Abstract
Emerging therapeutics that utilize RNA interference (RNAi) have the potential to treat broad classes of diseases due to their ability to reversibly silence target genes. In August 2018, the FDA approved the first siRNA therapeutic, called ONPATTRO™ (Patisiran), for the treatment of transthyretin-mediated amyloidosis. This was an important milestone for the field of siRNA delivery that opens the door for additional siRNA drugs. Currently, more than 20 small interfering RNA (siRNA)-based therapies are in clinical trials for a wide variety of diseases including cancers, genetic disorders, and viral infections. To maximize therapeutic benefits of siRNA-based drugs, a number of chemical strategies have been applied to address issues associated with efficacy, specificity, and safety. This review focuses on the chemical perspectives behind non-viral siRNA delivery systems, including siRNA synthesis, siRNA conjugates, and nanoparticle delivery using nucleotides, lipids, and polymers. Tracing and understanding the chemical development of strategies to make siRNAs into drugs is important to guide development of additional clinical candidates and enable prolonged success of siRNA therapeutics.
1. Introduction
The presence of endogenous RNA interference pathways in mammalian cells provides a powerful mechanism for the regulation of cellular signaling pathways by enabling precise modulation of gene expression1. As one component of the RNAi complex, siRNAs are able to “silence” expression of specific genes with complementary sequences. In contrast to traditional drugs, the sequence of a siRNA therapeutic can be identified based on knowledge of the sequence of the messenger RNA (mRNA) that would encode for the protein target. While siRNA has the potential to target essentially any gene, function requires methods to deliver these molecules inside of target cells safely and effectively. The FDA recently approved the first ever siRNA therapeutic, which is a lipid nanoparticle (LNP) called ONPATTRO™ (Patisiran) that encapsulates and delivers siRNA against mutant and wild type transthyretin to treat transthyretin-mediated amyloidosis2. We believe that ONPATTRO is the first of many RNAi medicines, but that additional development is required to achieve the broadest clinical application of siRNA. Below we describe how chemistry has played a critical role in improving the efficacy, specificity, and safety of siRNA therapeutics, since the initial description of RNAi in 19981,.
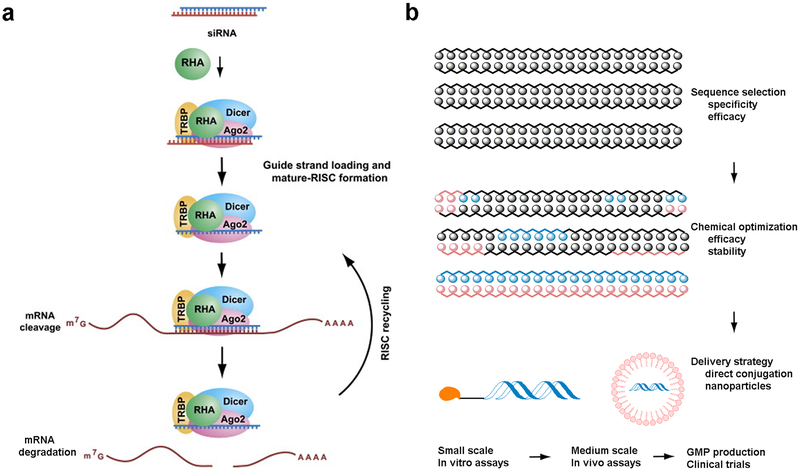
In 2006, the Nobel Prize was awarded for the discovery and characterization of RNA interference1. RNA interference is mediated by small double-stranded RNA molecules, or siRNAs, which associate with the RNA-Induced Silencing Complex (RISC) inside of cells. After loading, the strands are separated, leaving the anti-sense (or guide) strand to bind complementary mRNA (Fig. 1a)3–5. The Argonaute endoribonuclease cleaves the mRNA, which prevents it from being translated to protein5. The antisense strand stays bound to RISC, and is able to act catalytically to cleave additional mRNA strands. Although siRNA is generally double-stranded, it can be single-stranded, hairpin, or dumbbell shaped as long as the antisense strand can become loaded into the RNAi machinery6. For example, single stranded siRNA is able to activate RNAi in mice, albeit less efficiently than the canonical double stranded forms7,8.
Fig. 1. Mechanism of RNA interference and strategies for siRNA synthesis and delivery.
a. An illustration of RNA interference process3. b. Strategies and procedures for siRNA development and delivery.
Delivery of siRNA has been the major challenge to application of siRNA therapeutics in humans.4,9–12 One key challenge to delivery is the pharmacological properties of siRNA. For example, siRNAs are relatively large (~13 kDa) in molecular weight in comparison to small molecule drugs. They are highly anionic with approximately thirty eight to fifty phosphate groups, which make them difficult to diffuse across cellular membranes. Furthermore, unmodified siRNAs are unstable in the bloodstream and can induce immune responses through interaction with Toll-like receptors6. Intravenously administered siRNA must cross the vascular endothelial barrier and then diffuse through the extracellular matrix to function. They must also avoid filtration by kidneys and uptake by non-targeted cells. After cellular internalization, siRNAs need to be released from endosomal compartments, decomplex, and access the RNAi machinery. Throughout this whole process, siRNAs need to have sufficient resistance to nuclease degradation to enable function13. Therefore, siRNAs have significant delivery challenges that do not exist for small molecule drugs.5 Nevertheless, significant progress has been made, and there are currently more than 20 ongoing clinical trials with siRNA-based therapeutics14–17.
