cell pellets were collected at different time points of 1, 3, 6, 24, and 48 hours after radiation. Total cell lysate was collected from asynchronously proliferating cells in lysis buffer (ThermoFisher 89900) supplemented with 1% protease (ThermoFisher 78430) and 1% phosphatase (ThermoFisher 78420) inhibitor cocktails, resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and then immunoblotted using standard techniques. Transfer was conducted using 1x NU-Page transfer buffer (ThermoFisher NP0006–1) with 10% methanol. The protein samples were transferred to Immuno-Blot PVDF membranes (Bio-Rad 1620177) for 90 minutes at 40V for phospho-histone H2A.X (γH2AX) and β-actin or 120 minutes at 30V for phospho-Rad50 (pRad50) and Rad51. The membranes were blocked for 1 hour at room temperature in 5% skim milk in Tris-HCl buffered saline with 0.05% tween 20 (TBST), and then probed with primary antibodies (1:1000 dilution) for overnight at 4°C, washed three times in TBST, incubated with horseradish peroxidase-conjugated secondary antibody (1:2000 dilution), followed by washing in TBST and then in phosphate-buffered saline (PBS), and signals visualized by enhanced chemiluminescence (ECL) using 1:1 supersignal west pico plus substrate (ThermoFisher 34577) for pRad50 and Rad51, and 1:1 Pierce ECL western blotting substrate (ThermoFisher 32106) for γH2AX. The primary antibodies phospho-Rad50 (Ser635, #14223), Rad51 (D4B10, #8875), phospho-Histone H2A.X (Ser139, 20E3, #9718), and β-actin (13E5, #4970) and secondary antibody (Anti-Rabbit IgG, #7074P2) were obtained from Cell Signaling Technologies (Danvers, MA).
DNA repair assay
Green fluorescent protein (GFP)-reconstitution reporter cassettes for detection of homologous recombination (HR) and non-homologous end joining (NHEJ) have been previously reported (17). Plasmids containing HR or NHEJ reporter cassettes were linearized and transfected into cells to measure HR or NHEJ as a function of GFP expression. Transfections were performed using Lipofectamine 2000 (ThermoFisher 11668027). Cells with integrated reporter constructs were selected by adding 1mg/mL geneticin (ThermoFisher 10131–035). HR or NHEJ cassette expressing cells were treated with 6 μM of GSK-J4 for 72 hours, then transfected with a mixture of 5 μg ISceI expressing plasmid and 2 μg pDsRed2-N1 (Clonetech 632406). Four days following transfection, cells were harvested, suspended in PBS and placed on ice. Cells were then analyzed on FACS LSR Fortessa. Cells expressing either GFP, pDsRed2-N1, or no fluorescent protein were used as calibration controls. Data were analyzed using FlowJo software (FlowJo, LLC). DNA repair efficiency was determined as a ratio of GFP+ / DsRed+ cells normalized to 100% of vehicle control (DMSO).
Immunocytochemistry
50,000 cells were seeded into 8 well chamber slides and incubated with GSK-J4 (6 μM) for 72 hours and then irradiated with 6 Gy. 4% paraformaldehyde (PFA, Polysciences 18814) was used to fix tumor cells on cover slips at 1, 6, and 24 hours after radiation. Slips were then rinsed in PBS and blocked with 0.3% Triton X-100 and 5% normal goat serum (Cell Signaling 5425) in PBS for 1 hour at room temperature. Coverslips were incubated overnight at 4°C with γH2AX (Ser139, 20E3, Cell Signaling 9718) or 53BP1 (Cell Signaling 4937) antibody at 1:250 dilution or RAD51 (Abcam ab63801) antibody at 1:100 dilution in PBS containing 0.3% Triton X-100 and 5% normal goat serum followed by 50-minute incubation at room temperature in anti-rabbit antibody Alexa Fluor 488 (Cell Signaling 4412) or anti-rabbit antibody Alexa Fluor 555 (Cell Signaling 4413) at 1:1000 dilution in PBS with 5% normal goat serum for single reporter labeling. The coverslips were rinsed in PBS four times and nuclei were stained by incubating the coverslips at room temperature in the dark with 4’,6-diamidino-2-phenylindole (DAPI, Cell Signaling, 8961) diluted at 1:500 in PBS, followed by additional rinses in PBS and sterile water. The cover slips were then mounted on glass slides using Vectashield (Vector Laboratories H-1200) and analyzed with a Carl Zeiss Axioimager 2 microscope (Zeiss, Oberkochen, Germany). Foci were counted in 100 cells per treatment condition.
Clonogenic survival assay
Six-well tissue culture plates were seeded with 400–10,000 cells. The cells were allowed to adhere for 16 hours and treated with GSK-J4 alone at concentration of 0, 1, and 2 μM for 72 hours, and 2.5 μM of GSK-J4 for 24 or 72 hours, either before or after radiation at dose of 0.5, 1, 2, 3, 4, 6, 8, and 10 Gy. Radiation was delivered by gamma irradiator. Cells were incubated at 37°C for three weeks after which colonies were counted following staining with 0.05% crystal violet. Plating efficiencies were calculated as the ratio of the number of colonies formed to the number of cells seeded. Colonies of >50 cells were used to indicate surviving fractions. Surviving fractions were calculated as the plating efficiency of treated cells divided by the plating efficiency of control cells. Dose enhancement factors (DEFs) were calculated at 10% survival.
Cell cycle analysis
GSK-J4 effects on cell cycle distributions were determined by treating cells with 6 μM of GSK-J4 for 72 hours and/or followed by radiation (6Gy). DIPG cells (1.0 × 106) were plated into 10-cm dishes and allowed to adhere overnight. Samples were then treated with GSK-J4 only, radiation (RT) only, or with radiation and GSK-J4. GSK-J4 treatments were initiated 72 hours before RT (GSK-J4 + RT). Vehicle only (DMSO) was used as a control. At 24 hours after radiation, cells were fixed in 80% ethanol and stained with propidium iodide. Cells were then subjected to flow cytometric analysis using a BD FACSCalibur instrument and data were analyzed using FlowJo 8.8 software.
Xenograft studies
Six-week-old female athymic mice (rnu/rnu genotype, BALB/c background) were purchased from Envigo and housed under aseptic conditions. Pontine injection of tumor cells was performed as previously described (10, 13). Each mouse was injected with 1 μL of SF8628 DIPG cell suspension (100,000 cells/μL) into the pontine tegmentum at a depth of 5 mm from the inner base of the skull. Animals were randomized into four treatment groups: 1) control (DMSO, n=9), 2) GSK-J4 monotherapy (intraperitoneal injection of 100 mg/kg of GSK-J4 for 10 consecutive days, n=9), 3) radiation monotherapy (0.5 Gy, 3 times a week for two consecutive weeks for a total dose of 6 Gy, n=9), 4) GSK-J4 and radiation combination therapy (n=7). Biweekly bioluminescence imaging was used to monitor tumor growth and response to therapy as previously described (10, 13). Mice were monitored daily and euthanized at endpoint which included irreversible neurological deficit or body condition score less than 2.
