Abstract
Objective:
Epidemiological studies have suggested that the promoter region polymorphisms of interleukin-10 (IL-10) gene may be associated with an increased risk of lung cancer. However, those studies results are controversial. Thus, a comprehensive meta-analysis was performed to evaluate the association of promoter region polymorphisms of IL-10 gene with susceptibility to lung cancer.
Methods:
a comprehensive search of PubMed, EMBASE, and CNKI databases was performed to find all eligible studies up to September 15, 2018. The pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of such association.
Results:
A total number of 19 case-control studies with 4084 cases and 6,131 controls were selected. The overall meta-analysis results showed that the -592A>C polymorphism was significantly associated with lung cancer risk under four genetic models, i.e., allele (CT vs. TT: OR= 1.17, 95% CI 1.01-1.35, p=0.02), homozygote (CC vs. AA: OR= 1.64, 95% CI 1.29-2.02, p≤0.001), heterozygote (CA vs. AA: OR= 1.26, 95% CI 1.06-1.50, p≤0.001), and dominant (CC+CA vs. AA: OR= 1.31, 95% CI 1.11-1.54, p=0.001). However, there was no significant association between -819T>C and -1082A>G polymorphisms of IL-10 and lung cancer risk. Similarly, subgroup analyses by ethnicity detected significant association between IL-10 -592A>C and lung cancer among Asians and Caucasians.
Conclusions:
Our meta-analysis suggests that the IL-10 -592A>C polymorphism might be risk factor for lung cancer, especially among Asian and Caucasians. In contrast, the IL-10 -819T>C and -1082A>G polymorphisms are not significantly associated with increased risk of lung cancer.
Key Words: Lung cancer- interleukin 10- polymorphism- meta-analysis
Introduction
Lung cancer is one of the most commonly diagnosed cancer (12.7% of the total cancer diagnoses) as well as the leading cause of cancer death (18.2% of the total cancer deaths) among all cancer patients worldwide (Mehrabi et al., 2017; Zhong et al., 2012). Lung cancer critically can be divided into two groups include Non-Small Cell Lung Carcinoma (NSCLC) and Small cell lung carcinoma (SCLC), which the first one accounts for about 80% of all lung cancers. Although, the mean onset age for lung cancer has been estimated ranged 60 to 70 years, less than 10% of all cases occurred at an early age (Rosenberger et al., 2008). Although tobacco smoke is probably the predominant etiological risk factor for lung cancer, the pathoetiology of lung cancer is not fully understood. Interestingly, lung cancer develops only in a small proportion of chain smokers (less than 11%), which indicating that genetic factors might play a critical role in its carcinogenic mechanisms (Cavic et al., 2014).
Genetic factors involved in lung cancers have been extensively studied and to date several genetic polymorphisms have been identified as candidates by meta-analyses. Epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma viral oncogene homolog (KRAS) deleterious mutations occur mutually among 5-15% of lung cancer cases (Choughule et al., 2014). The promoter region polymorphisms of IL-10 gene has been associated with susceptibility to several cancers including lung cancer (Namazi et al., 2018; Sheikhpour et al., 2017). However, published data on the possible association of IL-10 polymorphism with lung cancer have generated inconclusive results. For example, Zhang et al found a significant association between the -819T>C and -592A>C polymorphisms of IL-10 and lung cancer in a Chinese population (Zhang et al., 2015), while Hsia et al., (2014) have reported that the genotypes of IL-10 -819TC polymorphism may have a protective effect on lung cancer risk in a Taiwanese population. Therefore, to estimate the effect of promoter region polymorphisms of IL-10 gene and lung cancer risk, as well as to quantify the potential between-study heterogeneity, we performed this meta-analysis based on published case-control studies.
Materials and Methods
Search strategies
We systematically searched several online databases including PubMed, EMBASE, Web of Science, Google Scholar, China Biological Medicine Database, and China National Knowledge Infrastructure comprehensively for all studies on the association of promoter region polymorphisms of IL-10 gene with lung cancer up to September 15, 2018. The combinations of following keywords and terms were used: (‘’lung carcinomas’’ or ‘’lung adenocarcinoma’’ or ‘’lung cancer’’ or ‘’small cell lung cancer’’ or ‘’Non-Small Cell Lung Cancer’’) and (‘’-1082 G>A’’ or ‘’rs1800896’’ or ‘’-819 T>C’’ or ‘’rs1800871’’ or ‘’-592A>C’’ or ‘’rs1800872’’) and (‘‘polymorphism’’ or ‘‘single nucleotide polymorphism’’ or ‘‘variation’’ or ‘‘mutation’’). Moreover, the reference lists of retrieved studies were hand-searched to find out the missing studies. All analyses of this meta-analysis were based on previous published studies, and this meta-analysis did not have original data. Thus, no ethical approval and patient consent are required.