Fig 1b describes a typical process for the development of siRNA therapeutics15. First, for a specific biological target, many RNA strands need to be designed and synthesized to identify the most potent sequences along the target mRNA strand. This is generally done both experimentally and computationally18. After validation of the in vitro potency and specificity, siRNA strands are further stabilized via optimization of chemically modified nucleotides. Finally, a variety of different delivery formulation strategies can be implemented, as described below. This article will discuss the strategies and design of non-viral siRNA therapeutics from a chemical perspective.
2. Synthesis and chemical modification of siRNA
Chemically modified nucleotides can improve chemical stability and efficacy, increase cell specificity, reduce immunological effects, and decrease off-target effects6. siRNA conjugation is one important method to achieve efficacious RNA interference both in vitro and in vivo. This section will focus on the chemical modification of nucleotides and synthesis of siRNA.
2.1. Nucleotides modification
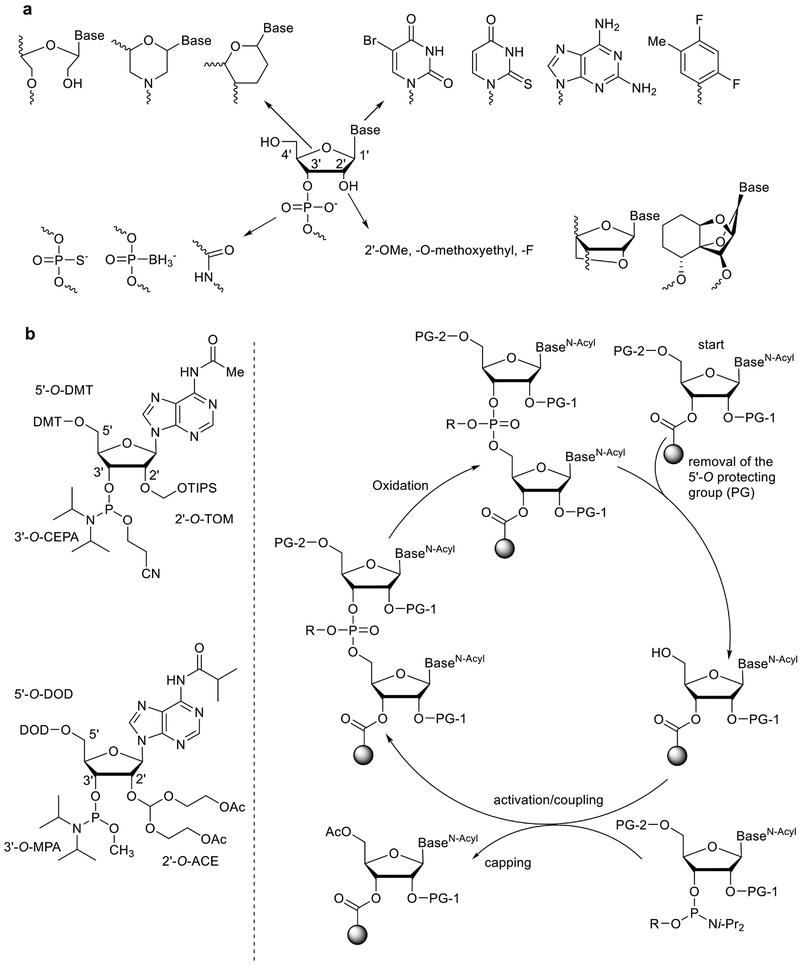
Nucleotides are the basic building blocks for both DNA and RNA. In general, natural nucleotides are composed of a ribose or 2’-deoxyribose sugar with 1’-nucleobase and 3’-phosphate groups (Fig. 2). Chemical modification of nucleotides dates back to the 1960s19. In recent years, the effects of modified nucleotides on siRNA activity have been extensively examined20,21. As illustrated in Fig. 2, four major sites have been explored for modification: the 2’-position, phosphate linkage, ribose, and nucleobase6.
Fig. 2. siRNA synthesis.
a. Chemical modification of nucleic acids including the 2’-position, phosphate linkage, ribose, and nucleobase. b. Solid-phase synthesis of RNA strands using automated RNA synthesizer49.
The 2’ position is the most common modification site in nucleotides. In 1959, Smith and Dunn isolated 2’-O-methyladenosine from wheat germ and rat liver22. The selective synthesis of this compound using diazomethane and 1,2-dimethoxyethane was described by Robins and co-workers23. Because chemical modification can potentially inhibit activity, Rana and co-workers performed a chemical modification analysis for siRNA function21. Their results indicated that 2’-OHs are not required for siRNA activity and that 2’ modification could significantly improve the stability of siRNA and extend their half-life21. The 2’ position has been modified with a number of residues including 2’-O-methyl, 2’-O-methoxyethyl, and fluoro (Fig. 2). Conformationally constrained nucleotides such as Locked Nucleic Acid (LNA) were developed by Imanishi and Wengel independently24,25. Introduction of LNA bases not only improves the stability of siRNA, but can also increase the binding affinity to RNA20. Recently, an α-L-tricyclic nucleic acid was developed, which is also highly constrained (Fig. 2)26.
