Abstract
Inositol hexakisphosphate kinases (IP6Ks) have been increasingly studied as therapeutically interesting enzymes. IP6K isoform specific knock-outs have been used to successfully explore inositol pyrophosphate physiology and related pathologies. A pan-IP6K inhibitor, N2-(m-trifluorobenzyl)-N6-(p-nitrobenzyl) purine (TNP), has been used to confirm phenotypes observed in genetic knock-out experiments; however, it suffers by having modest potency and poor solubility making it difficult to handle for in vitro applications in the absence of DMSO. Moreover, TNP’s pan-IP6K inhibitory profile does not inform which IP6K isoform is responsible for which phenotypes. In this report we describe a series of purine-based isoform specific IP6K1 inhibitors. The lead compound was identified after multiple rounds of SAR and has been found to selectively inhibit IP6K1 over IP6K2 or IP6K3 using biochemical and biophysical approaches. It also boasts increased solubility and IP6K1 potency over TNP. These new compounds are useful tools for additional assay development and exploration of IP6K1 specific biology.
Keywords: Inositol hexakisphosphate kinase, Inositol pyrophosphate, Selective enzyme inhibitors, Structure activity relationships, Kinases
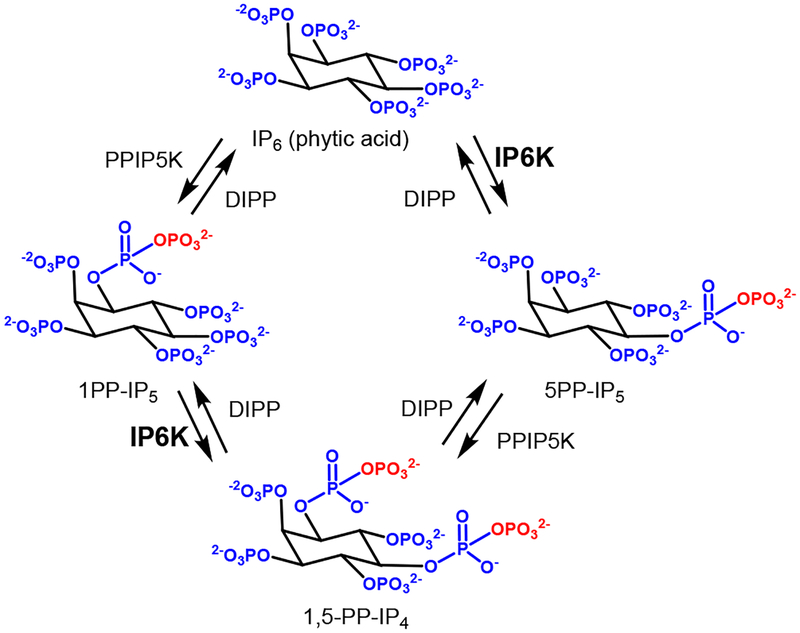
Inositol pyrophosphates (1PP-IP5, 5PP-IP5, 1,5PP-IP4, Figure 1) are becoming recognized as important signaling molecules that may help regulate cellular metabolism and energy levels.1–3 The intercellular pool of 5PP-IP5 is formed from inositol hexakisphosphate (IP6, phytic acid) by a family of three inositol hexakisphosphate kinases (IP6K1, IP6K2, and IP6K3). These kinases utilize ATP to form a high-energy pyrophosphate bond when they convert IP6 to 5PP-IP5. 5PP-IP5 is thought to integrate with a number of cellular processes including signal transduction, vesicular trafficking, apoptosis, and metabolism.4
Figure 1.
Biosynthesis of inositol pyrophosphates
All three IP6K isoforms catalyze the same reaction; however, global genetic knock out experiments have uncovered unique phenotypes associated with each isoform. IP6K1 knock out mice show metabolic changes and resistance to diet-induced obesity and fatty liver.5,6 IP6K2 knock out mice are sensitive to carcinogen-induced tumor formation,7 and IP6K3 knock out mice are resistant to age-induced weight gain and have an increased lifespan.8 IP6K1 (neuronal migration),9 IP6K2 (Purkinje cell morphology and cerebellar synapse formation),10 and IP6K3 (synapse formation)11 have also been implicated with neuronal functions. Each isoform has differential cell-type expression,8 sub-cellular localization,12 and protein-protein interactions13–16 that allow for the different phenotypes observed. Due to the importance of protein-protein interactions in maintaining normal cellular processes it is unclear if phenotypes observed in knock-out studies are fully driven by decreasing 5PP-IP5 production or if the loss of protein-protein interactions and scaffolding have a significant effect.
