Abstract
Aquaporins (AQPs) serve as water channel proteins and belong to major intrinsic proteins (MIPs) family, functioning in rapidly and selectively transporting water and other small solutes across biological membranes. Importantly, AQPs have been shown to play a critical role in abiotic stress response pathways of plants. As a species closely related to Arabidopsis thaliana, Eutrema salsugineum has been proposed as a model for studying salt resistance in plants. Here we surveyed 35 full-length AQP genes in E. salsugineum, which could be grouped into four subfamilies including 12 plasma membrane intrinsic proteins (PIPs), 11 tonoplast intrinsic proteins (TIPs), nine NOD-like intrinsic proteins (NIPs), and three small basic intrinsic proteins (SIPs) by phylogenetic analysis. EsAQPs were comprised of 237–323 amino acids, with a theoretical molecular weight (MW) of 24.31–31.80 kDa and an isoelectric point (pI) value of 4.73–10.49. Functional prediction based on the NPA motif, aromatic/arginine (ar/R) selectivity filter, Froger’s position and specificity-determining position suggested quite differences in substrate specificities of EsAQPs. EsAQPs exhibited global expressions in all organs as shown by gene expression profiles and should be play important roles in response to salt, cold and drought stresses. This study provides comprehensive bioinformation on AQPs in E. salsugineum, which would be helpful for gene function analysis for further studies.
Keywords: Eutrema salsugineum, Aquaporin, Abiotic stress, Gene structure, Expression pattern
Introduction
Water is the most abundant molecule in living cells, forming the basic medium in which all biochemical reactions take place (Dev & Herbert, 2018). Aquaporins (AQPs) belong to the major intrinsic proteins (MIPs) superfamily, which could selectively transport water molecules across the cell membrane. In addition, AQPs can also transport many small molecules, such as glycerol, urea, carbon dioxide (CO2), silicon, boron, ammonia (NH3) and hydrogen peroxide (H2O2) (Biela et al., 1999; Gerbeau et al., 1999; Uehlein et al., 2003; Ma et al., 2006; Takano et al., 2006; Loqué et al., 2005; Dynowski et al., 2008). AQP was first discovered in animals and subsequently found in almost all living organisms (Gomes et al., 2009). Compare to animals, plants have more robust and diverse AQPs. For instance, there are 35 AQPs in Arabidopsis thaliana, 33 in Oryza sativa, 40 in Sorghum bicolor, 72 in Glycine max, 47 in Cicer arietinum and 45 in Manihot esculenta (Johanson et al., 2001; Sakurai et al., 2005; Kadam et al., 2017; Zhang et al., 2013; Deokar & Tar’an, 2016; Putpeerawit et al., 2017).
Plant AQPs can be divided into seven subfamilies based on the protein sequence similarity analysis. Plasma membrane intrinsic proteins (PIPs) are the largest subfamily of plant AQPs. The most of the PIPs are commonly localized in the plasma membrane and are further divided into two phylogenetic groups PIP1 and PIP2. Tonoplast intrinsic proteins (TIPs) subfamily is usually localized in the tonoplast, which contain five classes TIP1, TIP2, TIP3, TIP4 and TIP5. NOD26-like intrinsic proteins (NIPs) named from NIP protein (Nodulin-26, GmNOD26), were discovered in the plasma membrane of soybean cells (Fortin, Morrison & Verma, 1987). Small basic intrinsic proteins (SIPs) are typically localized in the endoplasmic reticulum. X intrinsic proteins (XIPs) are present in some dicots but absent in Brassicaceae and monocots (Maurel et al., 2015). GlpF-like intrinsic proteins (GIPs) are found in moss (Physcomitrella patens) and similar to bacterial glycerol channels (Danielson & Johanson, 2008; Gustavsson et al., 2005). Hybrid intrinsic proteins (HIPs) are found in fern (Selaginella moellendorffii) and moss (Physcomitrella patens, Anderberg, Kjellbom & Johanson, 2012; Gustavsson et al., 2005). Therefore, some classes (such as XIPs, HIPs, or GIPs) are considered to be lost during the evolution of certain plant lineages due to function redundancies (Maurel et al., 2015).
AQPs are highly conserved in molecular structure, consisting of six transmembrane α-helical domains (TM1-TM6) linked by five loops (A-E), with both the N and C terminal having a cytoplasmic orientation. There are two highly conserved NPA (Asn-Pro-Ala) motifs in two half helices (HB and HE) of loopB and loopE at the center of the pore that have substrate selectivity (Taji et al., 2002). The narrow aromatic/arginine (ar/R) selectivity filter is formed with four residues from TM helix 2 (H2), TM helix 5 (H5), and loop E (LE1 and LE2), which has been shown to provide a size barrier for solute permeability (Bansal & Sankararamakrishnan, 2007). Froger’s position consists of five residues (P1-P5) that could transport two different types of molecules, water and glycerol (Froger et al., 1998). Moreover, it has been predicted that AQPs have nine specificity-determining positions (SDPs) for non-aqua substrates, such as ammonia, boron, carbon dioxide, hydrogen peroxide, silicon and urea, for each unique group (Hove & Bhave, 2011).
Salt cress previously named as Thellungiella halophila or Thellungiella salsuginea recently was corrected to Eutrema salsugineum based on taxonomy and systematics, which is a relative close to A. thaliana (Koch & German, 2013). As a salt-sensitive plant, Arabidopsis has certain limits to study the mechanism of salt and drought resistance. In contrast, E. salsugineum, with a small genome, is quite tolerant to salt, drought and low temperature stresses, being considered to be a halophyte model plant for investigating the mechanism of plant resistance to stress (Zhu, 2001; Inan et al., 2004). The E. salsugineum AQPs like TsTIP1;2, TsMIP6 and TsPIP1;1 have been found to play an important role in plant response to abiotic stress (Wang et al., 2014; Sun et al., 2015; Li et al., 2018). The E. salsugineum genome was sequenced in 2012 and 2013 at the chromosome level and scaffold level, respectively (Wu et al., 2012; Yang et al., 2013), promoting the bioinformatics analysis of whole aquaporin family.
In this study, a genome-wide analysis of AQP genes was carried out in E. salsugineum, a total of 35 full-length AQP genes were identified. Based on the phylogenetic analysis, we found that the identified EsAQPs were quite similar to AtAQPs. The EsAQPs could be grouped into four subfamilies, including PIPs, TIPs, NIPs and SIPs. Protein sequences, chromosome distributions, gene structures and putative functions were analyzed for each of these members. The expression level of EsAQP genes in different organs and the abundance change of EsAQP genes in response to salt, drought and cold stresses were also investigated.
Materials & Methods
Identification and chromosomal location of EsAQPs
The whole genome of E. salsugineum was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/12266, Wu et al., 2012; Yang et al., 2013). To identify E. salsugineum AQP candidate genes, a Hidden Markov Model (HMM) analysis was used. HMM profile of MIP (PF00230) was downloaded from Pfam protein family database (http://pfam.sanger.ac.uk/) and used as the query (P < 0.05) to search for AQP proteins in the E. salsugineum genome. To avoid missing potential AQP members, the NCBI BLAST tool was used to search E. salaugineum AQPs and known Arabidopsis AQP protein sequences as a query, and the top five aligned sequences were considered as candidates. After removing all of the redundant sequences, the sequences of putative EsAQP genes were loaded on relative chromosomes of E. salsugineum using the SnapGene tool. The map of the chromosome position of each EsAQP genes was drawn by MapInspect 1.0.
Classification, phylogenetic analysis and structural features
Multiple sequence alignments of putative AQP proteins were performed by ClustalW, and a phylogenetic tree was constructed using neighbor joining with MEGA 6.0 (Tamura et al., 2013). The transmembrane regions were detected using TOPCONS (http://topcons.cbr.su.se/pred/) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). Protein subcellular localization of E. salsugineum AQPs was predicted in Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) and WoLF PSORT (http://www.genscript.com/wolf-psort.html). Functional predictions, such as NPA motifs, ar/R filters (H2, H5, LE1 and LE2), Froger’s positions (P1-P5) and nine specificity-determining positions (SDP1-SDP9), were analyzed by the alignments with function known AQPs (Quigley et al., 2001; Park et al., 2010; Hove & Bhave, 2011). The gene structure for each EsAQP was illustrated with the Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). The conserved motifs of EsAQP proteins were analyzed by MEME suite (http://meme-suite.org/).
Plant materials and stress treatments
E. salsugineum seeds (ecotype Shandong, China) were provided by Prof. Hui Zhang (Shandong Normal University, Jinan, China). The seeds were plated on 1/2 MS medium and treated at 4 °C in dark for 7 days, then cultured in plant growth chamber with illumination of 150 µmol/m2/s, photoperiod 16/8 h of light/darkness at 25 °C and 60% relative humidity. After one week, the seedlings were transferred into a mixed medium with soil and vermiculite (3:1). Vernalization treatment for bolting was conducted in 4-week old seedlings at 4 °C for 4 weeks, and then they were moved back to the growth chamber until they grew flowers. Samples of roots, stems, leaves, flowers and siliques were collected, immediately frozen in liquid nitrogen and stored at −80 °C for further analysis.
For abiotic stress assays, the 4-week old seedlings were exposed to 300 mM NaCl for 24 h as a salt stress condition, treated at 4 °C for 24 h as cold stress, and not irrigated until the soil moisture content was less than 20% for 7 days as drought stress. The aerial part of seedling was collected for further analysis.
RNA extraction, cDNA synthesis and qRT-PCR
The total RNA was extracted using TRIzol reagent (Takara) following the manufacturer’s protocol. The quality of the RNA was determined using an ultraviolet spectrophotometer (BioMate 3S; Thermo Fisher). After removing genomic DNA contamination with DNase I, cDNA was synthesized by using the PrimeScript™ RT Reagent Kit (Takara). Three biological replicates of cDNA samples were used for qRT-PCR analysis with three technical replicates.
Primers of EsPIP genes were designed using Primer 3.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and the reference gene was taken from Wang et al. (2014). All of primers were listed in Table S1. The qRT-PCR analysis was conducted in Applied Biosystems 7500 Real-Time PCR System (ABI, USA) by using SYBR Premix Ex TaqTM II (Takara). Reaction system contained 10 µl SYBR Premix Ex Taq II, 2 µl 5-fold diluted cDNA, 0.8 µl of each primer (10 mM), and ddH2O to a final volume of 20 µl. The PCR program was set as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. Then, a melting curve was generated to analyze the specificity of each primer with a temperature shift from 60 to 95 °C. The fold changes of the EsAQP genes expression under abiotic stresses were calculated with the 2−ΔΔCt method, while the gene expressions level of EsAQP genes in each organ were calculated with the ΔCt method. The heat map of gene expression pattern was visualized using HemI software.
Subcellular localization of EsPIP1;2 and EsPIP2;1 proteins
The coding sequences of EsPIP1;2 and EsPIP2;1 were amplified using primers containing the XbaI/SalI restriction site (Table S2). The purified products were subcloned into a reconstructed pBI121 vector which was composed of the XbaI/SalI site, and GFP. pBI121-EsPIP1;2-GFP and pBI121-EsPIP2;1-GFP vectors were transformed into A. tumefaciens strain GV3101. Then transient transformation in onion epidermis according to the method of Xu et al. (2014) took place. Images of epidermal cells were taken by fluorescence microscope with a mirror unit (U-BW).