Inclusion and exclusion criteria
Inclusion criteria were: 1) studies with case-control or cohort design; 2) evaluation of association of promoter region polymorphisms of IL-10 gene with lung cancer risk; 3) provided sufficient data to calculate the odds ratios (ORs), 95% confidence intervals (CIs). Accordingly, exclusion criteria were: 1) linkage studies or family based studies; 2) studies did not provided genotype frequencies; 3) duplicates or overlapping data; and 4) abstracts, reviews, meta-analyses, case reports, editorials, and animal studies.
Data extraction
The identified studies were reviewed separately by two authors independently and carefully to extraction necessary data and then recorded in a standardized form. We have resolved any disagreement by a discussion with the senior investigator. We sought the following data from each study: first author’s name, year of publication, ethnicity of each study population, country, source of controls, genotyping method, number of cases and controls, as well as numbers of cases and controls for IL-10 polymorphisms, minor allele frequency (MAF) in healthy subjects, and evidence of Hardy-Weinberg equilibrium (HWE).
Statistical analysis
All of the calculations were performed using Comprehensive Meta-Analysis (CMA) version 2.0 (Biostat, USA). Two-sided P-values<0.05 were considered statistically significant. The strength of association of promoter region polymorphisms of IL-10 gene with lung cancer risk was assessed using ORs and 95%CIs. The Z-test was employed to determine the significance of the pooled ORs.
Pooled ORs were calculated under four genetic models, i.e., allele (B vs. A), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB+BA vs. AA), and recessive (BB vs. BA+AA), which a ‘‘A’’ denotes a major allele; ‘‘B’’ denotes a minor allele. Between studies heterogeneity was tested by the Cochran Q-test, in which p-value less than 0.05 showed presence of heterogeneity. In addition, we used the I2 to detect the degree of heterogeneity between the included studies (I2=0-25%, no heterogeneity; I2=25-50%, moderate heterogeneity; I2>50%, large heterogeneity). The study-specific ORs were pooled using a fixed-effect or random-effect model depending on the heterogeneity. When a Q test indicated P <0.05 or I2 >50%, a random-effect model was used; otherwise, a fixed-effect model was applied. The departure from Hardy-Weinberg equilibrium (HWE) in the controls was assessed using Pearson’s χ2 test. Subgroup analyses based on ethnicity, source of controls (SOC), and genotyping methods were also performed. The stability of pooled results or influence of individual studies on the pooled ORs was tested through sensitivity analysis by omitting each study sequentially. Publication bias was estimated using the egg’s funnel plot and Egger linear regression test. If the publication bias observed, the Duval and Tweedie ‘‘trim and fill’’ method was used to adjust the bias.
Results
After a comprehensive literatures search we have identified 81 articles, then we reviewed all retrieved articles in accordance with the defined criteria. Of those articles, 62 articles were excluded due to be reviews, previous meta-analyses, case reports, letters, no detailed genotyping data, and duplicate publication. Finally, a total of 19 case-control studies in ten publications (Colakogullari et al., 2008; Hao et al., 2009; Hart et al., 2011; Hsia et al., 2014; Liang et al., 2011; Peddireddy et al., 2016; Seifart et al., 2005; Shih et al., 2005; Vogel et al., 2008; Zhang et al., 2015) were included in the meta-analysis. The details of each study were shown in Table 1. Of those studies, seven case-control studies with 1839 cases and 2,613 controls were on -592A>C, five studies with 963 cases and 1558 controls on -819T>C, and seven case-control studies with 1,282 cases and 1,960 controls on -1082A>G. Among the included studies, eleven were preformed among the Asians and eight among the Caucasians. The sample size in cases ranged between 44 and 436. Three genotyping methods were utilized in the selected studies, including PCR-RFLP, PCR-SSP, and TaqMan.
Table 1.
