Abstract
For more than 40 years after its discovery, histone methylation was thought to be largely irreversible. However, the first histone demethylase (HDM) was identified in 2004, challenging this notion. Since that time, more than 20 HDMs have been identified and characterized, and many have been shown to have critical roles in organismal development, cell fate, and disease. Here, we highlight some of the recent advances in our understanding of the function of HDMs in the context of neuronal development, plasticity, and disease. We focus in particular on molecular genetic studies of LSD1, Kdm6b, and Kdm5c that have elucidated both enzymatic and non-enzymatic gene regulatory functions of these HDMs in neurons.
Keywords: Histone methylation, chromatin regulation, neuronal transcription, brain development, intellectual disability, mouse models
Introduction
Post-translational modifications to the N-terminal tails of the histone proteins play crucial roles in genome regulation [1]. These modifications (e.g. acetylation, methylation, phosphorylation) are deposited by so-called “writer” enzymes and dynamically removed by the action of “eraser” enzymes. Although histone methylation was long thought to be irreversible due to its chemical stability, the identification of a large group of enzymes with histone demethylase activity challenged this assumption. Histone demethylases (HDMs) can be broadly classified into two families based on their mechanisms of enzymatic action: the two amine oxidase demethylases (LSD1/KDM1A and LSD2/KDM1B) and the much larger Jumonji C domain (JmjC) family, which has more than 20 members [2]. The discovery of such a large set of HDMs, along with the observation of their specificity for demethylation of distinct residues on the histone tails, immediately raised the possibility that these enzymes dynamically regulate histone methylation-dependent cellular processes. Indeed, HDMs have been found to play crucial roles in development and contribute to pathological processes like cancer and aging.
Neurons are long-lived postmitotic cells that continuously adapt their gene expression programs to the environment; thus, these cells serve as a particularly good substrate for discovering the function of chromatin regulators including HDMs in genome dynamics. Here we review recent hallmark studies on the functions of HDMs in neurons, focusing in particular on biological and biochemical studies of three of the best studied neuronal HDMs (summarized in Fig. 1) that offer new insight into the roles of chromatin regulation in the brain.
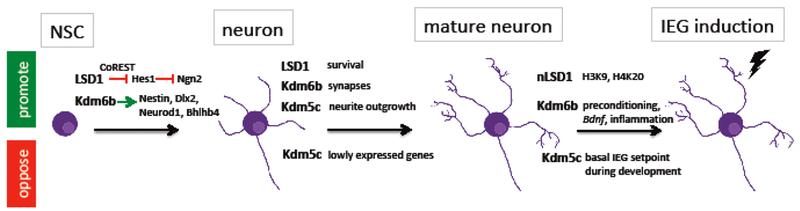
Figure 1:
Functions of HDMs in neuronal development and plasticity. Summary of the functions of LSD1, Kdm6b, and Kdm5c at different stages of neuronal development as described in the text. Green arrow or box indicates activation of a process or gene, Red bar or box indicates repression of a process or gene. NSC, neural stem cell. nLSD1, neuronal splice variant of LSD1. IEG, immediate early gene. The lightning bold represents synaptic activity and neuronal action potential firing.
LSD1: protein complexes and splicing determine target specificity
Lysine-specific demethylase 1 (LSD1) was the first HDM to be discovered [3] and has been one of the most widely studied. LSD1 was initially characterized as a transcriptional repressor that interacts with the CoREST complex and specifically demethylates the transcriptional activation-associated mark histone H3 mono or dimethylated at lysine 4 (H3K4me1 and H3K4me2) [4]. However, LSD1 was also co-purified with the androgen receptor and was found to act as a coactivator of transcription via its ability to demethylate the repressive marks H3K9me1 and H3K9me2 [5]. The association of LSD1 with distinct protein complexes appears to direct its specificity toward different histone modifications, allowing this single enzyme to have opposing functions in transcriptional regulation in distinct contexts [6]. Mutations that impair the enzymatic activity of LSD1 have been associated with intellectual disability (ID), suggesting the relevance of LSD1’s demethylase function in human brain development (Fig. 2A) [7].