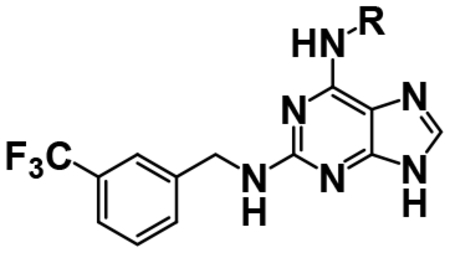
Nearly a decade ago N2-(m-trifluorobenzyl)-N6-(p-nitrobenzyl) purine (TNP, compound 1, Table 1) was discovered as a pan-IP6K inhibitor.17 Since then TNP has been used in a number of studies to examine the effects of 5PP-IP5 in various contexts (see4 for a comprehensive review). In particular, Ghoshal et al. demonstrated that mice treated with TNP were resistant to high-fat diet induced weight gain while remaining sensitive to glucose and insulin.18 While TNP has proved itself a useful tool, it can certainly be improved -- particularly in terms of potency, solubility, and isoform selectivity in order to better understand inositol pyrophosphate biology. TNP is reported to have low micromolar potency against the IP6K family,17,19,20 however, it is sparingly soluble in aqueous buffers and is thus difficult to work with in vitro without precipitation and difficult to formulate for in vivo animal studies. Finally, TNP does not preferentially inhibit one IP6K isoform over the others. Therefore, it is not possible to use TNP to study the effects of one IP6K isoform in a context where the others are also present.
Table 1.
Inhibitory activities of the first iteration of purine analogs with variation at the N6-position.
 | ||
|---|---|---|
| Compound | R | IP6K1 pIC50 |
| 1, TNP |  |
< 3.70* |
| 4 | 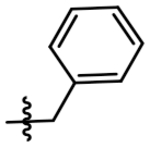 |
< 3.70 |
| 5 | 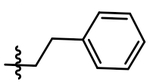 |
< 3.70 |
| 6 | 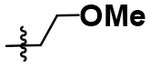 |
3.93 ± 0.10 |
Solubility issues preclude proper IC50 determination
Data represented as the mean ± SEM.
Puhl-Rubio and colleagues have identified a number of IP6K2 inhibitors after screening a library of kinase-focused compounds.20 Gu et al. have also recently reported a series of flavonoids as inositol pyrophosphate kinase inhibitors.21 However, we adopted a medicinal chemistry approach to identify IP6K1 specific inhibitors by systematically varying the N2 and N6 positions off of the purine core.
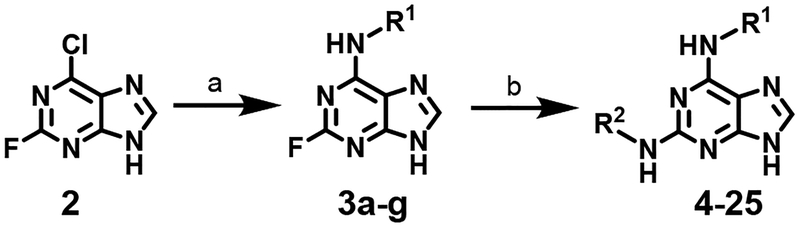
A simple two-step SNAr reaction depicted in Scheme 1 from 6-chloro-2-fluoro purine allows for rapid exploration of the 6- and 2- positions on the purine core. Initial efforts were spent exploring the N6 position to determine if the nitrobenzyl moiety present on TNP was essential for compound potency (Table 1). The nitrobenzyl group is undesirable because it is capable of forming toxic metabolites in vivo and reduces compound solubility.22 In this study, all compounds were tested in the Promega ADP-Glo Max assay optimized for IP6K performance as described previously.23 Briefly, the assay is conducted with physiologically relevant concentrations of ATP (1 mM) that is consumed as the kinase reaction progresses over time. After a time, the reaction is stopped, the remaining ATP is enzymatically depleted, and the ADP produced is converted back to ATP with a recycling reaction. This regenerated ATP reacts with luciferin/luciferase to produce a bioluminescent signal proportional to IP6K activity. In this assay, TNP performance is inconsistent with previously published work presumably due to the immense hydrophobic nature of the inhibitor causing it to precipitate out of the assay. Nitrobenzyl replacement with benzyl (4) or phenethyl (5) moieties caused a dramatic decrease in potency compared to the published value for TNP. A methoxyethyl (6) substitution produced a soluble compound capable of completely inhibiting IP6K1, albeit very weakly with a pIC50 of 3.93.