Characteristics of the Studies Included in Meta-Analysis
| First Author | Ethnicity | SOC | Genotyping | Case/Control | Cases | Control | MAFs | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Country) | Methods | Genotypes | Alleles | Genotypes | Alleles | |||||||||||
| -592A>C | AA | AC | CC | A | C | AA | AC | CC | A | C | ||||||
| Shih 2005 | Taiwan(Asian) | HB | PCR-RFLP | 154/205 | 66 | 70 | 18 | 202 | 106 | 116 | 76 | 13 | 308 | 102 | 0.24 | 0.9 |
| Colakogullari 2008 | Turkey(Caucasian) | HB | PCR-SSP | 44/59 | 2 | 23 | 19 | 27 | 61 | 7 | 25 | 27 | 39 | 79 | 0.66 | 0.74 |
| Vogel 2008 | Denmark(Caucasian) | HB | PCR | 403/744 | 13 | 149 | 241 | 175 | 631 | 42 | 250 | 452 | 334 | 1154 | 0.77 | 0.34 |
| Liang 2011 | China(Asian) | HB | PCR-RFLP | 116/120 | 69 | 36 | 11 | 174 | 58 | 69 | 44 | 7 | 182 | 58 | 0.24 | 0.99 |
| Hart 2011 | Norway(Caucasian) | HB | TaqMan | 434/433 | 15 | 175 | 243 | 205 | 661 | 26 | 144 | 264 | 196 | 672 | 0.77 | 0.28 |
| Hsia 2014 | Taiwan(Asian) | HB | PCR-RFLP | 358/716 | 173 | 145 | 40 | 491 | 225 | 368 | 277 | 71 | 1013 | 419 | 0.29 | 0.07 |
| Zhang 2015 | China(Asian) | HB | PCR-RFLP | 330/336 | 64 | 156 | 110 | 284 | 376 | 85 | 176 | 75 | 346 | 326 | 0.48 | 0.37 |
| -819T>C | TT | TC | CC | T | C | TT | TC | CC | T | C | ||||||
| Seifart 2005 | Germany(Caucasian) | HB | PCR-RFLP | 77/242 | 2 | 14 | 24 | 18 | 62 | 14 | 88 | 140 | 116 | 368 | 0.76 | 0.97 |
| Shih 2005 | Taiwan(Asian) | HB | PCR-RFLP | 154/205 | 66 | 58 | 30 | 190 | 128 | 104 | 86 | 15 | 294 | 116 | 0.28 | 0.62 |
| Colakogullari 2008 | Turkey(Caucasian) | HB | PCR-SSP | 44/59 | 2 | 23 | 19 | 27 | 61 | 7 | 26 | 26 | 40 | 78 | 0.66 | 0.89 |
| Hsia 2014 | Taiwan(Asian) | HB | PCR-RFLP | 358/716 | 212 | 128 | 18 | 552 | 164 | 372 | 265 | 79 | 1009 | 423 | 0.29 | 0 |
| Zhang 2015 | China(Asian) | HB | PCR-RFLP | 330/336 | 108 | 135 | 87 | 351 | 309 | 145 | 144 | 47 | 434 | 238 | 0.35 | 0.24 |
| -1082A>G | AA | AG | GG | A | G | AA | AG | GG | A | G | ||||||
| Seifart 2005 | Germany(Caucasian) | HB | PCR-RFLP | 39/243 | 6 | 21 | 12 | 33 | 45 | 86 | 115 | 42 | 287 | 199 | 0.4 | 0.73 |
| Shih 2005 | Taiwan(Asian) | HB | PCR-RFLP | 115/205 | 115 | 39 | 0 | 269 | 39 | 194 | 11 | 0 | 399 | 11 | 0.02 | 0.69 |
| Colakogullari 2005 | Turkey(Caucasian) | HB | PCR-SSP | 44/59 | 11 | 30 | 3 | 52 | 36 | 33 | 21 | 5 | 87 | 31 | 0.26 | 0.53 |
| Hao 2009 | China(Asian) | PB | TaqMan | 44/52 | 36 | 7 (AG+GG) | - | - | 46 | 6 (AG+GG) | - | - | - | |||
| Hart 2011 | Norway(Caucasian) | HB | TaqMan | 436/435 | 120 | 207 | 109 | 447 | 425 | 104 | 226 | 105 | 434 | 436 | 0.5 | 0.41 |
| Hsia 2014 | Taiwan(Asian) | HB | PCR-RFLP | 358/716 | 273 | 69 | 16 | 615 | 101 | 561 | 130 | 25 | 1252 | 180 | 0.12 | 0 |