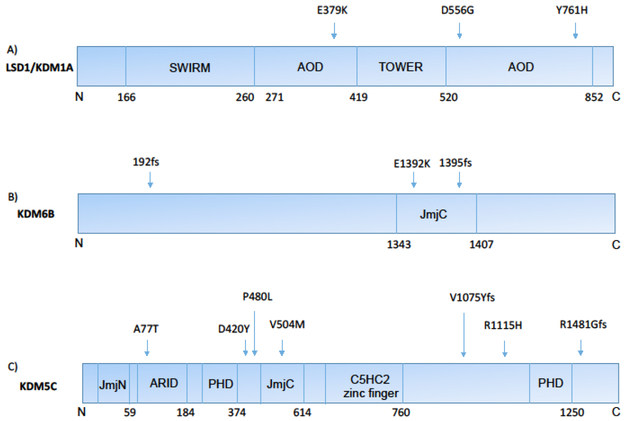
Figure 2:
Point mutations in KDM1A, KDM6B, and KDM5C associated with neurodevelopmental disorders. Protein domains are indicated by boxes and names, and the positions of disease associated mutations indicated by arrows. A) KDM1A mutations associated with cognitive impairment [7]. B) De novo KDM6B mutations identified in patients with ASD [28]. C) KDM5C mutations associated with XLID [34, 37] and one mutation (R1115H) associated with ASD/ID [39].
Kdm1a knockout mice die by embryonic day 6.5, precluding the study of brain development in these animals [8]. However, knockdown or inhibition of LSD1 in neuronal progenitors has revealed that it serves as a positive regulator of neuronal differentiation in the developing brain. In cortical progenitors, LSD1 forms a complex with the transcriptional corepressor CoREST and binds to the promoter of Hes1, a transcriptional repressor and key effector of the Notch signaling pathway [9]. Knockdown of LSD1 or CoREST by in utero electroporation leads to an increase in Hes1 expression and reduced expression of its proneural downstream target, the basic helix-loop-helix transcription factor, Ngn2. These data indicate that LSD1 opposes Notch to promote cortical neuronal differentiation. Similar functions for LSD1 were observed in retinal development, where peak LSD1 expression occurs during the period of rod photoreceptor differentiation [10]. Inhibition of LSD1 in cultured retinal explants was shown to increase expression and H3K4me2 deposition on promoters of progenitor-expressed genes, including Hes1, and block induction of terminal rod differentiation markers despite the normal expression of rod photoreceptor transcription factors.
LSD1 also contributes to the regulation of specific programs of gene expression in postmitotic, fate-committed neurons. Perhaps its most specialized role is in the complex series of epigenetic regulatory events that permit each olfactory sensory neuron to express one and only one of the thousands of possible olfactory receptor genes [11]. To more broadly study the function of LSD1 in the adult brain, Christopher et al. [12] conditionally deleted floxed Kdm1a in adult mice with a tamoxifen-inducible Cre transgene. Loss of LSD1 from the adult brain resulted in widespread neuronal death throughout the cortex and hippocampus. Gene expression analysis showed reactivation of stem cell genes in the hippocampus of LSD1 knockout mice and bioinformatics suggested an overall similarity of the knockout gene expression profile to that seen in cases of Alzheimer’s disease and frontotemporal dementia. Whether insults to LSD1 function directly cause neurodegeneration remains unknown; however, these data suggest that continuous expression of LSD1 in the adult brain is required for maintenance of proper neuronal function.
Interestingly, LSD1 has a neuronal-specific splice variant, which comprises about half of all LSD1 in the adult brain and is the predominant form of LSD1 expressed in the early postnatal brain [13]. Referred to as NeuroLSD1 or LSD1n, this variant is characterized by the inclusion of a 12-nucleotide microexon (E8a) that adds just four amino acids to the enzymatic amine oxidase domain [14]. Nonetheless, variant-specific knockout or knockdown of LSD1n has shown that this small change in the LSD1 sequence has major effects on LSD1 function. The brains of LSD1n knockout mice are hypoexcitable and show decreased seizure susceptibility [15]. LSD1n mutant mice also display impaired spatial learning and memory in the Barnes maze [16] and reduced stress-induced anxiety-like behavior [17]. These behavioral changes correlate with impaired activity-dependent induction of immediate-early genes (IEGs), which have been suggested to play important roles in the cellular and circuit adaptations that underlie learning and memory.
