Abstract
Introduction
Congenital anomalies of the kidney and urinary tract (CAKUT) are the most common cause of chronic kidney disease (~45%) that manifests before 30 years of age. The genetic locus containing COL4A1 (13q33–34) has been implicated in vesicoureteral reflux (VUR), but mutations in COL4A1 have not been reported in CAKUT. We hypothesized that COL4A1 mutations cause CAKUT in humans.
Methods
We performed whole exome sequencing (WES) in 550 families with CAKUT. As negative control cohorts we used WES sequencing data from patients with nephronophthisis (NPHP) with no genetic cause identified (n=257) and with nephrotic syndrome (NS) due to monogenic causes (n=100).
Results
We identified a not previously reported heterozygous missense variant in COL4A1 in three siblings with isolated VUR. When examining 549 families with CAKUT, we identified nine additional different heterozygous missense mutations in COL4A1 in 11 individuals from 11 unrelated families with CAKUT, while no COL4A1 mutations were identified in a control cohort with NPHP and only one in the cohort with NS. Most individuals (12/14) had isolated CAKUT with no extrarenal features. The predominant phenotype was VUR (9/14). There were no clinical features of the COL4A1-related disorders (e.g., HANAC syndrome, porencephaly, tortuosity of retinal arteries). Whereas COL4A1-related disorders are typically caused by glycine substitutions in the collagenous domain (84.4% of variants), only one variant in our cohort is a glycine substitution within the collagenous domain (1/10).
Conclusion
We identified heterozygous COL4A1 mutations as a potential novel autosomal dominant cause of CAKUT that is allelic to the established COL4A1-related disorders and predominantly caused by non-glycine substitutions.
Keywords: COL4A1, congenital anomalies of the kidney and urinary tract (CAKUT), whole-exome sequencing (WES)
INTRODUCTION
Congenital anomalies of the kidney and urinary tract (CAKUT) are the commonest cause of CKD in the first three decades of life, amounting to ~45% of cases (Chesnaye et al. 2014). CAKUT can present as an isolated renal condition or as part of a clinical syndrome (Soliman et al. 2015; van der Ven et al. 2018b; Vivante et al. 2014). To date, 42 monogenic causes of isolated CAKUT have been identified (Supplementary Table S1) (van der Ven et al. 2018a).
Collagen IV is a major component of the basement membrane and is part of a specialized extracellular matrix structure that influences cell behavior and signaling. While COL4A1 mutations have been known to cause autosomal dominant forms of porencephaly (OMIM# 175780), they have also been recognized to cause a broader spectrum of conditions summarized as “COL4A1-related disorders” with eye defects (e.g., tortuosity of retinal arteries, OMIM# 180000), brain small vessel disease with or without ocular anomalies (OMIM# 607595), and systemic defects (e.g., hereditary angiopathy with nephropathy, aneurysms, and muscle cramps; HANAC syndrome, OMIM# 611773) (Meuwissen et al. 2015). Notably, the clinical phenotype of patients with COL4A1 variants is noticed to be extremely variable, with broad intra- and interfamilial variation and evidence for reduced penetrance (Meuwissen et al. 2015). While the COL4A1 locus (13q34) has been implicated in vesicoureteral reflux (VUR) and other CAKUT phenotypes (Supplementary Table S2), no single nucleotide mutations of COL4A1 have yet been reported in isolated CAKUT (Vats et al. 2006; Wang et al. 2017).
We have previously shown that whole-exome sequencing (WES) provides an etiologic diagnosis in up to 14% of patients with CAKUT (Connaughton and Hildebrandt 2019; van der Ven et al. 2018a). By means of WES, we identified a heterozygous COL4A1 variant in three siblings with VUR and, hence, hypothesized that COL4A1 mutations may present a novel cause of non-syndromic CAKUT. To this effect, we evaluated WES data for mutations in COL4A1 from a cohort of 550 families, who had WES done because of a clinical diagnosis of CAKUT without detecting a mutation in one of the 42 known CAKUT genes. We identified 11 additional families with heterozygous variants in the COL4A1 gene. Interestingly, the individuals in our cohort had predominantly nonglycine substitutions, while COL4A1-related disorders are typically caused by glycine substitutions. We identified heterozygous COL4A1 mutations as a potential novel autosomal dominant cause of CAKUT that is allelic to the established COL4A1-related disorders and predominantly caused by non-glycine substitutions
MATERIALS AND METHODS
Study participants
The study was approved by the institutional review board of Boston Children’s Hospital as well as the institutional review boards of institutions where we recruited families. It was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments and comparable ethical standards. Following informed consent, 550 different families were enrolled and had WES performed on DNA samples. All patients with CAKUT were referred to us by their pediatric nephrologist or urologist who made a clinical diagnosis of CAKUT on the basis of renal imaging studies. CAKUT was defined as demonstration of any abnormality of number, size, shape, or anatomic position of the kidneys or other parts of the urinary tract and included at least one of the following: renal agenesis, renal hypo/dysplasia, multicystic dysplastic kidneys, hydronephrosis, ureteropelvic junction obstruction, hydroureter, vesicoureteral reflux, ectopic or horseshoe kidney, duplex collecting system, ureterovesical junction obstruction, epi/hypospadias, and posterior urethral valves.