Xenopus oocyte expression and osmotic water permeability assay
The coding regions of EsPIP1;2 and EsPIP2;1 were subcloned into pCS107 vector using the restriction sites BamHI and EcoRI (see primers in Table S2). After linearization, the cRNAs were synthesized in vitro using the Sp6 mMessage mMachine kit (Ambion). Oocyte preparation, injection, and expression were performed as described by Hu et al. (2012) with a little modification. A total of 10 nl water or cRNAs of EsPIP1;2 and EsPIP2;1 (one ng/nl) were injected into oocytes, respectively, and then the oocytes were incubated at 18 °C for 48 h in Oocyte Culture Medium (OCM, 50% L-15, 40% HEPES (pH 7.4), 10% calf serum, 0.5% penicillin and 10 mg/ml streptomycin).The osmotic water permeability coefficient of oocytes was determined as described by Zhang & Verkman (1991). To measure the osmotic water permeability coefficient, oocytes were transferred to 5-fold diluted OCM solution. Changes in the oocytes volume were monitored at room temperature with a microscope video system. Oocytes volumes (Vs) were calculated from the measured area of each oocyte. The osmotic Pf was calculated for the Erst 10 min using the formula Pf = V 0[d(V∕V 0)∕dt]∕[S0 × V w(Osmin–Osmout)]. V 0 and S0 are the initial volume and surface area of each individual oocyte, respectively; d(V/V0)/dt is the relative volume increase per unit time; Vw is the molar volume of water (18 cm3 mol−1); and Osmin–Osmout is the osmotic gradient between the inside and outside of the oocyte.
Results
Characterization, classification and chromosome localization of EsAQPs
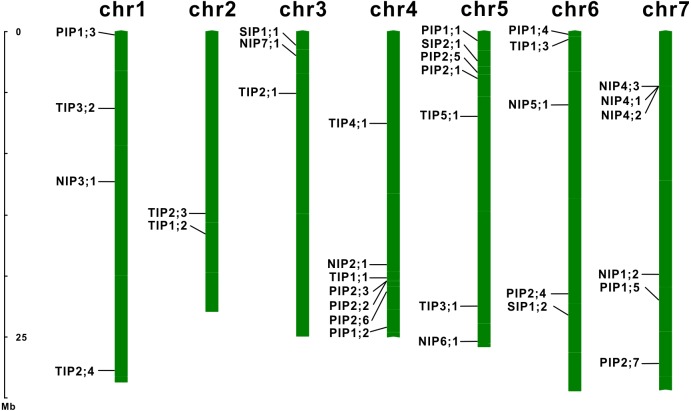
To extensively identify AQPs in E. salsguineum, HMM profile of the MIP domain (PF00230) was used. As a result, a total of 35 putative EsAQPs were identified for further analysis (Table 1). To classify the AQP members, a phylogenetic tree was constructed according to the similarity of AQP protein sequences in E. salsugineum and Arabidopsis through the neighbor-joining method (Fig. 1). Based on the phylogenetic analysis, we found that the identified EsAQPs have very high similarity with AtAQPs which can be grouped into four subfamilies, including 12 PIPs, 11 TIPs, nine NIPs and three SIPs. In addition, the EsPIP subfamily was further divided into two classes (five EsPIP1s and seven EsPIP2s), the EsTIP subfamily into five classes (three EsTIP1s, four EsTIP2s, two EsTIP3s, one EsTIP4s and one EsTIP5s), the EsNIP subfamily into seven classes (1 EsNIP1s, 1 EsNIP2s, 1 EsNIP3s, three EsNIP4s, one EsNIP5s, one EsNIP6s and one EsNIP7s), and the EsSIP subfamily into two classes (two EsSIP1s and one EsSIP2s). The nomenclature of E. salsugineum AQPs was based on their phylogenetic relationship with AtAQPs (Fig. 1). These results were also supported by the existing annotation for E. salsugineum obtained from Phytozyme (https://phytozome.jgi.doe.gov/pz/portal.html#!bulk?org=Org_Esalsugineum), most of our identified EsAQPs were matched to the existing annotations (Table S3). In addition, we have updated some annotations i.e., Thhalv10008397m and Thhalv10025910m, which were both annotated as PIP1;4 in Phytozyme, were renamed with EsPIP1;3 and EsPIP1;5, respectively, and other details were listed in Table S3. Compare to AtAQPs, PIP2;8 and NIP1;1 were missing in E. salsugineum. And TIP2;4 and NIP4;3 were identified in E.salsugineum, but not found in Arabidopsis, which were shared high similarity with their homologous genes. In Arabidopsis, 35 AQP genes were unevenly distributed on the five chromosomes Feng et al. (2017). As shown in Table 1 and Fig. 2, the chromosomal locations of 34 EsAQP genes were randomly assigned to all the seven chromosomes. However, chromosomal location of EsTIP2;2 could not be determined. Overall, AQPs from E. salsugineum had a very close relationship with those from Arabidopsis.
Table 1. Details of EsAQP genes identified from the genome-wide search analysis.
| Name | Chromosomal Localization | Scaffold | Coding sequence | Protein ID | Plant-mPLoc | WoLF PSORT | Plant species | Subcellular localization | Reference |
|---|---|---|---|---|---|---|---|---|---|
| EsPIP1;1 | Chr5;748,014∼746,287 | NW_006256838.1 | XM_006402419.1 | XP_006402482.1 | plas | plas | Oryza ativa | plas | Liu et al. (2013) |
| EsPIP1;2 | Chr4;24,198,933∼24,200,732 | NW_006256812.1 | XM_006397718.1 | XP_006397781.1 | plas | plas | Musa nana | plas | Sreedharan, Shekhawat & Ganapathi (2013) |
| EsPIP1;3 | Chr1;227,418∼229,068 | NW_006256612.1 | XM_006418376.1 | XP_006418439.1 | plas | plas | |||
| EsPIP1;4 | Chr6;182,520∼180,408 | NW_006256756.1 | XM_006396178.1 | XP_006396241.1 | plas | plas | Arabidopsis thaliana | plas | Li et al. (2015) |
| EsPIP1;5 | Chr7;21,955,256∼21,956,964 | NW_006256909.1 | XM_006413496.1 | XP_006413559.1 | plas | plas | |||
| EsPIP2;1 | Chr5;3,815,044∼3,817,131 | NW_006256858.1 | XM_006403628.1 | XP_006403691.1 | plas | plas | A. thaliana | plas | Li et al. (2011) |
| EsPIP2;2 | Chr4;20,408,518∼20,407,373 | NW_006256908.1 | XM_006410833.1 | XP_006410896.1 | plas | plas | Vitis vinifera | plas | Leitäo et al. (2012) |
| EsPIP2;3 | Chr4;20,411,864∼20,413,318 | NW_006256908.1 | XM_006410834.1 | XP_006410897.1 | plas | plas | |||
| EsPIP2;4 | Chr6;21,418,342∼21,416,629 | NW_006256829.1 | XM_006400761.1 | XP_006400824.1 | plas | plas | Zea mays | plas | Zelazny et al. (2009) |
| EsPIP2;5 | Chr5;3,318,416∼3,315,956 | NW_006256858.1 | XM_006403468.1 | XP_006403531.1 | plas | plas | Z. mays | plas | Zelazny et al. (2009) |
| EsPIP2;6 | Chr4;21,319,556∼21,322,584 | NW_006256908.1 | XM_006411061.1 | XP_006411124.1 | plas | plas | M. nana | plas | Sreedharan, Shekhawat & Ganapathi (2015) |
| EsPIP2;7 | Chr7;27,180,960∼27,182,785 | NW_006256909.1 | XM_006412089.1 | XP_006412152.1 | plas | plas | A. thaliana | plas | Hachez et al. (2014) |
| EsTIP1;1 | Chr4;20,182,942∼20,184,210 | NW_006256908.1 | XM_006410791.1 | XP_006410854.1 | vacu | cyto | A. thaliana | vacu | Ma et al. (2004) |
| EsTIP1;2 | Chr2;16,508,526∼16,506,789 | NW_006256547.1 | XM_006395487.1 | XP_006395549.1 | vacu | plas/vacu | Eutrema salsiguneum | vacu | Wang et al. (2014) |
| EsTIP1;3 | Chr6;663,103∼662,130 | NW_006256756.1 | XM_006396285.1 | XP_006396348.1 | vacu | cyto | |||
| EsTIP2;1 | Chr3;5,624,419∼5,626,413 | NW_006256885.1 | XM_006406794.1 | XP_006406857.1 | vacu | chlo/vacu | A. thaliana | vacu | Loque et al. (2005) |
| EsTIP2;2 | NA | NW_006256909.1 | XM_006414179.1 | XP_006414242.1 | vacu | vacu | Triticum aestivum | vacu | Chunhui et al. (2013) |
| EsTIP2;3 | Chr2;14,894,399∼14,893,306 | NW_006256828.1 | XM_006398375.1 | XP_006398438.1 | vacu | vacu | A. thaliana | vacu | Loque et al. (2005) |
| EsTIP2;4 | Chr1;27,709,976∼27,708,236 | NW_006256486.1 | XM_006392888.1 | XP_006392950.1 | vacu | vacu | |||
| EsTIP3;1 | Chr5;22,490,388∼22,491,488 | NW_006256342.1 | XM_006390520.1 | XP_006390582.1 | vacu | chlo/cyto/vacu | A. thaliana | plas/vacu | Gattolin, Sorieul & Frigerio (2011) |
| EsTIP3;2 | Chr1;6,309,744∼6,311,048 | NW_006256612.1 | XM_006416602.1 | XP_006416665.1 | vacu | chlo/mito/vacu | A. thaliana | plas/vacu | Gattolin, Sorieul & Frigerio (2011) |
| EsTIP4;1 | Chr4;7,484,947∼7,486,691 | NW_006256895.1 | XM_006408738.1 | XP_006408801.1 | vacu | vacu | |||
| EsTIP5;1 | Chr5;6,934,814∼6,933,858 | NW_006256858.1 | XM_006404316.1 | XP_006404379.1 | vacu / plas | chlo | A. thaliana | mito | Soto et al. (2010) |
| EsNIP1;2 | Chr7;19,890,089∼19,892,520 | NW_006256909.1 | XM_006413978.1 | XP_006414041.1 | plas | plas | A. thaliana | plas | Wang et al. (2017) |
| EsNIP2;1 | Chr4;19,043,681∼19,042,522 | NW_006256908.1 | XM_006410521.1 | XP_006410584.1 | plas | vacu: | A. thaliana | plas/E.R | Choi & Roberts (2007), Mizutani et al. (2006) |
| EsNIP3;1 | Chr1;12,292,410∼12,294,335 | NW_006256612.1 | XM_006415218.1 | XP_006415281.1 | plas | vacu | O. sativa | plas | Hanaoka et al. (2014) |
| EsNIP4;1 | Chr7;4,484,562∼4,482,986 | NW_006256877.1 | XM_006405767.1 | XP_006405830.1 | plas | plas | A. thaliana | plas/vacu | DiGiorgio et al. (2016) |
| EsNIP4;2 | Chr7;4,513,301∼4,511,485 | NW_006256877.1 | XM_006405768.1 | XP_006405831.1 | plas | plas | A. thaliana | plas/vacu | DiGiorgio et al. (2016) |
| EsNIP4;3 | Chr7;4,481,446∼4,479,745 | NW_006256877.1 | XM_006405766.1 | XP_006405829.1 | plas | plas | |||
| EsNIP5;1 | Chr6;6,005,178∼6,008,910 | NW_006256756.1 | XM_006397006.1 | XP_006397069.1 | plas | plas | A. thaliana | plas | Takano et al. (2006) |
| EsNIP6;1 | Chr5;25,383,958∼25,386,014 | NW_006256342.1 | XM_006389768.1 | XP_006389830.1 | plas | plas | A. thaliana | plas | Tanaka et al. (2008) |
| EsNIP7;1 | Chr3;1,929,290∼1,927,201 | NW_006256885.1 | XM_006407920.1 | XP_006407983.1 | plas | cyto | |||
| EsSIP1;1 | Chr3;1,105,251∼1,102,416 | NW_006256885.1 | XM_024159977.1 | XP_024015745.1 | plas | plas | A. thaliana | E.R | Ishikawa et al. (2005) |
| EsSIP1;2 | Chr6;23,161,081∼23,162,581 | NW_006256829.1 | XM_006400314.1 | XP_006400377.1 | vacu plas | vacu | A. thaliana | E.R | Ishikawa et al. (2005) |
| EsSIP2;1 | Chr5;2,401,441∼2,403,463 | NW_006256838.1 | XM_006402867.1 | XP_006402930.1 | plas | E.R | A. thaliana | E.R | Ishikawa et al. (2005) |
Notes.
