Abstract
Purpose
Paclitaxel is an integral component of primary therapy for breast and epithelial ovarian cancers, but less than half of these cancers respond to the drug. Enhancing the response to primary therapy with paclitaxel could improve outcomes for women with both diseases.
Experimental Design
Twelve kinases that regulate metabolism were depleted in multiple ovarian and breast cancer cell lines to determine whether they regulate sensitivity to paclitaxel in Sulforhodamine B assays. The effects of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 depletion on cell metabolomics, extracellular acidification rate, nicotinamide adenine dinucleotide phosphate, reactive oxygen species and apoptosis were studied in multiple ovarian and breast cancer cell lines. Four breast and ovarian human xenografts and a breast cancer patient-derived xenograft (PDX) were used to examine the knockdown effect of PFKFB2 on tumor cell growth in vivo.
Results
Knockdown of PFKFB2 inhibited clonogenic growth and enhanced paclitaxel sensitivity in ovarian and breast cancer cell lines with wtTP53. Silencing PFKFB2 significantly inhibited tumor growth and enhanced paclitaxel sensitivity in 4 xenografts derived from 2 ovarian and 2 breast cancer cell lines, and prolonged survival in a triple-negative breast cancer PDX. Transfection of siPFKFB2 increased the glycolysis rate, but decreased the flow of intermediates through the pentose-phosphate pathway in cancer cells with wtTP53, decreasing NADPH. ROS accumulated after PFKFB2 knockdown, which stimulated JNK and TP53 phosphorylation, and induced apoptosis that depended upon upregulation of p21 and Puma.
Conclusions
PFKFB2 is a novel target whose inhibition can enhance the effect of paclitaxel-based primary chemotherapy upon ovarian and breast cancers retaining wtTP53.
Keywords: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2); pentose phosphate pathway (PPP); reactive oxygen species (ROS); cell cycle arrest; apoptosis; p53; p21; paclitaxel sensitivity
Introduction
Paclitaxel has emerged as an important agent for adjuvant treatment of both ovarian and breast cancers. When used as a single agent, however, less than half of these cancers respond (1–3). Enhancing the response to primary therapy with paclitaxel could improve outcomes for women with both diseases. The tumor suppressor TP53 is frequently mutated in human cancers including breast and ovarian cancers. Somatic TP53 mutations occur in over 50% of all tumors (4,5), across nearly all types of cancer. TP53 mediates multiple processes of which the best understood include cell cycle arrest, DNA repair and apoptosis (6,7). Recent work indicates that TP53 has an important role in modulating metabolic processes including glycolysis, oxidative phosphorylation, and the pentose phosphate pathway (PPP) (8,9).
Cancer cells can exhibit enhanced glycolysis and lactate production even under normoxic conditions, i.e., the Warburg Effect (10,11). Cancer cells often exhibit increased expression of glycolytic enzymes and glucose uptake which result in markedly enhanced glycolytic rates. In addition to producing additional ATP, increased glycolysis also generates three-carbon building blocks for the biosynthesis of proteins, lipids and nucleic acids in rapidly growing cells (12). Many previous studies have shown that oncogenic signaling pathways regulate the activity of metabolic enzymes to support macromolecular synthesis in cancer cells which is required for their rapid proliferation. Enzymes involved in altered glucose metabolism might provide important targets for cancer therapy (13,14). The pentose phosphate pathway consists of oxidative and non-oxidative branches. The oxidative branch is a major source of reduced nicotinamide adenine dinucleotide phosphate (NADPH), a key reducing equivalent for lipid, nucleotide, and aromatic amino acid biosynthesis (15). NADPH also plays a key role in maintaining intracellular redox homeostasis by maintaining a pool of reduced glutathione which neutralizes reactive oxygen species (ROS). Cancer cells frequently have an increased burden of oxidative stress (16,17) and are likely to be more sensitive to the additional oxidative damage promoted by ROS. Multiple studies indicate that tumors are particularly dependent on redox-regulating pathways and that this vulnerability can be exploited by targeted therapeutics (18,19). In recent years, several kinases have been identified that regulate the sensitivity of cancer cells to paclitaxel by inhibiting centrosome splitting (20) or enhancing microtubule stability (21,22). Much less attention has been given to kinases that affect paclitaxel sensitivity by modulating cancer cell metabolism.
In earlier studies, we found that 20% of potential kinase targets whose knockdown modulates paclitaxel sensitivity participate in glucose and energy metabolism (23). In the present study, we targeted genes engaged in glucose and energy metabolism in ovarian and breast cancer cell lines and were able to validate 4 genes (CKMT1B, CKMT2, PFKFB2 and PFKFB4) whose knockdown reduced cancer cell survival and enhanced paclitaxel sensitivity in at least 3 of 6 cancer cell lines tested. CKMT1B and CKMT2 are mitochondrial creatine kinase isoenzymes which catalyze the reversible transfer of phosphate groups from phosphocreatine to ADP to yield ATP and creatine. CKMT1B and CKMT2 are the key molecule for oxidative phosphorylation and apoptosis in mitochondria. PFKFB2 and PFKFB4 belong to a family of bifunctional enzymes that control the levels of fructose 2,6-bisphosphate. Several studies have demonstrated that some cancer cell lines produce markedly elevated level of Fru-2,6 bisphosphate (24,25). Among these 4 genes, a leading candidate was 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), an isoform of the glycolytic enzyme phosphofructokinase (PFK2). Analysis of this gene’s function in ovarian and breast cancers revealed that PFKFB2 maintains intracellular redox balance which attenuates TP53-dependent apoptotic cell death. Knockdown of PFKFB2 increased the glycolysis rate, but decreased the flow of intermediates through the pentose-phosphate pathway in cancer cells with wtTP53, decreasing NADPH. ROS accumulated after PFKFB2 knockdown, which stimulated JNK and p53 phosphorylation, induced apoptosis that depended upon upregulation of p21 and Puma.
Materials and Methods
Cell lines and cell culture
Cell lines used in this manuscript are listed in Table S1. The identity of all cell lines was confirmed with STR DNA fingerprinting (12/18/2018) in the MDACC Characterized Cell Line core (supported by NCI P30CA016672). Cell line mycoplasma test was performed using an ATCC PCR kit on 11/20/2018.
Custom siRNA Screen and data analysis
A confirmatory screen with 12 positive hits (CKM, CKMT1B, CKMT2, GCK, PDK3, PDK4, PFKFB2, PFKFB3, PFKFB4, PFKL, PFKM and PGK10), which were selected from our initial screen, was conducted in 4 ovarian cancer (A2780, HeyA8, OC316 and OVCAR8) and 2 breast cancer (MCF-7 and ZR-75–1) cell lines. The same screening procedures were applied as in the primary screen (23) and reagents used in the screen were listed in Table S2.
Individual siRNA transfection
ON-TARGETplus PFKFB2 siRNA #6, 7, 8 and 9, TP53 siRNA #14, 15 and 16, SMARTpool G6PD siRNA and Non-targeting siRNA #2 were purchased from Dharmacon (Lafayette, CO). 30 nM of siRNA and 1.5‰ DharmaFECT 4 (Dharmacon, Lafayette, CO) were diluted in OPTI-MEM medium individually and then mixed together for 20 min at room temperature. Cells were laid on top of siRNA-DharmaFECT mixture in 6-well plates and incubated for 24, 48 or 72 hrs before analyses. We validated all siRNA oligos in multiple cell lines and discovered the knockdown efficiency of siPFKFB2 #6 and #7 is better than #8 and 9, and siTP53 #14 is better than #15 and 16.
Cell viability assays
3–8×103 Cells were reversely transfected with a mixture of 20 nM of siRNA and 1.5‰ DharmaFECT 4 in triplicate wells of 96-well plates. 24 hrs post transfection, cells were treated with a serial dilution of paclitaxel and incubated for another 72 hrs. Cells were fixed with 10% trichloroacetic acid at 4ºC for 30 min. Plates were rinsed with distilled H2O and 100uL of 0.4% sulforhodamine B (SRB) in 1% acetic acid was added to each well. Plates were incubated at room temperature for 30 min, and then rinsed with 1% acetic acid to remove excessive dye. SRB was solubilized with 100 μL of 10mM Tris buffer at pH8 for 5 min with shaking. Absorbance was determined at 570 nm on a Synergy 2 microplate reader from BioTek (Winooski, VT). Paclitaxel dose response curves were plotted and IC50 was determined by GraphPad Prism 7 (San Diego, CA).
Clonogenic assay
400–800 Cells were seeded in triplicate wells of 6-well plates 24 hrs post siRNA transfection. Cells were incubated for up to 14 days until colonies became visible. Cells were washed twice with PBS, fixed in 0.1% Brilliant Blue R with 10% v/v acetic acid and 30% v/v methanol for 1 min and washed with tap water until background was clear. Colonies with more than 50 cells were counted.
Reactive Oxygen Species (ROS) assays
1.5–3×105 Cells were transfected with control or PFKFB2 siRNA for 24–72 hrs and then incubated either with 10 μM CM-H2DCFDA (5- and 6-chloromethyl-2’,7’-dichlorodihydrofluorescein diacetate acetyl ester, Thermo Fisher, Waltham, MA) in PBS for 30 min or with 10 μM dihydroethidium (Thermo Fisher) in PBS for 1 hr in a 37°C incubator. Cells were then washed and suspended in 0.5 mL of PBS. Florescence was measured in a flow cytometer using the FITC and PE-Texas Red channels.