Scheme 1.
Preparation of N2- and N6- substituted purine analogs 2–25. Reagents and reaction conditions: (a) R1-NH2, DIPEA, nBuOH, 110°C (b) R2-NH2, DIPEA, DMSO, 110°C.
The N6-methoxyethyl substitution was initially retained for solubility while we attempted to regain potency at the 2-position (Table 2). Replacing the mCF3-benzyl (6) with phenethyl (7) resulted in a modest increase in potency. Removing electron density (8) from the phenyl ring did not increase potency while adding electron density (9) increased potency from pIC50 4.10 to 4.95. Large electron-rich aromatic rings (indole, 10) at the 2-position further increase potency to pIC50 5.08 while large hydrophobic aromatic rings are less active (napthyl, 11). A smaller imidazole group (12) similarly abolishes potency.
Table 2.
Inhibitory activities of the second iteration of purine analogs with variation at the N2-position.
 | ||
|---|---|---|
| Compound | R | IP6K1 pIC50 |
| 7 | 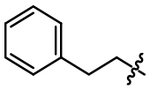 |
4.10 ± 0.12 |
| 8 | 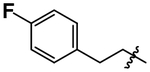 |
4.05 ± 0.04 |
| 9 | 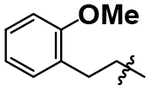 |
4.95 ± 0.19 |
| 10 | 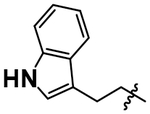 |
5.08 ± 0.10 |
| 11 | 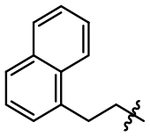 |
4.03 ± 0.09 |
| 12 | 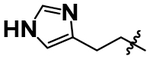 |
< 3.70 |
Data represented as the mean ± SEM.
Using compound 10 as a baseline, we set out to explore how essential the hydrogen bond donors in the molecule are by alkylating the NH groups with a methyl cap. We found that alkylating any of the exocyclic NHs abolished potency and 1-methyltrytamine dramatically reduced it (Supplementary Data Table 1). Likewise, purine N7 and N9 to carbon substitutions abolished potency. These data suggest that there is an intricate hydrogen bond network in the IP6K1 active site.
We next explored the chemical space surrounding the tryptamine (Table 3). Generally, substitutions at the 5 position on the indole are not tolerated (13–16). Substitutions at the 2 and 6 positions increase potency (17, 18). Increasing the linker length from 2 to 3 carbons (19) dramatically decreases potency and constraining free rotation of the tryptamine abolishes potency (data not shown).
Table 3.
Inhibitory activities of the third iteration of purine analogs with exploration around an N2-tryptamine.