Abbreviation
- plas
- plasma membrane
- cyto
- cytosol
- vacu
- tonoplast membrane
- chlo
- chloroplast
- mito
- mitochondria
- E.R
- endoplasmic reticulum
- NA
- not applicable
Figure 1. Phylogenetic tree of AQP amino acid sequences from E. salsugineum and A. thaliana.
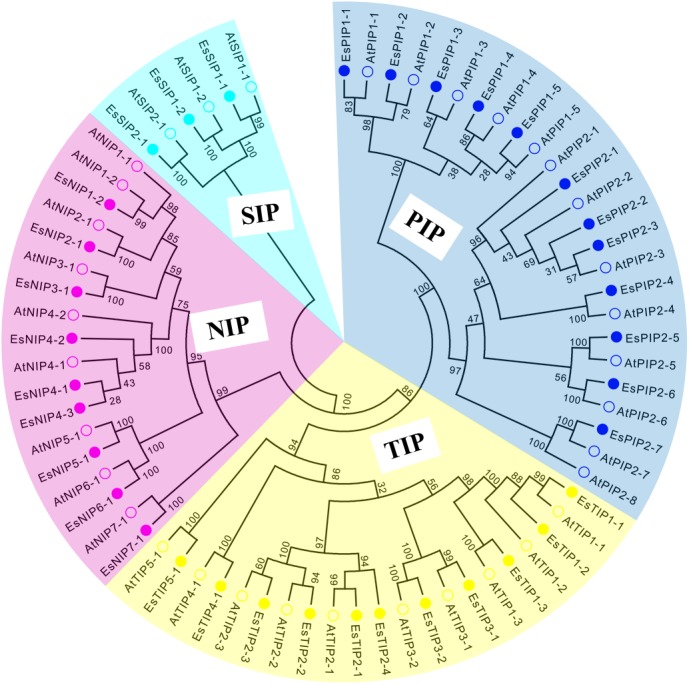
Alignments were performed using the default parameter of ClustalW and the phylogenetic tree was constructed using the Neighbor-Joining tree method with 1,000 bootstrap replicates in MEGA6.0 software. Each subfamily of AQPs was well separated in different clades and represented by different colors. The solid circle represents EsAQPs and the hollow circle represents AtAQPs.
Figure 2. Chromosomal localization of the EsAQP genes.
The diagram was drawn using the MapInspect software, and 34 out of 35 EsAQPs were located on seven chromosomes (except EsTIP2;2).
Gene structure and subcellular localization analysis of EsAQPs
Gene structure analysis of the 35 EsAQP genes was performed on the Gene Structure Display Server of NCBI. Based on their mRNA and genomic DNA sequences, we found exon lengths were mostly conserved in each subfamily of EsAQP gene with same exon number, but introns varied in both length and position (Fig. 3). All members of the EsPIP subfamily contained four exons with similar length (289–328, 296, 141 and 93–126 bp, respectively) and conserved sequences in the 2nd and 3rd exon, except for EsPIP2;4, which had a shorter 2nd and longer 3rd exon (307, 151, 286, and 111 bp). The majority members of the EsTIP subfamily contained three exons with similar lengths, and the other members had two exons with similar lengths, except for EsTIP1;3, which had only one exon without intron. In the EsNIP subfamily, some members exhibited five exons with similar lengths, while others had four exons with varied lengths. All EsSIP subfamily genes displayed three exons with similar lengths. This description of exon-intron structure provides additional evidence to support the classification results (Kong et al., 2017).
Figure 3. Gene structures of the EsAQP genes.
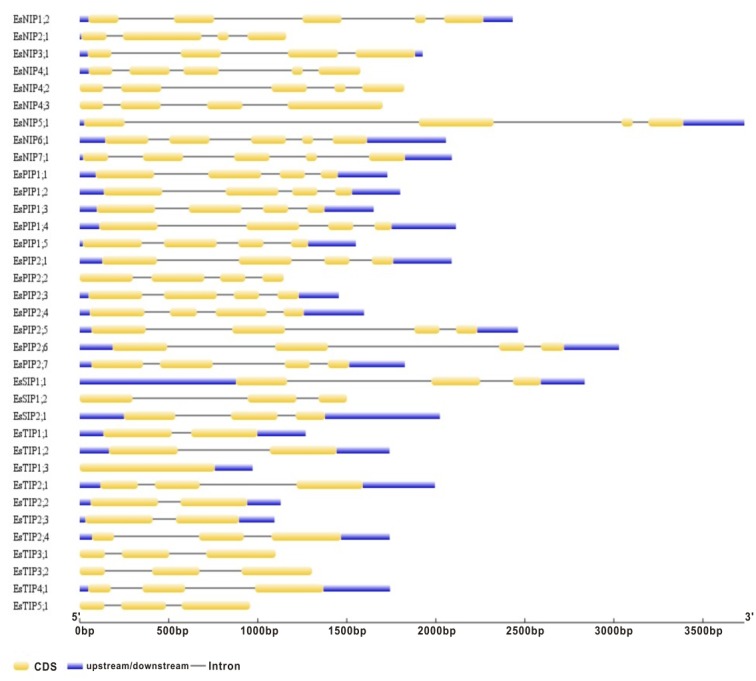
The blue rectangle, yellow rectangle and black line represent UTR, exon and intron, respectively.
The prediction of subcellular localization showed diverse results, not always in agreement with experimentally determined localizations (reviewed in Katsuhara et al., 2008). In summary, the prediction of EsAQP subcellular localization in Plant-mPLoc showed that EsPIP, EsNIP and EsSIP subfamilies were localized in plasma membrane, while EsTIP subfamily members were localized in tonoplast membrane. Among them, EsPIP1;2 and EsTIP5;1 were localized in both tonoplast membrane and plasma membrane (Table 1). Moreover, WoLF PSORT predicts different location for EsAQPs and assigns values for that location (Table S4). The highest values list in Table 1 showed that EsPIPs were predicted to localize in plasma membrane, which were consistent with the Plant-mPLoc prediction and many other reports (Cui et al., 2008; Hu et al., 2012; Xu et al., 2014). The majority of other AQP members were predicted to localize in plasma membrane or tonoplast membrane, except for EsTIP5;1, EsNIP7;1 and EsSIP2;1, which were predicted to be associated with chloroplast, cytosol and endoplasmic reticulum, respectively. Moreover, some members showed multiple type of localization; for example, EsTIP3;1 was predicted to be associated with chloroplast/cytosol/tonoplast membrane and EsTIP3;2 with chloroplast/mitochondria/tonoplast membrane. The subcellular localization of most published AQP homologous was consistent with the predicted results in E. salsugineum (Table 1). These observations demonstrated that the subcellular localization of AQPs may be complex and diverse.
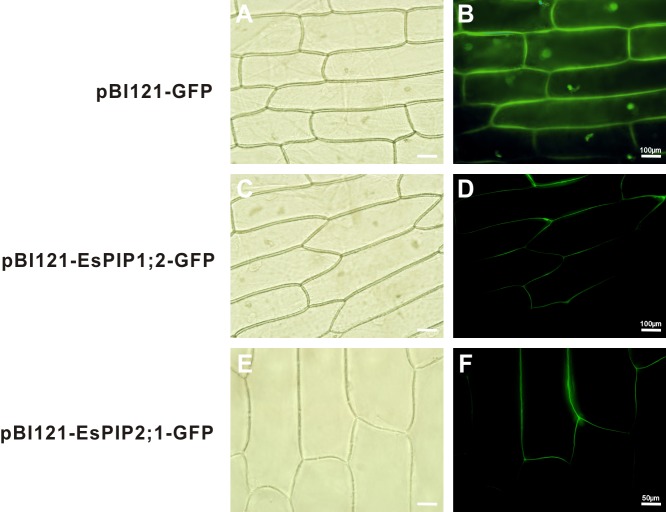
To verify the predictions, genes of EsPIP1;2 and EsPIP2;1 were cloned into the pBI121-GFP vector to create the 35S::EsPIP-GFP fusion proteins. The plasmid was transformed into onion epidermis by agrobacterium-mediated transformation. As shown in Fig. 4, the GFP fluorescence mainly exhibit in plasma membrane, indicated that EsPIP1;2 and EsPIP2;1 proteins were consistent with the predictions. Although not conclusive, the predicted localization could serve as a useful reference for further studies on EsAQPs protein functions in plants.
Figure 4. Subcelluar localizations of EsPIP1;2 and EsPIP2;1 proteins.
Onion epidermal cells transiently transformed with empty vector (A, B), EsPIP1;2-GFP (C, D) and EsPIP2;1-GFP (E, F), respectively. The images were visualized under fluorescence microscope. A, C, E: bright-field images; B, D, F: green fluorescence images.
Structure characteristics of EsAQPs
Sequence analysis showed that all EsAQPs contain six transmembrane domains (TMDs) comprising 237–323 amino acids, had theoretical molecular weights (MW) of 24.31–31.80 kDa and isoelectric point (pI) values of 4.73–10.49 (Table 2). The EsPIP subfamily had a similar molecular weight of approximately 30.84 kDa. Most members of the EsNIP subfamily exhibited a similar molecular weight and isoelectric point of EsPIP subfamily. The EsTIP and EsSIP subfamilies had lower MW among the EsAQPs, and the isoelectric points of these two subfamilies were acidic and alkaline, respectively (Fig. S1).