Apoptosis
Apoptosis was detected with the Alexa Fluor® 488 Annexin V/Dead Cell Apoptosis kit (Thermo Fisher). Cells were reversely transfected with siRNA for 24 hrs and treated with paclitaxel or diluent for 48 hrs. Cells were stained with 0.1 mL of Annexin binding buffer containing 5 μL of Annexin V Alexa 488 conjugate and 1μg/mL of PI for 15 min at room temperature (Thermo Fisher). Stained cells were read on Gallios analyzer (Beckman Coulter, Atlanta, GA) and 20,000 events were counted.
RNA extraction and RT-qPCR analysis
Forty eight hours post siRNA transfection, cells were lysed in QIAzol and total RNA was extracted using an RNeasy kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions.cDNA was synthesized from 1 μg of RNA using the Superscript II First Strand Synthesis Kit (Thermo Fisher). qPCR was performed using CFX Connect Real-time System (Bio-Rad, Hercules, CA) in a total volume of 20 μL, which included 10 μL of 2X iTaq Universal PCR master mix, 300 nM of each forward and reverse primers, and 40 ng of cDNA. Puma primers are forward 5’-CACCTAATTGGGCTCCATCTC-3’ and reverse 5’-GACGACCTCAACGCACAGTA-3’. GAPDH primers are forward 5’-TCGACAGTCAGCCGCATCTTCTTT-3’ and reverse 5’-ACCAAATCCGTTGACTCCGACCTT-3’.
ECAR measurements
Extracellular acidification rates (ECAR) were measured using a Seahorse XF24 analyzer (Agilent Technologies, Santa Clara, CA). 1.5–4×104 Cells were seeded into XF24 cell culture microplates and then transfected with siRNA at 24, 48 and 72 hrs prior to analysis. Cells were gently washed and incubated in XF assay media with pH 7.4 at 37°C without CO2 for 30 min. ECAR was detected under basal conditions followed by the sequential addition of 10 mM glucose, 2 μM oligomycin and 50 mM 2-deoxy-glucose (2-DG) (XF Glycolysis Stress Test kit, Agilent). pH levels were measured three times using XF24 Extracellular Flux Analyzer after the injection of stressing reagents. ECAR data were normalized to cell number of each sample and glycolysis rate was calculated with a formula: Glycolysis = ECAR after addition of glucose – ECAR after 2-DG treatment (26).
Metabolomics
Sample preparation and LC-MS/MS analysis followed a published protocol with modifications (27). Cells were transfected with control or PFKFB2 siRNA for 28 hrs and then washed twice with cold PBS. Cells were lysed on dry ice with 3 mL of cold extraction solvent (80% methanol containing 13C6-G6P and 13C3-pyruvate as internal standard) and were incubated at −80 °C for 30 min. Cells were then scraped and lysates were transferred to 15 mL pre-chilled conical tubes. Cells were pelleted at 2000 RPM for 5 min and the supernatant was transferred into pre-chilled 15 mL conical tubes. The pellet was washed with 1 mL of the cold extraction solvent and then centrifuged again at 2000 RPM for 5 min. Supernatant (1 mL) was transferred into Eppendorf tubes and dried down in a speed vacuum concentrator. The residues were reconstituted with 50% ACN/H2O (100 uL), vortexed, and centrifuged for 10 min at 15,000 rpm at 4°C. The supernatant (80 uL) was transferred into HPLC vials for LC-MS/MS analysis. Samples were analyzed on an Agilent 1290 infinity LC system coupled with an Agilent 6460 triple quadrupole mass spectrometer operated in a negative mode.
G6PD and NADPH/NADP+ measurements
The enzyme activity of glucose-6-phosphate dehydrogenase was measured using a G6PD activity colorimetric assay kit (BioVision, Milpitas, CA). Cells were homogenized in 500 μL of cold PBS. Cell lysates were assayed following the manufacturer’s instructions. The concentrations of NADP+ and NADPH were determined with a NADP / NADPH-Glo™ Assay (Promega, Madison, WI). Cells were transfected with siRNA in 96-well plates. 48 hrs post transfection, cells were lysed and treated under heat and acid conditions to separate NADPH and NADP+ components and lysates were incubated with luminescent substrate. NADPH and NADP+ concentrations were determined by NADPH and NADP+ standard curves, respectively. The NADPH/NADP+ ratio was calculated to represent cellular redox potential.
Immunohistochemistry
Tissue slides were subjected to immunohistochemical staining according to the manufacturer’s protocol (Biocare Medical, Concord, CA). Six-micron sections were cut. Slides were baked at 60°C overnight, and deparaffinized in two changes of xylene. After rehydration, antigen was retrieved in 10 mM sodium citrate buffer (pH 6.0) at 125 °C for 10 min. Slides were blocked with PeroxAbolish, avidin, biotin, 3% bovine serum albumin (BSA) in PBS and Rodent Blocking M for xenografts. Slides were incubated at 4°C overnight with primary antibodies indicated in figures (sources of antibodies were listed in Table S3), followed by biotinylated secondary antibody and streptavidin conjugated HRP. Slides were stained in DAB chromogen for 1 min and then counterstained with hematoxylin. Tissue microarray was scored and analyzed by two investigators (H.Y. and Z.L.) independently.
Establishment of MCF7 and HIM3 shPFKFB2 inducible cell lines
MCF7 and HIM3 breast cancer cells were infected with SMARTvector Tet-inducible shPFKFB2 lentivirus (Clone ID V3SH7669–224728719) and subsequently selected using 1 μg/mL of puromycin according to the manufacturer’s protocol (Dharmacon). Clonal populations were generated by limiting dilution under puromycin selection. Clones with best knockdown efficiency were selected by Western blotting under 1 μg/mL of doxycycline for 48 hrs. MCF7 and HIM3 inducible cells were maintained in DMEM (Corning, Corning, New York) supplemented with 10% FBS and 0.5 μg/mL of puromycin (Sigma, St. Louis, MO).
Growth of human ovarian and breast cancer xenografts in mice
Experiments with Hsd:Athymic nude-Foxn1nu homozygous mice (Envigo) and nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Charles River) were reviewed and approved by the Institutional Animal Care and Use Committee of M. D. Anderson Cancer Center (IACUC 00001052). For ovarian cancer xenografts, 60 female nude mice were injected intraperitoneally (i.p.) with 1×106 A2780 cells or 1×106 HeyA8 cells. After 7-day inoculation, mice were randomly assigned to 4 treatment groups (n = 10) as indicated. In the MCF7 breast cancer xenograft study, mice were implanted with 17β-estradiol pellets (0.5 mg per pellet, 60-day release; Innovative Research of America, Sarasota, FA) under 2% isofluorane. 3×106 MCF-7 cells suspended in 100 μL of equal part of PBS and matrigel with reduced growth factor (Corning) were injected into the fourth mammary fat pads three days after estradiol pellet implantation. When xenografts reached 50 mm3, tumor-bearing mice were randomly divided into 4 groups as indicated (n=7). In the HIM3 xenograft study, xenograft models were established according to published protocols (28). 2.5×105 Irradiated immortalized human mammary stromal fibroblasts were mixed with 2.5×105 non-irradiated cells. Fibroblasts were then mixed with 1×106 tumor cells and 1/3 volume of Matrigel. This mixture was injected into the fourth mammary fat pads of 4 week-old NOD/SCID mice. After xenografts reached 50 mm3, tumor-bearing mice were randomly divided into 4 groups as indicated (n=13; 8 for tumor growth and 5 for IHC). For IHC analysis, mice were sacrificed and tumors were obtained after one week of doxycycline treatment. Tumor sizes were measured every week in two dimensions using a caliper, and tumor volume was calculated with the following formula: tumor volume (mm3) = 0.5 × ab2 (a and b being the longest and the shortest diameters of the tumor, respectively). Mice were monitored until tumor burden reached 1000 mm3 for euthanasia.
Statistical analysis
Data are represented as means +/− standard deviations unless specified otherwise. Statistical significance was determined using an independent sample Mann-Whitney U test. Survival was analyzed by the Kaplan-Meier method and compared using Logrank (Mantel-Cox) test. All experiments were performed in triplicate and repeated at least 2–3 time. The minimal level of significance was p=0.05.
Results
PFKFB2 modulates paclitaxel sensitivity in ovarian and breast cancer cells with wild-type TP53 (wtTP53)
We previously performed siRNA kinome-screens to identify kinases that modulate paclitaxel resistance in ovarian cancer cells (23). To identify targetable molecules that alter paclitaxel sensitivity in ovarian and breast cancer cells, we conducted a confirmatory screen in multiple cancer cell lines, knocking down the 12 positive hits from our initial screen that were known to participate in glucose and energy metabolism. Each of the 12 was knocked down in 4 ovarian and 2 breast cancer cell lines to determine whether decreased expression of each kinase would enhance paclitaxel sensitivity (z-score <−0.5) in at least 3 of the cancer cell lines. Of the 12 kinases screened, knockdown of 4 kinases (CKMT1B, CKMT2, PFKFB2 and PFKFB4) with siRNAs were shown to have significant effects in at least 3 of 6 ovarian and breast cancer cell lines. Knockdown of these 4 kinases preferentially enhanced paclitaxel sensitivity in ovarian and breast cancer cell lines that preserve wtTP53 (HeyA8, A2780 and ZR-75–1) compared to cell lines that contained mutant or null TP53 (SKOv3ip, OC316 and BT-20) (Figure S1A and Table S4).