Control cohorts
Control cohort I consisted of 257 different families with a clinical diagnosis of Nephronophthisis (NPHP) with no genetic cause identified. The diagnosis of NPHP or an NPHP-related ciliopathy (NPHP-RC) was based on previously published clinical criteria (Chaki et al. 2011). As a first diagnostic step, homozygous deletions of NPHP1 were excluded in all patients by applying a multiplex PCR-based deletion analysis described elsewhere (Otto et al. 2008). Subsequently, WES was performed in all 257 patients, with no genetic cause identified after analysis of more than 90 NPHP-RC genes and screening for novel autosomal recessive causes. Control cohort II consisted of 100 families with steroid resistant nephrotic syndrome due to an underlying monogenic cause (Supplementary Table S3).
Whole exome sequencing and variant calling
WES was performed as previously described (Braun et al. 2016). In brief, genomic DNA was isolated from blood lymphocytes or saliva samples and subjected to exome capture using Agilent SureSelect human exome capture arrays (Life Technologies), followed by next generation sequencing on the Illumina HighSeq sequencing platform. Sequence reads were mapped to the human reference genome assembly (NCBI build 37/hg19) using CLC Genomics Workbench (version 6.5.2) software (CLC Bio, Aarhus, Denmark). After alignment to the human reference genome, variants were filtered for most likely deleterious variants as previously described. (Gee et al. 2014; Sadowski et al. 2015) Variants with minor allele frequencies >1% in the dbSNP (version 147) or the 1000 Genomes Project (1094 subjects of various ethnicities; May 2011 data release) databases were excluded, because they were unlikely to be deleterious. Synonymous and intronic variants that were not located within canonical splice site regions were excluded. Kept variants, which included nonsynonymous variants and splice site variants, were then analyzed.
Screening for mutations in known monogenic causes of CAKUT
We evaluated WES data for causative mutations in 42 monogenic genes for isolated CAKUT known at the time (Supplemental Table S1) (van der Ven et al. 2018a). To assess deleteriousness of potential mutations, remaining variants were ranked based on their probable effect on the function of the encoded protein considering evolutionary conservation among orthologs across phylogeny using ENSEMBL Genome Browser and assembled using Clustal Omega as well as the web-based prediction programs CADD, MutationTaster, PolyPhen-2, and SIFT. Variant filtering based on population frequency was performed using population databases (EVS server, ExAC, gnomAD, and 1000-genomes) to include only rare alleles (i.e., minor allele frequency <1%). Remaining variants were confirmed in original patient DNA by Sanger sequencing and ranked by deleteriousness using the ACMG criteria (Richards et al. 2015). Whenever familial DNA (parents or siblings) was available, segregation analysis was performed.
Variant filtering to identify novel monogenic causes of CAKUT
We used homozygosity mapping data to test for homozygosity by descent in all families (>100 Mb of cumulative homozygosity according to mapping) and performed trio-WES analysis to identify genomic candidate loci for the underlying mutation. The remaining calls (i.e., compound heterozygous variants or heterozygous variants only) were ranked for their predicted pathogenicity based on the following criteria (Lovric et al. 2016; Sadowski et al. 2015; Vivante and Hildebrandt 2016): a) protein-truncating or obligatory splice site mutation vs. missense mutation or in-frame deletion/insertion, b) evolutionary conservation, c) minor allele frequency in control databases (gnomAD, EVS), d) chemical difference between the wildtype and the altered amino acid residue, e) web-based mutation analysis prediction tools (SIFT, PolyPhen-2, MutationTaster).
Consideration of structural data and evolutionary conservation for variant evaluation
Protein domain structure cartoons and evaluation was based on the uniprot (Universal Protein Resource) database. Orthologous proteins used to evaluate evolutionary conservation were obtained from the Ensemble Genome Browser and were aligned using the Clustal Omega multiple sequence alignment tool (EMBL-EBI) (Sievers et al. 2011). The datasets generated and/or analyzed during the current study are available from the corresponding author on request.
Statistical analysis
Categorical variables were compared by using two-tailed Fisher’s exact test. A P-value <0.05 was considered statistically significant.