With regard to the phosphate linkage, chemical synthesis using nucleotide phosphorothioate (including the relevant diastereomers) was the earliest exploration of nucleotide modification19. These linkages significantly stabilize the RNA to nuclease degradation such as those found in human serum, but may lead to increased toxicity20. Interestingly, DNA phosphorothioation was discovered in 2007 as an endogenous process in bacteria27. Recently, Wu and co-workers explored different types of phosphorodithioate on siRNAs and their antitumor activity in mouse models28. In addition, a polyamide-based Peptide Nucleic Acid (PNA) was reported by Nielsen and co-workers in 199129. Later, PNA modification was installed at the end of RNA strands30. This PNA-siRNA was reported to inhibit the activity of telomerase and introduce cell death of human tumor cells. Boron-containing nucleotide analogues were reported in the early 1990s31. A recent study reports that boranophosphate siRNAs can be more potent than native siRNA and phosphorothioate siRNAs32. However, PNA and boranophosphate siRNAs have not been investigated as extensively in animal models as phosphorothioate modified siRNAs, which are in human clinical trials15.
Modification of the ribose ring has also been investigated (Fig. 2). A morpholino nucleoside was developed in 1989, which has been examined in a number of antisense applications33. Morpholino nucleotide oligomers are being examined clinically as a therapy for Duchenne muscular dystrophy34. Also of note, Wengel and co-workers developed Unlocked Nucleic Acid (UNA) in 200935. These acyclic UNA based siRNAs were shown to induce silencing in vitro and in vivo36. Moreover, UNA modification was reported to reduce off-target effects and improve biostability in mice. At the same time, hexitol and anitrol based nucleotides were reported, and show potential for use as siRNA modifications37. Bramsen and coworkers performed an analysis of chemically modified siRNA for their silencing activity, stability and toxicity in human tumor cells38. They report that the sense strand can tolerate diverse chemical modifications, while the antisense strand can be moderately modified under certain conditions. siRNA stabilization does not require the modification of the whole siRNA duplex, but only on a few selected positions. They also reported that UNA modification introduced less toxicity and retained the silencing activity38. In 2018, oxabicyclic nucleoside phosphonates were chemically synthesized39 and might be incorporated into siRNA sequences for gene silencing.
Finally, modification of the nucleobase has been examined to improve the nuclease resistance of siRNA duplexes in serum. For instance, siRNAs containing several 2-thiouracil modified units were thermally stable and showed similar silencing activity compared with the unmodified siRNAs40. Other representative examples include 5-bromouracil, 6-diaminopurine, and 2,4-difluorotoluene21,40–42. In general, base modifications are not widely applied for siRNA modification, but nevertheless offer an additional opportunity for chemical manipulation.
2.2. Synthesis of siRNA
The synthesis of siRNA is based on research that originated in 1950s43. In 1978, Zamecnik and Stephenson reported that a synthetic oligonucleotide complementary to Rous sarcoma virus 35S RNA could inhibit protein expression44,45. This work reported that a synthetic antisense oligonucleotide (ASO) could bind to a target mRNA through Watson-Crick base pairing led to substantial research development of ASOs as therapeutics. In 2013, the FDA approved mipomersen, an ASO against apolipoprotein B, for the treatment of homozygous familial hypercholesterolemia46. Nusinersen (Spinraza) was later approved by the FDA in 2016 for the indication of spinal muscular atrophy47. In general, ASOs do not function in vivo through the RNAi pathway and therefore are not considered siRNAs48. Nonetheless, research on ASOs provided the basis for much of the chemistry used in siRNA synthesis and chemical modification today.
Solid-phase synthesis is currently the primary approach used to make synthetic RNA49,50. As shown in Fig. 2b RNA synthesis is a repetitive chemical cycle in which each nucleotide is added on a solid support. This cycle starts with a deprotection step to remove the protective group on 5’-hydroxyl of the solid support bound nucleotide. The resulting 5’-hydroxyl is then coupled with an activated 3’-phosphorous ester, followed by a capping step to remove the unreacted nucleotides from the reaction system. The intermediate undergoes another step to oxidize phosphite to phosphorous ester. After the chain assembly, the oligomer is released from the solid support, deprotected, and purified by HPLC. Two types of building blocks were later developed for the efficient synthesis of RNA including 2’-O-TOM and 2’-O-ACE modified nucleotides (Fig. 1c)49. Both methods provide a coupling yield of over 99%49. The whole process has been successfully automated by utilizing oligonucleotide synthesizers49.