Immunohistochemistry
Brains were collected from the mice at 3 hours following completion of the last treatment (n=2 for each treatment). Paraformaldehyde-fixed brains were paraffin-embedded and sectioned (10 μm) for hematoxylin and eosin (HE). Immunohistochemistry (IHC) was performed using an automated processor (DISCOVERY ULTRA, Ventana Medical Systems, Inc.). The primary antibodies used for IHC are as follows; PCNA (PC10, ab29, Abcam, dilution 1:10,000), XRCC1 (33-2-5, ab1838, Abcam, dilution 1:50), PolD1 (A304–005A, Bethyl, dilution 1:500), JMJD3 (AP1022A, ABGENT, dilution 1:40), H3K27me3 (C36B11, #9733, Cell Signaling, dilution 1:200) and Ki67 (2 μg/mL, Ventana Inc.). To assay apoptotic response to treatment, TUNEL staining was performed using the DeadEnd Colorimetric TUNEL system (Promega) according to the manufacturer’s protocol. All images were taken at 40x magnification.
Statistics
Survival plots were generated and analyzed using the Kaplan-Meier method and Graph-Pad Prism v7.0 software (GraphPad Software, San Diego, CA). Differences between survival plots were estimated using a log-rank test with Holm adjustment (18, 19). For all other analyses, a two-tailed unpaired t-test was applied using the Prism software.
Study approval
All animal protocols were approved by the Northwestern University Institutional Animal Care and Use Committee.
RESULTS
Effect of GSK-J4 on genes associated with DNA damage repair in K27M DIPG cells
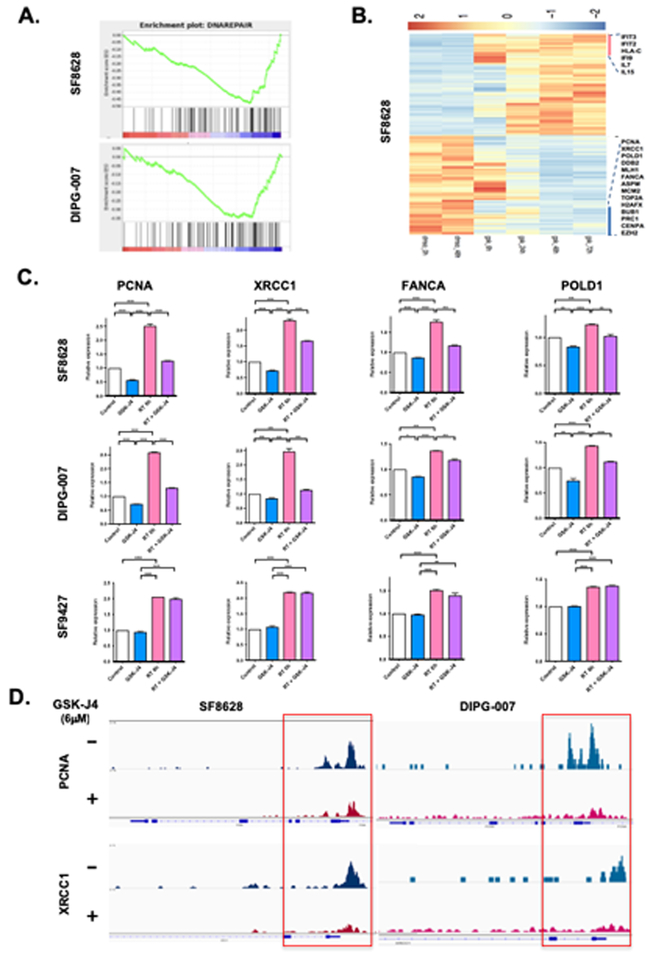
We have previously shown that GSK-J4 treatment causes a change in the expression of the genes that promote tumor growth in K27M DIPG (10). In our current RNA sequence analysis (RNA-Seq), we performed unsupervised Principal component analysis of multiple samples derived from two K27M DIPG cell lines (SF8628, DIPG-007) including DMSO treated samples collected at 0 and 48 hours and GSK-J4 treated samples collected at 0, 6, 24, 48, 72 hours following treatment. We found a global gene expression shift in GSK-J4 treated DIPG cells compared to DMSO treated samples (Supplementary Fig. S1A). In addition, we compared the RNA-Seq data between SF8628 and DIPG-007 cell lines. The direction of gene expression change between these two DIPG cell lines is highly correlated (2-sided Pearson correlation test p < 0.001***), suggesting the observed gene expression changes are shared among DIPG cell lines (Supplementary Fig. S1B). We performed GSEA pathway analysis and found that DNA DSB repair pathways were among the most significantly down-regulated in both SF8628 and DIPG-007 DIPG cell lines upon GSK-J4 treatment (Fig. 1A, B and Supplementary Table S1).
Figure 1: Histone H3 demethylase inhibition by GSK-J4 down-regulates DNA damage repair gene expression and decreases DNA accessibility in K27M DIPG cells.
(A) GSEA analysis generated from RNA-Seq data in SF8628 and DIPG-007 K27M DIPG cells, showing decrease gene sets associated with 6 μM of GSK-J4 treatment. Each gene set was permuted 1,000 times to calculate p value and FDR values. (B) Heat map showing differentially expressed genes in SF8628 K27M DIPG cell lines treated with DMSO or GSK-J4 (6 μM) for 6, 24, 48, and 72 hours. (C) Quantitative RT-PCRs (q-PCRs) showing the expression of DNA DSB repair genes (PCNA, XRCC1, FANCA, POLD1) in irradiated K27M DIPG cells (SF8628, DIPG-007) and H3 wildtype GBM cells (SF9427) with and without 6 μM of GSK-J4 treatment. Value shown are based on averages from triplicate samples and error bars represent SD (mean ± SD). Unpaired t-test values for comparisons between samples: ****P < 0.0001; ***P = 0.0002 for XRCC1 between control vs. GSK-J4, control vs. RT in DIPG-007; ***P = 0.0001 for XRCC1 between GSK-J4 vs. RT in DIPG-007; ***P = 0.0003 for XRCC1 between RT vs. RT + GSK-J4 in DIPG-007; ***P = 0.0002 for FANCA between RT vs. RT + GSK-J4 in SF8628, *P = 0.0111 for FANCA between control vs. GSK-J4 in DIPG-007; ***P = 0.0003 for FANCA between control vs. RT in DIPG-007; ***P = 0.0008 for FANCA between RT vs. RT + GSK-J4 in DIPG-007; **P = 0.0012 for FANCA between GSK-J4 vs. RT + GSK-J4 in SF9427; **P = 0.0034 for POLD1 between control vs. GSK-J4 in SF8628; ***P = 0.0007 for POLD1 between control vs. RT in SF8628; **P = 0.0020 for POLD1 between RT vs. RT + GSK-J4 in SF8628; **P = 0.0054 for POLD1 between control vs. GSK-J4 in DIPG-007. (D) Representative tracks of PCNA and XRCC1 peaks at promoter generated from ATAC-Seq data in SF8628 and DIPG-007 K27M DIPG cells. All panels are representative of two independent experiments.