Web Resources
CADD – Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu/
Clustal Omega, http://www.ebi.ac.uk/Tools/msa/clustal
Ensembl Genome Browser, http://www.ensembl.org
Exome Variant Server, http://evs.gs.washington.edu/EVS
Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org
HGMD Professional 2016.3, https://portal.biobase-international.com/hgmd
Homozygosity Mapper, http://www.homozygositymapper.org/
MutationTaster http://www.mutationtaster.org
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Polyphen2, http://genetics.bwh.harvard.edu/pph2
Sorting Intolerant From Tolerant (SIFT), http://sift.jcvi.org
UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
Uniprot Consortium, http://www.uniprot.org/
1000 Genomes Browser http://browser.1000genomes.org
RESULTS
Heterozygous COL4A1 missense variants identified in patients with CAKUT
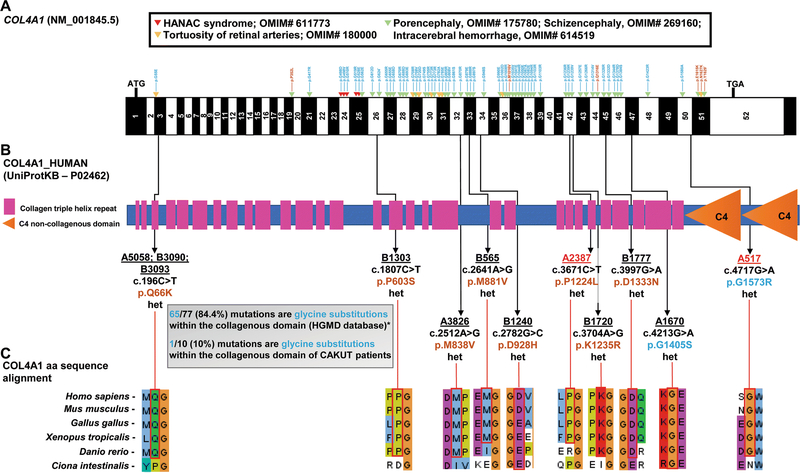
We performed WES in 550 families with CAKUT. First, we excluded disease-causing variants in 42 genes that are known to cause monogenic CAKUT, if mutated (Supplementary Table S1). In a non-consanguineous family from Serbia (A1670) with three siblings affected by CAKUT, and two siblings that were unaffected, we analyzed the genomic regions shared by all three affected individuals for homozygous, compound heterozygous, and heterozygous variants. Using this strategy, we identified a shared genetically strong heterozygous missense mutation in the gene COL4A1 (NM_001845.5), encoding the alpha-1 subunit of collagen type IV (Table1, Figure 1B). The mutation was inherited from the unaffected mother and was not present in the two healthy siblings. The identified variant causes a glycine to serine substitution in a highly conserved protein region (c. 4213G>A, p.G1405S; conserved to Ciona intestinalis) (Table1, Figure 1C).
Table 1.
Heterozygous likely disease-causing mutations of COL4A1 identified by WES in twelve families with CAKUT.
| Family-Individual | Nucleotide change | Aa change | Exon (zygosity, segregation) | PPH2 Score | SIFT | Mut. Taster | CADD Score | Aa conservation to species | gnomAD (hom/het/wildtype) | Sex | Ethnicity | Consanguinity | Extrarenal features | Renal phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A5058_21 | c.196C>A | p.Q66K | 3 (het, m) | 0.814 | D | DC | 24 | Danio rerio | 0/11/251484 | M | Albanian | No | none | Right hypo/dysplastic kidney |
| B3090_21 | c.196C>A | p.Q66K | 3 (het, p) | 0.814 | D | DC | 24 | Danio rerio | 0/11/251484 | M | Macedonia | no | none | Right sided ureter duplex with hydronephrosis |
| B3093_21 | c.196C>A | p.Q66K | 3 (het, m) | 0.814 | D | DC | 24 | Danio rerio | 0/11/251484 | M | Macedonia | no | none | Left UVJO with hydronephrosis |
| B1303_21 | c.1807C>T | p.P603S | 26 (het, m) | 0.563 | D | DC | 23.7 | Danio rerio | 0/3/276118 | F | Macedonia | no | none | MCDK |
| A3826_21 | c.2512A>G | p.M838V | 32 (het, n/d) | 0.603 | D | DC | 22.9 | Danio rerio | not reported | M | Indian | yes | none | Right VUR, bilateral hydronephrosis |
| B565_21 | c.2641A>G | p.M881V | 33 (het, m) | 0.698 | D | DC | 23.7 | Xenopus tropicalis | 0/19/282818 | M | Caucasian | no | none | Right VUR grade V, hydronephrosis, solitary kidney, PUV |
| B1240_21 | c.2782G>C | p.D928H | 34 (het, p) | 0.992 | D | DC | 24.5 | Mus musculus | not reported | M | Arabic | no | none | Right VUR grade IV, hydronephrosis, PUV |
| A2387_21 | c.3671C>T | p.P1224L | 42 (het, m) | 1 | T | DC | 22.5 | Xenopus tropicalis | 0/15/269928 | M | Albanian | no | Intellectual disability, facial dysmorphism, blue sclerae | Left VUR grade IV, ectopic dysplastic left kidney |
| B1720_21 | c.3704A>G | p.K1235R | 42 (het, p) | 0.998 | D | DC | 24 | Danio rerio | 0/3/234250 | F | Arabic | yes | none | Bilateral VUR grade III |
| B1777 | c.3997G>A | p.D1333N | 45 (het, n/d) | 0.999 | D | DC | 23.1 | Danio rerio | 0/19/282894 | M | Arabic | yes | none | Right VUR grade V, Left VUR grade IV |
| A1670_21 | c.4213G>A | p.G1405S | 47 (het, m) | 1 | D | DC | 24.8 | Ciona intestinalis | not reported | F | Serbian | no | none | Left VUR |
| A1670_22 | c.4213G>A | p.G1405S | 47 (het, m) | 1 | D | DC | 24.8 | Ciona intestinalis | not reported | F | Serbian | no | none | Left VUR |
| A1670_23 | c.4213G>A | p.G1405S | 47 (het, m) | 1 | D | DC | 24.8 | Ciona intestinalis | not reported | F | Serbian | no | none | Bilateral VUR |
| A517_21 | c.4717G>A | p.G1573R | 50 (het, m) | 1 | D | DC | 26.3 | Danio rerio | 0/3/251382 | M | Macedonia | no | Intestinal obstruction | Left ectopic kidney, cross ectopia |
Aa, amino acid; CADD, Combined Annotation Dependent Depletion (>20, predicted to be among the 1% most deleterious substitutions in the human genome); DC, predicted to be disease-causing (SIFT); D, predicted to be deleterious (MutationTaster); gnomAD, genome aggregation database; het, heterozygous; hom, homozygous; m, maternal; MCDK, multi-cystic dysplastic kidney; n/d, no data; p, paternal; PPH2 score, PolyPhen-2 prediction score (0.0 to 1.0, i.e., tolerated to deleterious, variants from 0.85 to 1 are more confidently predicted to be damaging); SIFT, Sorting intolerant from tolerant; T, tolerated; PUV, posterior urethral valve; UVJO, ureterovesical junction obstruction; VUR, vesicoureteral reflux.