PFKFB2 is overexpressed in a fraction of ovarian and breast cancers
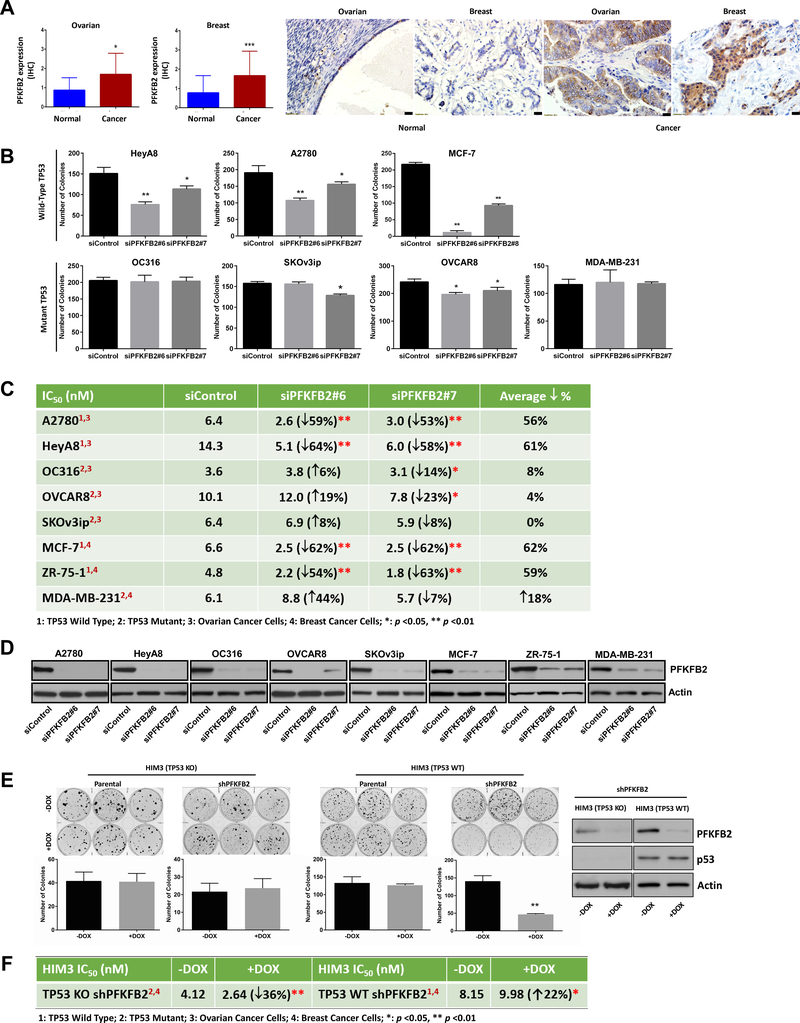
Gene expression of the 12 kinases was examined using the TCGA database. Only PFKFB2 was overexpressed in a fraction of both ovarian (10%) and breast cancers (26%) (Figure S1B) (29–31). Of the 10% ovarian cancer samples, 4.2% of samples are wtTP53 and of the 26% breast cancer samples, 49.4% of samples are wtTP53. PFKFB2 is a bifunctional glycolytic enzyme that regulates the level of fructose-2,6-bisphosphate (Fru-2,6-BP), a strong allosteric activator of phosphofructokinase 1 (PFK1) (32,33), one of the most important regulatory enzyme of glycolysis. Consequently, PFKFB2 was chosen for further investigation. To assess the prevalence of PFKFB2 overexpression in ovarian and breast cancers, PFKFB2 protein expression was measured by immunohistochemical analysis, in ovarian and breast cancer tissue microarrays (TMA) prepared from specimens obtained at the time of initial surgery. The expression of PFKFB2 was significantly upregulated in cancer cells compared to normal ovarian/fallopian (p=0.026) and breast epithelial cells (p<0.0001) (Figure 1A, S2 and Table S5–6). Intense staining (3–4+) was observed in 25% of ovarian cancers and 25% of breast cancers, but in 0% of normal ovarian/fallopian epithelium and 4.3% of normal breast epithelial cells. In addition, differential expression of PFKFB2 was shown between ovarian and breast cancers (intense staining) and all other normal organ tissues (Figure S3).
Figure 1. PFKFB2 protein is overexpressed in a fraction of ovarian and breast cancers and regulates tumor cell growth and paclitaxel sensitivity in ovarian and breast cancer cell lines.
(A) Expression of PFKFB2 was measured by immunohistochemical staining of two tissue microarray (TMA) sections that contained cores from 225 primary ovarian cancers and 142 primary breast cancers, respectively. The staining intensity was scored on a scale of 0 to 4 in comparison with staining observed in normal ovarian surface/fallopian epithelial cells and normal mammary epithelial cells (Figure S1). The detailed patient information was listed in Table S5 (ovarian) and S6 (breast). The columns indicate the mean of expression, and the bars indicate the S.D. (* p<0.05; *** p<0.001 cancer versus normal). Representative normal and cancer images are presented next to the bar graphs. Scale bars: 20 μm. (B) Clonogenic assays were performed in ovarian and breast cancer cells with or without PFKFB2 siRNAs. Cells were transfected with two different PFKFB2 siRNAs (#6 and #7) for 24 hrs, and then re-plated in 6-well plates, at a density of 400–800 cells/well. Data were obtained from three independent experiments. The columns indicate the mean of colony numbers, and the bars indicate the S.D. (* p<0.05; ** p<0.01). (C) Paclitaxel sensitivity was measured in ovarian and breast cancer cells with or without PFKFB2 siRNAs. Cells were transfected with PFKFB2 or control siRNA for 24 hrs followed by treatment with serial diluted paclitaxel for an additional 72 hrs. Cell viability was measured with SRB assays. The IC50 value of paclitaxel for each cell line was determined using GraphPad Prism. Means of IC50 were obtained from three independent experiments, (* p<0.05; ** p<0.01). (D) siRNA knockdown efficiency for paclitaxel sensitivity experiments described in (C) was measured by Western blot analysis. (E) Clonogenic assays were performed in HIM3 TP53 isogenic PDX cell lines. shRNA was induced by addition of DOX. Knockdown of PFKFB2 was shown in the Western Blots to the right. The columns indicate the mean and the bars indicate the S.D. (** p<0.01). Data were obtained from three independent experiments. (F) Paclitaxel sensitivity represented by IC50 values for HIM3 cell lines were calculated as described in (C).
Silencing PFKFB2 inhibits cancer cell growth and increases sensitivity to paclitaxel
To validate the siRNA screen results and address whether high expression of PFKFB2 is one of the important factors contributing to tumorigenic phenotype in ovarian and breast cancers, we performed clonogenic assays after PFKFB2 siRNA or control siRNA transfection. PFKFB2 siRNA significantly inhibited cell growth, when compared to control siRNA. Notably, the most marked effects of silencing PFKFB2 on cell proliferation were seen in HeyA8, A2780 and MCF-7 ovarian and breast cancer cell lines that express wtTP53, with more modest inhibition observed in the OC316, SKOv3ip, OVCAR8 and MDA-MB-231 cells that express mutant or null p53 (Figure 1B and S4A). Moreover, the ability of PFKFB2 knockdown to sensitize ovarian and breast cancer cells to paclitaxel was greater in HeyA8, A2780, MCF-7 and ZR-75–1 than in OC316, OVCAR8, SKOv3ip and MDA-MB-231 cells. When the concentrations of paclitaxel required to inhibit cancer cell growth by 50% (IC50) were compared with and without PFKFB2 knockdown, silencing PFKFB2 using two individual PFKFB2 siRNAs in the HeyA8, A2780 MCF-7 and ZR-75–1 cell lines decreased cell viability and reduced the average paclitaxel IC50 by 2.5 fold (range = 2.3 – 2.6 fold) (Figure 1C and S4B). However, when the IC50 of paclitaxel was measured in OC316, OVCAR8, SKOv3ip and MDA-MB-231 cells with mutant or null TP53, silencing PFKFB2 with a single siRNA reduced the average paclitaxel IC50 by only 0.95 fold (range = 0.8 – 1 fold) (Figure 1C and S4B). Similar knockdown efficiencies were observed across both sets of cell lines. (Figure 1D). In addition, we used paired isogenic patient derived xenograft (PDX) lines differing only in TP53 (ER-PR-HER2-TP53 wild-type and a TP53 deficient subline) (28), to evaluate the cancer cell growth and paclitaxel sensitivity in cell culture. Consistent with the above data, we observed significant growth inhibition (Figure 1E) and paclitaxel sensitization (Figure 1F) with the wtTP53 parental cell line HIM3 TP53 WT, but not with the TP53-deficent subline HIM3 TP53 KO by addition of doxycycline (DOX) to silence PFKFB2 in cultured cells. Conversely, overexpression of PFKFB2 increased cell growth and reduced sensitivity to paclitaxel, increasing the IC50 of paclitaxel (p<0.05 in HeyA8 and p<0.01 in MCF-7), when HeyA8 and MCF-7 cells were transiently transfected with a PFKFB2 plasmid and selected with G418 (Figure S4C). Thus, these observations support the hypothesis that high expression of PFKFB2 can facilitate cell proliferation and paclitaxel resistance in ovarian and breast cancer cells that express wtTP53.
Knockdown of PFKFB2 increases glycolysis and reduces shunting of glycolytic intermediates to the oxidative pentose phosphate pathway (PPP) only in ovarian and breast cancer cells with wtTP53
Increased glucose metablism in cancer cells is required to meet their high energetic and biosynthethic demands. Many glycolytic enzymes, including PFKFB2, are induced in malignant cells (34) and PFKFB2 is throught to play an important role in regulating carbon flux through the glycolytic and pentose phosphate pathways (PPP) (Figure 2A) (35,36). To understand the metabolic basis for the therapeutic effect of silencing PFKFB2 on tumor cell growth and sensitivity to paclitaxel, we have studied the mechanism(s) by which silencing PFKFB2 alters glycolysis and the PPP.