We verified the change in the DNA DSB repair gene expression by qPCR in irradiated K27M DIPG (SF8628, DIPG-007) and H3 wild-type GBM cells (SF9427) with and without GSK-J4 treatment (Fig. 1C). Radiation treatment (RT) increases the expression of DNA DSB repair genes including PCNA, XRCC1, FANCA, and POLD1 while GSK-J4 decreases the expression of these genes in K27M DIPG cells. Furthermore, radiation-induced DNA DSB repair gene expression was significantly reduced by combination treatment with GSK-J4 and RT. In contrast to K27M DIPG cell lines, the DNA DSB repair gene expression was not changed by GSK-J4 treatment in irradiated SF9427 H3 wild-type GBM cells (Fig.1C).
To investigate the influence of GSK-J4 on genome-wide DNA accessibility, we performed the assay transposase accessible chromatin sequencing (ATAC-Seq) and found an overall concordance between ATAC-Seq and RNA-Seq data. Based on ATAC-Seq, we sought to compare the overall accessibility between GSK-J4 and DMSO treatment samples based on the identified binding peaks. We found a trend towards reduced DNA accessibility as indicated by the long-tail of marginally down-regulated binding sites in SF8628 DIPG cells treated with GSK-J4 when compared with DMSO treated cells (Supplemental Fig. S1C). This is consistent with our previous study showing the chromatin-repressive role of GSK-J4 through increased H3K27 tri- and di-methylation in K27M DIPG cells (10). To investigate the enrichment of transcriptional factor among the decreased ATAC-Seq binding sites, we created a union of differential peaks between DMSO and GSK-J4 treated samples (p < 0.05). We performed HOMER motif analysis in SF8628 DIPG cells and found a significant enrichment of DNA sequencing motifs involving neuron developmental transcriptional factors such as GLIS2, PITX1 (Supplemental Table S2). Interestingly, the promoters of two DNA repair genes, PCNA and XRCC1, have diminished binding in the GSK-J4 treated SF8628 and DIPG-007 cells (Fig. 1D). Taken together, our results suggest that GSK-J4 promotes the transcriptionally silence of chromatin state through an increase in H3K27 methylation and represses the genes involving DNA DSB repair in K27M DIPG cells.
GSK-J4 enhances radiation-induced DNA damage
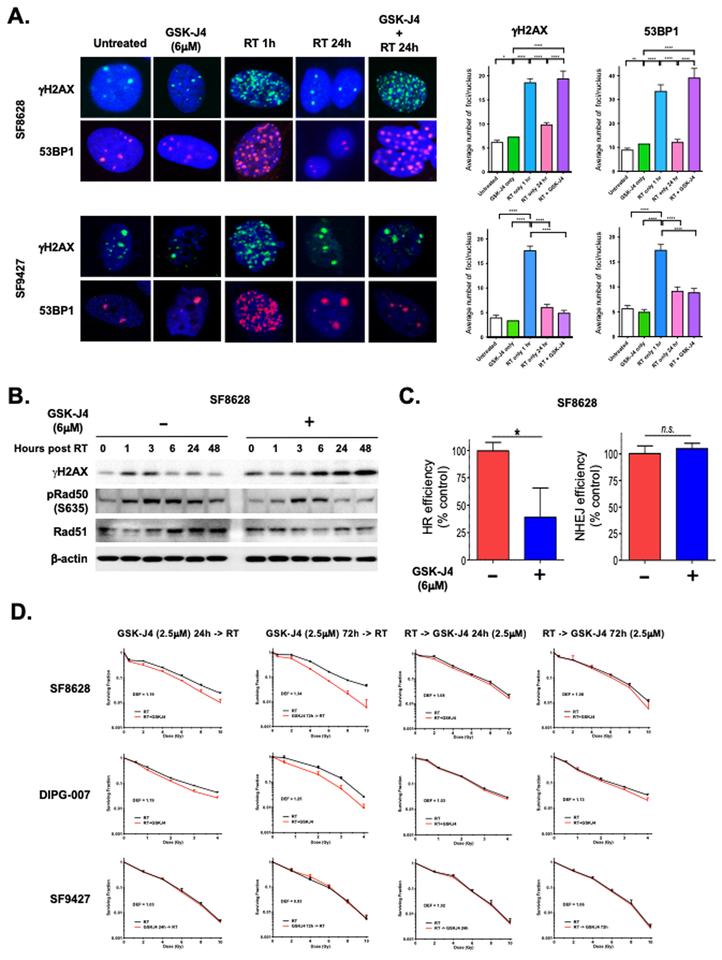
To determine the effect of GSK-J4 on radiation-induced DNA damage, fluorescence immunocytochemistry of DSB markers γH2AX and 53BP1 was examined to quantitate the extent of DNA damage in irradiated K27M DIPG cells (SF8628) and H3 wild-type GBM cells (SF9427) either in the presence or absence of GSK-J4. SF8628 K27M DIPG cells treated with GSK-J4 showed increase γH2AX and 53BP1 foci relative to untreated cells. However, GSKJ-4 did not cause DNA damage in SF9427 wild-type GBM cells. In both SF8628 K27M DIPG and SF9427 wild-type GBM cells, γH2AX and 53BP1 foci increased one hour following radiation therapy, indicating increased DNA damage by irradiation. At 24 hours after irradiation, γH2AX and 53BP1 foci were largely reduced in those cells due to successful repair of the previously noted DNA damage. However, irradiated SF8628 K27M DIPG cells treated with GSK-J4 sustained high levels of γH2AX and 53BP1 at 24 hours compared to irradiated SF9427 wild-type GBM cells treated with GSK-J4 (Fig. 2A). This might be due to the lack of response to GSK-J4 inhibitory effect in SF9427 wild-type GBM cells, as previously reported (10). Furthermore, western blotting revealed that radiation-induced γH2AX was increased and sustained until 48 hours in SF8628 K27M DIPG cells subjected to combination of GSK-J4 and radiation, unlike cells treated with radiation alone (Fig. 2B). These findings suggest that GSK-J4 uniquely inhibits DNA damage repair in irradiated K27M DIPG cells to sustain radiation-induced DNA damage.