Table 2. Structural characteristics of the EsAQPs.
| Name | AA | TM | MW(KD) | pI | NPA motif | ar/R selectivity filter | Froger’s positions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LB | LE | H2 | H5 | LE1 | LE2 | P1 | P2 | P3 | P4 | P5 | |||||
| PIPs | |||||||||||||||
| EsPIP1;1 | 286 | 6 | 30.77 | 9.14 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP1;2 | 286 | 6 | 30.60 | 9.16 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP1;3 | 286 | 6 | 30.62 | 9.02 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP1;4 | 286 | 6 | 30.56 | 9.02 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP1;5 | 287 | 6 | 30.61 | 9.00 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;1 | 287 | 6 | 30.48 | 6.95 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;2 | 284 | 6 | 30.21 | 6.50 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;3 | 285 | 6 | 30.31 | 6.51 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;4 | 285 | 6 | 30.12 | 7.62 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;5 | 286 | 6 | 30.57 | 8.82 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;6 | 290 | 6 | 31.11 | 7.69 | NPA | NPA | F | H | T | R | Q | S | A | F | W |
| EsPIP2;7 | 281 | 6 | 29.82 | 9.11 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| TIPs | |||||||||||||||
| EsTIP1;1 | 251 | 6 | 25.62 | 6.03 | NPA | NPA | H | I | A | V | T | A | A | Y | W |
| EsTIP1;2 | 253 | 6 | 25.70 | 5.32 | NPA | NPA | H | I | A | V | T | A | A | Y | W |
| EsTIP1;3 | 252 | 6 | 25.85 | 5.10 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| EsTIP2;1 | 277 | 6 | 28.32 | 7.80 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| EsTIP2;2 | 250 | 6 | 25.02 | 4.87 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| EsTIP2;3 | 243 | 6 | 24.31 | 4.73 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| EsTIP2;4 | 254 | 6 | 25.85 | 5.43 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| EsTIP3;1 | 265 | 6 | 27.94 | 7.17 | NPA | NPA | H | T | A | R | T | A | A | Y | W |
| EsTIP3;2 | 267 | 6 | 28.29 | 6.58 | NPA | NPA | H | M | A | R | T | T | A | Y | W |
| EsTIP4;1 | 249 | 6 | 26.16 | 5.49 | NPA | NPA | H | I | A | R | T | S | A | Y | W |
| EsTIP5;1 | 257 | 6 | 26.70 | 7.72 | NPA | NPA | N | V | G | C | V | A | A | Y | W |
| NIPs | |||||||||||||||
| EsNIP1;2 | 297 | 6 | 31.80 | 8.83 | NPA | NPA | W | V | A | R | F | S | A | Y | L |
| EsNIP2;1 | 286 | 6 | 30.56 | 6.78 | NPA | NPG | W | V | A | R | F | S | A | Y | L |
| EsNIP3;1 | 323 | 6 | 34.46 | 5.94 | NPA | NPA | W | I | A | R | F | S | A | Y | L |
| EsNIP4;1 | 283 | 6 | 30.49 | 8.73 | NPA | NPA | W | V | A | R | F | S | A | Y | L |
| EsNIP4;2 | 284 | 6 | 30.34 | 8.80 | NPA | NPA | W | V | A | R | F | S | A | Y | L |
| EsNIP4;3 | 283 | 6 | 30.30 | 8.98 | NPA | NPA | W | V | A | R | F | S | A | Y | L |
| EsNIP5;1 | 301 | 6 | 31.20 | 8.31 | NPS | NPA | A | I | G | R | F | T | A | Y | L |
| EsNIP6;1 | 305 | 6 | 31.78 | 8.57 | NPA | NPA | A | I | A | R | F | T | A | Y | L |
| EsNIP7;1 | 275 | 6 | 28.62 | 6.12 | NPS | NPA | A | V | G | R | Y | S | A | Y | L |
| SIPs | |||||||||||||||
| EsSIP1;1 | 238 | 6 | 25.41 | 9.89 | NPT | NPA | I | V | P | I | I | A | A | Y | W |
| EsSIP1;2 | 242 | 6 | 25.96 | 9.83 | NPC | NPA | V | F | P | I | I | A | A | Y | W |
| EsSIP2;1 | 237 | 6 | 25.85 | 9.64 | NPL | NPA | S | H | G | A | F | V | A | Y | W |
Notes.
Abbreviation
- AA
- amino acids length
- TM
- transmembrane domain
- MW
- molecular weight
- pI
- isoelectricpoint NPA Asn-Pro-Ala motif
- ar/R
- aromatic/arginine
NPA motifs, ar/R selectivity filters and Froger’s positions of AQP protein sequences play critical role in channel selectivity. The sequence alignment between AtAQPs and GhAQPs was carried out to analyze the conserved domains (Quigley et al., 2001; Park et al., 2010). The results in Table 2 showed that all EsPIP subfamily members had two typical NPA motifs in loop B and loop E, with a water transport ar/R filter with amino acid of F-H-T-R. Froger’s position consists of Q-S-A-F-W in most cases, except for EsPIP2;7, which had an M at the P1 position. All EsTIP subfamily had two typical NPA motifs. The ar/R was composed of H–I-A-V in EsTIP1s, H-I-G-R in EsTIP2s and H-T/M/I-A-R in other EsTIP members, while in EsTIP5;1, it was composed of N-V-G-C. Froger’s position consists of T-A/S-A-Y-W, except for EsTIP5;1 and EsTIP3;2, which had a V at the P1 position and a T at the P2 position respectively. Most members of EsNIP subfamily had two typical NPA motifs, not in EsNIP2;1 (with an NPG in LE), EsNIP5;1 and EsNIP7;1 (with an NPS in LB). The ar/R filter consists of residues like W/A-V/I-A/G-R, and Froger’s position consists of F-S-A-Y-L, except for EsNIP7;1, which had a Y at the P1 position, and for EsNIP5;1 and EsNIP6;1 had a T at the P2 position. The EsSIP subfamily showed a variable site in the first NPA, the alanine (A) was replaced by threonine (T), cysteine (C) or leucine (L). The ar/R filter was also inconsistent with each other: I-V-P-I in EsSIP1;1, V-F-P-I in EsSIP1;2 and S-H-G-A in EsSIP2;1. The Forger’s position was composed of I-A-A-Y-W in EsSIP1s, while it was F-V-A-Y-W in EsSIP2;1.
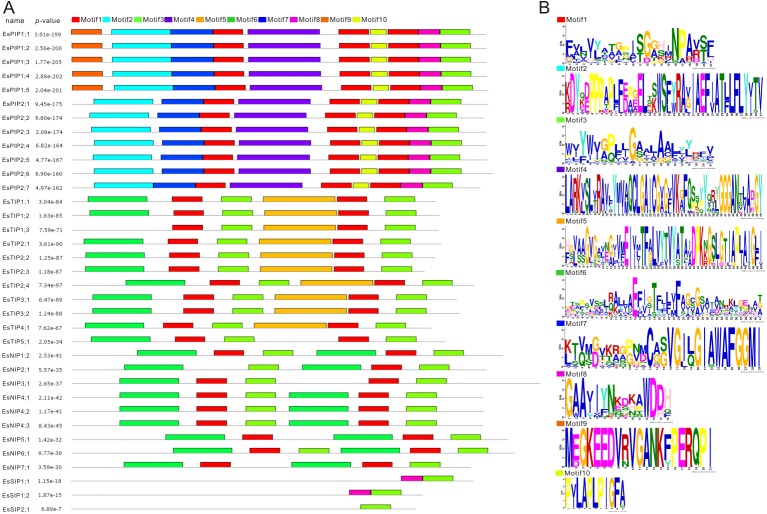
MEME (Multiple EM for Motif Elicitation) is one of the most widely used tools for searching for novel “signals” in sets of biological sequences, include the discovery of new transcription factor binding sites and protein domains (Bailey et al., 2006). Conserved motifs of EsAQP proteins were predicted by MEME suite (Fig. 5). The result showed that motif 1, 2, 3, 4, 7, 8, and 10 were same in all EsPIPs, and motif 2, 4, 7, and 10 were unique. In addition, motif 9 was unique in EsPIP1s and can be used to distinguish EsPIP1s from EsPIP2s. This pattern of conserved motifs in the PIP subfamily also occurs in other plants and PIP1s contain one unique motif (Tao et al., 2014; Yuan et al., 2017). In the EsTIP subfamily, almost all EsTIPs had two motif 1, two motif 3, one motif 5 and one motif 6. Except for EsTIP1;3, which had no motif 6. Motif 5 could be an identifier of EsTIPs among the AQPs of E. salsugineum except for EsTIP5;1. Most of members in NIP subfamily had two motif 1, two motif 3, and two motif 6, except for EsNIP2;1 (lose one motif 1), EsNIP3;1 (lose one motif 6) and EsNIP5;1 (lose one motif 3). The two motif 6 might be used to distinguish EsNIPs with other EsAQPs. All EsSIP subfamily carried motif 3. Motif 8 appeared in EsSIP1s but not in EsSIP2;1, so it might be an specific trait of this group. This is a common phenomenon in plant SIP subfamily contains less motifs (Tao et al., 2014; Reddy et al., 2015; Yuan et al., 2017; Kong et al., 2017). Based on these analysis, it was evident that there were structural differences in various EsAQP subfamilies, but conserved in their own subfamily.
Figure 5. Conversed motif analysis in EsAQPs.
The conversed motif prediction was identified using MEME motif search analysis, and the maximum number parameter was set to 10. Different motifs were represented by different colors. (A) Conversed motifs of 35 EsAQP proteins correspond to p-values. (B) Motif consensus sequences.
Expression pattern of EsAQPs
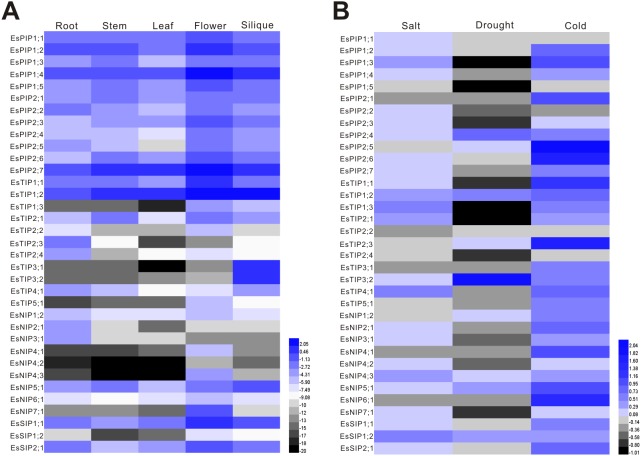
The expression of EsAQP genes in different organs, including root, stem, leaf, flower and silique, was analyzed by RT-qPCR. The results showed that 35 EsAQP genes were detected in all the organs (Fig. 6A). Almost all EsPIP genes were highly expressed in all organs, except for EsPIP2;5 in leaf. In addition, the EsPIP genes, EsTIP1;1, EsTIP1;2, EsNIP1;2, EsNIP5;1, EsSIP1;1 and EsSIP2;1 were also highly expressed in all organs. Some EsAQP genes, such as EsTIP2;3, EsTIP2;4, EsNIP2;1 and EsNIP3;1, were specifically highly expressed in root. Two EsTIPs (EsTIP2;2 and EsTIP5;1), three EsNIPs (EsNIP4;1, EsNIP4;3 and EsNIP7;1) and EsSIP1;2 were highly expressed only in flower. Two EsTIPs (EsTIP3;1 and EsTIP3;2) were expressed in silique with relative high abundance. Compared analysis of each EsAQP gene between different organs revealed that most EsAQP genes showed higher expression level in flower than in other organs.
Figure 6. Expression profiles of the EsAQP genes.
(A) EsAQP genes expression in response to abiotic stress. The color scale represents the 2−ΔΔCt value normalized to untreated controls and log2 transformed counts, where green indicates downregulated expression and red indicates upregulated expression. (B) Expression of EsAQP genes in various organs of E. salsugineum. Color scales represent 2ΔCt values normalized to actin and log2 transformed counts, where green indicates low expression and red indicates high expression.
Abiotic stresses are the main limiting factors for plants during environmental conditions that induce osmotic stress and disturb water balance. AQPs play major roles in maintaining water homeostasis and responding to environmental stresses in plants. Therefore, we further investigated the expression patterns of EsAQP genes under salt, drought and cold stress by qRT-PCR. The results showed that most of the EsAQP genes were up-regulated under salt and cold stress but down-regulated under drought stress (Fig. 6B). We found that five EsAQP genes were up-regulated under all the types of abiotic stresses, including EsPIP2;4, EsTIP1;2, EsNIP4;3, EsNIP5;1 and EsSIP1;2, while three EsAQP genes were down-regulated under all the types of abiotic stresses, including EsPIP1;5, EsTIP2;2 and EsTIP2;4. In addition, EsPIP1;1 and EsPIP2;2 were specifically up-regulated under salt stress, and EsPIP2;1, EsTIP2;1, EsTIP5;1, EsNIP4;1 and EsNIP6;1 were up-regulated only under cold stress.