3. siRNA-ligand conjugates
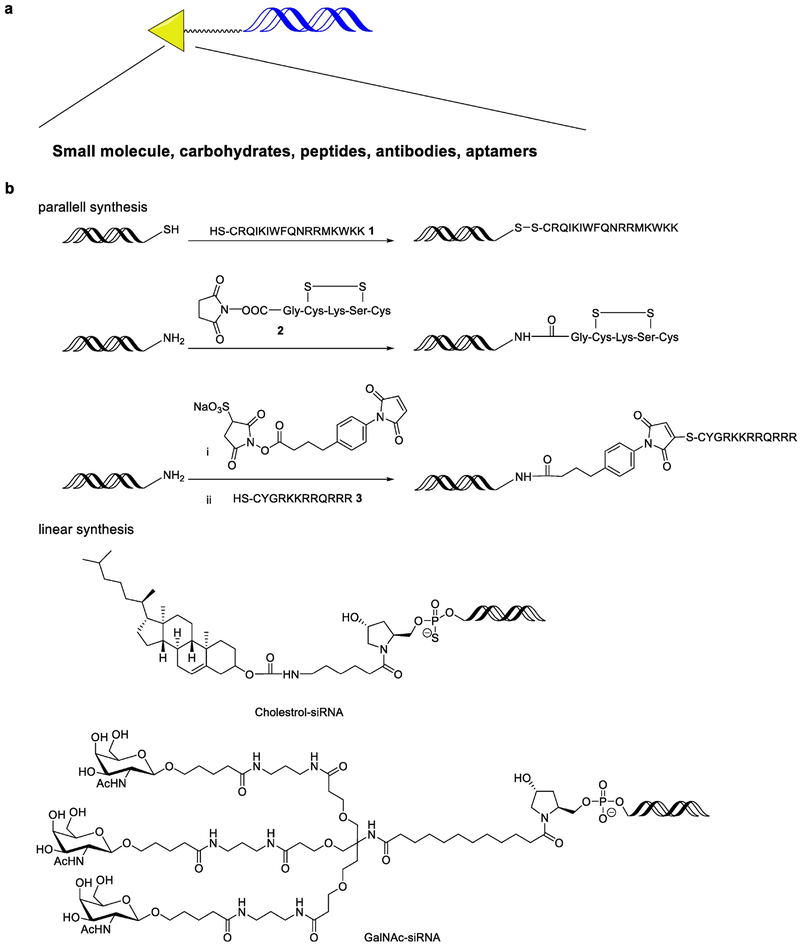
Direct ligand conjugation to siRNA is a promising delivery strategy. Diverse ligands including small molecules, carbohydrates, aptamers, peptides and antibodies have been covalently linked to siRNA in order to improve cellular uptake and target specific cell types (Fig. 3a)51–55. An advantage of siRNA conjugates is that they reduce the need for extra delivery materials, and may thereby improve the tolerability and safety profile of the delivery formulation56. Because the 5’ end of the antisense strand is required for silencing activity, conjugation is typically performed on the sense strand or 3’ end of antisense strand51. Two synthetic approaches have been applied: parallel synthesis and linear synthesis.
Fig. 3. Chemical strategies for synthesis of siRNA conjugates.
a. siRNA conjugates with ligands including small molecules, carbohydrates peptides, antibodies andaptamers. b. Parallel and linear synthesis siRNA-peptide and -cholesterol conjugates. c. An example of linear synthesis of GalNAc-siRNA conjugates.
For parallel synthesis (Fig. 3b), siRNA and its relevant conjugate ligand are synthesized in separate synthetic routes and then are conjugated with each other usually through biodegradable bonds51. For example, conjugation of membrane permanent peptides (MPPs, 1) and anti-GFP siRNA was achieved through a disulfide bond using a diamide oxidizing reagent57. Rana and co-workers applied a similar strategy to conjugate TAT (2), a cell penetrating peptide and siRNA through a succinimidyl 4-[pmaleimidophenyl]butyrate (SMPB) based linker58. Condensation of insulin receptor substrate 1 (IRS1, 3) and siRNA was also achieved through an amide linkage59. The above siRNA conjugates were reported to show silencing effects in different human cell lines.
Linear synthesis (functional groups are added sequentially) is also widely used for a variety of chemical conjugations to siRNA (Fig. 3b). In 1989, Letsinger and co-workers reported the synthesis of amide-linked, cholesterol-modified oligonucleotides60. In 2004, cholesterol and lipid modified siRNAs were created using a pyrrolidine-based linkage61. These lipophilic siRNA conjugates were shown to silence apolipoprotein B through intravenous injection in mice62, via a lipoprotein-dependent mechanism. Both low-density lipoprotein receptor and scavenger receptor class B type I are required for the uptake of siRNA conjugates by the liver and other tissues.