Figure 2: GSK-J4 enhances radiation-induced DNA damage.
(A) Effect of GSK-J4 (6 μM) on γH2AX and 53BP1 foci formation in irradiated SF8628 K27M DIPG and SF9427 H3 wild-type GBM cells. Left: Representative images of nuclei from each treatment, showing γH2AX (upper) and 53BP1 (lower) foci. Right: Mean and SD values for γH2AX and 53BP1 foci/nucleus are shown, and are based on foci counts from three independent experiments. Unpaired t-test values for γH2AX comparisons between treatments: **** indicate p < 0.0001. * indicated p = 0.0126 between untreated vs. GSK-J4 only for SF8628. Unpaired t-test values for 53BP1 comparisons between treatments: **** indicates p < 0.0001. ** indicated p = 0.0047 between untreated vs. GSK-J4 only for SF8628. (B) Western blot results showing effects of GSK-J4 (6 μM) on expression of γH2AX and homologous recombination (HR) repair pathway proteins, phospho-Rad50 and total Rad51 in SF8628 K27M DIPG cells. (C) DNA repair assay showing effect of GSK-J4 (6 μM) on HR and nonhomologous end-joining (NHEJ) pathways in SF8628 K27M DIPG cells. Repair efficiency represents the ratio of GFP+ to DsRed+ cells normalized to 100% of vehicle control (DMSO). Value shown are based on averages from duplicate or triplicate samples and error bars represent SD (mean ± SD). Unpaired t-test values for comparisons between samples: * indicates P = 0.0205, ** indicates P = 0.0089. (D) Clonogenic survival results for K27M DIPG cells (SF8628, DIPG-007) and H3 wild type glioma (SF9427) cells treated with GSK-J4 (2.5 μM) for 24 or 72 hours before or after radiation (RT alone: black line, RT and GSK-J4: red line). Survival fractions, shown as mean ± SD based on averages from triplicate samples, were normalized to plating efficiency for each cell lines. Dose Enhancement Factor (DEF) for each cell line was calculated at 10% survival level.
GSK-J4 inhibits DNA damage repair through homologous recombination
DNA DSBs are repaired by two major pathways; via homologous recombination (HR) in late S and G2 phase and via nonhomologous end-joining (NHEJ) throughout the cells cycle (20, 21). During HR, the missing information is copied into the DSB site from a homologous sequence, whereas during NHEJ joins the broken ends are joined without homology (22). In order to quantify the DSB repair pathways, we employed green fluorescent protein (GFP)-reconstitution reporter cassettes for detection of HR and NHEJ (16). Using reporter constructs for each DSB repair pathway, we studied the DNA repair ability of SF8628 K27M DIPG cells and human astrocytes expressing K27M (Astro KM) and wild-type (Astro WT) H3F3A genes, in the presence and absence of GSK-J4. GSK-J4 exerted an inhibitory effect on the cells’ ability to repair DNA via the HR pathway while largely sparing the NHEJ pathway (Fig. 2C, Supplementary Fig. S2A). This inhibitory effect of GSK-J4 on DNA damage repair was observed in the cells expressing K27M H3F3A gene (SF8628 DIPG and Astro KM) but not in the cells expressing H3 wild-type H3F3A gene (Astro WT) (Fig. 2C, Supplementary Fig. S2A).
To better understand the effect of GSK-J4 on DNA repair dynamics, the cell cycle distribution was evaluated in SF8628 K27M DIPG, Astro KM, and Astro WT cells treated with GSK-J4, radiation, and the combination of GSK-J4 and radiation (Supplementary Fig. S2B). In the SF8628 K27M DIPG and Astro KM cells, GSK-J4 treatment increased the S phase fraction (SF8628: 28.8%, Astro KM: 25.9%) relative to control (SF8628: 12.2%, Astro KM: 12.0%). Radiation increased G2/M cell fraction (SF8628: 28.9%, Astro KM: 25.5%) relative to control (SF8628: 21.8%, Astro KM: 19.3%) or GSK-J4 alone (SF8628: 4.5%, Astro KM: 9.3%). Combination treatment of GSK-J4 and radiation also increased S phase fraction (SF8628: 21.2%, Astro KM: 22.4%) relative to radiation alone (SF8628: 11.2%, Astro KM: 17.4%). Thus, GSK-J4 maintained a high proportion of S phase fraction during response to radiation in SF8628 K27M DIPG and Astro KM cells. In the Astro WT cells, GSK-J4 showed the least increase S phase fraction (13.4%) relative to control (11.2%).
Western blot analysis revealed that treatment with GSK-J4 prior to radiation causes a time-dependent reduction in the level of phosphorylated RAD50 and total RAD51 protein, which both play a major role in HR DNA DSB repair pathway, in SF8628 K27M DIPG cells (Fig. 2B). Fluorescence immunocytochemistry of RAD51 in SF8628 K27M DIPG cells showed that RAD51 foci formation increased in a time dependent manner with peak at 24 hours following radiation (Supplementary Fig. S2C). GSK-J4 inhibited RAD51 foci formation at each time point, indicating inhibition of HR DNA damage repair by GSK-J4.
Effect of GSK-J4 on cell proliferation in combination with radiation
Next, we examined the effect of GSK-J4 on radiation response in K27M DIPG cells. Clonogenic survival assay was carried out using two K27M DIPG cell lines (SF8628 and DIPG-007) and H3 wild-type GBM cell line (SF9427). Cells were treated with GSK-J4 administered at either 24 or 72 hours before or after radiation. GSK-J4 treatment for 72 hours before radiation resulted in the largest enhancement of radiation effect compared to that of radiation only treatment in K27M DIPG cells (SF8628: DEF of 1.34, DIPG-007: 1.25, Fig. 2D). GSK-J4 treatment for 24 hours before radiation was intermediate in its radiation-enhancing effect (SF8628: DEF of 1.19, DIPG-007: DEF of 1.19, Fig. 2D). Exposure of DIPG cells to GSK-J4 for either 24 or 72 hours after radiation showed the least radiation dose enhancement (SF8628: DEF of 1.08 for 24 hours and 1.08 for 72 hours, DIPG-007: DEF of 1.03 for 24 hours and 1.13 for 72 hours, Fig. 2D). There is a minor effect of GSK-J4 on enhancing the response to radiation in SF9427 (DEF between 0.93 and 1.06). GSK-J4 treatment for 72 hours in the absence of radiation inhibited the clonal growth of K27M DIPG cells, but had no effect on the clonal growth of H3 wild-type GBM cells (Supplementary Fig. S2D).