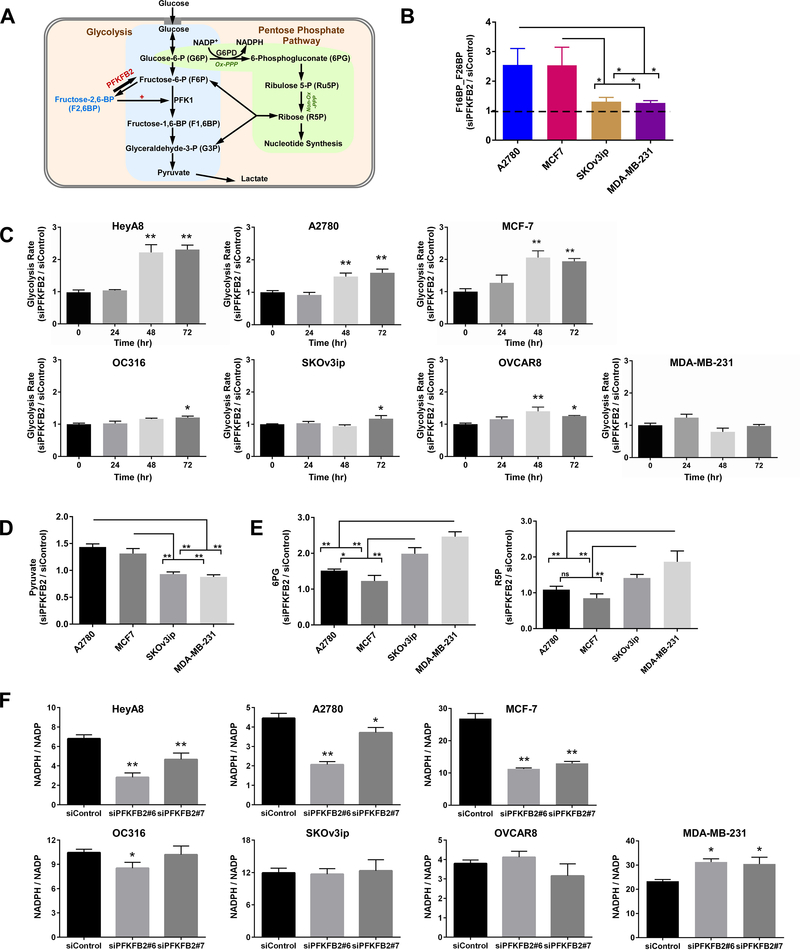
Figure 2. Identification of the mechanism(s) by which silencing PFKFB2 alters glycolysis and the pentose phosphate pathway (PPP).
(A) Working model demonstrating the regulation of glycolysis and pentose phosphate pathway by PFKFB2 in cancer cells. (B) Measurement of F1,6BP and F2,6BP. The concentrations of metabolites were measured by LC-MS in ovarian and breast cancer cell lines with or without depletion of PFKFB2. Cells were transfected with PFKFB2 siRNA for 28 hrs and then cell lysates were collected for the analysis. Because of technical limitations, we measured the mixture of F1,6BP and F2,6BP. The columns indicate the relative concentrations, and the bars indicate the S.D. (* p<0.05). Data were obtained from two independent experiments. (C) Measurement of glycolytic flux. The glycolytic flux was detected in 7 ovarian and breast cancer cell lines with or without depletion of PFKFB2 using a Seahorse glycolysis stress assay. The columns indicate the mean of lactate excretion, and the bars indicate the S.D. (* p<0.05 compared with “0” time point). Data were obtained from two independent experiments. (D and E) Measurement of pyruvate, 6PG and Ru5P. The concentrations of metabolites were measured by LC-MS in ovarian and breast cancer cell lines with or without depletion of PFKFB2. Cells were transfected with PFKFB2 siRNA for 28 hrs and then cell lysates were collected for analysis. The columns indicate the concentrations, and the bars indicate the S.D. (* p<0.05). Data were obtained from two independent experiments. (F) Determination of the ratios of NADPH/NADP+. NADPH and NADP+ were quantified with a Promega NADP/NADPH-Glo™ Kit in 7 ovarian and breast cancer cell lines with or without trasfection of siPFKFB2 for 48 hrs. The columns indicate the mean, and the bars indicate the S.D. (* p<0.05 compared with siControl). Data were obtained from two independent experiments.
Initially, we measured the concentration of fructose-2,6-bisphosphate in two ovarian cancer cell lines and two breast cancer cell lines by liquid chromatography-mass spectrometry (LC-MS) with or without silencing PFKFB2. Knockdown of PFKFB2 increased fructose-2,6-bisphosphate and fructose-1,6-bisphosphate (F1,6BP / F2,6BP) (siPFKFB2 / siControl > 1) in all the cell lines tested (Figure 2B), consistent with increased glycolysis. The relative concentrations of F1,6BP + F2,6BP were considerably higher in the A2780 and MCF-7 ovarian and breast cancer cell lines that express wtTP53 than those in the SKOv3ip and MDA-MB-231 cell lines that express mutant TP53 (Figure 2B). Consistent with these results, studies with the Seahorse XF glycolysis stress assay demonstrated a higher glycolytic flux in HeyA8, A2780 and MCF-7 cells than in OC316, SKOv3ip and MDA-MB-231 cells (Figure 2C and S5A). Moreover, pyruvate, the end product of glycolysis, was significantly higher in wtTP53 cell lines than that in TP53 mutant or null cell lines after depletion of PFKFB2 (Figure 2D and S5B). In contrast, we measured two oxidative PPP products by LC-MS (6-phosphoglyconate, 6PG, and ribulose 5-phosphate, Ru5P) and found that silencing PFKFB2 increased the concentrations of both (Figure 2E) in SKOv3ip, and MDA-MB-231 cells compared to A2780 and MCF-7 cell lines, suggesting that knockdown of PFKFB2 resulted in the shunting of glycolytic intermediates to the oxidative PPP in cancer cells with mutant TP53.
One possible consequence of increasing fructose-2,6-bisphophate (F1,6BP/F2,6BP) would be an increase in PFK1 activity. It would be predicted to increase glycolytic flux and decrease the amount of glucose-6-phosphate (G6P) entering the PPP which would reduce the biosynthesis of NADPH. On the other hand, higher levels of 6PG and Ru5P would imply higher oxidative PPP flux and increased production of NADPH. To test this, we measured NADPH and NADP+ using cancer cell lysates and found that silencing PFKFB2 decreased the ratio of NADPH/NADP+ (Figure 2F and S5C) in A2780, HeyA8 and MCF-7 cells with wtTP53. In contrast, silencing PFKFB2 produced minimal changes in the NADPH to NADP+ ratio in OC316, SKOv3ip, OVCAR8 and MDA-MB-231 cancer cells with mutant TP53. Thus, oxidative PPP flux was significantly upregulated in TP53 mutant cell lines, however it was downregulated in wtTP53 cell lines after depletion of PFKFB2 by siRNA.
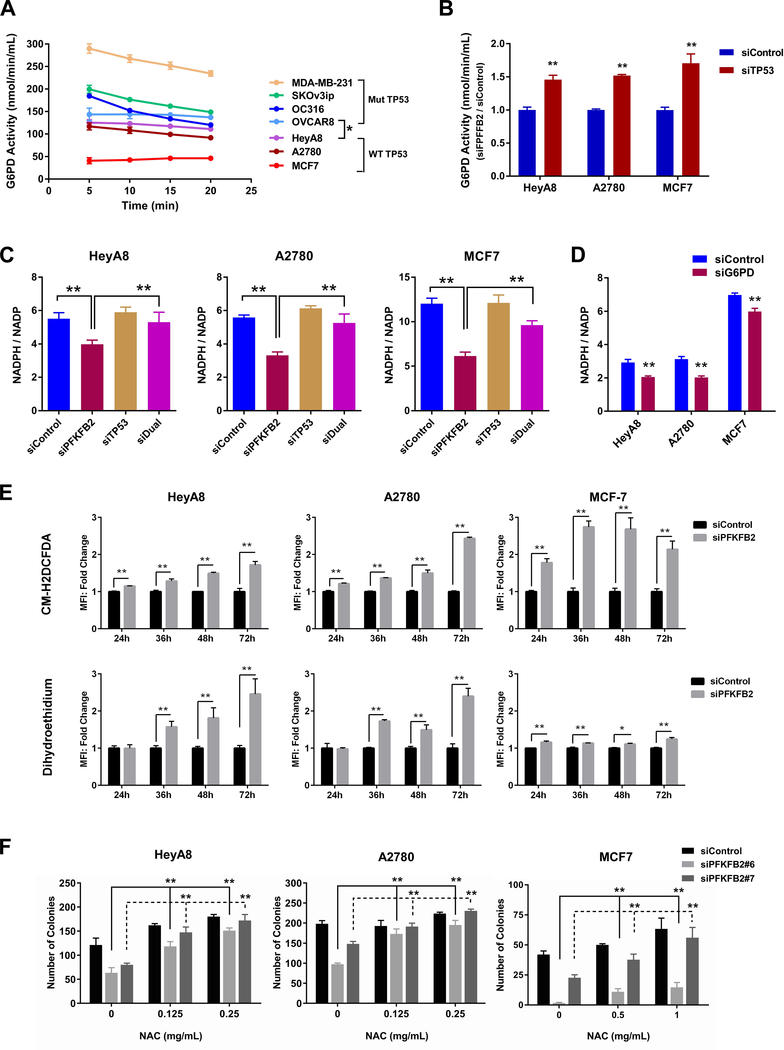
Downregulation of PPP by silencing PFKFB2 depends upon suppression of G6PD activity by wtp53
To document differences in response to depletion of PFKFB2 in TP53 wild-type and TP53 mutant cells, the basal activity of G6PD was measured in HeyA8, A2780, MCF-7, OC316, SKOv3ip, OVCAR8 and MDA-MB-231 cells. We found that basal activity of G6PD was much lower in A2780, HeyA8 and MCF-7 cancer cells with wtTP53 (Figure 3A) than in OC316, SKOv3ip, OVCAR8 and MDA-MB-231 with mutant TP53 (Figure 3A). This low basal activity was significantly increased after knockdown of TP53 in TP53 wild-type cell lines (Figure 3B and S6A). In line with these results, decreased NADPH was observed in wtTP53 cancer cells following knockdown of PFKFB2, and the reduction in NADPH after PFKFB2 knockdown was rescued by knockdown of TP53 (Figure 3C and S6B) and decreased NADPH was also observed in TP53 wild-type cancer cells following knockdown of G6PD (Figure 3D and S6C). These results are consistant with our observation that silencing PFKFB2 in wtTP53 cell lines resulted in increased glycolysis (Figure 2D) and decreased PPP flux (Figure 2E). Taken together, these results suggest that tumor-associated TP53 mutants in high grade ovarian cancer which lack G6PD-inhibitory activity may promote tumor cell proliferation by directing glucose towards PPP flux and NADPH production. In contrast, ovarian cancers with wtTP53 may attenuate PPP activity and instead channel glucose through glycolysis to generate ATP and pyruvate for TCA cycle and OxPhos metabolism (Figure S6D).