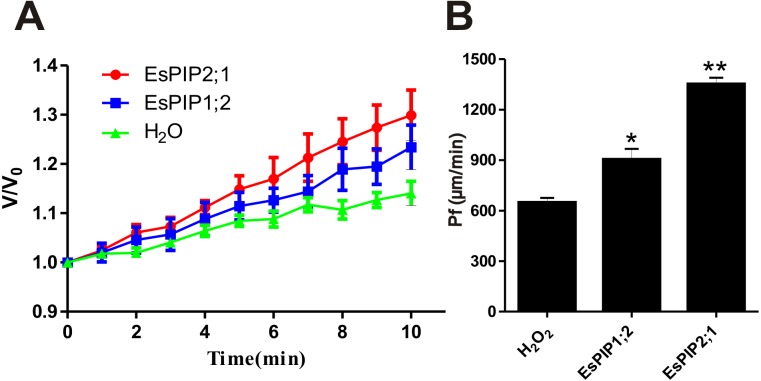
Water permeability of EsPIP1;2 and EsPIP2;1
Previously, AtPIP2;1 has been reported that is an integral membrane protein that facilitates water transport across plasma membrane while AtPIP1;2 has no function (Li et al., 2011; Heckwolf et al., 2011). To determine the water channel activity of EsPIP1;2 and EsPIP2;1, proteins were tested in the Xenopus oocyte system. After two days of cRNA or water injection, the change rate in oocyte volume (Fig. 7A) and the osmotic water permeability coefficient (Pf) (Fig. 7B) were calculated. Expression of EsPIP2;1 conferred a rapid osmotically driven increase in relative volume, while expression of EsPIP1;2 enabled an increase in relative volume at a slower rate than the water-injected oocytes. Compared with water-injected control, the oocytes expressing EsPIP1;2 and EsPIP2;1 showed 1.39-fold and 2.08-fold increase in Pf, suggesting that both EsPIP1;2 and EsPIP2;1 are functional AQP with water channel activity. Meanwhile, our result is consistent with the known information that PIP2s have high efficiency water transfer activity but PIP1s have little or no increase in the Pf (Chaumont & Tyerman, 2014).
Figure 7. Water channel activity appraisals of EsPIP1;2 and EsPIP2;1.
(A) The swelling rates of Xenopus oocytes injected with H2O, or cRNA encoding EsPIP1;2 and EsPIP2;1, respectively. The rate of oocyte swelling upon immersion in hypo-osmotic medium is drawn as V/V0, where V is the volume at a given time point and V0 is the initial volume. (B) Water permeability codfficient (Pf) of oocytes injected with cRNA encoding H2O, or EsPIP1;2, or EsPIP2;1. The Pf values were calculated from the rate of oocyte swelling. Vertical bars indicate the SE. Asterisks indicate significant differences in comparison with oocytes injected with water. Statistical analysis were performed by SPSS 16.0 using one-way ANOVA and Least Significant Difference (LSD) test to detect significant differences (*p < 0.05, **p < 0.01).
Discussion
Gene duplication is a ubiquitous event that plays an important role in biological evolution, which may also contribute to stress tolerance via gene dosage increasing, avoiding some deleterious mutations and creating the opportunity for new function emergence (Innan & Kondrashov, 2010). AQPs are abundant, diverse and widely distributed in plants and involved in regulation of plant growth and development. From algae (two in Thalassiosira pseudonana and five in Phaeodactylum tricornutum) (Armbrust et al., 2004; Bowler et al., 2008) to fern (19 in Selaginella moellendorffii) (Danielson & Johanson, 2008) and moss (23 in Physcomitrella patens) (Anderberg, Kjellbom & Johanson, 2012) to higher plants (35 AQPs in Arabidopsis, 33 in Oryza sativa, 72 in Glycine max) (Johanson et al., 2001; Sakurai et al., 2005; Zhang et al., 2013), the number of AQPs has largely increased with evolution. Here, we provide a genome-wide information of AQP family of E. salsugineum.
In previous studies, it was shown that more than 95% gene families are shared in T. salsuginea (the former name of E. salsugineum, Koch & German, 2013) with A. thaliana (Wu et al., 2012) or more than 80% E. salsugineum genes had high homology orthologs in A. thaliana (Yang et al., 2013). The number of AQPs identified in E. salsugineum is the same as that in A. thaliana, and their protein sequences have very high similarities. No homologies of AtAQPs, PIP2;8 and NIP1;1 were not identified in E. salsugineum, while another two AQPs, TIP2;4 and NIP4;3 were found instead, which were not existed in Arabidopsis. These differences may not be directly illustrated the superiority of E. salsugineum in stress resistance, the functions of EsAQPs in resistance need to be further studied.
Structural analysis and functional inference of EsAQPs
Exon-intron structural divergence commonly happened in duplicate gene evolution and even in sibling paralogs; these changes occurred through the mechanisms of gain/loss, exonization/pseudoexonization and insertion/deletion (Xu et al., 2012). In common bean (Phaseolus vulgaris L.), each aquaporin subfamily are completely conserved in number, order and length of exons but varies in introns (Ariani & Gepts, 2015). The MEME motifs of the AQPs were conserved in all subfamilies, while a few were deleted, unique or family-specific, and a previous report also found this pattern in ZmPIPs (Bari et al., 2018). In our study, the exon-intron structure of EsAQP genes and the conserved MEME motifs of EsAQP protein sequences showed some common patterns (Figs. 3 and 5). These results indicated that the gene structure and the conserved motifs of EsAQPs shown subfamily-specific, these traits may provide new evidence to support the classification.
High conservation of signature sequences or residues was shown in plant PIP proteins. In our study (Table 2), EsPIPs showed a typical NPA motif, a highly conserved ar/R selectivity filter and Froger’s position of F-H-T-R and Q/M-S-A-F-W, these characteristics are correlated with water transport activity (Quigley et al., 2001). In addition to water transport, plant PIPs also could transfer carbon dioxide, hydrogen peroxide, boric acid, and urea (Gaspar et al., 2003; Bienert et al., 2014; Heckwolf et al., 2011). According to the SDP analysis proposed by Hove & Bhave (2011), all EsPIPs had H2O2-type and urea-type SDPs (Table 3, Fig. S2). In addition, all EsPIP1s and EsPIP2;5 had boric acid-type SDPs, and all EsPIP1s had CO2-type SDPs, including two novel types of SDP showed in EsPIP1;3 and EsPIP1;4 which have an M in place of I in SDP2, it also have been found in RcPIPs, JcPIPs and BvPIPs (Zou et al., 2015; Zou et al., 2016; Kong et al., 2017). In addition, EsPIP2;4 owned another novel CO2-type SDPs (V-I-C-A-V-E-W-D-W), with E replaced by D in SDP6. These results showed the conservation of plant PIPs in the transport of urea and hydrogen peroxide (Gaspar et al., 2003; Bienert et al., 2014), and PIP1s not PIP2s are main CO2 and boric acid channels (Heckwolf et al., 2011).
Table 3. Identified typical SDPs in EsAQPs.
The red font represent novel site.
| Aquaporin | Specificity-determining positions | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SDP1 | SDP2 | SDP3 | SDP4 | SDP5 | SDP6 | SDP7 | SDP8 | SDP9 | |
| Ammonia Transporters | F/T | K/L/N/V | F/T | V/L/T | A | D/S | A/H/L | E/P/S | A/R/T |
| EsTIP2;1 | T | L | T | V | A | S | H | P | A |
| EsTIP3;1 | T | L | G | T | A | S | H | P | A |
| EsNIP1;2 | F | K | F | T | G | D | L | E | T |
| EsNIP4;1 | F | T | F | T | A | D | L | E | T |
| EsNIP4;3 | F | T | F | T | A | D | L | E | T |
| Boric Acid transporter | T/V | I/V | H/I | P | E | I/L | I/L/T | A/T | A/G/P/K |
| EsPIP1;1 | T | I | H | P | E | L | L | T | P |
| EsPIP1;2 | T | I | H | P | E | L | L | T | P |
| EsPIP1;3 | T | I | H | P | E | L | L | T | P |
| EsPIP1;4 | T | I | H | P | E | L | L | T | P |
| EsPIP1;5 | T | I | H | P | E | L | L | T | P |
| EsPIP2;5 | T | I | H | P | E | L | L | T | P |
| EsNIP5;1 | T | I | H | P | E | L | L | A | P |
| EsNIP6;1 | T | I | H | P | E | L | L | A | P |
| EsNIP7;1 | V | I | H | P | E | L | L | T | P |
| CO2transporter | I/L/V | I | C | A | I/V | D | W | D | W |
| EsPIP1;1 | L | I | C | A | I | D | W | D | W |
| EsPIP1;2 | V | I | C | A | I | D | W | D | W |
| EsPIP1;3 | V | M | C | A | I | D | W | D | W |
| EsPIP1;4 | V | M | C | A | I | D | W | D | W |
| EsPIP1;5 | V | I | C | A | I | D | W | D | W |
| EsPIP2;4 | V | I | C | A | V | E | W | D | W |
| H2O2transporters | A/S | A/G | L/V | A/F/L/V/T | I/L/V | H/I/L/Q | F/Y | A/V | P |
| EsPIP1;1 | A | G | V | F | I | H | F | V | P |
| EsPIP1;2 | A | G | V | F | I | H | F | V | P |
| EsPIP1;3 | A | G | V | F | I | H | F | V | P |
| EsPIP1;4 | A | G | V | F | I | H | F | V | P |
| EsPIP1;5 | A | G | V | F | I | H | F | V | P |
| EsPIP2;1 | A | G | V | F | I | H | F | V | P |
| EsPIP2;2 | A | G | V | F | I | H | F | V | P |
| EsPIP2;3 | A | G | V | F | I | H | F | V | P |
| EsPIP2;4 | A | G | V | F | I | Q | F | V | P |
| EsPIP2;5 | A | G | V | F | I | H | F | V | P |
| EsPIP2;6 | A | G | V | F | I | Q | F | V | P |
| EsPIP2;7 | A | G | V | F | I | H | F | V | P |
| EsTIP1;1 | S | A | L | A | I | H | Y | A | P |
| EsTIP1;2 | S | A | L | A | I | H | Y | A | P |
| EsTIP1;3 | A | A | L | S | I | H | Y | V | P |
| EsTIP2;1 | S | A | L | V | I | H | Y | V | P |
| EsTIP2;2 | S | A | L | V | I | I | Y | V | P |
| EsTIP2;3 | S | A | L | V | I | I | Y | V | P |
| EsTIP3;2 | A | A | L | A | I | H | Y | V | P |
| EsTIP4;1 | S | A | L | L | T | H | Y | V | P |
| EsNIP1;2 | S | A | L | L | V | I | Y | V | P |
| EsNIP3;1 | S | A | L | V | I | L | Y | V | P |
| EsNIP5;1 | S | A | L | V | V | L | Y | V | P |
| Silicic acid transporters | C/S | F/Y | A/E/L | H/R/Y | G | K/N/T | R | E/S/T | A/K/P/T |
| Not found | |||||||||
| Urea Transporters | H | P | F/I/L/T | A/C/F/L | L/M | A/G/P | G/S | G/S | N |
| EsPIP1;1 | H | P | F | F | L | P | G | G | N |
| EsPIP1;2 | H | P | F | F | L | P | G | G | N |
| EsPIP1;3 | H | P | F | F | L | P | G | G | N |
| EsPIP1;4 | H | P | F | F | L | P | G | G | N |
| EsPIP1;5 | H | P | F | F | L | P | G | G | N |
| EsPIP2;1 | H | P | F | F | L | P | G | G | N |
| EsPIP2;2 | H | P | F | F | L | P | G | G | N |
| EsPIP2;3 | H | P | F | F | L | P | G | G | N |
| EsPIP2;4 | H | P | F | F | L | P | G | G | N |
| EsPIP2;5 | H | P | F | F | L | P | G | G | N |
| EsPIP2;6 | H | P | F | F | L | P | G | G | N |
| EsPIP2;7 | H | P | F | F | L | P | G | G | N |
| EsTIP1;1 | H | P | F | F | L | A | G | S | N |
| EsTIP1;2 | H | P | F | F | L | A | G | S | N |
| EsTIP1;3 | H | P | F | F | L | A | G | S | N |
| EsTIP2;1 | H | P | F | A | L | P | G | S | N |