Figure 1. WES detects in 12 out of 550 CAKUT families with isolated CAKUT heterozygous COL4A1 mutations, that are mostly non-glycine substitutions, as opposed to the established COL4A1-related disorders that mostly report glycine substitutions.
A) Exon structure of human COL4A1 cDNA is denoted in black and white; ATG, start codon; TGA, stop codon; arrow heads denote missense mutations in COL4A1-related disorders found in the HGMD® database: blue denotes glycine substitutions, while brown denotes non-glycine substitutions. Note that 65 out of 77 (84.4%) mutations are glycine substitutions.
*Only 64 of the 77 mutations of the HGMD® database are depicted due to space limitations.
B) Protein domain structure of human COL4A1 showing the domain position of each of the missense mutations identified in ten families with CAKUT. Family ID is underlined; family ID is highlighted in red if extra-renal clinical manifestations are noted, p.change is in brown for non-glycine substitutions and in blue for glycine substitutions. Note that only one out of ten mutations (10%) is a glycine substitution within the collagenous domain.
C) Clustal alignment of amino acid sequences of COL4A1 demonstrating evolutionary conservation from mammalia to ascidiacea for each amino acid residue.
Subsequently, we evaluated WES data from a cohort of affected individuals from the remaining 549 families with CAKUT for variants in the COL4A1 gene. We additionally identified nine different heterozygous mutations in COL4A1 in 11 individuals from 11 unrelated families, for whom an evaluation of the known CAKUT genes and an evaluation for novel autosomal-recessive genes did not reveal an underlying cause (Figure 2). The family structures are suggestive of an autosomal dominant mode of inheritance with reduced penetrance. An autosomal dominant mode of inheritance is further supported by the observation that COL4A1 demonstrates extreme loss-of-function intolerance (pLI = 1; observed variants/expected variants = 0.06, 95% CI = 0.03 – 0.013; gnomAD) (Lek et al. 2016). Notably, individuals from family A5058, B3090, and B3093 all shared the same variant (c.196C>A, p.Q66K). All three families were originally from Macedonia, suggesting the possibility of a founder effect due to a common ancestor. All variants showed strong evolutionary conservation (Table1, Figure 1C).
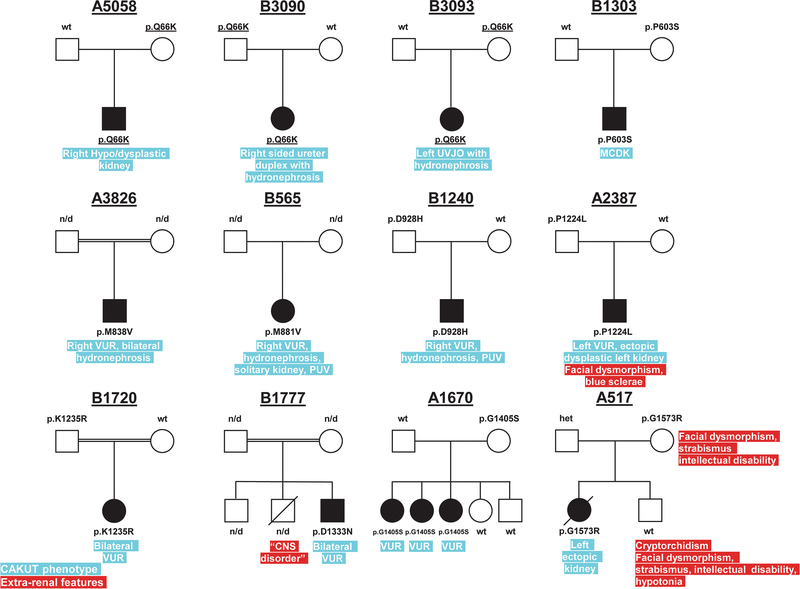
Figure 2. Pedigrees of families with CAKUT and COL4A1 mutations.
In index family A1670, the variant identified in COL4A1 (p.G1405S) segregated to all three affected siblings, while being absent in the unaffected siblings. The variant was present in the mother for whom, no clinical information was available. Families A5058, B3090, and B3093 carry the same rare COL4A1 variant (p.Q66K), which was inherited from a parent, and all three families are from Macedonia. This suggests the possibility of a founder effect due to a common ancestor. The observation that all mutations are inherited from an unaffected parent suggests incomplete penetrance and variable expressivity, which has been well documented in CAKUT.