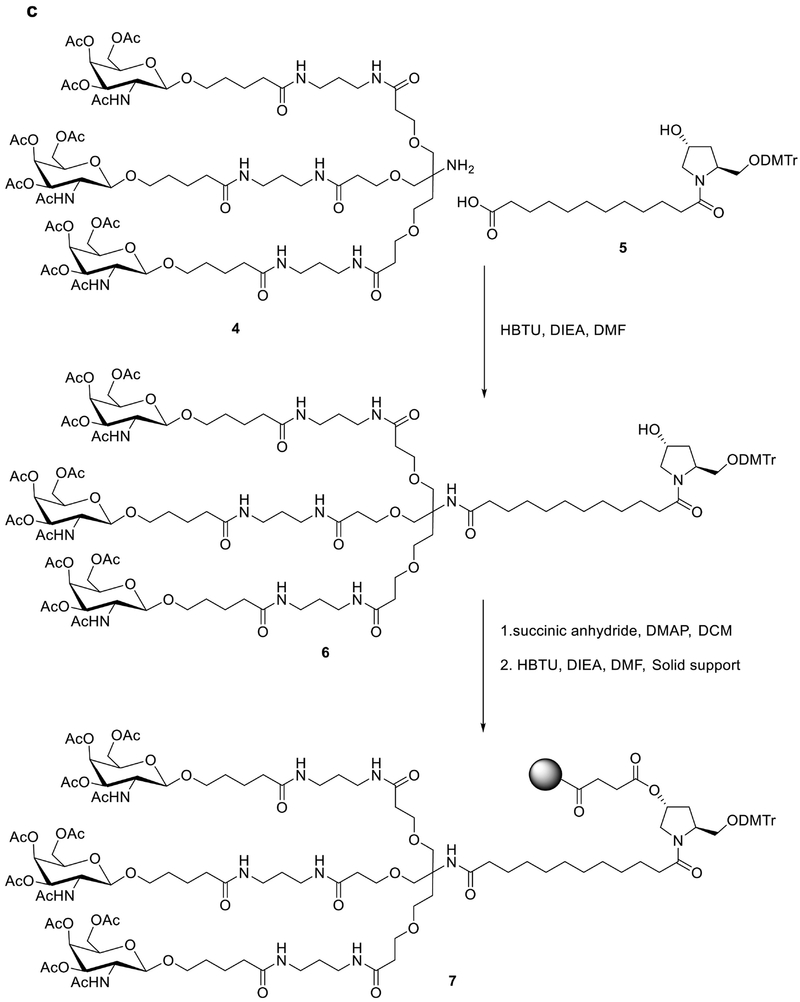
A similar conjugation strategy was applied to develop hepatocyte-targeted delivery using asialoglycoprotein receptor (ASGPR) targeted ligand, N-Acetyl-D-galactosamine (GalNAc)63. As shown in Fig. 3c, pyrrolidine derivative 4 and GalNAc derivative 5 were condensed to afford an intermediate 663. A solid support was installed on the intermediate through a succinic acid linker and gave the substrate 7 that could then be used to generate the siRNA strand (Fig. 3c). Finally, the GalNAc-siRNA strand was synthesized by adding the nucleotide one by one through the solid phase-based approach as discussed above. This GalNAc-siRNA conjugate is able to significantly silence target gene in hepatocytes via subcutaneous administration at a dose of single digit mg/kg in mice56. Results from a clinical trial of this conjugate demonstrate significant reduction of serum transthyretin (TTR) protein for treating TTR mediated amyloidosis64. Currently, a series of these GalNAc-siRNA conjugates are in early or late stage of clinical trials2,17. Cemdisiran is in phase II clinical investigation for patients with complement-mediated diseases2,17. In addition, several promising candidates including Vutrisiran (indication: TTR-mediated amyloidosis), Fitusiran (indication: hemophilia and rare bleeding disorders), Inclisiran (indication: hypercholesterolemia), and Lumasiran (indication: primary hyperoxaluria type 1) are in Phase III clinical trials2,17. In addition to the clinical advance, a wide variety of new chemical approaches were reported to optimize the GalNAc-siRNA conjugate. For example, GalNAc can be conjugated on the 2’-position of the ribose65. Matsuda el al systematically explored the effects of GalNAc on different sites of siRNA strands and identified several potent sequences65. Most recently, Parmar and co-workers incorporated (E)-Vinylphosphonate at the 5’- end of the antisense strand, which stabilized the siRNA and improved its potency66. These findings provide new insights on next generation siRNA conjugates.
4. Nucleotides derived nanoparticles
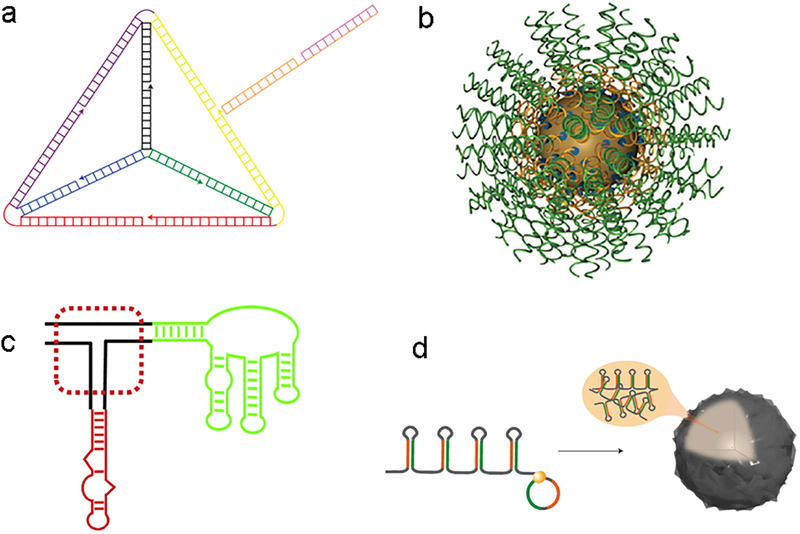
Nucleotides have long been utilized as building blocks to assemble a wide variety of nanoparticles67. For example, DNA nanostructures, also called DNA origami, have been explored for over 30 years67. A number of two-dimensional and three-dimensional nanostructures have been self-assembled through branched DNA motifs and crystalized for characterization and visualization. In 2012, self-assembled DNA-siRNA tetrahedral nanoparticles were developed for siRNA delivery68. The 28.6 nm tetrahedron nanoparticles were composed of 186 Watson-Crick base pairs. Each edge is 30 base pairs long and contains a nick in the middle (Fig. 4). This nick is complementary to the overhang of siRNA strands to serve as a siRNA carrier, which was also applied for aptamer-based siRNA delivery55. In order to differentiate tumor cells from normal cells, a cancer-targeting ligand, folate, was installed on the nanoparticle surface. The targeted siRNA-DNA origami showed significant silencing in tumor cells at a dose of 2.5 mg/kg (anti-luciferase siRNA) in a mouse xenograft model. It also displayed a longer blood circulation time (t1/2 ≈ 24.2 min) compared with free siRNA (t1/2 ≈ 6 min). Different from the DNA origami, Mirkin and co-workers first reported spherical nucleic acid (SNA) conjugates in 1996, which were made with gold cores and DNA shells69. SNA nanostructures are determined by the shape of the cores and the shells can accommodate both single- and double-stranded nucleic acids with sequences of interest. To construct functional SNA, three components were necessary: a particle attachment moiety, a spacer region, and a programmable recognition region69. In the past decade, a number of inorganic cores and nucleic acids shells have been investigated for diverse applications including diagnosis, small molecular drug delivery, and DNA and siRNA