Figure 3. PFKFB2 promotes ovarian and breast cancer survival through a p53-dependent pathway.
(A and B) Measurement of G6PD in ovarian and breast cancer cells. The basal activity of G6PD (A) was measured at different incubation times. The enzymatic reaction was saturated after 20 mins. Each point represents the mean ± S. D. of product concentration (* p<0.05 compared to TP53 wild-type cell lines). G6PD Activity with or without TP53 siRNA was measured in (B) at 15-min incubation. (** p< 0.01 compared to siControl). Data were obtained from two independent experiments. (C and D) NADPH/NADP+ measurements were conducted in ovarian and breast cancer cells with PFKFB2 and/or TP53 siRNA (C) and with G6PD siRNA (D). Cells were transfected with targeting or control siRNA for 48 hrs prior to analyses. (E) Detection of ROS in ovarian and breast cancer cells. Cells were transfected with PFKFB2 siRNA for different time intervals as indicated, then cells were harvested and incubated with ROS detection dyes (CM-H2DCFDA and dihydroethidium). Cells were then washed prior to flow cytometric analysis. Mixture of siPFKFB2 #6 and #7 was used in this experiment. The columns indicate the fold change, and the bars indicate the S.D. (** p<0.01). Data were obtained from three independent experiments. (F) Clonogenic assays were conducted in ovarian (HeyA8 and A2780) and breast (MCF7) cancer cells with or without PFKFB2 siRNA in the absence or presence of the ROS inhibitor (NAC). Cells were transfected with PFKFB2 siRNA for 24 hrs, and then re-plated into 6-well plates with culture medium supplemented with increasing concentrations of NAC as indicated. The medium supplemented with NAC was refreshed daily. Cells were incubated for up to 14 days and viability was measured by colony counts. Data were obtained from three independent experiments. The columns indicate the mean of colony numbers, and the bars indicate the S.D. (** p<0.01).
ROS accumlates in ovarian and breast cancer cells with wtTP53, suggesting a mechanism by which PFKFB2 siRNA induces growth inhibition
To explore whether decreased levels of NADPH following PFKFB2 silencing result in ROS accumulation and decreased viability in ovarain and breast cancer cells, ROS was measured using two fluorescent dyes, CM-H2DCFDA and dihydroethidium. Fluoresence intensity of the two dyes was enhanced by silencing PFKFB2, indicating the accumulation of ROS. However, more significent accumulation of ROS was obeserved in HeyA8, A2780, and MCF-7 cells with wtTP53 (Figure 3E) than that in OC316, SKOv3ip and MDA-MB-231 cells with mutant TP53 (Figure S7A). These results are consistent with the hypothesis that ROS accumulation may result from diminshed pools of intracellular NADPH in TP53 wild-type ovarian and breast cancer cell lines. To measure the effect of increasd ROS on ovarian cancer growth, we performed clonogenic assays in the presence and absence of N-acetyl-L-cysteine (NAC), an ROS inhibitor. NAC rescued PFKFB2 knockdown-mediated growth inhibition in HeyA8 and A2780 cancer cells (Figure 3F and S7B). In contrast, NAC had little effect on OC316, SKOv3ip and OVCAR8 cells (Figure S7C). These studies support the interpretation that PFKFB2 knockdown affects the proliferation of ovarian cancer cells with wtTP53 in an ROS-dependent manner.
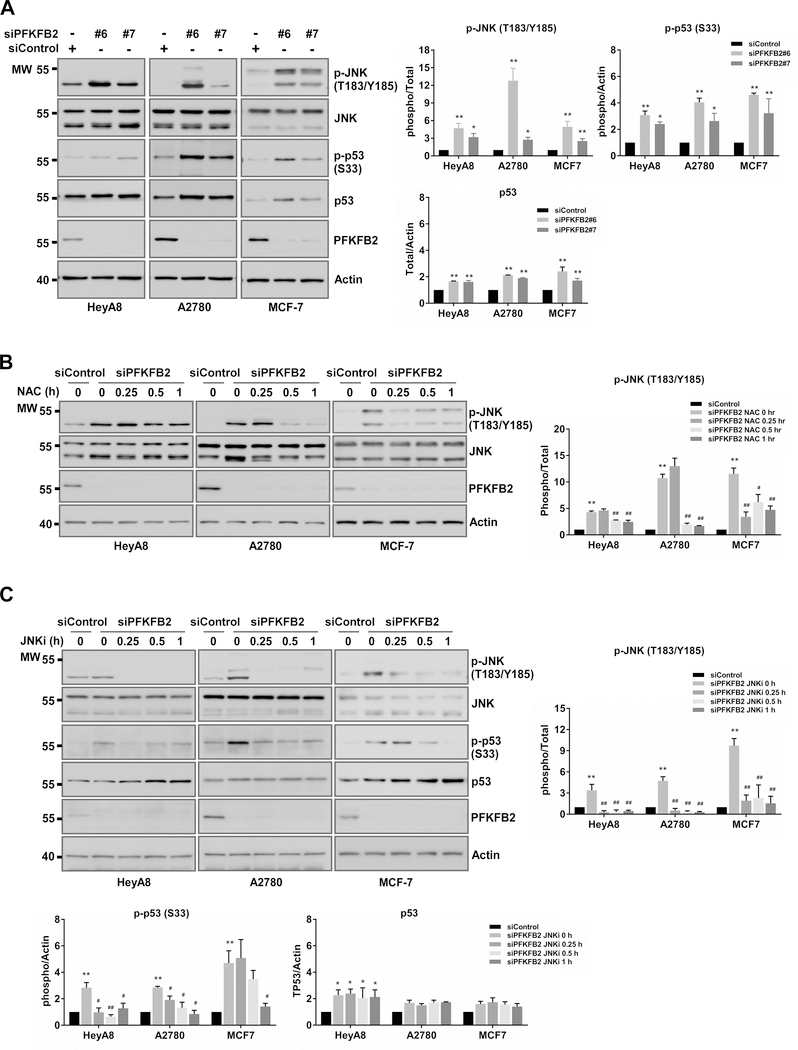
ROS-dependent phosphorylation of Jun N-terminal kinase (JNK) leads to p53 activation in ovarian and breast cancer cells after knockdown of PFKFB2
Previous studies indicate that oxidative stress can trigger the JNK pathway and the phosphorylation of p53 (37,38). Therefore, we examined phosphorylation of JNK and phosphorylation of p53 by Western blot analysis in HeyA8, A2780, and MCF-7 cells using siRNA to silence PFKFB2. We found that silencing PFKFB2 induced expression of p53 and, more importantly, significantly increased the phosphorylation of both JNK and p53, suggesting that ROS induced-JNK phosphorylation led to p53 activation (Figure 4A). To confirm that p53 activation is triggered by ROS-mediated JNK phosphorylation, we investigated the effect of ROS and JNK inhibitors on p53 phosphorylation (Figure 4B–C). We observed that NAC (Figure 4B) and JNK inhibitors (Figure 4C) substantially impeded JNK and p53 phosphorylation after PFKFB2 silencing in HeyA8, A2780 and MCF-7 cells. This result confirms that JNK is a mediator of p53 activation in response to oxidative stress resulting from downregulation of PFKFB2. Collectively, our data suggest that p53 is a crucial node downstream of ROS / JNK in the molecular pathway, inhibiting growth in ovarian and breast cancers.
Figure 4. After knockdown of PFKFB2, ROS-dependent phosphorylation of Jun N-terminal kinase (JNK) leads to p53 activation in ovarian and breast cancer cells.
(A) PFKFB2 siRNAs increases phosphorylation of JNK and p53. HeyA8, A2780 and MCF-7 cells were transfected with two different PFKFB2 siRNA (#6 and #7) for 48 hrs. Cell lysates were analyzed by Western blots with indicated antibodies. (B) N-Acetyl cysteine (NAC), an ROS scavenger, reduces JNK phosphorylation levels. 48 hrs post PFKFB2 siRNA transfection, cells were treated with 1 mg/mL of NAC for different time intervals as indicated. Mixture of siPFKFB2 #6 and #7 was used in this experiment. Cell lysates were analyzed on Western blots. (C) JNK inhibitor (JNKi) decreases p53 phosphorylation levels. Cells were transfected as described in (B) and then treated with 10 μM of JNKi (SP600125, Selleck Chemicals). The density of protein bands in A-C was determined by Image J (developed by NIH) and was plotted next to the blots. Data are represented as mean ± S. D. from three individual experiments (* p<0.05 and ** p<0.01 compared to siControl).