CNS, central nervous system; MCDK, multi-cystic dysplastic kidney; n/d, no data; PUV, posterior urethral valve; UVJO, ureterovesical junction obstruction; VUR, vesicoureteral reflux.
All families were screened for genes known to cause non-syndromic isolated CAKUT. However, no likely causative heterozygous or biallelic variants were detected in those genes, except for a genetically strong SIX2 mutation in family B1720.
Individuals in our cohort demonstrated a spectrum of CAKUT phenotypes (Table 1), with VUR being the predominant phenotype, present in nine out of 14 affected individuals. Two patients had extra-renal features (Table 1). Notably, none of the patients had features found in the established COL4A1-related disorders.
Individuals with COL4A1 mutations and CAKUT have a preponderance of non-glycine substitutions, in contrast to patients with the currently established COL4A1-related disorders
The COL4A1 mutations that we identified in 14 individuals with CAKUT from 12 different families appeared to be randomly distributed throughout the gene, with four of the ten variants present in amino acid sequence stretches of low complexity, i.e., collagen triple helix repeats, while one variant was present in the globular non-collagenous C4 domain (Figure 1B). While the currently established COL4A1-related disorders are typically caused by glycine substitutions within the collagenous domain (65 out of 77 (84.4%) missense mutations reported in the HGMD® database), nine out of ten mutations (90%) of our 12 families are not glycine substitutions within the collagenous domain (Figure 1B).
Heterozygous COL4A1 missense variants are enriched in patients with CAKUT when compared to patients with a clinical diagnosis of nephronophthsis (NPHP) and nephrotic syndrome (NS)
In total, WES was performed in 550 families with a CAKUT phenotype as defined above (see Methods). As a negative control, we screened two different control cohorts for heterozygous COL4A1 variants. Control cohort I had WES performed because of a clinical diagnosis of NPHP with no causative mutation identified in any of the almost 100 known NPHP genes (n=257 families) (Connaughton and Hildebrandt 2019). Control cohort II consisted of 100 families who had WES performed because of NS, and who were found to have a causative mutation in one of the 55 established NS genes (Warejko et al. 2018). COL4A1 variants with a minor allele frequency <1% were analyzed in these control cohorts using predefined criteria as described in the Methods section. Following this step-wise approach, no COL4A1 variants remained in control cohort I, while one variant remained in control cohort II. The overall burden of COL4A1 variants meeting all predefined screening criteria was statistically significantly increased by almost eight-fold in families with CAKUT when compared to families in both control cohorts combined (2.2% vs 0.28%, respectively; OR = 7.9, P = 0.0201; Table 2). For a detailed analysis of all variants identified in all three cohorts, see Supplementary Table S4, and Supplementary table S5.
Table 2.
Overview of COL4A1 variant evaluation from WES data in 550 families with CAKUT, and negative control families represented by 257 families with NPHP (control I), and 100 families with NS (control II).
| Cohort | CAKUT N (%) | NPHP (control I) N (%) | NS (control II) N (%) | Control I+II N (%) |
|---|---|---|---|---|
| 1) Total # of families with WES data a | 550 | 257 | 100 | 357 |
| 2) Total # of rare heterozygous COL4A1 variants (MAF<1%) b | 30 (5.5%) | 10 (3.9%) | 8 (8%) | 18 (5%) |
| 3) Remaining # of families after evaluation by predefined criteria c | 12 (2.2%) | 0 (0%) | 1 (1%) | 1 (0.28%) |
CAKUT, congenital anomalies of the kidney and urinary tract; MAF, minor allele frequency; NPHP, nephronophthisis; NS, nephrotic syndrome
Whole exome sequencing was performed in 550 families with CAKUT, 257 families with NPHP, and 100 families with NS.
Non-synonymous and splice-site variants were filtered for a MAF<1%.
Variants with a MAF<1% were analyzed for three filter criteria: Filter criterion 1, evolutionary conservation (moderate to strong; i.e., at least to Xenopus tropicalis); Filter criterion 2, severity (i.e., predicted to be deleterious by two out of three in-silico prediction tools and CADD score >20); and Filter criterion 3, allele frequency (i.e., variant present in less than 20 heterozygous alleles) in a healthy control cohort (gnomAD) (Lek et al. 2016). Remaining variants had to meet all three filter criteria (Supplemental table S4 and Supplemental table S5).
Discussion
While COL4A1 mutations have been confirmed to cause ocular, cerebral, renal (i.e., in form of a cystic nephropathy) and muscular defects, we here show for the first time heterozygous missense mutations in the COL4A1 gene as a potential novel cause of non-syndromic CAKUT. The genomic locus of COL4A1 (13q34) has previously been implicated in CAKUT/VUR by deletion mapping on chromosome 13q, in at least ten children with genomic heterozygous microdeletions and syndromic CAKUT (Supplemental Table S2) (Kaylor et al. 2014; Luo et al. 2000; Mimaki et al. 2015; Turleau et al. 1978; Vats et al. 2006; Walczak-Sztulpa et al. 2008; Walsh et al. 2001). Furthermore, Wnt5a−/− knockout mice have been shown to have an abnormal ureteric tree development with reduced Col4a1 expression levels due to Wnt5a deficiency (Pietila et al. 2016). Interestingly, more recent evidence in a cohort of 49 children with VUR (stage II-IV) demonstrated the association of the rs565470 COL4A1 polymorphism with VUR (Petritsa 2017). Notably, VUR was the predominant phenotype in our cohort, present in nine out of 14 CAKUT patients with heterozygous missense mutations in COL4A1.