delivery69. By conjugating different siRNA sequences, SNA achieved gene silencing of a variety of biological targets including luciferase, epidermal growth factor receptor (EGFR), and Bcl2Like1270. Currently, this platform is in the clinical trial for treating glioblastoma. In addition, RNA nanostructures were also applied to siRNA delivery71,72. For example, Guo and co-workers constructed multifunctional RNA nanoparticles based on RNA three-way junctions, which showed effective delivery of siRNA targeting survivin73. Later on, a diverse set of RNA based nanomaterials were created for gene silencing and other applications71,74. In 2012, Hammond and co-workers developed self-assembled RNA interference microsponges via rolling circle replication (RCT), which was applied by viruses to amplify their genes75. To achieve this process, they constructed a linear DNA strand encoding the antisense and sense sequences. Also, their ends were partially complementary to the T7 promoter. After hybridization with a T7 promoter, this linear DNA formed a circular DNA with its nick closed by a T4 DNA ligase. During the RNA transcription, T7 RNA polymerase can produce multiple copies of antisense and sense sequences to form hairpin RNA structures. As the concentration of circular DNA increased from 3 nM to 100 nM, RNA products grow from fiber-like structures to sponge-like structures. A single sponge contains approximately five hundred thousand copies of siRNA. After condensation with polyethyleneimine (PEI), the microsponge size reduced from 2 μm to 200 nm75.
Fig. 4. Nucleotides derived nanoparticles.
a. Tetrahedron DNA origami. b. Spherical nucleic acid (SNA) conjugates with a gold core and siRNA shell. c. Three-way junction RNA nanoparticles. d. RNAi microsponges with multiple copies of shRNA.
5. Lipid-based delivery systems
Phospholipids are natural components of cell membranes that form lipid bilayers76. Liposomes have been developed as drug delivery carriers using a variety of synthetic lipids76. They have been widely used to encapsulate small molecule drugs for treating diseases in humans, most notably Doxil for breast cancer and AmBisome for fungal infection76. For example, the formulation of Doxil is composed of doxorubicin, methoxypolyethylene glycol 2000 −1,2- distearoyl-sn-glycero-3 phosphoethanolamine (MPEG-DSPE), hydrogenated soy phosphatidylcholine (HSPC), and cholesterol76. These previous studies provide important guidance for the development of lipid-based siRNA delivery systems.
5.1. Lipid analogs with cationic head groups and hydrophobic tails
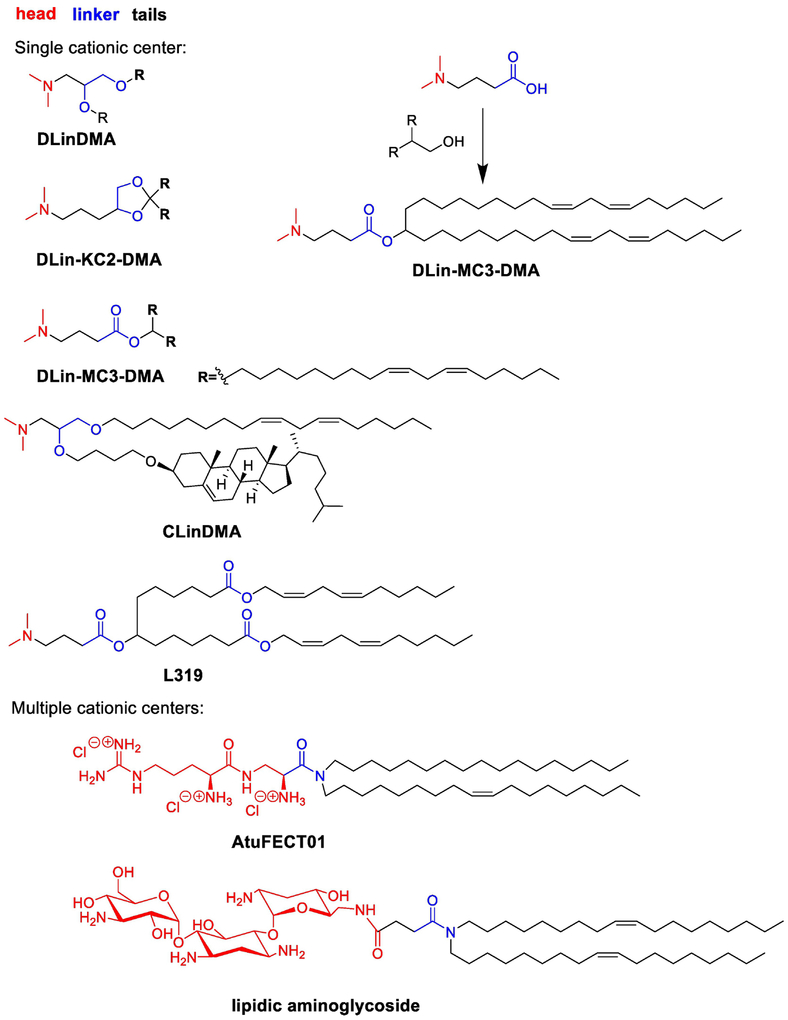
Lipid-based nanoparticles (LPNs), particularly lipids with single or multiple cationic centers (Fig. 5), are highly effective carriers of siRNA77. In 2005, Stabilized Nucleic Acid Lipid Particles (SNALPs) for intracellular delivery of siRNA were reported78. SNALPs are typically composed of a formulation consisting of an amine-based lipid, cholesterol, a PEG-lipid, as well as helper phospholipids78. Early SNALPs were formulated from DlinDMA, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) as a helper lipid, mPEGC-DMA, and cholesterol (DlinDMA:DSPC:Chol:PEG-C-DMA 30:20:48:2 molar percent)78. DlinDMA is composed of an ionizable amino head group, a glycerol-based ether linker, and two unsaturated carbon tails (Fig. 5)78. Amino lipids are central components of SNALPs, as they play a role in the assembly of the nanoparticles, by binding the siRNA through electrostatic interactions79. These amino groups also facilitate endosomal escape, through interaction with endosomal components during acidification80. The common structure of this type of lipids includes a cationic head group, a linker, and two long hydrophobic domains (Fig. 5).
Fig. 5. Lipid analogs with cationic head groups, linker and hydrophobic tails and a representative synthetic route to DLin-MC3-DMA.