| Peddireddy 2016 | India(Asian) | HN | PCR-RFLP | 246/250 | 156 | 69 | 21 | 381 | 111 | 130 | 84 | 36 | 344 | 156 | 0.31 | 0 |
Quantitative Synthesis
IL-10 -592A>C Polymorphism
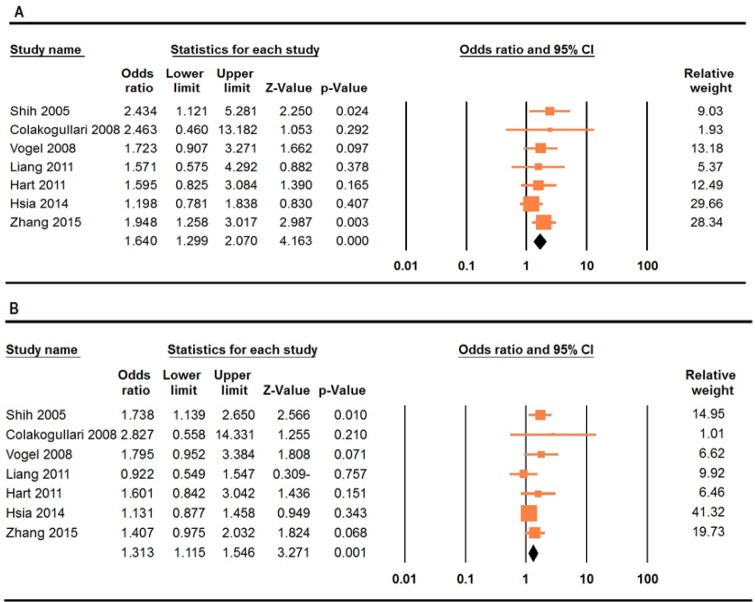
The summary of the meta-analysis of the association between IL-10 -592A>C polymorphism and lung cancer risk were listed in Table 2. There was a significant association between the -592A>C polymorphism and lung cancer risk under four genetic models, i.e., allele (CT vs. TT: OR= 1.14, 95% CI 1.03-1.25, p=0.007), homozygote (CC vs. AA: OR= 1.64, 95% CI 1.29-2.02, p≤0.001, Figure 1A), heterozygote (CA vs. AA: OR= 1.26, 95% CI 1.06-1.50, p≤0.001), and dominant (CC+CA vs. AA: OR= 1.31, 95% CI 1.11-1.54, p=0.001, Figure 1B). Stratified analysis by ethnicity revealed that there was a significant association between IL-10 -592A>C polymorphism and lung cancer among Asians under four genetic models, i.e., allele (C vs. A: OR= 1.26, 95% CI 1.11-1.43, p≤0.001), homozygote (CC vs. AA: OR= 1.64, 95% CI 1.22-2.21, p=0.001), dominant (CC+CA vs. AA: OR= 1.25, 95% CI 1.04-1.49, p≤0.001), and recessive (CC vs. CA+AA: OR= 1.52, 95% CI 1.19-1.93, p=0.001), and Caucasians under four genetic models, i.e., homozygote (CC vs. AA: OR= 1.70, 95% CI 1.09-2.65, p=0.01), heterozygote (CA vs. AA: OR= 2.08, 95% CI 1.32-3.27, p=0.001), dominant (CC+CA vs. AA: OR= 1.76, 95% CI 1.13-2.71, p=0.01), and recessive (CC vs. CA+AA: OR= 1.52, 95% CI 1.19-1.93, p=0.001).
Table 2.
Summary Risk Estimates for Association between IL-10 Polymorphism and Risk of Lung Cancer
| Subgroup | Genetic Model | Type of Model | Heterogeneity | Odds Ratio (OR) | Publication Bias | ||||
|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | ZOR | POR | PBeggs | |||
| -592A>C | C vs. A | Fixed | 49.9 | 0.06 | 1.14 | 1.03-1.25 | 2.68 | 0.007 | 1 |
| CC vs. AA | Fixed | 0 | 0.68 | 1.64 | 1.29-2.02 | 4.16 | ≤0.001 | 0.54 | |
| CA vs. AA | Fixed | 37.21 | 0.14 | 1.26 | 1.06-1.50 | 2.68 | ≤0.001 | 0.22 | |
| CC+CA vs. AA | Fixed | 15.63 | 0.31 | 1.31 | 1.11-1.54 | 3.27 | 0.001 | 0.36 | |
| CC vs. CA+AA | Random | 62.3 | 0.01 | 1.16 | 0.89-1.52 | 1.12 | 0.26 | 0.54 | |
| Asians | C vs. A | Fixed | 43.73 | 0.14 | 1.26 | 1.11-1.43 | 3.61 | ≤0.001 | 0.73 |
| CC vs. AA | Fixed | 17.91 | 0.3 | 1.61 | 1.22-2.21 | 3.43 | 0.001 | 0.73 | |
| CA vs. AA | Fixed | 20.93 | 0.28 | 1.16 | 0.96-1.40 | 1.58 | 0.11 | 1 | |
| CC+CA vs. AA | Fixed | 35.76 | 0.19 | 1.25 | 1.04-1.49 | 2.49 | 0.01 | 1 | |
| CC vs. CA+AA | Fixed | 0 | 0.4 | 1.52 | 1.19-1.93 | 3.41 | 0.001 | 0.73 | |
| Caucasians | C vs. A | Fixed | 0 | 0.67 | 0.99 | 0.85-1.15 | -0.09 | 0.92 | 1 |