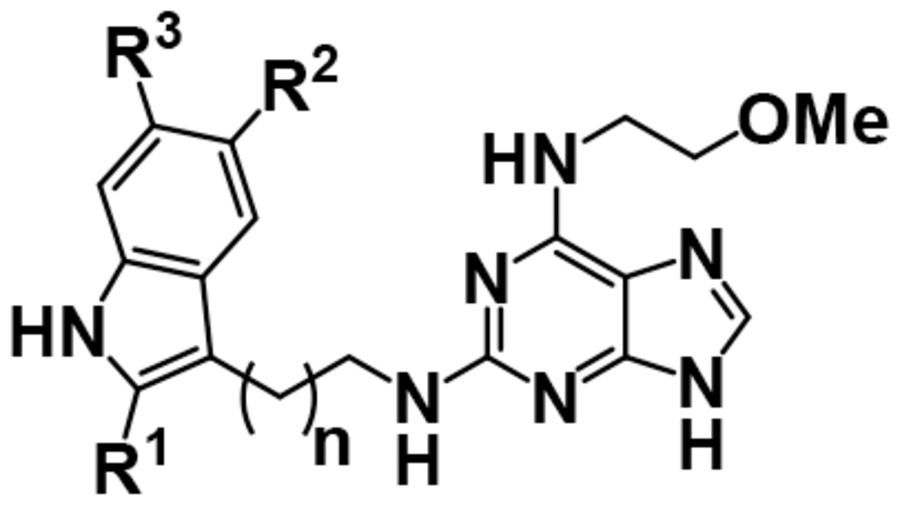 | |||||
|---|---|---|---|---|---|
| Compound | R1 | R2 | R3 | n | IP6K1 pIC50 |
| 13 | H | OMe | H | 1 | 3.86 ± 0.08 |
| 14 | H | OH | H | 1 | 4.50 ± 0.14 |
| 15 | H | Cl | H | 1 | 4.57 ± 0.20 |
| 16 | H | Me | H | 1 | 3.91 ± 0.08 |
| 17 | H | H | OMe | 1 | 5.56 ± 0.09 |
| 18 | Me | H | H | 1 | 5.71 ± 0.14 |
| 19 | H | H | H | 2 | 4.16 ± 0.01 |
Data represented as the mean ± SEM.
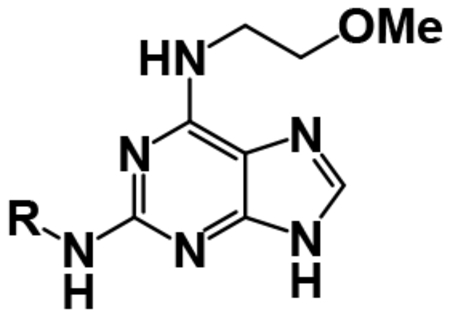
After achieving low micromolar potency with compounds 17 and 18 by optimizing the N2 position off of the purine core, we returned to the N6 position to see if nitro replacements (relative to TNP) exist which are more potent than methoxyethylamine (Table 4). Benzoxadiazole (20), 4-cyanobenzyl (22), 4- and 5-methyl-2-pyridone (21, 23) derivatives were synthesized and tested. Compound 23 increased potency from pIC50 5.08 to 5.76 over the parent compound 10. This 5-methyl-2-pyridone substitution was then combined with the more potent N2–6-methoxytryptamine or N2–2-methyltryptamine derivatives to yield compounds 24 and 25. N2–2-Methyltryptamine (25) is equipotent to unsubstituted tryptamine (10); however, N2–6-methoxytryptamine (24) is additive with 5-methyl-2-pyridone yielding for the first time a sub-micromolar IP6K1 inhibitor with a pIC50 of 6.13.
Table 4.
Inhibitory activities of the fourth iteration of TNP analogs with nitro replacements at N6.