| EsTIP2;2 | H | P | L | A | L | P | G | S | N |
| EsTIP2;3 | H | P | L | A | L | P | G | S | N |
| EsTIP2;4 | H | P | F | V | L | P | G | S | N |
| EsTIP3;1 | H | P | F | L | L | P | G | S | N |
| EsTIP3;2 | H | P | L | L | L | P | G | S | N |
| EsTIP4;1 | H | P | I | L | L | A | G | S | N |
| EsTIP5;1 | H | P | F | A | L | P | G | S | N |
| EsNIP1;2 | H | P | I | A | L | P | G | S | N |
| EsNIP2;1 | H | P | I | A | L | E | G | S | N |
| EsNIP3;1 | H | P | I | A | L | P | G | S | N |
| EsNIP4;1 | H | P | V | A | L | P | G | S | N |
| EsNIP4;2 | H | P | F | A | L | P | G | S | N |
| EsNIP4;3 | H | P | I | A | L | P | G | S | N |
| EsNIP5;1 | H | P | I | A | L | P | G | S | N |
| EsNIP6;1 | H | P | I | A | L | P | S | S | N |
| EsNIP7;1 | H | P | I | A | V | P | G | S | N |
Compared to PIPs, TIPs are more diverse which have a variety of selectivity filters. Two typical NPA motifs were found in all the EsTIPs, and the ar/R filters and Froger’s position were conserved in the EsTIP1s and EsTIP2s classes, but different with other classes. All the EsTIPs showed urea-type SDPs, and most of them had H2O2-type SDPs (except for EsTIP3;1 and EsTIP5;1). EsTIP2;1 had an NH3-type SDPs, as confirmed in Arabidopsis TIP2;1 (Loque et al., 2005). EsTIP3;1 possessed a novel NH3-type SDPs (T-L-G-T-A-S-H-P-A) with F/T replaced by G in SDP3. The NIP subfamily has low intrinsic water permeability and the ability to transport solutes like glycerol and ammonia (Choi & Roberts, 2007). Most EsNIPs held two typical NPA motifs, but some varied at the third residue in the first or second NPA motif. All EsNIPs had urea-type SDPs, EsNIP1;2, EsNIP3;1 and EsNIP5;1 had H2O2-type SDPs. EsNIP5;1, EsNIP6;1 and EsNIP7;1 had boric acid-type SDPs, which have been found in Arabidopsis (Takano et al., 2006). EsNIP1;2 possessed a novel NH3-type SDPs with a substitution of G for A at SDP4. In addition, EsNIP4;1 and EsNIP4;3, which both had the substitution of T for K/L/N/V at SDP2. EsSIPs varied in the third residue of the first NPA motif, with diverse ar/R filters and Froger’s positions. However, the residues were consistent with the corresponding SIP in Arabidopsis. AtSIP1;1 and AtSIP1;2 could transport water in the ER. AtSIP2;1 might act as an ER channel for other small molecules or ions (Ishikawa et al., 2005), and their similarity in these motifs suggests that these EsSIPs may have similar function. These results indicate that the diversity of AQPs in E. salsugineum may have crucial role in response to environmental stress.
Distinct expression profiles of EsAQP genes in various organs
Previous studies have shown that many AQPs show similar expression patterns, suggesting that they may act synergistic in some organs. For instance, PIPs and TIPs are abundant in all organs in many plant species (Quigley et al., 2001; Venkatesh, Yu & Park, 2013; Reuscher et al., 2013; Zou et al., 2015; Yuan et al., 2017). The qRT-PCR results showed that the transcripts of EsAQP genes could be detected in all organs, but their expression levels were diverse (Fig. 6A). Among them, the most abundant transcripts were EsPIPs and a few EsTIPs (EsTIP1;1 and EsTIP1;2), which were consistent with previous studies, especially with Arabidopsis AQP genes (Jang et al., 2004). The high expression of these AQP genes may be related to their effective water channel function that mediates water uptake in plant (Jang et al., 2004; Gomes et al., 2009). Moreover, EsTIP3;1 and EsTIP3;2 were highly expressed in silique specifically. It has been reported that seed-specific TIP3;1 and TIP3;2 play a role in maintaining seed longevity, and as target genes of ABI3 transcription factor which known to be involved in seed desiccation tolerance and seed longevity (Mao & Sun, 2015). It suggested that TIP3s may be involve in cellular osmoregulation and maturation of the vacuolar apparatus to support optimal water uptake and growth of the embryo during seed development and germination (Shivaraj et al., 2017). In general, the transcript level of NIP subfamily is lower than others. However, the EsNIP5;1 was high abundant in all organs, and some of them showed organ specific. For example, EsNIP2;1 and EsNIP3;1 were predominant expression in root, EsNIP4;1, EsNIP4;1 and EsNIP7;1 were predominantly expressed in flower. These may rely on their transport function of diverse substrates (Mitani-Ueno et al., 2011). Strikingly, the SIP1;1 and SIP2;1 exhibited higher expression than many TIPs and NIPs in both E. salsugineum (this study) and Arabidopsis (Alexandersson et al., 2005). Compared with different organs, many AQP genes are mainly expressed in roots and flowers, whereas no AQP isoform is leaf specific in Arabidopsis (Alexandersson et al., 2005). These results were also observed in our investigation. Above all, the parallel expression patterns of AQP genes in different organs between E. salsugineum and Arabidopsis may further indicated their similarity.
Stress responsive AQP genes in E. salsugineum
Environmental stress factors such as salt, drought and low temperature can quickly reduce water transport rates (Javot & Maurel, 2002), thus the maintenance of osmotic potential is a major challenge for plants. Since AQPs are known to be involved in the maintenance of water balance in the plant, we investigated the expression of EsAQP genes at aerial parts of seedlings under various abiotic stresses including salt, drought and cold. In Arabidopsis, most AQP genes are down-regulated upon drought stress in leaves, with the exception of AtPIP1;4 and AtPIP2;5, which are up-regulated (Alexandersson et al., 2005). Besides, the expression analysis of AtPIPs at aerial parts show that only the PIP2;5 was up-regulated by cold treatment, and most of the AtPIP genes were down-regulated by cold stress whereas less-severely modulated by high salinity (Jang et al., 2004). In our data (Fig. 6B), major AQP genes of E. salsugineum were down-regulated expression to drought treatment, however, nine genes (EsPIP2;4, EsPIP2;5, EsTIP1;2, EsTIP2;3, EsTIP3;2, EsNIP1;2, EsNIP4;3, EsNIP5;1 and EsSIP1;2) were up-regulated. Among these, the level of EsTIP3;2 was most significantly increased after drought treatment, which has low abundance in leaf (Fig. 6A). It is suggested that EsTIP3;2 may play a unique role under drought stress. While most of AQP genes were up-regulated under salt stress, it is consistent with those in barley and bamboo (Hove et al., 2015; Sun et al., 2016). Contrary to Arabidopsis, most of AQP genes in E. salsugineum were up-regulated under cold stress. This type of expression pattern has been reported in Sorghum bicolor (Reddy et al., 2015), to improve water transport efficiency and enhance cold tolerance (Li et al., 2008). Moreover, EsPIP1;5 was down-regulated under abiotic stresses but highly abundant in all organs, the EsTIP1;2 and EsNIP5;1 were highly abundant in all organs and up-regulated under various stresses. These AQP genes were induced by external stimuli, and implied to play role in maintaining water homeostasis during environmental stress (Jang et al., 2004).
Conclusions
In our study, a genome-wide information of E. salsugineum AQP gene family was provided. 35 EsAQP s, located in seven chromosomes, were identified and divided into four subfamilies based on phylogenetic analysis, which was also supported by the subfamily-specific gene structure and MEME motifs analysis. Furthermore, functional properties were investigated through the analysis of ar/R filters, Froger’s positions and SDPs, which have potential outputs for the widely function of EsAQPs. Moreover, the expression analysis was performed by qRT-PCR, showing AQP genes were widely involved in E. salsugineum organs development and abiotic stress response, and may have the potentially important roles in E. salsugineum. Our work not only provided a full-scale bioinformation of E. salsugineum AQP genes, but also offered a positive assessment for the underlying candidate EsAQPs in abiotic stress response.
Supplemental Information
Mismatched names highlighted in Yellow.
Multiple alignments were performed using ClustalX. The SDPs are highlighted in yellow, mismatch site highlighted in red and the representative sequences are marked in blue. The Genbank accession numbers: AtPIP1;2 (Q06611), AtPIP2;1 (P43286), AtPIP2;4 (Q9FF53), AtTIP1;1 (P25818), AtTIP1;2 (Q41963), AtTIP1;3 (NP_192056), AtTIP2;1 (Q41951), AtTIP2;3 (Q9FGL2), AtTIP4;1 (O82316), AtTIP5;1 (NP_190328), AtNIP1;2 (Q8LFP7), AtNIP5;1 (NP_192776), AtNIP6;1 (NP_178191), CpNIP1 (CAD67694), GmNOD26 (P08995), HvPIP1;3 (BAA23745), HvPIP1;4 (BAF33068), HvPIP2;1 (BAA23744), NtAQP1 (O24662), NtTIPa (Q9XG70), OsNIP2;1 (Q6Z2T3), TaTIP2;1 (AAS19468), TaTIP2;2 (AAS19469), ZmPIP1;1 (Q41870), ZmPIP1;5 (Q9AR14).
Acknowledgments
The authors appreciate those contributors who make the Eutrema salsugineum genome data accessible in public databases.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31570396) and the Fundamental Research Funds for the Central Universities (2572016DA05). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Weiguo Qian conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Xiaomin Yang, Jiawen Li and Rui Luo performed the experiments.
Xiufeng Yan conceived and designed the experiments, contributed reagents/materials/analysis tools, approved the final draft.
Qiuying Pang conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The primers used in qRT-PCR are available in Table S1. The specificity determining positions (SDPs) analysis of E.salsugineum AQPs from alignments with putative amino acid sequences of AQPs transporting non-aqua substrates are available in Fig. S2.