| CC vs. AA | Fixed | 0 | 0.89 | 1.7 | 1.09-2.65 | 2.36 | 0.01 | 1 | |
| CA vs. AA | Fixed | 0 | 0.85 | 2.08 | 1.32-3.27 | 3.18 | 0.001 | 0.29 | |
| CC+CA vs. AA | Fixed | 0 | 0.81 | 1.76 | 1.13-2.71 | 2.54 | 0.01 | 1 | |
| CC vs. CA+AA | Fixed | 0 | 0.4 | 1.52 | 1.19-1.93 | 3.41 | 0.001 | 0.73 | |
| -819T>C | T vs. C | Random | 95.76 | ≤0.001 | 0.98 | 0.86-1.11 | -0.25 | 0.8 | 0.8 |
| TT vs. CC | Random | 87.95 | ≤0.001 | 1.53 | 0.57-4.06 | 0.85 | 0.39 | 0.8 | |
| TC vs. CC | Fixed | 19.96 | 0.28 | 1.01 | 0.84-1.22 | 0.16 | 0.87 | 0.8 | |
| TT+TC vs. CC | Random | 94.81 | ≤0.001 | 0.71 | 0.30-1.71 | -0.74 | 0.45 | 0.8 | |
| TT vs. TC+CC | Random | 92.33 | ≤0.001 | 0.98 | 0.33-2.41 | -0.04 | 0.96 | 0.8 | |
| -1082A>G | G vs. A | Random | 90.8 | ≤0.001 | 1.51 | 0.95-2.38 | 1.76 | 0.07 | 0.13 |
| GG vs. AA | Random | 71.79 | ≤0.001 | 1.15 | 0.63-2.08 | 0.46 | 0.64 | 0.46 | |
| GA vs. AA | Random | 88.58 | ≤0.001 | 1.67 | 0.92-3.04 | 1.7 | 0.08 | 0.45 | |
| GG+GA vs. AA | Random | 89.81 | ≤0.001 | 1.76 | 0.97-3.20 | 1.86 | 0.06 | 0.76 | |
| GG vs. GA+AA | Fixed | 53.87 | 0.07 | 1.02 | 0.81-1.30 | 0.23 | 0.81 | 0.8 | |
| Asians | G vs. A | Random | 95.29 | ≤0.001 | 1.64 | 0.63-4.22 | 1.03 | 0.3 | 1 |
| GG vs. AA | Random | 80.06 | 0.02 | 0.79 | 0.29-2.10 | -0.46 | 0.64 | NA | |
| GA vs. AA | Random | 92.73 | ≤0.001 | 1.56 | 0.60-4.03 | 0.91 | 0.36 | 1 | |
| GG+GA vs. AA | Random | 92.97 | ≤0.001 | 1.68 | 0.65-4.32 | 1.07 | 0.28 | 0.73 | |
| GG vs. GA+AA | Random | 73.27 | 0.05 | 0.83 | 0.36-1.91 | -0.42 | 0.67 | NA | |
| Caucasians | G vs. A | Random | 82.69 | 0.003 | 1.46 | 0.82-2.59 | 1.31 | 0.18 | 1 |
| GG vs. AA | Random | 73.42 | 0.02 | 1.72 | 0.59-5.00 | 1 | 0.31 | 1 | |
| GA vs. AA | Random | 87.66 | ≤0.001 | 1.95 | 0.61-6.24 | 1.13 | 0.25 | 1 | |
| GG+GA vs. AA | Random | 87.64 | ≤0.001 | 1.99 | 0.65-6.08 | 1.21 | 0.22 | 1 | |
| GG vs. GA+AA | Fixed | 36.07 | 0.2 | 1.14 | 0.86-1.51 | 0.93 | 0.35 | 1 | |
Figure 1.
Forest Plots Showed Significant Association between IL-10 -592A>C Polymorphism and Lung Cancer. A, homozygote model (AA vs. CC); B, dominant model (CC+CA vs. AA)
IL-10 -1082A>G and -819T>C Polymorphisms
The summary of the meta-analysis of the association between -1082A>G and -819T>C polymorphisms of IL-10 gene and lung cancer risk were listed in Table 2. The pooled results showed that there was no significant association between -1082A>G and -819T>C polymorphisms of IL-10 gene and lung cancer in overall population under all five genetic models. Moreover, stratified analysis by ethnicity also revealed that there was not a significant association between -1082A>G and -819T>C polymorphisms of IL-10 gene and lung cancer among Asians and Caucasians (Table 2).