GSK-J4 enhances radiation antitumor effect in human DIPG xenografts
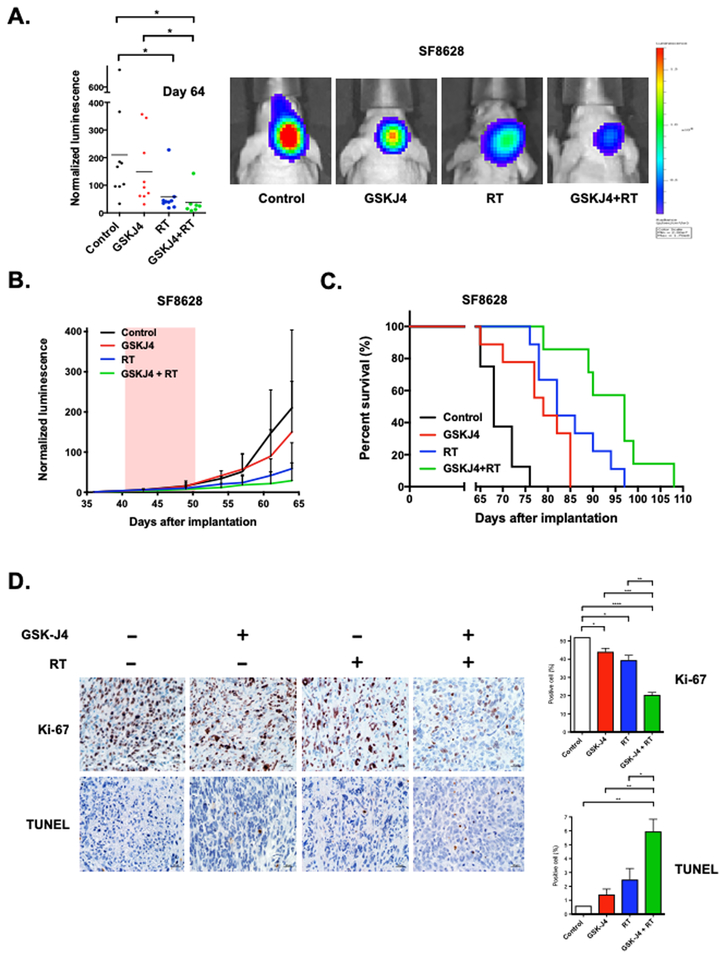
Based on the effects of GSK-J4 on radiation-induced DNA DSB repair and clonogenic survival in irradiated DIPG cells, we hypothesized that GSK-J4 treatment would enhance radiation anti-tumor activity to suppress tumor growth and increase survival benefit in mice with orthotopic human DIPG xenografts. To address this, mice with SF8628 K27M DIPG xenografts were treated with a daily dose of 100mg/kg of GSK-J4 for 10 days in the presence or absence of radiation treatment (RT) at total dose of 3 Gy (0.5 Gy per day for 3 days a week for 2 weeks). GSK-J4 monotherapy resulted in reduced tumor growth rate and conferred substantial survival benefit against human DIPG xenograft model, as indicated by bioluminescence monitoring (Fig. 3A, B) and survival of treated mice (**P = 0.0021, Fig. 3C), consistent with previous study (10). Similarly, radiation monotherapy provided a significant benefit (*P = 0.0416 in Fig. 3A, ***P < 0.0001 in Fig. 3C). We found that combination treatment of DIPG xenografts with GSK-J4 and RT significantly increase therapeutic benefit, outperforming either monotherapy (GSK-J4 vs. GSK-J4 + RT: **P = 0.0429 in Fig. 3A, **P = 0.0014 in Fig. 3C; RT vs. GSK-J4 + RT: *P = 0.0348 in Fig. 3C). This DIPG xenograft experiment included mice that were euthanized at the end of treatment to obtain intracranial tumor samples to analyze the expression level of K27me3, JMJD3, DNA damage repair (PCNA, XRCC1, POLD1), tumor cell proliferation (Ki-67), and apoptosis (TUNEL). Immunohistochemistry (IHC) analysis of tumors obtained from mice treated with GSK-J4 showed that GSKJ4 treatment significantly increased tumor cell K27me3 positivity (***P = 0.0002) and decreased tumor cells JMJD3 positivity (**P = 0.0022, Supplementary Fig. S3A). IHC analysis of DNA damage repair revealed that RT significantly increased the expression of DNA damage repair proteins including PCNA (****P < 0.0001), XRCC1 (**P = 0.0046), and POLD1 (*P = 0.0102) relative to control, while GSK-J4 decreased the expression level of these proteins. In addition, the expression level of DNA damage repair proteins were significantly reduced by combination treatment with GSK-J4 and RT in compared to RT alone (PCNA: **P = 0.0024, XRCC1: ****P < 0.0001, POLD1: *P = 0.0291). Analysis of intra-tumor Ki-67 staining showed all therapies significantly reduced DIPG xenograft cell proliferation relative to the control group (GSK-J4: *P = 0.0174, RT: *P = 0.0107, GSK-J4 + RT: ****P < 0.0001, Fig. 3D). There were significantly less Ki-67 positive cells in the samples treated with combination therapy compared to either monotherapy (***P = 0.0005 in relation to GSK-J4 alone, **P = 0.0031 in relation to RT alone). TUNEL staining results showed the highest proportion of positive cells in tumors from mice receiving combination therapy of GSK-J4 and RT relative to either monotherapy (**P = 0.0092 relative to GSK-J4 alone, *P = 0.0433 relative to RT alone). No TUNEL positivity was evident in normal brain surrounding tumor in mice receiving any of the combination therapy treatments (Fig. 3D). In contrast to K27M tumors, xenografts established from GBM43 cells expressing wild-type H3 showed no response to GSK-J4 treatment and no therapeutic benefit from combination therapy of GSK-J4 and radiation relative to radiation monotherapy.
Figure 3: GSK-J4 enhances radiation antitumor activity in vivo.