The inheritance pattern in all our families suggests an autosomal dominant mode of inheritance with reduced penetrance (Figure 2; Supplemental Figure S6). Reduced penetrance (45–66%) and variable expressivity has been well documented for CAKUT (Chapman et al. 1985; Noe et al. 1992). However, it is likely that for many risk genes the actual penetrance is even lower, given that penetrance estimates are likely confounded by ascertainment bias. For example, BMP4 missense variants are frequently found to be inherited from a mildly or unaffected parent (Reis et al. 2011; Suzuki et al. 2009; Weber et al. 2008), while mice with heterozygous ROBO2 mutations demonstrate a 15% penetrance of the CAKUT phenotype (Lu et al. 2007).
In index family A1670, the COL4A1 variant (p.G1405S) was present in all three siblings with VUR but absent in the two unaffected siblings. The variant was inherited from the mother for whom no clinical information was available. In family A517, the mother carried the COL4A1 variant (p.G1573R) and was reported to have intellectual disability, strabismus, and facial dysmorphic features. While strabismus has been described in one patient with brain small vessel disease (OMIM#175780) none of these features are thought of as characteristic clinical features of the COL4A1-related disorders (https://www.ncbi.nlm.nih.gov/books/NBK7046/#col4a1-dis.Clinical_Characteristics), making them likely to be due to a different undiagnosed genetic etiology. Notably, individual A517_II-2 was found to have similar clinical features as the mother (e.g., intellectual disability, strabismus, and facial dysmorphic features), but did not carry the COL4A1 variant. However, the COL4A1 variant was inherited by the first-born child (A517_II-1), who presented with left ectopic kidney, renal failure of unclear etiology, and died in the intensive care unit due to a suspected bowel obstruction. In family B1720, the index patient carried a competing SIX2 variant, which was inherited from the unaffected father. The clinical relevance of variants in the SIX2 gene in the development of CAKUT is still unclear (Faguer et al. 2012). Families A5058, B3090, and B3093 carried the same rare COL4A1 variant (p.Q66K), which was inherited from a parent, and all three families are from Macedonia. This suggests the possibility of a founder effect due to a common ancestor.
COL4A1 is highly conserved across species and is present in almost all basement membranes (Kuo et al. 2012). As a component of the extra-cellular matrix, the basement membrane is important to provide physical support for tissue architecture, but moreover, it also serves as a reservoir for growth factors and other signaling molecules by binding them through receptors, such as integrins (Kim and Nelson 2012). In the kidney in particular, the basement membrane plays an important role in ureteric-bud (UB) branching morphogenesis. Multiple growth factors, such as FGF and TGFβ, have been identified as important modulators of UB branching. However, also structural molecules such proteoglycans, which are an integral part of the basement membrane, have been shown to affect UB morphogenesis in rat kidney development, by regulating growth factor activity along the branching UB and in the developing metanephric mesenchyme (Shah et al. 2011). While it is conceivable that COL4A1 may play a similar role during kidney development, one is mostly left to speculate on the possible underlying pathological mechanism.
The currently established COL4A1-related disordersare typically caused by glycine substitutions within the collagenous domain (65/77, 84.4% of the missense mutations in the HGMD® database). Interestingly, only one of our twelve families had a glycine substitution within the collagenous domain. Glycine residues are required at every third position of the collagenous domain, since the absence of a side chain allows glycine to fit into the core of the triple helix (Kuo et al. 2012; Prockop and Kivirikko 1995). Both the position of the mutation and the residue replacing the glycine can influence the biosynthetic consequence of the mutation (Baker et al. 2007). HANAC syndrome (OMIM# 611773) is typically caused by glycine substitutions in close proximity to exons 24 and 25 (Figure 1A), which are thought to encode integrin binding sites (Alamowitch et al. 2009; Meuwissen et al. 2015; Plaisier et al. 2010; Plaisier et al. 2007). Only family B1303 of our cohort had a non-glycine variant close to this region in exon 26 (p.P603S). The proband had no clinical findings of HANAC syndrome (Table 1). The glycine mutations in our two families are close to the C-terminus of the protein, one in the collagen domain (A1670, p.G1405S; Figure 1B) and one in the non-collagenous C4 domain (A517, p.G1573R; Figure 1B). Non-glycine mutations are generally thought to be less severe than glycine mutations. Two of our patients had proline substitutions (B1303, p.P603S and A2387, p.P1224L; Figure 1B) and hydroxylation of proline has been shown to be critical for triple helix stabilization (Kuo et al. 2012). Family B1720 had a mutation affecting a lysine residue (p.K1235R; Figure 1B). Lysine residues of type IV collagens are hydroxylated and glycosylated by lysyl-hydroxylases and pathogenic lysine substitutions have been identified in other collagenopathies (Kuo et al. 2012). Families A3826 and B565 had methionine substitutions (p.M838V and p.M881V, respectively; Figure 1B) and families A5058, B3090, and B3093 all carried the same glutamine substitution (p.Q66K; Figure 1B). While both methionine and glutamine substitutions have been demonstrated in COL41-related disorders, there is less knowledge regarding their biochemical implications (Kuo et al. 2012).