Heterogeneity test and Sensitivity analysis
There was a significant in almost genetic models for IL-10 -592A>C and -819T>C polymorphisms. We have performed sensitivity analysis to detect the influence of each study on the pooled OR by deleting the single study and by deleting those studies did not accordance with HWE. However, sensitivity analysis suggested that a single study did not significantly change the pooled ORs, which indicated our meta-analysis results were robust and stable.
Publication bias
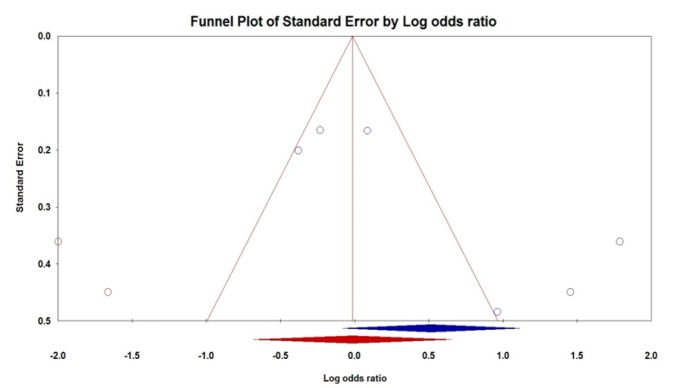
To determine the possible publication bias of the literature, we used the Begg’s funnel plot and Egger test. The shapes of Begg’s funnel plot did not show evidence of obviously asymmetrical for IL-10 -592A>C and -819T>C polymorphisms under all five genetic models and further confirmed by Egger test (Table 2). Although, Egger’s test revealed a significant publication bias under the heterozygote genetic model (GA vs. AA: PBeggs = 0.45 and PEggers = 0.04, Figure 2) for IL-10 -1082A>G polymorphism. Therefore, the Duval and Tweedie non-parametric ‘‘trim and fill’’ method was applied to the publication bias result IL-10 -1082A>G polymorphism. However, the outcomes showed that the current meta-analysis with and without ‘‘trim and fill’’ did not draw different results, indicating that our results were statistically reliable.
Figure 2.
Begg’s Funnel Plots of IL-10 -1082A>G Polymorphism and Lung Cancer Risk for Publication Bias Test under Heterozygote Model (GA vs. AA)
Discussion
To date, several case-control studies that have been performed to evaluate the association of IL-10 polymorphisms with the risk of lung cancer. However, the results are controversial due to the relatively small sample size of individual study, the different distributions of cases or healthy subjects, different lung cancer types, and the different genotyping methods. Meta-analysis as a powerful tool can provide more reliable results than a single study especially in explaining controversial conclusions. Thus, we performed this meta-analysis with larger sample size and subgroup to achieve a better understanding of the association of promoter region polymorphisms of IL-10 gene with lung cancer risk. To the best of our knowledge, our meta-analysis, on the basis of 19 case-controls 4,084 cases and 6,131 controls is the largest and most comprehensive assessment to evaluate association of IL-10 with lung cancer and the final results showed that -592A>C polymorphism was associated with an increased lung cancer risk.
In 2012, Wang et al., (2012) in a meta-analysis based on three studies have evaluated the association of IL-10 - 819C>T polymorphism and lung cancer. Their results showed that IL-10 - 819C>T polymorphism was significantly associated with increased risk of lung cancer. However, Yu et al., (2013) in a meta-analysis of four case-controls studies found that the IL-10 -819 C>T polymorphism was not significantly associated with lung cancer. In other meta-analysis, Peng et al., have reported that the IL-10 1082G>A, 819C>T and 592C>A polymorphisms were significantly associated with risk of lung cancer (Peng et al., 2012). Although the previous meta-analyses have reported positive association between IL-10 gene polymorphisms and lung cancer, the number of studies that they have included considerably smaller than that needed to receive the reliable results. Thus, those meta-analyses results should be interpreted with caution. Because, the number of studies included considerably smaller than that needed to receive the reliable results. However, our meta-analysis could offer adequate power to detect the association between IL-10 gene polymorphisms and lung cancer. In addition, in this meta-analysis we have performed subgroup analysis by ethnicity and the significant associations were only found for IL-10 -592A>C polymorphism among Asians and Caucasians. Therefore, IL-10 -592A>C polymorphism may be important factor contributed to lung cancer development.