How does LSD1n promote IEG induction? Phosphorylation of the LSD1n tetrapeptide has been suggested to inhibit association with CoREST and to block the repressive H3K4 demethylase activity of LSD1 [18]. Given evidence that LSD1n binds to at least some of the same target genes as LSD1, one possibility is that LSD1n functions as a dominant negative, blocking the repressive actions of LSD1 [13]. Alternatively, one group found that LSD1n binds a co-regulatory protein called supervillin, which directs the complex to demethylate H3K9me2, thus promoting gene activation [19]; another group proposed a model in which LSD1n acquires specificity to demethylate H4K20, promoting transcriptional elongation [16].
Kdm6b: Relief of polycomb-mediated repression
Kdm6b/Jmjd3 is one of a small family of two HDMs (Kdm6a/Utx is the other HDM) that has specificity for removal of the H3K27me2/3 marks laid down by the polycomb repressive complex. Over the course of neuronal differentiation, Kdm6b is important for removing H3K27me3 in the context of bivalent chromatin marks. Bivalency describes regulatory elements that are associated with both H3K4me3 and H3K27me3, which are histone modifications normally correlated with gene activation and repression, respectively [20]. During the early stages of cell fate commitment, bivalent chromatin is found at promoters of cell-type specific genes that are poised to turn either on or off depending on the fate of the cell. For example, Kdm6b-dependent resolution of bivalency is required for proper activation of Nestin expression in embryonic stem cell-derived neural progenitor cells during commitment to the neuronal lineage [21].
In vivo, conditional knockout or knockdown of Kdm6b impairs neurogenesis in the olfactory bulb (OB) [22], the spinal cord [23], and the retina [24]. These studies have revealed diversity in the action of Kdm6b. In the spinal cord, Kdm6b is bound to a set of gene promoters that are activated during TGFβ-dependent neural differentiation, and it collaborates with Smad3 to induce promoter demethylation and transcriptional activation [23]. By contrast, although Kdm6b is required for the expression of the transcription factor Dlx2 in OB progenitors, it acts by regulating H3K27me3 at a distal enhancer of this gene rather than at the gene promoter [22]. In the retina, although Kdm6b is also pro-differentiation, Kdm6b is expressed most highly in young postmitotic neurons, and loss of Kdm6b function impairs the development of only a subset of retina cell types that undergo developmental loss of H3K27me3 at the Bhlhb4 promoter [24]. Finally, given that substantial developmental loss of H3K27me3 during neural commitment can still occur in the absence of the Kdm6 family [25], it remains important to test which of the functions of Kdm6b in neural differentiation require the action of its histone demethylation function.
Despite these data implicating Kdm6b as a positive regulator of neuronal differentiation, and surprisingly given the evidence that germline knockouts die perinatally due to an inability to breathe, the brains of Kdm6b knockout mice are remarkably normal in gross morphology [26,27]. Interestingly, Kdm6b acts in postmitotic neurons to regulate synapse function. Burgold et al. (2012) found that Kdm6b is required to maintain functionality of the Pre-Bötzinger complex (PBC), the neural pacemaker that controls the respiratory rhythm. In the absence of Kdm6b, although the neurons of the PBC are present, the circuit fails to functionally mature into a network state that can control proper breathing by the time of birth. Clues to the potential mechanisms of these circuit defects are suggested by studies of Kdm6b-dependent gene expression in developing cerebellar granule neurons (CGNs) [27]. CGN differentiation provides an effective means to identify gene expression that changes across the full time course of differentiation including postmitotic stages of neuronal and synaptic maturation. Conditional knockout of Kdm6b in fate-committed CGN progenitors did not affect the expression of early neuronal marker genes and did not disrupt cerebellar morphology. However loss of Kdm6b expression did impair the late upregulation of gene products including specific GABA and glutamate receptor subunits that confer mature properties upon synaptic function [27]. These data are intriguing in light of the discovery of de novo KDM6B mutations in patients diagnosed with autism spectrum disorder (ASD), which is thought to be a primary disorder of synapses (Fig. 2B) [28].