We concluded that rare COL4A1 mutations constitute a potential novel autosomal dominant cause of CAKUT that is allelic to HANAC syndrome and other COL4A1related disorders and distinct by a preponderance of non-glycine mutations. This is further supported by the observation of an almost eight-fold increase of COL4A1 variants by predefined evaluation criteria when compared to two control cohorts (Table2). Hence, we propose heterozygous COL4A1 mutations as a potential novel autosomal dominant cause of CAKUT.
Supplementary Material
Acknowledgements
We would like to thank the families and study individuals for their contribution.
Sequencing and data processing was performed by the Broad and Yale Centers for Mendelian Genomics funded by the National Human Genome Research Institute (UM1 HG008900 to DGM and HLR and U54 HG006504 to RPL).
This research was supported by grants from the National Institutes of Health to F.H. (DK076683).
A.M. is supported by a Research Training in Pediatric Nephrology grant (T32-DK007726) and the Harvard Stem Cell Institute Kidney Inter-lab Fellowship Award at Harvard Medical School (F-KP-0003-17-00).
C.H.W.W. is supported by funding from the National Institutes of Health T32-GM007748 grant.
D.M.C. is funded by the Health Research Board, Ireland (HPF-206-674), the International Pediatric Research Foundation Early Investigators’ Exchange Program and the Amgen Irish Nephrology Society Specialist Registrar Research Bursary.
N.M. is supported by funding from the National Institute of Health (T32-DK007726) grant at Boston Children’s Hospital.
T.M.K. is supported by a Post-Doctoral Fellowship award from the KRESCENT Program, a national kidney research training partnership of the Kidney Foundation of Canada, the Canadian Society of Nephrology, and the Canadian Institutes of Health Research.
F.H. and S.S. are supported by the Begg Family Foundation.
Footnotes
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Alamowitch S et al. (2009) Cerebrovascular disease related to COL4A1 mutations in HANAC syndrome Neurology 73:1873–1882 doi: 10.1212/WNL.0b013e3181c3fd12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NL et al. (2007) Molecular consequences of dominant Bethlem myopathy collagen VI mutations Ann Neurol 62:390–405 doi: 10.1002/ana.21213 [DOI] [PubMed] [Google Scholar]
- Braun DA et al. (2016) Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome Nat Genet 48:457–465 doi: 10.1038/ng.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M et al. (2011) Genotype-phenotype correlation in 440 patients with NPHP-related ciliopathies Kidney Int 80:1239–1245 doi: 10.1038/ki.2011.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman CJ, Bailey RR, Janus ED, Abbott GD, Lynn KL (1985) Vesicoureteric reflux: segregation analysis Am J Med Genet 20:577–584 doi: 10.1002/ajmg.1320200403 [DOI] [PubMed] [Google Scholar]
- Chesnaye N et al. (2014) Demographics of paediatric renal replacement therapy in Europe: a report of the ESPN/ERA-EDTA registry Pediatr Nephrol 29:2403–2410 doi: 10.1007/s00467-014-2884-6 [DOI] [PubMed] [Google Scholar]
- Connaughton DM, Hildebrandt F (2019) Personalized medicine in chronic kidney disease by detection of monogenic mutations Nephrol Dial Transplant doi: 10.1093/ndt/gfz028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faguer S, Chassaing N, Bandin F, Prouheze C, Chauveau D, Decramer S (2012) Should SIX2 be routinely tested in patients with isolated congenital abnormalities of kidneys and/or urinary tract (CAKUT)? Eur J Med Genet 55:688–689 doi: 10.1016/j.ejmg.2012.06.003 [DOI] [PubMed] [Google Scholar]
- Gee HY et al. (2014) Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies Kidney Int 85:880–887 doi: 10.1038/ki.2013.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaylor J, Alfaro M, Ishwar A, Sailey C, Sawyer J, Zarate YA (2014) Molecular and cytogenetic evaluation of a patient with ring chromosome 13 and discordant results Cytogenet Genome Res 144:104–108 doi: 10.1159/000368649 [DOI] [PubMed] [Google Scholar]
- Kim HY, Nelson CM (2012) Extracellular matrix and cytoskeletal dynamics during branching morphogenesis Organogenesis 8:56–64 doi: 10.4161/org.19813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo DS, Labelle-Dumais C, Gould DB (2012) COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets Hum Mol Genet 21:R97–110 doi: 10.1093/hmg/dds346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans Nature 536:285–291 doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovric S, Ashraf S, Tan W, Hildebrandt F (2016) Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol Dial Transplant 31:1802–1813 doi: 10.1093/ndt/gfv355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W et al. (2007) Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux Am J Hum Genet 80:616–632 doi: 10.1086/512735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Balkin N, Stewart JF, Sarwark JF, Charrow J, Nye JS (2000) Neural tube defects and the 13q deletion syndrome: evidence for a critical region in 13q33–34 Am J Med Genet 91:227–230 [DOI] [PubMed] [Google Scholar]