Mice with SF8628 intracranial tumor were randomized to four treatment groups: control (DMSO, n=9), GSK-J4 alone (100mg/kg/day for 10 consecutive days, n=9), radiation treatment (RT) alone (0.5 Gy, 3 times a week for two consecutive weeks for a total dose of 3 Gy, n=9), and GSK-J4 + RT (n=7). (A) Left: Dot plot representation of tumor bioluminescence values in mice at day 64 post-tumor cell injection. Horizontal bars indicate the mean value for each treatment group. Unpaired t-test values for comparisons between treatments: control vs. RT, *P = 0.0416; control vs. GSK-J4 + RT, *P = 0.0389; GSK-J4 vs. GSK-J4 + RT, *P = 0.0429. Right: Tumor bioluminescence overlay images showing relative bioluminescence intensities. (B) Growth plots for intracranial tumors. Tumor bioluminescence values show mean and standard deviation (SD) for normalized against bioluminescence values obtained at day 36 post-tumor cell injection. (C) Corresponding survival plots for each experiment. Statistical analysis was performed using a log-rank test with Holm adjustment: control vs. GSK-J4, **P = 0.0021; control vs. RT, ***P < 0.0001; control vs. GSK-J4 + RT, ***P = 0.0001; GSK-J4 vs. GSK-J4 + RT, **P = 0.0014; RT vs. GSK-J4 + RT, *P = 0.0348. (D) Left: Images of representative Ki-67 and TUNEL staining for intracranial tumor from mice euthanized at the end of treatment. Right: Mean and SD values representing the average number of positive cells in four high-powered fields in each tumor. Unpaired t-test values for comparisons between treatments. Ki-67: control vs. GSK-J4, *P = 0.0174; control vs. RT, *P = 0.0107; control vs. GSK-J4 + RT, ****P < 0.0001; GSK-J4 vs. GSK-J4 + RT, ***P = 0.0005. TUNEL: control vs. GSK-J4 + RT, **P = 0.0074; GSK-J4 vs. GSK-J4 + RT, **P = 0.0092; RT vs. GSK-J4 + RT, *P = 0.0433.
DISCUSSION
DNA damage is thought to be the most deleterious effect caused by radiation therapy. Treatment of DIPG with fractionated focal radiation therapy often provides transient relief from tumor symptoms, however, tumor recurrence and patient demise is inevitable. We and others indicated that the majority of the DNA DSB caused by radiation is repaired within 24 hour of treatment (Fig. 2A) (23–25). This DNA damage repair is possibly responsible for the tumor progression observed in DIPG after completion of radiation therapy, thereby ultimately conveying no survival benefits to the patients. It is essential to identify therapeutic agents that inhibit DNA DSB repair to enhance the anti-tumor effects of radiation, based on the understanding of the molecular mechanisms associated with chromatin modification in K27M DIPG.
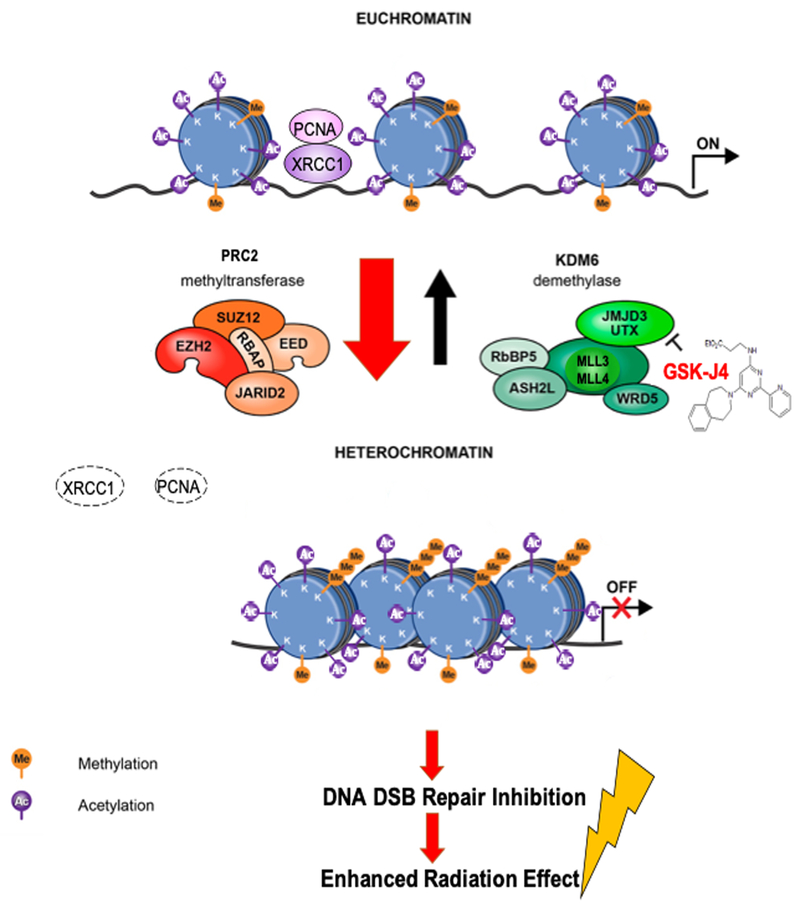
We, as well as others, have identified the gain-of-function mechanism for the K27M mutation. The mutant protein sequesters and functionally inactivates EZH2, a catalytic subunit of the K27 histone methyltransferase, polycomb repressive complex 2 (PRC2), leading to a dramatic reduction in global methylation at K27 residues in the K27M DIPG (7,8). This reduction in H3K27 methylation leads to opening of the chromatin providing access for active transcription (26, 27). We have previously shown that inhibition of the H3K27 demethylase JMJD3 by GSK-J4 acts to restore K27 methylation in DIPG cells (10). Increased H3K27 methylation promotes heterochromatin which is transcriptionally silent and decreases gene expression (27). In our current study, GSK-J4-mediated H3K27 methylation significantly decreased DNA accessibility at both promoter and distal binding sites (Supplementary Fig. S1C) and decreased expression of the genes involved in DNA damage repair pathways in K27M DIPG cells (Fig. 1A, B, C, Supplementary Table S1). GSK-J4 indirectly modifies chromatin structure to reduce chromatin accessibility, thereby providing a plausible mechanism to prevent access of DNA repair proteins to the sites of DNA damage induced by radiation (Fig. 1D). Interestingly, GSK-J4 decreased enrichment of DNA sequencing motifs involving neuron developmental genes, while this observation is not direct relevance to the effect of GSK-J4 on DNA damage repair. The significant enrichment of genes involved in neuronal development has been identified by another group (9). Further investigations would be suitably addressed in future studies.