Finally, like LSD1n, Kdm6b is also implicated in activity-dependent gene regulation in the mature nervous system. However, unlike LSD1n, which likely acts in a constitutive manner to set the responsiveness of IEGs to synaptic stimuli, Kdm6b itself is a direct target of regulation by synaptic activity. Among all of the HDMs, the expression of Kdm6b is by far the most strongly induced by synaptic activity, showing over a 20-fold increase in mRNA expression in neurons of the hippocampus following pilocarpine-induced seizures [29]. Kdm6b is also induced in neurons of the prefrontal cortex following cocaine exposure and withdrawal [30]. Whether there are direct synaptic activity-regulated targets of Kdm6b remains to be fully understood, although one study showed that pre-treatment of hippocampal neurons with the Kdm6b inhibitor GSK-J4 impaired NMDA-receptor dependent transcription of Brain-Derived Neurotrophic Factor (Bdnf) [31]. In hippocampal neurons Kdm6b is required for a form of synaptic activity-dependent survival called preconditioning. Surprisingly, in this context, Kdm6b is required for the induction of a program of inflammatory gene expression rather than traditional pro-survival genes like Bdnf [29]. These data raise the possibility that the pervasive function of Kdm6b in the control of inflammation [32] may also contribute to its cell-type specific functions in activity-dependent neuronal adaptations.
Kdm5c: Mutations in X-linked intellectual disability
As mentioned above, targeted exome sequencing studies are rapidly revealing de novo mutations in a number of chromatin regulators associated with neurodevelopmental disorders including ID and ASD [33,34]. In addition, familial mutations in the H3K4-selective demethylase KDM5C (SMCX/JARID1C) have been identified as one of the more frequent causes of X-linked intellectual disability (XLID) (Fig. 2C)[35]. In either case, the genetic association between any chromatin regulator and disease is only the beginning of the story, and recent efforts have focused on elucidating the cellular mechanisms that link chromatin dysregulation through gene expression and circuit formation to impaired brain function.
Mice with germline deletion of Kdm5c recapitulate many of the cognitive, adaptive and social abnormalities seen in patients with KDM5C mutations and, thus, offer a model for studying the neurological effects of Kdm5c disruption [36]. Behaviorally, Kdm5c knockout mice exhibit increased aggression, decreased anxiety, and impaired social behaviors as well as defects in learning and memory. At the cellular level, neurons in the knockout mice have defects in dendritic branching and spine morphology. Expression of disease-associated KDM5C mutations (KDM5CH514A, KDM5CY751C and KDM5CF642L) in the Neuro2A cell line suppress retinoic acid-induced neurite growth, consistent with these point mutations being KDM5C loss-of-function at least for this cellular phenotype [37].
In Kdm5c germline and forebrain conditional knockout mice, RNA sequencing showed the upregulation of a large set of genes, suggesting that Kdm5c functions primarily as a transcriptional repressor [36,38]. While no changes in global H3K4me3 levels were observed in the knockout neurons, there were thousands of differentially methylated peaks, primarily exhibiting local increases in H3K4me3. De novo H3K4me3 peaks were most common at intergenic regions that are marked by H3K4me1 in wildtype, suggesting that enhancers more than promoters are particularly sensitive to the loss of Kdm5c [38]. Despite the fact that Kdm5c is broadly bound at the promoters of expressed genes, genes that showed the highest induction of expression and the greatest increase in H3K4me3 at their promoters in the absence of Kdm5c were those that were lowly expressed in wildtype [36,38]. These data suggest that Kdm5c is primarily required for fine-tuning gene expression by suppressing the accumulation of H3K4me3 at active regulatory elements. Notably, although most ID-associated mutations in KDM5C disrupt the histone demethylase activity of this enzyme, a point mutant was recently identified that neither disrupts protein expression or enzyme activity (Fig. 2C) [39]. Thus Kdm5c may also have non-enzymatic activities that contribute to brain development.
Other HDMs in neurodevelopmental disorders
In addition to the HDMs highlighted here, many other HDMs are expressed in neurons [29], and mutations in several of these have been found in neurodevelopmental syndromes, highlighting their importance human brain development. For example, microdeletion of 12a24.31, which is a genomic region that contains both KDM2B and the histone methyltransferases SETD1B, causes an ID syndrome [40], and multiple mutations in the H3K9 demethylase JMJD1C (KDM3C) were found upon targeted resequencing of a set of patients clinically diagnosed with ASD, ID, or Rett Syndrome without an identified genetic defect [41].