Both p53 and Puma and p21 are required for silencing PFKFB2 induced-apoptosis in ovarian and breast cancers
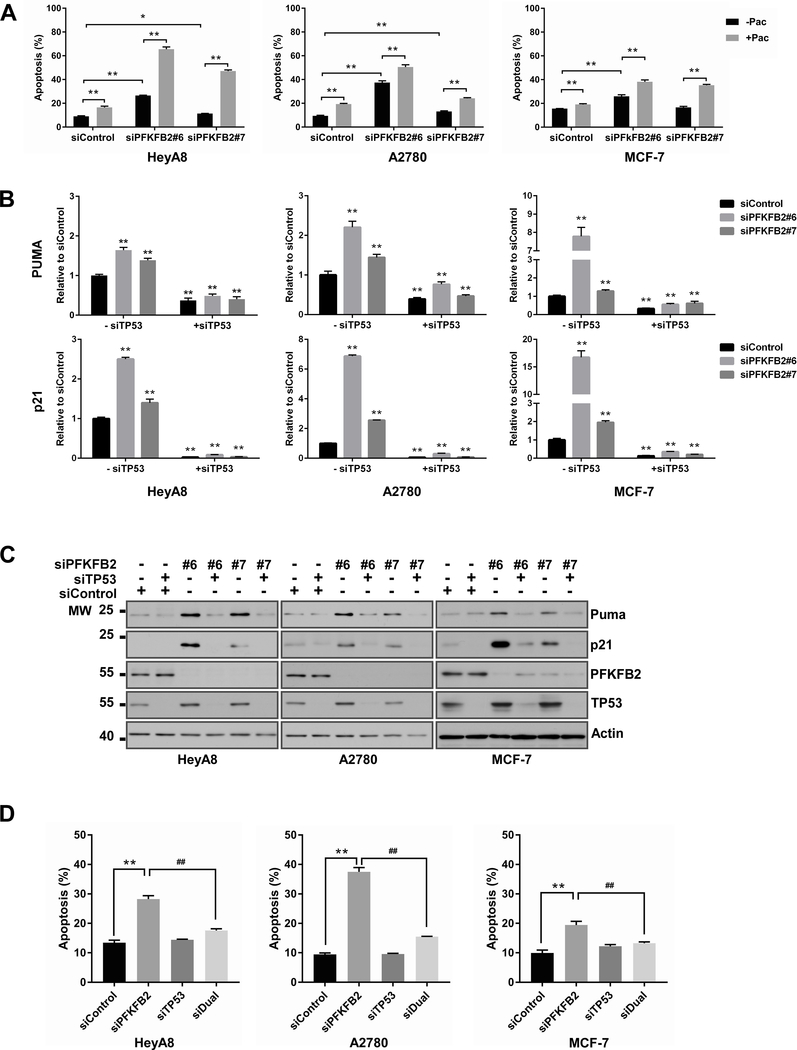
To identify possible mechanism(s) by which PFKFB2 knockdown enhances the cytotoxic activity of paclitaxel in ovarian and breast cancer cells, we first examined the effect of PFKFB2 siRNA on apoptosis in the presence and absence of paclitaxel. Flow cytometry revealed that PFKFB2 siRNA induced a higher percentage of annexin V positive cells, and the percentage was further increased when combined with paclitaxel (Figures 5A). We then tested whether p53 is a critical mediator in response to PFKFB2 knockdown-induced apoptosis. We measured the expression of Puma and p21, which are downstream targets of TP53 in HeyA8, A2780 and MCF-7 cells. The mRNA and protein levels of Puma and p21 were increased by silencing PFKFB2 (Figure 5B–C and S8), implying that PFKFB2 siRNA induced apoptosis in HeyA8, A2780 and MCF-7 cells is mediated by p53 activation. Moreover, TP53 knockdown decreased the mRNA and protein levels of Puma and p21 (Figure 5B–C and S8) and rescued cells from apoptosis induced by PFKFB2 knockdown (Figure 5D), confirming that p53, Puma and p21 are critical mediators of ROS/JNK induced apoptosis.
Figure 5. PFKFB2 inhibits p53-dependent apoptosis in ovarian and breast cancers.
(A) Apoptosis was measured in ovarian and breast cancer cells with or without PFKFB2 siRNA in the presence or absence of paclitaxel. HeyA8, A2780 and MCF-7 cells were transfected with PFKFB2 siRNA or control siRNA for 24 hrs followed by paclitaxel treatment (7 nM for A2780, 10 nM for HeyA8 and 8 nM for MCF7) for an additional 48 hrs (apoptosis) or 24 hrs (cell cycle). Cells were stained with Annexin V-Alexa 488 prior to flow cytometric analysis. Data were obtained from three independent experiments. The columns indicate the mean, and the bars indicate the S.D. (** p<0.01). (B) mRNA and (C) protein of Puma and p21 in HeyA8, A2780 and MCF-7 cells with or without depletion of PFKFB2 and/or TP53. Cells were transfected with siRNA for 48 hrs prior to RT-qPCR and Western blot analyses. Experiments were repeated three times. Quantification of Western blots are presented in supplementary Figure S8. (D) Measurement of apoptosis was conducted in ovarian and breast cancer cells with or without depletion of PFKFB2 and/or TP53. HeyA8, A2780 and MCF-7 cells were transfected with siRNA for 48 hrs. Cells were stained as described in (A) for flow cytometric analysis. Mixture of siPFKFB2 #6 and #7 was used in this experiment. Data were obtained from three independent experiments. The columns indicate the mean, and the bars indicate the S.D. (** p<0.01 compared to siControl; ## p<0.01 compared to siPFKFB2).
PFKFB2 siRNA or shRNA inhibits tumor growth and enhances sensitivity to paclitaxel in human ovarian and breast cancer xenografts and PFKFB2 shRNA prolongs survival in a triple negative breast patient-derived xenograft (PDX)
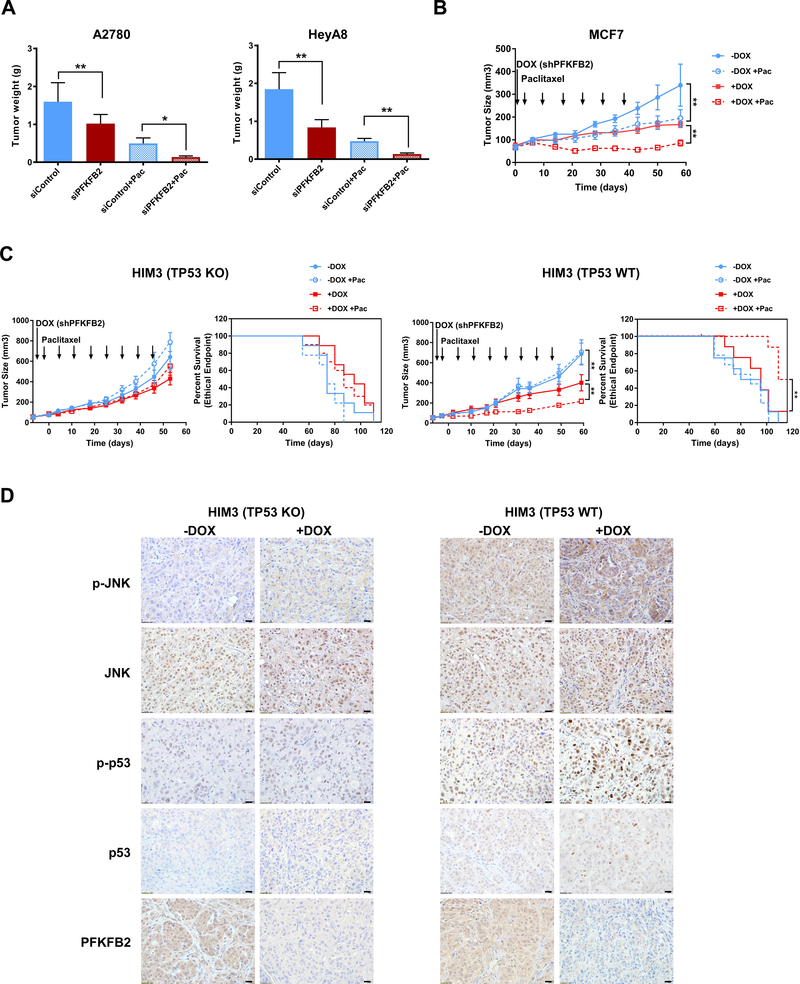
Given the outcome of cell culture studies, we asked whether depletion of PFKFB2 affects the ability of TP53 wild-type ovarian and breast cancer cells to form tumors in immunodeficient mice. Orthotopic human ovarian and breast xenograft models were used to determine the potential of PFKFB2 as a target for clinical therapy. We used a well-characterized DOPC (1,2-dioleoyl-sn-glycero-3-phosphatidylcholine) nanoliposomal system for in vivo siRNA delivery in mouse ovarian cancer models (20). For each cancer cell line, 4 groups of 10 mice were treated for 4 weeks as follows: 1) control siRNA-DOPC, 2) PFKFB2 siRNA-DOPC, 3) a combination of control siRNA-DOPC plus paclitaxel, 4) a combination of PFKFB2 siRNA-DOPC plus paclitaxel. Liposome encapsulated PFKFB2 siRNA significantly inhibited tumor growth in both ovarian cancer xenograft models (HEYA8 and A2780). Greater growth inhibition was observed when PFKFB2 siRNA was combined with paclitaxel than that with either single treatment (p<0.05) (Figure 6A). In a murine human breast cancer xenograft model, MCF-7, a lentiviral vector-mediated doxycycline (+DOX) inducible shRNA expression system was used to silence PFKFB2. After addition of DOX to drinking water, PFKFB2 was silenced and the inhibitory effect on tumor growth as a single agent or in combination with paclitaxel treatment was examined. shPFKFB2 (+DOX) or paclitaxel alone produced significant inhibition of tumor growth (p<0.01), compared to control (-DOX), moreover, shPFKFB2 plus paclitaxel produced greater inhibition of tumor growth than either single agent in this orthotopic breast cancer mouse model (p< 0.01) which is consistent with the ovarian cancer data (Figure 6B). To further evaluate the role of TP53 in PFKFB2 depletion mediated growth inhibition in vivo, we used paired isogenic patient derived xenograft (PDX) lines and assessed the outcome in xenograft models by inducing shPFKFB2 expression using DOX. DOX induction (silencing PFKFB2) plus paclitaxel significantly inhibited tumor growth, and prolonged survival of mice bearing HIM3 wtTP53 tumor (p=0.008), compared to mice bearing HIM3 TP53 Knockout (KO) tumor (Figure 6C). Moreover, the concentration of paclitaxel used in the experiments remained identical (5 mg/kg) in MCF-7 and HIM3 breast cancer xenograft studies. In the MCF-7 model, paclitaxel alone showed statistically significant inhibition of tumor growth as described above. However in the HIM3 wtTP53 model, which is a triple negative breast cancer, paclitaxel alone did not inhibit tumor growth; but when combined with PFKPB2 shRNA (+DOX), tumor growth was significantly inhibited with prolonged survival. As expected, p-JNK and p-p53 increased in HIM3 wtTP53 xenografts with depletion of PFKFB2 (+DOX) by immunohistochemistry (Figure 6D). In addition, HIM3 TP53 KO xenografts showed minimal changes in p-JNK and p-p53 with PFKFB2 depletion (Figure 6D). Taken together, these results demonstrate an important effect of knockdown of PFKFB2 and identify a potential therapeutic target to enhance paclitaxel sensitivity in breast cancers that retain functional TP53.