References
- Alexandersson et al. (2005).Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P. Whole gene family expression and drought stress regulation of aquaporins. Plant Molecular Biology. 2005;59:469–484. doi: 10.1007/s11103-005-0352-1. [DOI] [PubMed] [Google Scholar]
- Anderberg, Kjellbom & Johanson (2012).Anderberg HI, Kjellbom P, Johanson U. Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Frontiers in Plant Science. 2012;3 doi: 10.3389/fpls.2012.00033. Article 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariani & Gepts (2015).Ariani A, Gepts P. Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris, L.) Molecular Genetics & Genomics. 2015;290:1771–1785. doi: 10.1007/s00438-015-1038-2. [DOI] [PubMed] [Google Scholar]
- Armbrust et al. (2004).Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS. The Genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Bailey et al. (2006).Bailey TL, Williams N, Misleh C, Li WW. Meme: discovering and analyzing dna and protein sequence motifs. Nucleic Acids Research. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal & Sankararamakrishnan (2007).Bansal A, Sankararamakrishnan R. Homology modeling of major intrinsic proteins in rice, maize and Arabidopsis: comparative analysis of transmembrane helix association and aromatic/arginine selectivity filters. BMC Structural Biology. 2007;7:27–27. doi: 10.1186/1472-6807-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari et al. (2018).Bari A, Farooq M, Hussain A, Muhammad TUQ, Abbas MW, Mustafa G, Karim A, Ahmed I, Hussain T. Genome-wide bioinformatics analysis of aquaporin gene family in maize (Zea mays L.) Journal of Phylogenetics & Evolutionary Biology. 2018;6 doi: 10.4172/2329-9002.1000197. Article 197. [DOI] [Google Scholar]
- Biela et al. (1999).Biela A, Grote K, Otto B, Hoth S, Hedrich R, Kaldenhoff R. The Nicotiana tabacum plasma membrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. Plant Journal. 1999;18:565–570. doi: 10.1046/j.1365-313X.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- Bienert et al. (2014).Bienert GP, Heinen RB, Berny MC, Chaumont F. Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochimica et Biophysica Acta. 2014;1838:216–222. doi: 10.1016/j.bbamem.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Bowler et al. (2008).Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Chaumont & Tyerman (2014).Chaumont F, Tyerman SD. Aquaporins: highly regulated channels controlling plant water relations. Plant Physiology. 2014;164:1600–1618. doi: 10.1104/pp.113.233791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi & Roberts (2007).Choi WG, Roberts DM. Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. Journal of Biological Chemistry. 2007;282:24209–24218. doi: 10.1074/jbc.M700982200. [DOI] [PubMed] [Google Scholar]
- Chunhui et al. (2013).Chunhui X, Meng W, Li Z, Taiyong Q, Guangmin X, Jauhar A. Heterologous expression of the wheat aquaporin gene TaTIP2;2 compromises the abiotic stress tolerance of Arabidopsis thaliana. PLOS ONE. 2013;8:e79618. doi: 10.1371/journal.pone.0079618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui et al. (2008).Cui XH, Hao FS, Chen H, Chen J, Wang XC. Expression of the Vicia faba VfPIP1gene in Arabidopsis thaliana plants improves their drought resistance. Journal of Plant Research. 2008;121:207–214. doi: 10.1007/s10265-007-0130-z. [DOI] [PubMed] [Google Scholar]
- Danielson & Johanson (2008).Danielson JÅ, Johanson U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biology. 2008;8:45. doi: 10.1186/1471-2229-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deokar & Tar’an (2016).Deokar AA, Tar’an B. Genome-wide analysis of the aquaporin gene family in Chickpea (Cicer arietinum L.) Frontiers in Plant Science. 2016;7 doi: 10.3389/fpls.2016.01802. Article 1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev & Herbert (2018).Dev TB, Herbert JK. From aquaporin to ecosystem: plants in the water cycle. Journal of Plant Physiology. 2018;227:1–2. doi: 10.1016/j.jplph.2018.06.008. [DOI] [PubMed] [Google Scholar]
- DiGiorgio et al. (2016).DiGiorgio JA, Bienert GP, Ayub ND, Yaneff A, Barberini ML, Mecchia MA, Amodeo G, Soto GC, Muschietti JP. Pollen-specific aquaporins NIP4;1 and NIP4;2 are required for pollen development and pollination in Arabidopsis thaliana. The Plant Cell. 2016;28:1053–1077. doi: 10.1105/tpc.15.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynowski et al. (2008).Dynowski M, Schaaf G, Loque D, Moran O, Ludewig U. Plant plasma membrane water channels conduct the signaling molecule H2O2. Biochemical Journal. 2008;414:53–61. doi: 10.1042/BJ20080287. [DOI] [PubMed] [Google Scholar]
- Feng et al. (2017).Feng ZJ, Xu SC, Liu N, Zhang GW, Hu QZ, Xu ZS, Gong YM. Identification of the AQP members involved in abiotic stress responses from Arabidopsis. Gene. 2017;646:64–73. doi: 10.1016/j.gene.2017.12.048. [DOI] [PubMed] [Google Scholar]
- Fortin, Morrison & Verma (1987).Fortin MG, Morrison NA, Verma DPS. Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Research. 1987;15:813–824. doi: 10.1093/nar/15.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froger et al. (1998).Froger A, Tallur B, Thomas D, Delamarche C. Prediction of functional residues in water channels and related proteins. Protein Science. 1998;7:1458–1468. doi: 10.1002/pro.5560070623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar et al. (2003).Gaspar M, Bousser A, Sissoëff I, Roche O, Hoarau J, Mahé A. Cloning and characterization of ZmPIP1-5b, an aquaporin transporting water and urea. Plant Science. 2003;165:21–31. doi: 10.1016/j.jinsphys.2013.08.013. [DOI] [Google Scholar]
- Gattolin, Sorieul & Frigerio (2011).Gattolin S, Sorieul M, Frigerio L. Mapping of tonoplast intrinsic proteins in maturing and germinating Arabidopsis seeds reveals dual localization of embryonic TIPs to the tonoplast and plasma membrane. Molecular Plant. 2011;4:180–189. doi: 10.1093/mp/ssq051. [DOI] [PubMed] [Google Scholar]
- Gerbeau et al. (1999).Gerbeau P, Guclu J, Ripoche P, Maurel C. Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. Plant Journal. 1999;18:577–587. doi: 10.1046/j.1365-313x.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- Gomes et al. (2009).Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochimica et Biophysica Acta. 2009;1788:1213–1228. doi: 10.1016/j.bbamem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Gustavsson et al. (2005).Gustavsson S, Lebrun AS, Kristina N, François C, Johanson U. A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiology. 2005;139:287–295. doi: 10.1104/pp.105.063198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez et al. (2014).Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, Rycke RD, Inzé D, Blatt MR, Russinova E, Chaumont F. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. The Plant Cell. 2014;26:3132–3147. doi: 10.1105/tpc.114.127159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka et al. (2014).Hanaoka H, Uraguchi S, Takano J, Tanaka M, Fujiwara T. OsNIP3;1, a rice boric acid channel, regulates boron distribution and is essential for growth under boron-deficient conditions. The Plant Journal. 2014;78:890–902. doi: 10.1111/tpj.12511. [DOI] [PubMed] [Google Scholar]
- Heckwolf et al. (2011).Heckwolf M, Pater D, Hanson DT, Kaldenhoff R. The Arabidopsis thaliana aquaporin AtPIP1;2 is a physiologically relevant CO2 transport facilitator. Plant Journal. 2011;67:795–804. doi: 10.1111/j.1365-313X.2011.04634.x. [DOI] [PubMed] [Google Scholar]
- Hove & Bhave (2011).Hove RM, Bhave M. Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Molecular Biology. 2011;75:413–430. doi: 10.1007/s11103-011-9737-5. [DOI] [PubMed] [Google Scholar]
- Hove et al. (2015).Hove RM, Mark Z, Mrinal B, Ricardo A. Identification and expression analysis of the barley (Hordeum vulgare L.) aquaporin gene family. PLOS ONE. 2015;10:e0128025. doi: 10.1371/journal.pone.0128025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2012).Hu W, Yuan Q, Wang Y, Cai R, Deng X, Wang J, Zhou S, Chen M, Chen L, Huang C, Ma Z, Yang G, He G. Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic Tobacco. Plant and Cell Physiology. 2012;53:2127–2141. doi: 10.1093/pcp/pcs154. [DOI] [PubMed] [Google Scholar]
- Inan et al. (2004).Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, Shi H, Damsz B, Charbaji T, Gong Q, Ma S, Fredricksen M, Galbraith DW, Jenks MA, Rhodes D, Hasegawa PM, Bohnert HJ, Joly RJ, Bressan RA, Zhu JK. Salt Cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiology. 2004;135:1718–1737. doi: 10.1104/pp.104.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan & Kondrashov (2010).Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nature Reviews Genetics. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Ishikawa et al. (2005).Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M. Novel type aquaporin SIPs are mainly localized to the er membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Letters. 2005;579:5814–5820. doi: 10.1016/j.febslet.2005.09.076. [DOI] [PubMed] [Google Scholar]
- Jang et al. (2004).Jang JY, Kim DG, Kim YO, Kim JS, Kang H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Molecular Biology. 2004;54:713–725. doi: 10.1023/b:plan.0000040900.61345.a6. [DOI] [PubMed] [Google Scholar]
- Javot & Maurel (2002).Javot H, Maurel C. The role of aquaporins in root water uptake. Annals of Botany. 2002;90:301–313. doi: 10.1093/aob/mcf199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson et al. (2001).Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology. 2001;126:1358–1369. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam et al. (2017).Kadam S, Abril A, Dhanapal AP, Koester RP, Vermerris W, Jose S, Fritschi FB. Characterization and regulation of aquaporin genes of sorghum [Sorghum bicolor (L.) Moench] in response to waterlogging stress. Frontiers in Plant Science. 2017;8 doi: 10.3389/fpls.2017.00862. Article 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuhara et al. (2008).Katsuhara M, Hanba YT, Shiratake K, Maeshima M. Expanding roles of plant aquaporins in plasma membranes and cell organelles. Functional Plant Biology. 2008;35:1–14. doi: 10.1071/FP07130. [DOI] [PubMed] [Google Scholar]