Between studies heterogeneity is one of the challenging issues in a meta-analysis (Aslebahar et al., 2019; Yazdi et al., 2017). In a meta-analysis, heterogeneity could explain by sample study design, size, genotyping methods, source of controls, cancer types, life style, and so on (Jafari Nedooshan et al., 2017; Mehdinejad et al., 2017; Sobhan et al., 2017a; Sobhan et al., 2017b). In the current meta-analysis there was significant heterogeneity for -819T>C and -1082A>G polymorphisms under almost genetic models. Thus, we conducted subgroup analyses based on ethnicity and the types of control groups. However, the results showed that the heterogeneity may have resulted due to something more than ethnicity and the types of control groups. Moreover, publication bias is another key factor that might affect the quality and reliability of a meta-analysis (Sadeghiyeh et al., 2017). There was publication bias for IL-10 -1082A>G polymorphism under heterozygote genetic model might be due insufficient size of sample. Moreover, the ‘‘trim and fill’’ method results did not draw different results, indicating that the results were statistically reliable.
Although we conducted the most comprehensive meta-analysis based on all eligible studies, some limitations of this meta-analysis should be acknowledged. First, the number of eligible case-control studies included in this meta-analysis was small which limited statistical power to detect a potential association for those polymorphisms. Second, the sample size of some subgroups in the stratified analyses was limited, which may have reduced the statistical power to explore the association of the polymorphism with lung cancer susceptibility. Third, we have only included published studies in English in the meta-analysis. It is possible that some related unpublished studies and in other languages were missed; therefore, publication bias may have been present, even though statistical analysis indicated this not to be the case. Finally, lung cancer as most malignancies is a multifactorial disease that results from complex interactions between genetic and environmental factors. Our results were based on unadjusted estimates and a more precise analysis could be conducted if other covariates such as age, gender, smoking status, type of lung cancer, environmental factors, and lifestyle are available. Further evaluation of lung cancer risk should pay more attention to the potential interactions between gene-gene, gene-environment, and even between IL-10 -592A>C, -819T>C and -1082A>G polymorphisms.
In summary, our findings suggest that IL-10 -592A>C polymorphism might be risk factor for lung cancer, especially among Asians and Caucasians. However, IL-10 -819T>C and -1082A>G polymorphisms did not significantly associated with increased risk of lung cancer. Moreover, further studies with large sample sizes and well-designed multicenter analyses are required to clarify the association of IL-10 polymorphisms with susceptibility of lung cancer.
References
- Aslebahar F, Neamatzadeh H, Meibodi B, et al. Association of Tumor Necrosis Factor-α (TNF-α) -308G>A and -238G>A polymorphisms with recurrent pregnancy Loss risk, A meta-analysis. Int J Fertil Steril. 2019;12:ePUB. doi: 10.22074/ijfs.2019.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavic M, Krivokuca A, Spasic J, et al. The influence of methylenetetrahydrofolate reductase and thymidylate synthetase gene polymorphisms on lung adenocarcinoma occurrence. J BUON. 2014;19:1024–8. [PubMed] [Google Scholar]
- Choughule A, Sharma R, Trivedi V, et al. Coexistence of KRAS mutation with mutant but not wild-type EGFR predicts response to tyrosine-kinase inhibitors in human lung cancer. Br J Cancer. 2014;111:2203–4. doi: 10.1038/bjc.2014.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colakogullari M, Ulukaya E, Yilmaztepe Oral A, et al. The involvement of IL-10, IL-6, IFN-gamma, TNF-alpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct. 2008;26:283–90. doi: 10.1002/cbf.1419. [DOI] [PubMed] [Google Scholar]
- Hao M, Wang W, Lu Y, et al. Association of genetic polymorphism within promoter region of TNF-α, IL-1β and IL-10 with the susceptibility to lung cancer. [Article Chinese] Huan Jing Yu Zhi Ye Yi Xue. 2009;26:24–7. [Google Scholar]
- Hart K, Landvik NE, Lind H, et al. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer. 2011;71:123–9. doi: 10.1016/j.lungcan.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Hsia T-C, Chang W-S, Liang S-J, et al. Interleukin-10 (IL-10) promoter genotypes are associated with lung cancer risk in Taiwan males and smokers. Anticancer Res. 2014;34:7039–44. [PubMed] [Google Scholar]