We observed that GSK-J4 increased radiation-induced DNA damage (Fig. 2A, B) by inhibiting HR DNA DSB repair pathway (Fig. 2C, Supplementary Fig. S2A) and enhancing radiosensitivity of K27M DIPG cell lines (Fig. 2D). These GSK-J4-mediated radiosensitizing effects were observed in two K27M DIPG cell lines (SF8628 and DIPG-007) but not in H3 wild-type GBM cell line (SF9427). This might be due to the global H3K27 hypomethylation in K27M DIPG cells and GSK-J4’s inhibitory effect on histone demethylase increasing H3K27 methylation, as previously reported (10). Given that HR occurs in late S/G2 phase while NHEJ repairs DSB throughout the cell cycle (20, 21), our results suggest that GSK-J4 maintains a high proportion of S phase fraction to inhibits the HR DNA repair pathway. These GSK-J4 effects on DNA damage repair were observed specifically in tumor cells expressing K27M mutant H3F3A (SF8628 and Astro KM) (Fig. 2C, Supplementary Fig. S2A). GSK-J4-mediated H3K27 methylation may function as an important mechanism to coordinate chromatin accessibility and transcription both at damage sites and in genes, and possibly to regulate repair of DNA lesions while also orchestrating transcriptional responses to DNA damage (Fig. 4). The role of H3K27 methylation in radiation-induced DNA damage and repair has been investigated but remains unclear. Previous studies have shown that radiation-induced DNA damage recruits EZH2 to DNA DSB sites leading to accumulation of H3K27me3 at the damage sites (28–31). Genetic and pharmacologic inhibitions of EZH2 increased radiosensitivity in human bone osteosarcoma epithelial cells (U2OS) (28) and atypical teratoid rhabdoid tumor cells (32), whereas it had no effect on the glioma cells (33). On the other hand, the expression of JMJD3 has been reported to increase in response to radiation-induced DNA damage (34).
Figure 4. Working model. Chromatin modification by GSK-J4 in K27M DIPG inhibits radiation-induced DNA double strand break (DSB) repair.
K27M mutation results in markedly reduced K27 methylation leading to open and transcriptionally active chromatin formation. Homologous recombination (HR) DNA repair proteins (PCNA, XRCC1) can be access to the DNA damage site induced by radiation. GSK-J4 treatment restores K27 methylation to promote compact and transcriptionally repressed chromatin state, in turn, down-regulates the expression of the genes involving with DNA DSB repair, limits to access HR DNA repair proteins (PCNA, XRCC1), and enhances radiation anti-tumor activity.
Consistent with in vitro results, our animal studies, using intracranial SF8628 K27M DIPG xenografts, demonstrated that GSK-J4 increased the genotoxic activity of RT, and that GSK-J4 and RT combination treatment has substantial growth inhibition and survival benefit that surpassed either agent (GSK-J4 or RT) administrated as a monotherapy (Fig. 3A, B, C). This antitumor activity of GSK-J4 + RT combination therapy inhibited radiation-induced DNA damage repair, by showing significant decrease in PCNA, XRCC1, POLD1 positive cells compared to radiation alone (Supplementary Fig. S3B). GSK-J4 + RT combination therapy also promotes apoptosis and cell death as evidenced by significantly increasing in TUNEL positive cell and decreasing Ki-67 positive cells in the combination therapy group compared to the monotherapy groups (Fig. 3D). The radiation dose used in the combination treatment with GSK-J4 was determined by a dose escalation study. We determined to use the fractionated radiation dose of 0.5 Gy/day for 3 days a week in 2 weeks showed intermediate survival benefit (**p = 0.0016 relative to control, median survivals extended by 64 days form 56.5 days of control group median survivals: Supplementary Fig. S3D).
GSK-J4 has been used to treat many kinds of tumors such as T-cell acute lymphoblastic leukemia (T-ALL), acute myeloid leukemia (AML), diffuse large B-cell lymphoma, DIPG, neuroblastoma, prostate cancer, and gastric cancer (10, 35–41). However, GSK-J4 is not yet in clinical development. The major challenge of GSK-J4 in clinical development is that GSK-J4 is a prodrug and is rapidly converted to active drug GSK-J1 in vivo (10, 42). The highly polar GSK-J1 compound has restricted cellular permeability (41). Further development for stabilizing GSK-J4 in vivo would be required for clinical application for this compound. In addition to stabilizing GSK-J4, local drug delivery systems such as convection-enhanced delivery (43) would provide an alternative strategy for reducing drug metabolism and potentially increasing the efficacy of GSK-J4 in vivo.
As mentioned above, children with DIPG inevitably die of their disease. The only therapeutic modality that is used routinely for the treatment of DIPG is radiation. Yet, even with radiotherapy the median overall survival is approximately 10 months (3–5). Increased morbidity from RT (44–46), and the poor long-term outcomes of this cancer have necessitated the frequent inclusion of focal radiation in young patients. Given this reality, there is a critical need to identify new therapeutics that enhance RT antitumor activity and have the potential to enable the use of lower doses and/or volumes of radiation to limit long-term side effects in young patients. Present findings here support the possibility of using GSK-J4 with RT for increased antitumor effect, that could potentially lessen the amount of RT while still providing substantial relief from disease for patients.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Diffuse intrinsic pontine glioma (DIPG) is one of the most devastating childhood cancers that limited response to radiation therapy (RT), resulting in median overall survival of approximately 10 months. There is a critical need for new therapeutics that enhance the radiation effect while enabling the use of lower dose of radiation to limit long-term side effects in young patients. DIPG pathogenesis involves histone H3 gene mutations and reduction of global H3K27 methylation that promotes tumor growth. We found that the JMJD3 demethylase inhibitor, GSK-J4, which restores H3K27 methylation, decreases expression of the genes involved in DNA damage repair, and enhances radiation-induced DNA damage by inhibiting homologous recombination repair, mediated by chromatin modification. The combination of GSK-J4 and RT, in human DIPG xenografts, had substantial growth inhibition and survival benefit, compared to either alone, supporting the possible use of GSK-J4 to increase RT anti-tumor effect while using lower doses of radiation.
ACKNOWLEDGMENTS
We thank Dr. Yoshihiro Tanaka from the Department of Preventive Medicine, Northwestern University (Chicago, USA) for statistical comment. This work was supported by grants from the Bear Necessities Pediatric Cancer Foundation and Rally Foundation for Childhood Cancer Research (R.H.), the Matthew Larson Foundation (R.H.), St. Baldrick’s Foundation (R.H.), Alex’s Lemonade Stand Foundation for Childhood Cancer (R.H.), John McNicholas Pediatric Brain Tumor Foundation (R.L., A.S. R.H.), and the U.S. National Institutes of Health grants: R01NS093079 (R.H.), F32CA216996 (D.U.), R01CA214035–15 (A.S.), and R35CA197569 (A.S.). Histology and fluorescent microscopy services were provided by the Mouse Histology and Phenotyping Laboratory and the Center for Advanced Microscopy/Nikon Imaging Center at Northwestern University, respectively.