The most common forms of ID are X-linked, adding urgency to the study of HDMs that localize to the X chromosome. The H3K9/H4K20 demethylase PHF8 (KDM7B) is associated with both syndromic and non-syndromic forms of X-linked ID [42]. Chen et al. (2018) generated Phf8 knockout mice as an animal model of these disorders and showed the knockouts exhibited learning and memory impairments in behavioral assays and long-term potentiation (LTP) deficits in slice electrophysiology [43]. A bioinformatics approach suggested the upregulation of the mTOR pathway in these mice and, interestingly, an inhibitor of mTOR (rapamycin) was able to rescue the learning/memory as well as LTP defects in these mice. Tang et al. (2017) conducted a similar study on a different X chromosome HDM, Kdm6a (Utx) [44]. They generated forebrain-specific Kdm6a knockout mice, which exhibited deficits in learning and memory, as well as morphological and functional abnormalities of hippocampal synapses [44]. Among the genes downregulated in the conditional knockouts, the authors focused on the serotonin receptor Htr5b and showed that overexpression could rescue dendritic defects in Kdm6a knockout neurons. These findings have potential relevance for the cognitive symptoms in Kabuki Syndrome, a congenital craniofacial disorder for which KDM6A mutations are one genetic cause. Although conditional genetic deletion of Kdm6a in the neural crest of mice recapitulates dysmorphic features of Kabuki syndrome, craniofacial development is mostly independent of the demethylase activity of Kdm6a because mice bearing a demethylase-dead knockin mutation of Kdm6a have normal facial morphology [45]. These data reiterate that the connections between the biological and biochemical functions of the HDMs remain to be fully explored, and they show that understanding the relevant actions of the HDMs in neurons will be required to develop meaningful potential therapeutics.
Concluding Remarks
The discovery of HDMs not only opened the door to understanding the roles of dynamic histone methylation but also gave us new genetic tools to explore the mechanisms and consequences of gene regulation in the brain. The identification of disease-associated mutations in HDMs and the study of mouse knockout models have revealed fundamental insights about the biological requirements for these enzymes. Future studies will reveal mechanistic insights about recruitment of HDMs to their target genes, the domains in HDMs that are critical for their function, as well as non-enzymatic functions of this family.
Highlights.
Neuronal functions of the histone demethylases LSD1, Kdm6b, and Kdm5c
Evidence for potential non-enzymatic functions of the histone demethylases
Summarize neurological disease-associated mutations in HDMs
Acknowledgements
We thank Urann Chan for assistance with the figures. This work was supported in part by National Institutes of Health grant 1R01NS098804 (A.E.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors have nothing to declare.
References
- 1.Strahl BD, Allis CD: The language of covalent histone modifications. Nature 2000, 403:41–45. [DOI] [PubMed] [Google Scholar]
- 2.Greer EL, Shi Y: Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012, 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA: Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119:941–953. [DOI] [PubMed] [Google Scholar]
- 4.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y: Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005, 19:857–864. [DOI] [PubMed] [Google Scholar]
- 5.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R: LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437:436–439. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. : Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007, 446:882–887. [DOI] [PubMed] [Google Scholar]
- 7.Pilotto S, Speranzini V, Marabelli C, Rusconi F, Toffolo E, Grillo B, Battaglioli E, Mattevi A: LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum Mol Genet 2016, 25:2578–2587.**This study uses structural modeling and biochemistry to determine the consequences of disease associated mutations on LSD1 stability, protein interactions, and enzymatic function.