Cationic head groups can be single or multiple cationic centers. Linkers span from ester, amide to ketal. Hydrophobic tails can accommodate unsaturated bonds, cholesterol, and ester groups.
In vivo, SNALPs have shown the ability to deliver siRNA to tumor tissues. A phase I clinical trial using DlinDMA based ALN-VSP to target vascular endothelial growth factor (VEGF) and kinesin family member 11 (KIF11 or KSP) demonstrated antitumor activity in patients with advanced solid tumors (dose >0.7 mg/kg). To improve the delivery efficiency, the ether linker in DlinDMA was replaced by a ketal linker to afford DLin-KC2-DMA81. This chemical alteration reduces the transition temperatures of DLin-KC2-DMA and facilitates its ability to form hexagonal structures when it interacts with naturally occurring anionic phospholipids in the endosomal membrane. This process is believed to promote endosomal release76. DLin-KC2-DMA showed improved delivery efficiency with an efficacious dose of 0.01 mg/kg for hepatocyte silencing of Factor VII in mice. Recently it was reported that DLin-KC2-DMA SNALPs can transfect leukemia cells in vivo as well82.
The structure-activity relationship for DLin-KC2-DMA derivatives has been investigated systematically (Fig. 5)79. Head groups were substituted by amino groups with different size and ring structures. SAR studies of 56 amino lipids in vivo indicate that a dimethyl substitution on the amine head group was preferred to diethyl, diisopropyl, and ringed structures. The linkers ranged from ester, amide, ketal, ether, and carbamate. Efficacy was generally retained for ester, ketal, and carbamate linkers, though the length and functional groups of the linker could significantly affect the activity. For example, both DLin-KC2-DMA and DLin-MC3-DMA displayed significant silencing of FVII in mice83. Lipids with amide and ether linkers possessed reduced delivery performance when formulated. The authors show that pKa is an important factor for delivery efficiency and an optimum pKa of the nanoparticles was between 6.2–6.5. Consistent with this report, a recent study investigated the correlation between nanoparticle properties and siRNA delivery efficiency84. In this study, pKa was also identified as a key determinant of nanoparticle delivery efficacy84. Hydrophobic tails can accommodate diverse functional groups, including unsaturated carbon bonds and small molecules85. Maier and coworkers report that an ester bond can be installed in the middle of hydrophobic tails (Fig. 5, L319), which retain delivery efficiency and improve biodegradability86. In this collection, DLin-MC3-DMA was reported to be the most potent lipid with siRNA formulations having an ED50 around 0.005 mg/kg in mice (Fig. 5)79. ONPATTRO™ (Patisiran) formulated from DLin-MC3-DMA was approved by FDA for the treatment of transthyretin-mediated amyloidosis in August 20182,87. A simple substitution reaction was used to prepare the DLin-MC3-DMA (Fig. 5)85.
In addition to lipids with single cationic center, numerous lipid derivatives and lipid-like materials with multiple cationic centers have been developed88. Here, we discuss two representative examples: aminoglycoside and amino acid derivatives. Aminoglycoside-based lipids are composed of an aminoglycosides head, an amide linker, and two unsaturated tails89 (Fig. 5) Formulated with 1,2-dioleoylsn-glycero-3-phosphoethanolamine (DOPE), lipidic aminoglycoside derivatives are capable of delivering siRNA in various human tumor cell lines89. In 2006, an arginine based lipid, AtuFECT01 was reported, which consists of arginine derived head, an amide linker, and two different carbon tails90. AtuFECT01 siRNA-lipoplexes were reported to silence in the vasculature of mice via systemic administration. Recently, a similar arginine based liposomal delivery system was developed for hepatic silencing91. The lead material was reported to have dose dependent silencing with an ED50 of 0.1 mg/kg in mice92. Both SNALP and AtuFECT01 based siRNA delivery systems were in clinical trials for treating solid tumors16.