Footnotes
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
REFERENCES
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KJ, Cullen J, Barnholtz-Sloan JS, Ostrom QT, Langer CE, Turner MC, et al. Childhood Brain Tumor Epidemiology: A Brain Tumor Epidemiology Consortium Review. Cancer Epidemiol Biomarkers Prev 2014;23(12):2716–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren KE. Diffuse intrinsic pontine glioma: poised for progress. Front Oncol 2012;2:205. doi: 10.3389/fonc.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robison NJ, Kieran MW. Diffuse intrinsic pontine glioma: a reassessment. J Neurooncol 2014;119(1):7–15. doi: 10.1007/s11060-014-1448-8. [DOI] [PubMed] [Google Scholar]
- 5.Roos DE, Smith JG. Randomized trial on radiotherapy for paediatric diffuse intrinsic pontine glioma (DIPG). Radiother Oncol 2014;113(3):425. [DOI] [PubMed] [Google Scholar]
- 6.Fontanilla HP, Pinnix CC, Ketonen LM, Woo SY, Vats TS, Rytting ME, et al. Palliative Reirradiation for Progressive Diffuse Intrinsic Pontine Glioma. Am J Clin Oncol 2012;35(1):51–57. [DOI] [PubMed] [Google Scholar]
- 7.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes and development 2013;27(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013;340(6134):857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 2013;24(5):660–672. [DOI] [PubMed] [Google Scholar]
- 10.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 2014;20(12):1394–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashizume R, Smirnov I, Liu S, Phillips JJ, Hyer J, McKnight TR, et al. Characterization of a diffuse intrinsic pontine glioma cell line: implications for future investigations and treatment. J Neurooncol 2012;110(3):305–313. [DOI] [PubMed] [Google Scholar]
- 12.Hashizume R, Gupta N. Patient-derived tumor models for diffuse intrinsic pontine gliomas. Current Neuropharmacol 2017;15:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki Y, Hashizume R, Ozawa T, Banarjee A, Prados M, James CD, et al. An experimental xenograft mouse model of diffuse pontine glioma designed for therapeutic testing. J Neurooncol J Neurooncol 2012;108(1):29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gilette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seluanov A, Mao Z, Gorbunova V. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J Vis Exp 2010;(43). doi: 10.3791/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bewick V, Cheek L, Ball J. Statistic review 12: survival analysis. Crit Care 2004;8:389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm S A simple sequentially rejective multiple test procedure. Scand J Stat 1979;6:65–70 [Google Scholar]
- 20.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008;77:229–57. [DOI] [PubMed] [Google Scholar]
- 21.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem 2008:283:1–5. [DOI] [PubMed] [Google Scholar]
- 22.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA Double-Strand Break Repair during the Mammalian Cell Cycle. Mol Cell Biol 2003;23(16):5706–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashizume R, Zhang A, Mueller S, Prados MD, Lulla RR, Goldman S, et al. Inhibition of DNA damage repair by the CDK4/6 inhibitor palbociclib delays irradiated intracranial atypical teratoid rhabdoid tumor and glioblastoma xenograft regrowth. Neuro Oncol 2016;18:1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin Oncol 2013;25(10):578–585. [DOI] [PubMed] [Google Scholar]
- 25.Mueller S, Hashizume R, Yang X, Kolkowitz I, Olow AK, Phillips J, et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol 2014;16:352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012;13(5):343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maury E, Hashizume R. Epigenetic modification in chromatin machinery and its deregulation in pediatric brain tumors: Insight into epigenetic therapies. Epigenetics 2017;12(5):353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 2013;12:2675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 2008;4:e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke ac, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage, Proc Natl Acad Sci U S A 2010;107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 2010;20:606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alimova I, Birks DK, Harris PS, Knipstein JA, Venkataraman S, Marquez VE, et al. Inhibition of EZH2 suppresses self-renewal and induces radiation sensitivity in atypical rhabdoid teratoid tumor cells. Neuro Oncol 2013;15:149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gursoy-Yuzugullu O, Carman C, Serafim RB, MyronakisM, Valente V, Price BD. Epigenetic therapy with inhibitors of histone methylation suppresses DNA damage signaling and increases glioma cell radiosensitivity. Oncotarget 2017;8:24518–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams K, Christensen J, Rappsilber J, Nielsen AL, Johansen JV, Helin K. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS ONE 2014;9:e96545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukemia. Nature 2014;514:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boila LD, Chatterjee SS, Banerjee D, Sengupta A. KDM6 and KDM4 histone lysine demethylase s emerge as molecular therapeutic targets in human acute myeloid leukemia. Exp Hematol 2018;58:44–51. [DOI] [PubMed] [Google Scholar]
- 37.Mathur R, Sehgal L, Havranek O, Köhrer S, Khashab T, Jain N, et al. Inhibition of demethylase KDM6B sensitizes diffuse large B-cell lymphoma to chemotherapeutic drugs. Haematologica 2017;102:373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 2015;21:555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lochmann TL, Powell KM, Ham J, Floros KV, Heisey DAR, Kurupi RIJ, et al. Targeted inhibition of histone H3K27 demethylation is effective in high-risk neuroblastoma. Sci Transl Med 2018;10(441). pii: eaao4680. doi: 10.1126/scitranslmed.aao4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morozov VM, Li Y, Clowers MM, Ishov AM. Inhibitor of H3K27 demethylase JMJD3/UTX GSK-J4 is a potential therapeutic option for castration resistant prostate cancer. Oncotarget 2017;8:62131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Xia Y, Xiao Z, Jia Y, Li L, Jin Y, et al. Comprehensive profiling of JMJD3 in gastric cancer and its influence on patient survival. Sci Rep 2019;9:868. doi: 10.1038/s41598-018-37340-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012;488:404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louis N, Liu S, He X, Drummond DC, Noble CO, Goldman S, et al. New therapeutic approaches for brainstem tumors: a comparison of delivery routes using nanoliposomal irinotecan in an animal model. J Neurooncol 2018;136:475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappaport R, Brauner R. Growth and Endocrine Disorders Secondary to Cranial Irradiation. Pediatr Res 1989;25:561–7. [DOI] [PubMed] [Google Scholar]
- 45.Moore BD, Copeland DR, Ried H. Neurophysiological Basis of Cognitive Deficits in Long-term Survivors of Childhood Cancer. Arch Neurol 1992;49:809–817. [DOI] [PubMed] [Google Scholar]
- 46.Palmer SL, Reddick WE, Glass JO, Gajjar A, Goloubeva O, Mulhern RK. Decline in Corpus Callosum Volume among Pediatric Patients with Medulloblastoma: Longitudinal MR Imaging Study. AJNR Am J Neuroradiol 2002;23:1088–94. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.