- Koch & German (2013).Koch MA, German DA. Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Frontiers in Plant Science. 2013;4 doi: 10.3389/fpls.2013.00267. Article 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong et al. (2017).Kong W, Yang S, Wang Y, Bendahmane M, Fu X. Genome-wide identification and characterization of aquaporin gene family in Beta vulgaris. PeerJ. 2017;5:e3747. doi: 10.7717/peerj.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitäo et al. (2012).Leitäo L, Prista C, Moura TF, Loureiro-Dias MC, Soveral G. Grapevine aquaporins: gating of a tonoplast intrinsic protein (TIP2;1) by cytosolic pH. PLOS ONE. 2012;7:e33219. doi: 10.1371/journal.pone.0033219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2008).Li GW, Peng YH, Yu X, Zhang MH, Cai WM, Sun WN, Su WA. Transport functions and expression analysis of vacuolar membrane aquaporins in response to various stresses in rice. Journal of Plant Physiology. 2008;165:1879–1888. doi: 10.1016/j.jplph.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Li et al. (2018).Li W, Qiang XJ, Han XR, Jiang LL, Zhang SH, Han J, He R, Cheng XG. Ectopic expression of a Thellungiella salsuginea aquaporin gene, TsPIP1;1, increased the salt tolerance of Rice. International Journal of Molecular Science. 2018;19 doi: 10.3390/ijms19082229. Article 2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2015).Li L, Wang H, Gago J, Cui H, Qian Z, Kodama N, Ji H, Tian S, Shen D, Chen Y, Sun F, Xia Z, Ye Q, Sun W, Flexas J, Dong H. Harpin hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Scientific Reports. 2015;5 doi: 10.1038/srep17207. Article 17207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2011).Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu DT, Maurel C, Lin J. Single-molecule analysis of PIP2;1 dynamics and partitioning reveals, multiple modes of Arabidopsis plasma membrane aquaporin regulation. The Plant Cell. 2011;23:3780–3797. doi: 10.1105/tpc.111.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2013).Liu C, Fukumoto T, Matsumoto T, Gena P, Frascaria D, Kaneko T, Katsuhara M, Zhong S, Sun X, Zhu Y, Iwasaki I, Ding X, Calamita G, Kitagawa Y. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiology and Biochemistry. 2013;63:151–158. doi: 10.1016/j.plaphy.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Loqué et al. (2005).Loqué D, Ludewig U, Yuan L, Wiren NV. Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiology. 2005;137:671–680. doi: 10.1104/pp.104.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2006).Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M. A silicon transporter in rice. Nature. 2006;440:688–691. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- Ma et al. (2004).Ma S, Quist TM, Ulanov A, Joly R, Bohnert HJ. Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. The Plant Journal. 2004;40:845–859. doi: 10.1111/j.1365-313X.2004.02265.x. [DOI] [PubMed] [Google Scholar]
- Mao & Sun (2015).Mao Z, Sun W. Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of abscisic acid insensitive 3. Journal of Experimental Botany. 2015;66:4781–4794. doi: 10.1093/jxb/erv244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel et al. (2015).Maurel C, Boursiac Y, Luu DT, Santoni V, Shahzad Z, Verdoucq L. Aquaporins in plants. Physiological Reviews. 2015;95:1321–1358. doi: 10.1152/physrev.00008.2015. [DOI] [PubMed] [Google Scholar]
- Mitani-Ueno et al. (2011).Mitani-Ueno N, Yamaji N, Zhao FJ, Ma JF. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. Journal of Experimental Botany. 2011;62:4391–4398. doi: 10.1093/jxb/err158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani et al. (2006).Mizutani M, Watanabe S, Nakagawa T, Maeshima M. Aquaporin NIP2;1 is mainly localized to the ER membrane and shows root-specific accumulation in Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:1420–1426. doi: 10.1093/pcp/pcl004. [DOI] [PubMed] [Google Scholar]
- Park et al. (2010).Park W, Scheffler BE, Bauer PJ, Campbell BT. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biology. 2010;10:142. doi: 10.1186/1471-2229-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putpeerawit et al. (2017).Putpeerawit P, Sojikul P, Thitamadee S, Narangaiavana J. Genome-wide analysis of aquaporin gene family and their responses to water-deficit stress conditions in cassava. Plant Physiology and Biochemistry. 2017;121:118–127. doi: 10.1016/j.plaphy.2017.10.025. [DOI] [PubMed] [Google Scholar]
- Quigley et al. (2001).Quigley F, Rosenberg JM, Shacharhill Y, Bohnert HJ. From genome to function: the Arabidopsis aquaporins. Genome Biology. 2001;3:1–17. doi: 10.1186/gb-2001-3-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy et al. (2015).Reddy PS, Rao TSRB, Sharma KK, Vadez V. Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L.) Plant Gene. 2015;1:18–28. doi: 10.1016/j.plgene.2014.12.002. [DOI] [Google Scholar]
- Reuscher et al. (2013).Reuscher S, Akiyama M, Mori C, Aoki K, Shibata D, Shiratake K. Genome-wide identification and expression analysis of aquaporins in tomato. PLOS ONE. 2013;8:e79052. doi: 10.1371/journal.pone.0079052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai et al. (2005).Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiology. 2005;46:1568–1577. doi: 10.1093/pcp/pci172. [DOI] [PubMed] [Google Scholar]
- Shivaraj et al. (2017).Shivaraj SM, Deshmukh RK, Rai R, Bélanger R, Agrawal PK, Dash PK. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum) Scientific Reports. 2017;7:46137. doi: 10.1038/srep46137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto et al. (2010).Soto G, Fox R, Ayub N, Alleva K, Muschietti J. TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. The Plant Journal. 2010;64:1038–1047. doi: 10.1111/j.1365-313X.2010.04395.x. [DOI] [PubMed] [Google Scholar]
- Sreedharan, Shekhawat & Ganapathi (2013).Sreedharan S, Shekhawat UKS, Ganapathi TR. Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1;2 display high tolerance levels to different abiotic stresses. Plant Biotechnology Journal. 2013;11:942–952. doi: 10.1111/pbi.12086. [DOI] [PubMed] [Google Scholar]
- Sreedharan, Shekhawat & Ganapathi (2015).Sreedharan S, Shekhawat UKS, Ganapathi TR. Constitutive and stress-inducible overexpression of a native aquaporin gene (MusaPIP2;6) in transgenic banana plants signals its pivotal role in salt tolerance. Plant Molecular Biology. 2015;88:41–52. doi: 10.1007/s11103-015-0305-2. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2016).Sun HY, Li LC, Lou YF, Zhao HS, Gao ZM. Genome-wide identification and characterization of aquaporin gene family in moso bamboo (Phyllostachys edulis) Molecular Biology Report. 2016;43:437–450. doi: 10.1007/s11033-016-3973-3. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2015).Sun LL, Yu GH, Han XR, Xin SC, Qiang XJ, Jiang LL, Zhang SH, Cheng XG. TsMIP6 enhances the tolerance of transgenic rice to salt stress and interacts with target proteins. Journal of Plant Biology. 2015;58:285–292. doi: 10.1007/s12374-015-0069-x. [DOI] [Google Scholar]
- Taji et al. (2002).Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Kanako I, Narusaka Y, Narusaka M, Tajkhorshid E, Nollert P, Jensen MØ, Miercke LJ, O’Connell J, Stroud RM, Schulten K. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science. 2002;296:525–530. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- Takano et al. (2006).Takano J, Wada M, Ludewig U, Schaaf G, Von Wiren N, Fujiwara T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. The Plant Cell. 2006;18:1498–1509. doi: 10.1105/tpc.106.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka et al. (2008).Tanaka M, Wallace IS, Takano J, Roberts DM, Fujiwara T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. The Plant Cell. 2008;20:2860–2875. doi: 10.1105/tpc.108.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao et al. (2014).Tao P, Zhong X, Li B, Wang W, Yue Z, Lei J, Guo W, Huang X. Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis) Molecular Genetics and Genomics. 2014;289:1131–1145. doi: 10.1007/s00438-014-0874-9. [DOI] [PubMed] [Google Scholar]
- Uehlein et al. (2003).Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature. 2003;425:734–737. doi: 10.1038/nature02027. [DOI] [PubMed] [Google Scholar]
- Venkatesh, Yu & Park (2013).Venkatesh J, Yu JW, Park SW. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiology & Biochemistry. 2013;73:392–404. doi: 10.1016/j.plaphy.2013.10.025. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2014).Wang LL, Chen AP, Zhong NQ, Liu N, Wu XM, Wang F, Yang CL, Romero MF, Xia GX. The Thellungiella salsuginea tonoplast aquaporin TsTIP1;2 functions in protection against multiple abiotic stresses. Plant & Cell Physiology. 2014;55:148–161. doi: 10.1093/pcp/pct166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang Y, Li R, Li D, Jia X, Zhou D, Li J, Lyi SM, Hou S, Huang Y, Kochian LV, Liu J. NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:5047. doi: 10.1073/pnas.1618557114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2012).Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, Wang YN, Yang Z, Liu Y, Zhang W, Zhang H, Chu J, Yan C, Fang S, Zhang J, Wang Y, Zhang F, Wang G, Lee SY, Cheeseman JM, Yang B, Li B, Min J, Yang L, Wang J, Chu C, Chen SY, Bohnert HJ, Zhu JK, Wang XJ, Xie Q. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12219–12224. doi: 10.1073/pnas.1209954109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2012).Xu GX, Guo CC, Shan HY, Kong HZ. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2014).Xu Y, Hu W, Liu J, Zhang J, Jia C, Miao H, Xu B, Jin Z. A banana aquaporin gene, MaPIP1;1 is involved in tolerance to drought and salt stresses. BMC Plant Biology. 2014;14:59. doi: 10.1186/1471-2229-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2013).Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, Schmutz J, Wing RA, Mitchell-Olds T, Schumaker KS, Wang X. The reference genome of the halophytic plant Eutrema salsugineum. Frontiers of Plant Science. 2013;4 doi: 10.3389/fpls.2013.00046. Article 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan et al. (2017).Yuan D, Li W, Hua YP, King GJ, Xu FS, Shi L. Genome-wide identification and characterization of the aquaporin gene family and transcriptional responses to boron deficiency in Brassica napus. Frontiers of Plant Science. 2017;8 doi: 10.3389/fpls.2017.01336. Article 1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny et al. (2009).Zelazny E, Miecielica U, Borst JW, Hemminga MA, Chaumont F. An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. The Plant Journal. 2009;57:346–355. doi: 10.1111/j.1365-313x.2008.03691.x. [DOI] [PubMed] [Google Scholar]
- Zhang & Verkman (1991).Zhang RB, Verkman AS. Water and urea permeability properties of Xenopus oocytes: expression of mRNA from toad urinary bladder. American Journal of Physiology. 1991;260:C26–C34. doi: 10.1152/ajpcell.1991.260.1.C26. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2013).Zhang DY, Ali Z, Wang CB, Xu L, Yi JX, Xu ZL, Liu XQ, He XL, Huang YH, Khan IA, Trethowan RM, Ma HX. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.) PLOS ONE. 2013;8:e56312. doi: 10.1371/journal.pone.0056312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu (2001).Zhu JK. Plant salt tolerance. Trends in Plant Science. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- Zou et al. (2015).Zou Z, Gong J, Huang QX, Mo YY, Yang LF, Xie GS. Gene structures, evolution, classification and expression profiles of the aquaporin gene family in castor bean (Ricinus communis L.) PLOS ONE. 2015;10:e0141022. doi: 10.1371/journal.pone.0141022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou et al. (2016).Zou Z, Yang L, Gong J, Mo Y, Wang J, Cao J, An F, Xie G. Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Frontiers of Plant Science. 2016;7:395. doi: 10.3389/fpls.2016.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mismatched names highlighted in Yellow.
Multiple alignments were performed using ClustalX. The SDPs are highlighted in yellow, mismatch site highlighted in red and the representative sequences are marked in blue. The Genbank accession numbers: AtPIP1;2 (Q06611), AtPIP2;1 (P43286), AtPIP2;4 (Q9FF53), AtTIP1;1 (P25818), AtTIP1;2 (Q41963), AtTIP1;3 (NP_192056), AtTIP2;1 (Q41951), AtTIP2;3 (Q9FGL2), AtTIP4;1 (O82316), AtTIP5;1 (NP_190328), AtNIP1;2 (Q8LFP7), AtNIP5;1 (NP_192776), AtNIP6;1 (NP_178191), CpNIP1 (CAD67694), GmNOD26 (P08995), HvPIP1;3 (BAA23745), HvPIP1;4 (BAF33068), HvPIP2;1 (BAA23744), NtAQP1 (O24662), NtTIPa (Q9XG70), OsNIP2;1 (Q6Z2T3), TaTIP2;1 (AAS19468), TaTIP2;2 (AAS19469), ZmPIP1;1 (Q41870), ZmPIP1;5 (Q9AR14).
Data Availability Statement
The following information was supplied regarding data availability:
The primers used in qRT-PCR are available in Table S1. The specificity determining positions (SDPs) analysis of E.salsugineum AQPs from alignments with putative amino acid sequences of AQPs transporting non-aqua substrates are available in Fig. S2.