- Jafari Nedooshan J, Forat Yazdi M, Neamatzadeh H, et al. Genetic association of XRCC1 Gene rs1799782, rs25487 and rs25489 polymorphisms with risk of thyroid cancer, A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:263–70. doi: 10.22034/APJCP.2017.18.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Zhao-Ran F, Xi-Ming Q. Relationship between the genetic polymorphism in Interleukin-10-592C/A and susceptibility to non-small cell lung cancer. [Article Chinese] Zhongguo Xian Dai Yi. 2011;21:189–93. [Google Scholar]
- Mehdinejad M, Sobhan MR, Mazaheri M, et al. Genetic association between ERCC2, NBN, RAD51 gene variants and osteosarcoma risk, A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:1315–21. doi: 10.22034/APJCP.2017.18.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi N, Moshtaghioun SM, Neamatzadeh H. Novel mutations of the CHRNA3 gene in non-small cell lung cancer in an Iranian population. Asian Pac J Cancer Prev. 2017;18:253–5. doi: 10.22034/APJCP.2017.18.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namazi A, Forat-Yazdi M, Jafari M, et al. Association of interleukin-10 -1082 A/G (rs1800896) polymorphism with susceptibility to gastric cancer, meta-analysis of 6,101 cases and 8,557 controls. Arq Gastroenterol. 2018;55:33–40. doi: 10.1590/S0004-2803.201800000-18. [DOI] [PubMed] [Google Scholar]
- Peddireddy V, Badabagni SP, Sulthana S, et al. Association of TNFα−308, IFNγ+874, and IL10−1082 gene polymorphisms and the risk of non-small cell lung cancer in the population of the South Indian state of Telangana. Int J Clin Oncol. 2016;21:843–52. doi: 10.1007/s10147-016-0972-2. [DOI] [PubMed] [Google Scholar]
- Peng W, He Q, Yang J, et al. Meta-analysis of association between cytokine gene polymorphisms and lung cancer risk. Mol Biol Rep. 2012;39:5187–94. doi: 10.1007/s11033-011-1315-z. [DOI] [PubMed] [Google Scholar]
- Rosenberger A, Illig T, Korb K, et al. Do genetic factors protect for early onset lung cancer? A case control study before the age of 50 years. BMC Cancer. 2008;8:60. doi: 10.1186/1471-2407-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghiyeh T, Hosseini Biouki F, Mazaheri M, et al. Association between Catechol-O-Methyltransferase Val158Met (158G/A) polymorphism and suicide susceptibility, A meta-analysis. J Res Health Sci. 2017;17:e00383. [PubMed] [Google Scholar]
- Seifart C, Plagens A, Dempfle A, et al. TNF-alpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21:157–65. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikhpour E, Noorbakhsh P, Foroughi E, et al. A survey on the role of interleukin-10 in breast cancer, A narrative. Reports Biochem Mol Biol. 2017;7:169–80. [PMC free article] [PubMed] [Google Scholar]
- Shih C-M, Lee Y-L, Chiou H-L, et al. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer. 2005;50:291–297. doi: 10.1016/j.lungcan.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Sobhan MR, Mehdinejad M, Jamaladini MH, et al. Association between aspartic acid repeat polymorphism of the asporin gene and risk of knee osteoarthritis, A systematic review and meta-analysis. Acta Orthop Traumatol Turc. 2017a;51:409–15. doi: 10.1016/j.aott.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhan MR, Forat Yazdi M, Mazaheri M, Zare Shehneh M, Neamatzadeh H. Association between the DNA Repair Gene XRCC3 rs861539 polymorphism and risk of osteosarcoma, a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017b;18:549–55. doi: 10.22034/APJCP.2017.18.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U, Christensen J, Wallin H, et al. Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat Res Mol Mech Mutagen. 2008;639:89–100. doi: 10.1016/j.mrfmmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Wang J, Ding Q, Shi Y, et al. The interleukin-10-1082 promoter polymorphism and cancer risk, a meta-analysis. Mutagenesis. 2012;27:305–12. doi: 10.1093/mutage/ger078. [DOI] [PubMed] [Google Scholar]
- Yazdi MM, Jamalaldini MH, Sobhan MR, et al. Association of ESRα Gene Pvu II T>C, XbaI A>G and BtgI G> A polymorphisms with knee osteoarthritis susceptibility, A systematic review and meta-analysis based on 22 case-control studies. Arch bone Jt Surg. 2017;5:351–62. [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Liu Q, Huang C, Wu M, Li G. The Interleukin 10 −819C/T polymorphism and cancer risk, A HuGE review and meta-analysis of 73 studies including 15,942 cases and 22,336 controls. Omi A J Integr Biol. 2013;17:200–14. doi: 10.1089/omi.2012.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D, Li G, Long J, Wu J, Hu Y. The hOGG1Ser326Cys polymorphism and increased lung cancer susceptibility in caucasians, An updated meta-analysis. Sci Rep. 2012;2:548. doi: 10.1038/srep00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-M, Mao Y-M, Sun Y-X. Genetic polymorphisms of IL-6 and IL-10 genes correlate with lung cancer in never-smoking Han population in China. Int J Clin Exp Med. 2015;8:1051–8. [PMC free article] [PubMed] [Google Scholar]