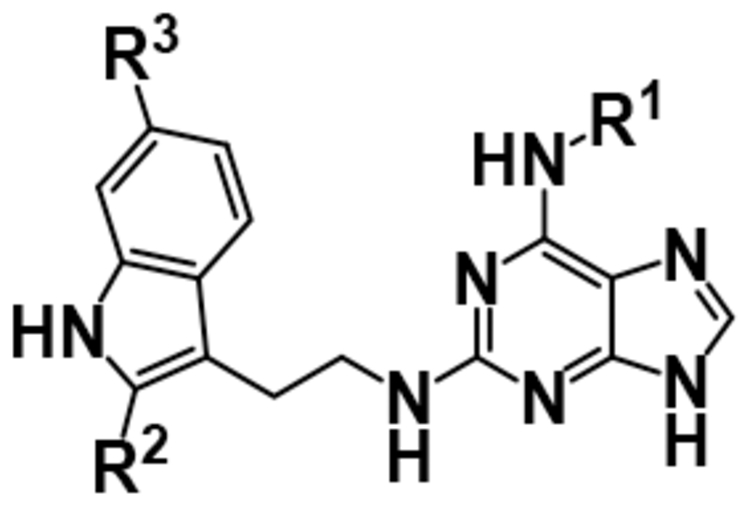 | ||||
|---|---|---|---|---|
| Compound | R1 | R2 | R3 | IP6K1 pIC50 |
| 20 | 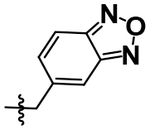 |
H | H | 3.82 ± 0.00 |
| 21 | 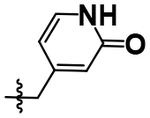 |
H | H | 5.00 ± 0.11 |
| 22 | 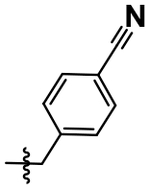 |
H | H | 4.33 ± 0.17 |
| 23 | 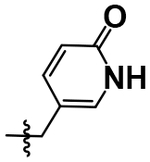 |
H | H | 5.76 ± 0.14 |
| 24 | 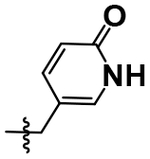 |
H | OMe | 6.13 ± 0.08 |
| 25 | 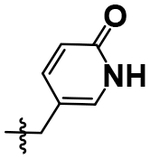 |
Me | H | 5.52 ± 0.07 |
Data represented as the mean ± SEM.
Compounds 1 (TNP), 10, 17, and 24 were selected for confirmation of binding to IP6K1 in an orthogonal biophysical thermal melt assay. In this assay purified IP6K1 was incubated over a temperature gradient in the presence of different concentrations of purine analogs. Compounds that bind tightly to IP6K1 will increase the protein melting temperature to a greater degree. Here, we observe good correlation between IC50 values obtained in the biochemical assay and an increase in melting temperature. As expected, compound 24 was able to increase the melting temperature to a greater degree than the other compounds at 11.1μM (Table 5). Moreover, compound 24 has a greater ΔTmMax than the other compounds tested, including TNP, indicating greater stabilization. Taken together with the biochemical data, these data suggest that compound 24 inhibits IP6K1 both enzymatically as well physically interacts with the enzyme in a meaningful way.
Table 5.
Compound 24 thermally stabilizes IP6K1 to a greater degree than other purine analogs.
| Compound | ΔTm at 11.1μM (°C) | ΔTmMax (°C) |
|---|---|---|
| 24 | 5.5 | 6.6 ± 0.6 |
| 17 | 3.6 | 4.7 ± 0.6 |
| 10 | 2.4 | 4.3 ± 0.9 |
| 1, TNP | 1.8 | 3.3 ± 0.6 |
Data represented as the mean ± SEM.
In order to examine IP6K isoform selectivity we tested compound 24 against purified IP6K2 and IP6K3 in the ADP-Glo Max assay (Table 6). We were gratified to find that compound 24 was ~25 and 50 fold more selective for IP6K1 over IP6K2 and IP6K3, respectfully, after converting observed IC50 values into Ki values with the Cheng-Prusoff equation.24 These results suggest that there are distinct enough differences in the IP6K family to allow for subtype specific inhibitors to be developed.
Table 6.
Selectivity of compound 24 against IP6K isoforms
| Compound 24 | |||
|---|---|---|---|
| IC50 (μM) | Ki (μM) | Selectivity (Ki/Ki IP6K1) | |
| IP6K1 | 0.75 | 0.20 | - |
| IP6K2 | 20 | 4.88 | 24.4 |
| IP6K3 | 15 | 9.62 | 48.1 |
Compound 24 was broadly profiled for off-target kinase inhibition in a diversity panel at Eurofins. This screen includes a number of kinases from different families and signaling pathways (see Supplementary Data Table 2). Compound 24 was tested at 10μM against 58 kinases and found to potently inhibit 4 kinases, 3 of which are part of the RAF/MAPK pathway in addition to casein kinase γ. The remaining 54 kinases were not significantly inhibited by compound 24. Importantly, this panel included a number of metabolically important enzymes including GSK3β, AKT, mTOR, and PI3K. 5PP-IP5 is thought to interact intimately with the PI3K/AKT/GSK3β pathway.5 As such, it is important that tools that manipulate 5PP-IP5 concentration do not inhibit these enzymes or the results may be confounded and difficult to interpret. These profiling results suggest that, surprisingly, the purine-based compound 24 is a relatively selective IP6K1 inhibitor against the rest of the kinome.