- 8.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. : The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 2009, 41:125–129. [DOI] [PubMed] [Google Scholar]
- 9.Lopez CI, Saud KE, Aguilar R, Berndt FA, Canovas J, Montecino M, Kukuljan M: The chromatin modifying complex CoREST/LSD1 negatively regulates notch pathway during cerebral cortex development. Dev Neurobiol 2016, 76:1360–1373. [DOI] [PubMed] [Google Scholar]
- 10.Popova EY, Pinzon-Guzman C, Salzberg AC, Zhang SS, Barnstable CJ: LSD1-Mediated Demethylation of H3K4me2 Is Required for the Transition from Late Progenitor to Differentiated Mouse Rod Photoreceptor. Mol Neurobiol 2016, 53:4563–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S: An epigenetic trap stabilizes singular olfactory receptor expression. Cell 2013, 154:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christopher MA, Myrick DA, Barwick BG, Engstrom AK, Porter-Stransky KA, Boss JM, Weinshenker D, Levey AI, Katz DJ: LSD1 protects against hippocampal and cortical neurodegeneration. Nat Commun 2017, 8:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusconi F, Grillo B, Toffolo E, Mattevi A, Battaglioli E: NeuroLSD1: Splicing-Generated Epigenetic Enhancer of Neuroplasticity. Trends Neurosci 2017, 40:28–38. [DOI] [PubMed] [Google Scholar]
- 14.Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E: Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci 2010, 30:2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusconi F, Paganini L, Braida D, Ponzoni L, Toffolo E, Maroli A, Landsberger N, Bedogni F, Turco E, Pattini L, et al. : LSD1 Neurospecific Alternative Splicing Controls Neuronal Excitability in Mouse Models of Epilepsy. Cereb Cortex 2015, 25:2729–2740. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Telese F, Tan Y, Li W, Jin C, He X, Basnet H, Ma Q, Merkurjev D, Zhu X, et al. : LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci 2015, 18:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rusconi F, Grillo B, Ponzoni L, Bassani S, Toffolo E, Paganini L, Mallei A, Braida D, Passafaro M, Popoli M, et al. : LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A 2016, 113:3651–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, et al. : Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem 2014, 128:603–616. [DOI] [PubMed] [Google Scholar]
- 19.Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, et al. : A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell 2015, 57:957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. : A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125:315–326. [DOI] [PubMed] [Google Scholar]
- 21.Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G: The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 2008, 3:e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, Testa G, Matzuk MM, Iwamori N, Lim DA: Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep 2014, 8:1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estaras C, Akizu N, Garcia A, Beltran S, de la Cruz X, Martinez-Balbas MA: Genome-wide analysis reveals that Smad3 and JMJD3 HDM co-activate the neural developmental program. Development 2012, 139:2681–2691. [DOI] [PubMed] [Google Scholar]
- 24.Iida A, Iwagawa T, Kuribayashi H, Satoh S, Mochizuki Y, Baba Y, Nakauchi H, Furukawa T, Koseki H, Murakami A, et al. : Histone demethylase Jmjd3 is required for the development of subsets of retinal bipolar cells. Proc Natl Acad Sci U S A 2014, 111:3751–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T: KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet 2014, 10:e1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, Spreafico F, Bevengut M, Gestreau C, Buontempo S, et al. : The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep 2012, 2:1244–1258. [DOI] [PubMed] [Google Scholar]
- 27.Wijayatunge R, Liu F, Shpargel KB, Wayne NJ, Chan U, Boua JV, Magnuson T, West AE: The histone demethylase Kdm6b regulates a mature gene expression program in differentiating cerebellar granule neurons. Mol Cell Neurosci 2018, 87:4–17.** This study demonstrates that knockdown of Kdm6b in cerebellar granule neurons prevents the induction of a late neuronal gene expression program that is enriched for gene products that confer mature function on synapses.
- 28.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. : Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wijayatunge R, Chen LF, Cha YM, Zannas AS, Frank CL, West AE: The histone lysine demethylase Kdm6b is required for activity-dependent preconditioning of hippocampal neuronal survival. Mol Cell Neurosci 2014, 61:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YX, Akumuo RC, Espana RA, Yan CX, Gao WJ, Li YC: The histone demethylase KDM6B in the medial prefrontal cortex epigenetically regulates cocaine reward memory. Neuropharmacology 2018, 141:113–125.*This study reports a function for Kdm6b in drug-induced behavioral plasticity in the adult nervous system.