Figure 6. Knockdown of PFKFB2 significantly increases paclitaxel sensitivity in ovarian and breast cancer xenografts.
(A) Tumor growth of ovarian cancer cells in female athymic nude mice after treatment with PFKFB2 siRNA-DOPC with or without paclitaxel. 1 million HeyA8 or A2780 cells were injected intraperitoneally (ip). After 7-day inoculation, mice (n=10) were treated with siRNA-DOPC at 5 ug per mouse twice per week and/or paclitaxel at 30 ug per mouse once a week for 4 weeks. All mice were sacrificed and tumors were weighed when mice in control group became moribund. Tumor growth by weight under different treatments was plotted as mean ± S.D. (*p<0.05; ** p<0.01). (B) Tumor growth of MCF-7 breast cancer cells in female athymic nude mice. 3 × 106 cells were injected into the fourth mammary fat pads three days after estradiol pellet implantation. Tumor-bearing mice were randomized into 4 treatment groups (n=7) when tumors reached 50 mm3. Mice were treated with 1. Sucrose water (-DOX); 2. Sucrose water and 5 mg/kg paclitaxel (-DOX + Pac); 3. Doxycycline water (+DOX) and 4. Doxycycline water and 5 mg/kg paclitaxel (+DOX + Pac). All mice were sacrificed after 6 weeks. (** p<0.01). (C) Tumor growth of HIM3 isogenic breast cancer cells in NOD/SCID mice. 1X106 tumor cells was injected into the fourth mammary fat pads of mice. After tumors reached 50 mm3, tumor-bearing mice were randomized into 4 groups (n=8). Mice were treated as described in Fig. 7B for 8 weeks. Animal survival was evaluated from the start of treatment until tumors reached 1000 mm3. Survival curves were generated by GraphPad Prism 7. (*p<0.05; ** p<0.01). (D) Representative images of IHC with indicated antibodies from HIM3 TP53 KO tumor tissue and HIM3 TP53 WT tumor tissue. –DOX: wild-type PFKPB2, +DOX: shPFKFB2, scale bar: 20μM.
Discussion
In this report we document that the PFK-2/FBPase-2 isoenzyme (PFKFB2) drives growth of ovarian cancer cells with wtTP53 and regulates paclitaxel sensitivity. While wtTP53 is found primarily in the small subset of low grade serous ovarian cancers, we found that PFKFB2 plays a similar role in the more prevalent TP53 wild-type breast cancers, making it an attractive target for drug development. Growth inhibition and cell death induced by silencing PFKFB2 have been shown to depend on dysregulation of the oxidative PPP, failure to detoxify ROS, activation of JNK and p53, upregulation of p21, Puma and the induction of apoptosis in wtTP53 cancer cells.
Cancer cells express altered levels of the different PFK-2/FBPase-2 isoenzymes and even modulate their relative kinase and bisphosphatase activities according to metabolic needs in a spatial and/or temporal manner (36). The high biosynthetic demand of rapidly proliferating tumor cells also requires the production of large amounts of NADPH and the bisphosphatase activity may be indispensable to limiting glycolytic flux and diverting glucose-6-phosphate into the oxidative PPP for NADPH production (39). It is therefore possible that cancer cells tightly regulate the activity of different PFK-2/FBPase-2 enzymes to meet their metabolic demands in order to maintain viability. In response to depletion of PFKFB2, the levels of NADPH and 6PG were decreased in cancer cells that harbored wtTP53, suggesting that PFKFB2 is acting primarily as a phosphatase in those cells. Consequently, silencing PFKFB2 results in ROS accumulation, oxidative stress and apoptosis in ovarian and breast cancers. A recent publication from Schulze et al reported that PFKFB4 isoform of PFK2 plays a role in prostate cancer cell survival (35). PFKFB4 was found to be required to maintain redox balance and to support tumor growth, highlighting the importance of metabolic regulation of biosynthesis and antioxidant production in prostate cancer, which is consistent with PFKFB2 function that we have observed in ovarian and breast cancers.
Our data have shown that silencing PFKFB2 only inhibits growth and increases paclitaxel sensitivity in cancer cell lines and xenografts that contain wtTP53, consistent with the context that p53 plays an important role in PPP flux and redox balance. TP53 is frequently mutated in many human malignancies including ovarian and breast cancers. Low grade ovarian cancers generally have wtTP53, but 99% of the more common high grade serous ovarian cancers exhibit TP53 mutations. More than half of breast cancers retain wtTP53 which decreases activity of the oxidative pentose phosphate pathway by binding to G6PD, the first and rate-limiting enzyme of the PPP (40). Wtp53 binding to G6PD prevents the formation of the active dimer and decreasing the production of NADPH for ROS detoxification and biosynthesis. Therefore, cancer cells with mutant TP53 that lack G6PD-inhibitory activity are prone to enhanced PPP glucose flux and NADPH production, resulting in reduced oxidative stress and decreased apoptosis. Silencing PFKFB2 in ovarian and breast cancers that harbor wtTP53 causes ROS/JNK-mediated activation of p53, implying the presence of a positive feedback loop which can block further PPP through induced expression of p53.
Elevated levels of ROS have been detected in almost all cancers, where they promote many aspects of tumor development and progression (41). However, tumor cells also express increased levels of antioxidant proteins to detoxify ROS (41,42), suggesting that careful regulation of intracellular ROS levels is required for cancer cell survival. Our study revealed that the ROS accumulation in ovarian and breast cancer cells in response to PFKFB2 depletion induces p53-dependent apoptosis and causes irreversible cellular damage, suggesting the importance of ROS metabolism in mutant TP53-driven cell proliferation and cancer development. Importantly, because metabolic reprogramming in cancer drives tumor cells to be highly dependent on ROS metabolism, disrupting the production of antioxidants could provide an effective treatment strategy (43). Chemotherapeutic agents that induce oxidative stress could be combined with strategies to block NADPH production for achieving synergistic effects. Our study confirmed that PFKFB2 modulates ROS production and activity of p53, resulting in growth inhibition and increased sensitivity to paclitaxel. It is, however, difficult to conclude that TP53 has a direct role as a determinant or biomarker of cell sensitivity to paclitaxel, particularly considering the conflicting reports on this theme in the literature (44–48).
In summary, PFKFB2 silencing upregulated glycolysis and attenuated PPP flux in cancer cells with wtTP53 function. Silencing PFKFB2 induces apoptotic cell death and paclitaxel sensitivity in ovarian cancer and breast cancer cells that harbored wtTP53 in vitro and in vivo. Our data support the importance of PFKFB2 as a potential predictive biomarker and target for molecular therapy. As PFKFB2 inhibitors become available, trials can be conducted to enhance the initial response of Type I low grade ovarian cancers to paclitaxel therapy. A majority of breast cancers have wtTP53 and even a small fraction of triple negative breast cancers retain p53 function (Figure S9A–B), making this approach applicable for several distinct cancer types. Although it remains to be determined if a particular subtype of breast cancer (e.g. Luminal A, Luminal B and Basal / triple negative) would be particularly susceptible to the combination treatment, our data suggest that those tumors with elevated PFKFB2 expression and functional TP53 would likely benefit from knockdown of PFKFB2 to enhance paclitaxel sensitivity. Furthermore the data is consistent with TCGA analysis which indicates an inverse correlation between high expression of PFKFB2 and poor overall survival (p=0.02389) in luminal A tumors where the majority are wtTP53 (Figure S9B–C). Finally, it will be interesting to evaluate whether depletion of PFKFB2 will provide antitumor efficacy and enhance chemosensitivity in other types of cancer that have retained wtTP53.