In summary, given the increasing body of evidence that IP6Ks and inositol pyrophosphates play an important role in cellular processes and pathologies, we became interested in developing IP6K specific inhibitors to serve as pharmacological compliments to the published genetic knock out models. These chemical tools will help deconvolute inositol pyrophosphate signaling and inform what each IP6K isoform is contributing to a particular phenotype. Our approach to systematically vary the N2 and N6 positions of TNP have uncovered compound 24 which boasts a number of improvements over TNP (Table 7). Primarily, compound 24 has sub-micromolar potency against IP6K1, it is 25 to 50-fold selective against IP6K2 and IP6K3, respectively, in addition to being inactive against 54/58 kinases tested in a diversity panel. Moreover, compound 24 eschews the nitrobenzyl moiety and possesses meaningful aqueous solubility compared to TNP, thus enabling in vitro experiments and in vivo formulations. We propose that compound 24 will be a useful tool to evaluate IP6K1-specific activity in physiological and pathological models.
Table 7.
Comparison of compound 24 with TNP
| Parameter | Compound 24 | TNP, 1 |
|---|---|---|
| Nitrobenzyl | Removed | Present |
| Solubility | 51μg/mL (PBS pH 7.4) | Mostly insoluble in 2% DMSO |
| Potency | 0.20μM Ki | 0.24μM Ki (published value)* |
| IP6K selectivity | 24.4 fold over K2 48.1 fold over K3 |
Not selective* |
| Microsome stability (t1/2) | Rat 38 min | Mouse 9 min** |
| Human 42 min | Human 32 min** |
published in Padmanabhan, et al. J Biol Chem 2009
published in Ghoshal, et al. Mol Metab. 2016
Supplementary Material
Highlights:
Structure-activity relationships based on a nonselective lead revealed compound 24 as a selective inhibitor of IP6K1.
Compound 24 has good selectivity over closely related kinases IP6K2 and IP6K3 as well as a panel of other diverse kinases.
Compound 24 is a useful tool to investigate the role of IP6K1 selective inhibitors.
Acknowledgements
The authors declare no competing financial interests. This research was supported by National Institutes of Health grant U01 MH112658 and the Lieber Institute for Brain Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334(6057):802–805. doi: 10.1126/science.1211908 [DOI] [PubMed] [Google Scholar]
- 2.Shears SB. Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol. 2009;76(2):236–252. doi: 10.1124/mol.109.055897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson MSC, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem J. 2013;452(3). [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty A The inositol pyrophosphate pathway in health and diseases. Biol Rev. December 2017. doi: 10.1111/brv.12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty A, Koldobskiy MA, Bello NT, et al. Inositol Pyrophosphates Inhibit Akt Signaling, Thereby Regulating Insulin Sensitivity and Weight Gain. Cell. 2010;143(6):897–910. doi: 10.1016/j.cell.2010.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Q, Ghoshal S, Rodrigues A, et al. Adipocyte-specific deletion of Ip6k1 reduces diet-induced obesity by enhancing AMPK-mediated thermogenesis. J Clin Invest. 2016;126(11):4273–4288. doi: 10.1172/JCI85510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28(25):2383–2392. doi: 10.1038/onc.2009.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moritoh Y, Oka M, Yasuhara Y, et al. Inositol Hexakisphosphate Kinase 3 Regulates Metabolism and Lifespan in Mice. Sci Rep. 2016;6:32072. doi: 10.1038/srep32072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu C, Xu J, Cheng W, et al. Neuronal migration is mediated by inositol hexakisphosphate kinase 1 via α-actinin and focal adhesion kinase. Proc Natl Acad Sci U S A. 2017;114(8):2036–2041. doi: 10.1073/pnas.1700165114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagpal L, Fu C, Snyder SH. Inositol Hexakisphosphate Kinase-2 in Cerebellar Granule Cells Regulates Purkinje Cells and Motor Coordination via Protein 4.1N. J Neurosci. 