- 31.Palomer E, Carretero J, Benvegnu S, Dotti CG, Martin MG: Neuronal activity controls Bdnf expression via Polycomb de-repression and CREB/CBP/JMJD3 activation in mature neurons. Nat Commun 2016, 7:11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G: The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007, 130:1083–1094. [DOI] [PubMed] [Google Scholar]
- 33.Duffney LJ, Valdez P, Tremblay MW, Cao X, Montgomery S, McConkie-Rosell A, Jiang YH: Epigenetics and autism spectrum disorder: A report of an autism case with mutation in H1 linker histone HIST1H1E and literature review. Am J Med Genet B Neuropsychiatr Genet 2018, 177:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwase S, Berube NG, Zhou Z, Kasri NN, Battaglioli E, Scandaglia M, Barco A: Epigenetic Etiology of Intellectual Disability. J Neurosci 2017, 37:10773–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al. : Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 2005, 76:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwase S, Brookes E, Agarwal S, Badeaux AI, Ito H, Vallianatos CN, Tomassy GS, Kasza T, Lin G, Thompson A, et al. : A Mouse Model of X-linked Intellectual Disability Associated with Impaired Removal of Histone Methylation. Cell Rep 2016, 14:1000–1009.**This study reports the behavioral and cellular phenotype of Kdm5c knockout mice and performs an initial molecular analysis of Kdm5c-dependent gene transcription.
- 37.Wei G, Deng X, Agarwal S, Iwase S, Disteche C, Xu J: Patient Mutations of the Intellectual Disability Gene KDM5C Downregulate Netrin G2 and Suppress Neurite Growth in Neuro2a Cells. J Mol Neurosci 2016, 60:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scandaglia M, Lopez-Atalaya JP, Medrano-Fernandez A, Lopez-Cascales MT, Del Blanco B, Lipinski M, Benito E, Olivares R, Iwase S, Shi Y, et al. : Loss of Kdm5c Causes Spurious Transcription and Prevents the Fine-Tuning of Activity-Regulated Enhancers in Neurons. Cell Rep 2017, 21:47–59.**Using the Kdm5c knockout mice reported in Ref. 36 as well as a forebrain conditional knockout line, this study conducts a detailed analysis of gene transcription and chromatin regulation that is disrupted by loss of Kdm5c.
- 39.Vallianatos CN, Farrehi C, Friez MJ, Burmeister M, Keegan CE, Iwase S: Altered Gene-Regulatory Function of KDM5C by a Novel Mutation Associated With Autism and Intellectual Disability. Front Mol Neurosci 2018, 11:104.*This study describes a new disease-associated KDM5C mutation with a distinct effect on KDM5C function that does not disrupt protein stability or histone demethylase activity.
- 40.Labonne JD, Lee KH, Iwase S, Kong IK, Diamond MP, Layman LC, Kim CH, Kim HG: An atypical 12q24.31 microdeletion implicates six genes including a histone demethylase KDM2B and a histone methyltransferase SETD1B in syndromic intellectual disability. Hum Genet 2016, 135:757–771. [DOI] [PubMed] [Google Scholar]
- 41.Saez MA, Fernandez-Rodriguez J, Moutinho C, Sanchez-Mut JV, Gomez A, Vidal E, Petazzi P, Szczesna K, Lopez-Serra P, Lucariello M, et al. : Mutations in JMJD1C are involved in Rett syndrome and intellectual disability. Genet Med 2016, 18:378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vallianatos CN, Iwase S: Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 2015, 7:503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Wang S, Zhou Y, Han Y, Li S, Xu Q, Xu L, Zhu Z, Deng Y, Yu L, et al. : Phf8 histone demethylase deficiency causes cognitive impairments through the mTOR pathway. Nat Commun 2018, 9:114.*This study reports learning and memory defects in Phf8 knockout mice. The authors use gene expression data from the knockouts to implicate the mTOR pathway and then use drugs targeting this pathway to improve LTP and learning in the mice.
- 44.Tang GB, Zeng YQ, Liu PP, Mi TW, Zhang SF, Dai SK, Tang QY, Yang L, Xu YJ, Yan HL, et al. : The Histone H3K27 Demethylase UTX Regulates Synaptic Plasticity and Cognitive Behaviors in Mice. Front Mol Neurosci 2017, 10:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shpargel KB, Starmer J, Wang C, Ge K, Magnuson T: UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc Natl Acad Sci U S A 2017, 114:E9046–E9055.* This study deletes Utx specifically in the neural crest lineage, which recapitulates key facial features of human Kabuki syndrome. Their data also predict that pathogenesis of this syndrome could be related to functions of Utx that are independent of its demethylase activity.