Supplementary Material
Translational Relevance.
Breast cancer is the most common cause of cancer death and epithelial ovarian cancer the fourth most common cause among women in the developed world. Paclitaxel-based chemotherapy continues to be an integral component in the treatment of breast and ovarian cancer, but less than half of these cancers respond to the drug. Enhancing the response to primary paclitaxel therapy could substantially improve outcomes for women with both diseases. Our group has found that silencing several different glycolytic enzymes can enhance paclitaxel sensitivity. The most significant enhancement of paclitaxel sensitivity has been observed by knocking down PFKFB2 in multiple ovarian and breast cancer cell lines and xenografts that expressed wtTP53. PFKFB2 is a novel target to enhance paclitaxel-based primary chemotherapy in ovarian cancers and breast cancers which retain wtTP53.
ACKNOWLEDGEMENT
This work was supported by the Cancer Prevention and Research Institute of Texas RP110595-P1, the MD Anderson SPOREs in Ovarian Cancer NCI P50 CA 083639 and CA 217985 the Shared Resources of the MD Anderson CCSG grant NCI P30 CA016672, R35 CA209904, the American Cancer Society Research Professor Award, the Frank McGraw Memorial Chair in Cancer Research, The National Foundation for Cancer Research, the philanthropic support from generous donations from Stuart and Gaye-Lynn Zarrow, the Mossy foundation and the Roberson endowment. AA is supported by grants from the NIHR National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC) and Ovarian Cancer Action. We also acknowledge R21CA181994 and R25T CA557730.
Footnotes
The authors have declared that no conflict of interest exists
REFERENCES
- 1.Ganz PA, Goodwin PJ. Breast Cancer Survivorship: Where Are We Today? Adv Exp Med Biol 2015;862:1–8 doi 10.1007/978-3-319-16366-6_1. [DOI] [PubMed] [Google Scholar]
- 2.Anampa J, Makower D, Sparano JA. Progress in adjuvant chemotherapy for breast cancer: an overview. BMC Med 2015;13:195 doi 10.1186/s12916-015-0439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muggia FM. Sequential single agents as first-line chemotherapy for ovarian cancer: a strategy derived from the results of GOG-132. Int J Gynecol Cancer 2003;13 Suppl 2:156–62. [DOI] [PubMed] [Google Scholar]
- 4.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2(1):a001008 doi 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silwal-Pandit L, Langerod A, Borresen-Dale AL. TP53 Mutations in Breast and Ovarian Cancer. Cold Spring Harb Perspect Med 2017;7(1) doi 10.1101/cshperspect.a026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408(6810):307–10 doi 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 7.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009;137(3):413–31 doi 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 8.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science 2006;312(5780):1650–3 doi 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 9.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer 2009;9(10):691–700 doi 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 10.Goetze K, Fabian CG, Siebers A, Binz L, Faber D, Indraccolo S, et al. Manipulation of tumor metabolism for therapeutic approaches: ovarian cancer-derived cell lines as a model system. Cell Oncol (Dordr) 2015;38(5):377–85 doi 10.1007/s13402-015-0237-5. [DOI] [PubMed] [Google Scholar]
- 11.Lincet H, Icard P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene 2015;34(29):3751–9 doi 10.1038/onc.2014.320. [DOI] [PubMed] [Google Scholar]
- 12.Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res 2018;28(3):265–80 doi 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keijer J, van Dartel DA. Reprogrammed metabolism of cancer cells as a potential therapeutic target. Curr Pharm Des 2014;20(15):2580–94. [DOI] [PubMed] [Google Scholar]
- 14.Teicher BA, Linehan WM, Helman LJ. Targeting cancer metabolism. Clin Cancer Res 2012;18(20):5537–45 doi 10.1158/1078-0432.CCR-12-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 2014;510(7504):298–302 doi 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez R, Lopez-Grueso MJ, Muntane J, Barcena JA, Padilla CA. Redox regulation of metabolic and signaling pathways by thioredoxin and glutaredoxin in NOS-3 overexpressing hepatoblastoma cells. Redox Biol 2015;6:122–34 doi 10.1016/j.redox.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucarelli G, Galleggiante V, Rutigliano M, Sanguedolce F, Cagiano S, Bufo P, et al. Metabolomic profile of glycolysis and the pentose phosphate pathway identifies the central role of glucose-6-phosphate dehydrogenase in clear cell-renal cell carcinoma. Oncotarget 2015;6(15):13371–86 doi 10.18632/oncotarget.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009;8(7):579–91 doi 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 19.Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol 2006;16(6):420–6 doi 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed AA, Lu Z, Jennings NB, Etemadmoghadam D, Capalbo L, Jacamo RO, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell 2010;18(2):109–21 doi 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, Bolanos-Garcia VM. The dynamics of signal amplification by macromolecular assemblies for the control of chromosome segregation. Front Physiol 2014;5:368 doi 10.3389/fphys.2014.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed AA, Wang X, Lu Z, Goldsmith J, Le XF, Grandjean G, et al. Modulating microtubule stability enhances the cytotoxic response of cancer cells to Paclitaxel. Cancer Res 2011;71(17):5806–17 doi 10.1158/0008-5472.CAN-11-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le XF, Mao W, He G, Claret FX, Xia W, Ahmed AA, et al. The role of p27(Kip1) in dasatinib-enhanced paclitaxel cytotoxicity in human ovarian cancer cells. J Natl Cancer Inst 2011;103(18):1403–22 doi 10.1093/jnci/djr280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nissler K, Petermann H, Wenz I, Brox D. Fructose 2,6-bisphosphate metabolism in Ehrlich ascites tumour cells. J Cancer Res Clin Oncol 1995;121(12):739–45. [DOI] [PubMed] [Google Scholar]
- 25.Miralpeix M, Azcon-Bieto J, Bartrons R, Argiles JM. The impairment of respiration by glycolysis in the Lewis lung carcinoma. Cancer Lett 1990;50(3):173–8. [DOI] [PubMed] [Google Scholar]
- 26.TeSlaa T, Teitell MA. Techniques to monitor glycolysis. Methods Enzymol 2014;542:91–114 doi 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 2012;7(5):872–81 doi 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell E, Shao J, Yuan Y, Chen HC, Cai S, Echeverria GV, et al. p53 deficiency linked to B cell translocation gene 2 (BTG2) loss enhances metastatic potential by promoting tumor growth in primary and metastatic sites in patient-derived xenograft (PDX) models of triple-negative breast cancer. Breast Cancer Res 2016;18(1):13 doi 10.1186/s13058-016-0673-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486(7403):346–52 doi 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474(7353):609–15 doi 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479 doi 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mor I, Cheung EC, Vousden KH. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb Symp Quant Biol 2011;76:211–6 doi 10.1101/sqb.2011.76.010868. [DOI] [PubMed] [Google Scholar]
- 33.Funato Y, Hayashi T, Irino Y, Takenawa T, Miki H. Nucleoredoxin regulates glucose metabolism via phosphofructokinase 1. Biochem Biophys Res Commun 2013;440(4):737–42 doi 10.1016/j.bbrc.2013.09.138. [DOI] [PubMed] [Google Scholar]
- 34.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev 2009;23(5):537–48 doi 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov 2012;2(4):328–43 doi 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- 36.Ros S, Schulze A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab 2013;1(1):8 doi 10.1186/2049-3002-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li P, Zhao QL, Wu LH, Jawaid P, Jiao YF, Kadowaki M, et al. Isofraxidin, a potent reactive oxygen species (ROS) scavenger, protects human leukemia cells from radiation-induced apoptosis via ROS/mitochondria pathway in p53-independent manner. Apoptosis 2014;19(6):1043–53 doi 10.1007/s10495-014-0984-1. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y, Nikulenkov F, Zawacka-Pankau J, Li H, Gabdoulline R, Xu J, et al. ROS-dependent activation of JNK converts p53 into an efficient inhibitor of oncogenes leading to robust apoptosis. Cell Death Differ 2014;21(4):612–23 doi 10.1038/cdd.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011;27:441–64 doi 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 40.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol 2011;13(3):310–6 doi 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res 2010;44(5):479–96 doi 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdul-Aziz A, MacEwan DJ, Bowles KM, Rushworth SA. Oxidative stress responses and NRF2 in human leukaemia. Oxid Med Cell Longev 2015;2015:454659 doi 10.1155/2015/454659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 2015;11(1):9–15 doi 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumontet C, Sikic BI. Mechanisms of action of and resistance to antitubulin agents: microtubule dynamics, drug transport, and cell death. J Clin Oncol 1999;17(3):1061–70 doi 10.1200/JCO.1999.17.3.1061. [DOI] [PubMed] [Google Scholar]
- 45.Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW, et al. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat Med 1996;2(1):72–9. [DOI] [PubMed] [Google Scholar]
- 46.Debernardis D, Sire EG, De Feudis P, Vikhanskaya F, Valenti M, Russo P, et al. p53 status does not affect sensitivity of human ovarian cancer cell lines to paclitaxel. Cancer Res 1997;57(5):870–4. [PubMed] [Google Scholar]
- 47.Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ Jr., Kohn KW, et al. An information-intensive approach to the molecular pharmacology of cancer. Science 1997;275(5298):343–9. [DOI] [PubMed] [Google Scholar]
- 48.O’Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res 1997;57(19):4285–300. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.