2018;38(34):7409–7419. doi: 10.1523/JNEUROSCI.1165-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu C, Xu J, Li R-J, et al. Inositol Hexakisphosphate Kinase-3 Regulates the Morphology and Synapse Formation of Cerebellar Purkinje Cells via Spectrin/Adducin. J Neurosci. 2015;35(31):11056–11067. doi: 10.1523/JNEUROSCI.1069-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem. 2001;276(42):39179–39185. doi: 10.1074/jbc.M106842200 [DOI] [PubMed] [Google Scholar]
- 13.Chakraborty A, Koldobskiy MA, Sixt KM, et al. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc Natl Acad Sci U S A. 2008;105(4):1134–1139. doi: 10.1073/pnas.0711168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo HR, Saiardi A, Nagata E, et al. GRAB: A Physiologic Guanine Nucleotide Exchange Factor for Rab3a, which Interacts with Inositol Hexakisphosphate Kinase. Neuron. 2001;31(3):439–451. doi: 10.1016/S0896-6273(01)00384-1 [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty A, Latapy C, Xu J, Snyder SH, Beaulieu J-M. Inositol hexakisphosphate kinase-1 regulates behavioral responses via GSK3 signaling pathways. Mol Psychiatry. 2013;19(November 2012):1–10. doi: 10.1038/mp.2013.21 [DOI] [PubMed] [Google Scholar]
- 16.Morrison BH, Bauer JA, Lupica JA, et al. Effect of inositol hexakisphosphate kinase 2 on transforming growth factor beta-activated kinase 1 and NF-kappaB activation. J Biol Chem. 2007;282(21):15349–15356. doi: 10.1074/jbc.M700156200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases. Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284(16):10571–10582. doi: 10.1074/jbc.M900752200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghoshal S, Zhu Q, Asteian A, et al. TNP [N2-(m-Trifluorobenzyl), N6-(p-nitrobenzyl)purine] ameliorates diet induced obesity and insulin resistance via inhibition of the IP6K1 pathway. Mol Metab. 2016;5(10):903–917. doi: 10.1016/j.molmet.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wundenberg T, Grabinski N, Lin H, Mayr GW. Discovery of InsP 6 -kinases as InsP 6 -dephosphorylating enzymes provides a new mechanism of cytosolic InsP 6 degradation driven by the cellular ATP/ADP ratio. Biochem J. 2014;462:173–184. doi: 10.1042/BJ20130992 [DOI] [PubMed] [Google Scholar]
- 20.Puhl-Rubio AC, Stashko MA, Wang H, et al. Use of Protein Kinase–Focused Compound Libraries for the Discovery of New Inositol Phosphate Kinase Inhibitors. SLAS Discov Adv Life Sci R&D. 2018;23(9):982–988. doi: 10.1177/2472555218775323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu C, Stashko MA, Puhl-Rubio AC, et al. Inhibition of Inositol Polyphosphate Kinases by Quercetin and Related Flavonoids: A Structure–Activity Analysis. J Med Chem. 2019;62(3):1443–1454. doi: 10.1021/acs.jmedchem.8b01593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macherey A-C, Dansette PM. Biotransformations Leading to Toxic Metabolites: Chemical Aspects. Pract Med Chem. January 2015:585–614. doi: 10.1016/B978-0-12-417205-0.00025-0 [DOI] [Google Scholar]
- 23.Wormald M, Liao G, Kimos M, Barrow J, Wei H. Development of a homogenous high-throughput assay for inositol hexakisphosphate kinase 1 activity. Song C, ed. PLoS One. 2017;12(11):e0188852. doi: 10.1371/journal.pone.0188852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y-C, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.