- Meuwissen ME et al. (2015) The expanding phenotype of COL4A1 and COL4A2 mutations: clinical data on 13 newly identified families and a review of the literature Genet Med 17:843–853 doi: 10.1038/gim.2014.210 [DOI] [PubMed] [Google Scholar]
- Mimaki M et al. (2015) Holoprosencephaly with cerebellar vermis hypoplasia in 13q deletion syndrome: Critical region for cerebellar dysgenesis within 13q32.2q34 Brain Dev 37:714–718 doi: 10.1016/j.braindev.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Noe HN, Wyatt RJ, Peeden JN Jr., Rivas ML (1992) The transmission of vesicoureteral reflux from parent to child J Urol 148:1869–1871 [DOI] [PubMed] [Google Scholar]
- Otto EA et al. (2008) Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing Hum Mutat 29:418–426 doi: 10.1002/humu.20669 [DOI] [PubMed] [Google Scholar]
- Petritsa (2017)
- Pietila I et al. (2016) Wnt5a Deficiency Leads to Anomalies in Ureteric Tree Development, Tubular Epithelial Cell Organization and Basement Membrane Integrity Pointing to a Role in Kidney Collecting Duct Patterning PLoS One 11:e0147171 doi: 10.1371/journal.pone.0147171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier E et al. (2010) Novel COL4A1 mutations associated with HANAC syndrome: a role for the triple helical CB3[IV] domain Am J Med Genet A 152A:2550–2555 doi: 10.1002/ajmg.a.33659 [DOI] [PubMed] [Google Scholar]
- Plaisier E et al. (2007) COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps N Engl J Med 357:2687–2695 doi: 10.1056/NEJMoa071906 [DOI] [PubMed] [Google Scholar]
- Prockop DJ, Kivirikko KI (1995) Collagens: molecular biology, diseases, and potentials for therapy Annu Rev Biochem 64:403–434 doi: 10.1146/annurev.bi.64.070195.002155 [DOI] [PubMed] [Google Scholar]
- Reis LM et al. (2011) BMP4 loss-of-function mutations in developmental eye disorders including SHORT syndrome Hum Genet 130:495–504 doi: 10.1007/s00439-011-0968-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology Genet Med 17:405–424 doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski CE et al. (2015) A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome J Am Soc Nephrol 26:1279–1289 doi: 10.1681/ASN.2014050489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MM, Sakurai H, Gallegos TF, Sweeney DE, Bush KT, Esko JD, Nigam SK (2011) Growth factor-dependent branching of the ureteric bud is modulated by selective 6-O sulfation of heparan sulfate Dev Biol 356:19–27 doi: 10.1016/j.ydbio.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega Mol Syst Biol 7:539 doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman NA, Ali RI, Ghobrial EE, Habib EI, Ziada AM (2015) Pattern of clinical presentation of congenital anomalies of the kidney and urinary tract among infants and children Nephrology (Carlton) 20:413–418 doi: 10.1111/nep.12414 [DOI] [PubMed] [Google Scholar]
- Suzuki S et al. (2009) Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip Am J Hum Genet 84:406–411 doi: 10.1016/j.ajhg.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turleau C, Seger J, de Grouchy J, Dore F, Job JC (1978) [Del (13) (q33). Exclusion of esterase D (ESD) from 13q33 and q34] Ann Genet 21:189–192 van der Ven AT et al. (2018a) Whole-Exome Sequencing Identifies Causative Mutations in Families with Congenital Anomalies of the Kidney and Urinary Tract J Am Soc Nephrol 29:2348–2361 doi: 10.1681/ASN.2017121265 [DOI] [Google Scholar]
- van der Ven AT, Vivante A, Hildebrandt F (2018b) Novel Insights into the Pathogenesis of Monogenic Congenital Anomalies of the Kidney and Urinary Tract J Am Soc Nephrol 29:36–50 doi: 10.1681/ASN.2017050561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats KR et al. (2006) A locus for renal malformations including vesico-ureteric reflux on chromosome 13q33–34 J Am Soc Nephrol 17:1158–1167 doi: 10.1681/ASN.2005040404 [DOI] [PubMed] [Google Scholar]
- Vivante A, Hildebrandt F (2016) Exploring the genetic basis of early-onset chronic kidney disease Nat Rev Nephrol 12:133–146 doi: 10.1038/nrneph.2015.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivante A, Kohl S, Hwang DY, Dworschak GC, Hildebrandt F (2014) Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans Pediatr Nephrol 29:695–704 doi: 10.1007/s00467-013-2684-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak-Sztulpa J et al. (2008) Chromosome deletions in 13q33–34: report of four patients and review of the literature Am J Med Genet A 146A:337–342 doi: 10.1002/ajmg.a.32127 [DOI] [PubMed] [Google Scholar]
- Walsh LE, Vance GH, Weaver DD (2001) Distal 13q Deletion Syndrome and the VACTERL association: case report, literature review, and possible implications Am J Med Genet 98:137–144 [DOI] [PubMed] [Google Scholar]
- Wang YP, Wang DJ, Niu ZB, Cui WT (2017) Chromosome 13q deletion syndrome involving 13q31qter: A case report Mol Med Rep 15:3658–3664 doi: 10.3892/mmr.2017.6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warejko JK et al. (2018) Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome Clin J Am Soc Nephrol 13:53–62 doi: 10.2215/CJN.04120417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S et al. (2008) SIX2 and BMP4 mutations associate with anomalous kidney development J Am Soc Nephrol 19:891–903 doi: 10.1681/ASN.2006111282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.