5.2. Combinatorial and high throughput strategies of lipid-like materials
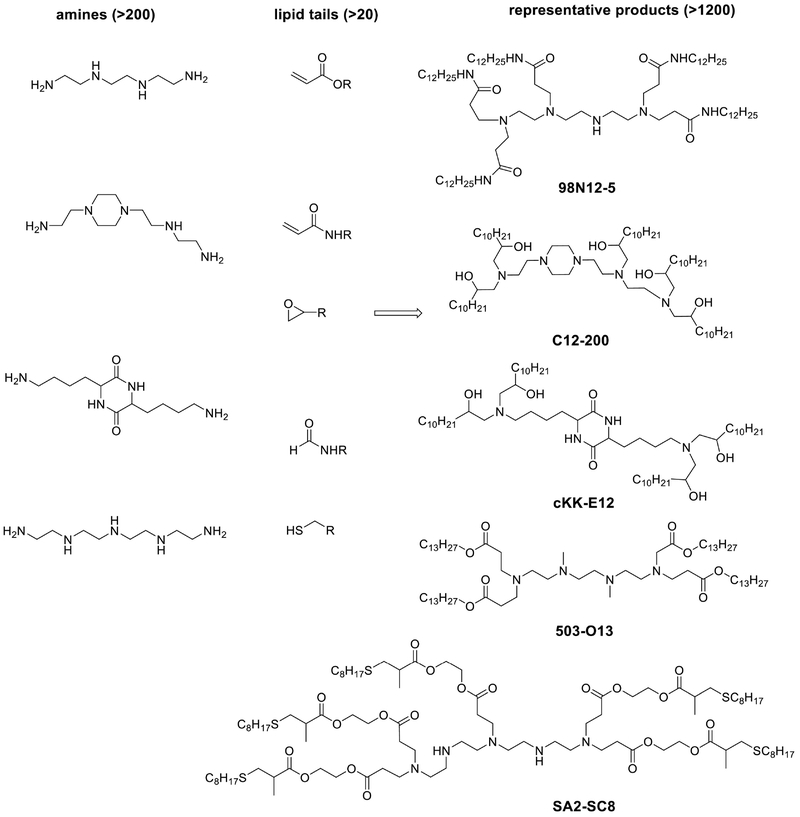
In 2008, a library of lipid-like molecules, termed lipidoids, was developed for siRNA delivery93 (Fig. 6). Lipidoids are composed of one or more amine centers and multiple hydrophobic tails. Over 1200 diverse lipidoids were synthesized with a range of functional amines and acrylates or acrylamides through a one-step Michael addition reaction without the need of catalysts or solvents. The amino groups function similarly to the amino head group in the lipid nanoparticles mentioned above, by neutralizing negative charges of siRNA and facilitating cytoplasmic release of siRNA. Lipidoids were formulated using a method similar to SNALPs. In general, lipidoids with amide linkages and more than two alkyl tails (8–12 carbons) had the most activity when formulated with siRNA. One lipidoid termed 98N12–5 (cite figure 6) in particular was investigated for its potential to deliver siRNA94. 98N12–5 was capable of silencing both APoB and FVII in mice and non-human primates.
Fig. 6. Combinatorial and high throughput strategies of lipid-like materials.
Thousands of lipid-like compounds were synthesized through Michael addition reactions, epoxide ring-opening reactions, reductive amination reactions and thiol-ene reactions. Materials were screened with high throughput bioassays both in vitro and in vivo.
In an effort to improve the delivery efficiency, a library of amino alcohol-based lipidoids was developed using epoxide-based chemistry95. A one-step ring-opening reaction between functional amines and epoxides afforded over 100 new lipidoids. The lead material, named C12–200 (Fig. 6), was composed of a piperazine ring and five lipid tails. The ED50 of siRNA formulated with C12–200 was reported as low as 0.01 mg/kg for hepatocytes in mice. The potency of this formulation also allows the potential for simultaneous gene silencing. C12–200 formulations containing five different siRNAs at once were shown capable of silencing their target genes in mice. This material was also shown capable of silencing certain immune cells in mice and primates96. More recently, several combinatorial libraries have been constructed using structural information gained in these earlier studies97. In particular, a lipopetide material termed cKK-E12, (Fig. 6), was developed and show to be the most potent and selective siRNA delivery systems for gene silencing reported thus far for hepatocytes98. Meanwhile, Whitehead and co-workers made a series of biodegradable lipidoids such as 503-O13 (Fig. 6)99. In 2016, Siegwart et al applied a two-step synthetic route, Michael addition and thiol-ene reaction, to prepare degradable dendrimers (Fig. 6), of which 5A2-SC8 induced strong gene silencing effects and extended overall survival of mice with liver cancer100. More and more lipid analogs and derivatives are under development, which may facilitate siRNA or other types RNA therapeutics.
6. Polymer-based siRNA delivery
The utility of polymers to function as intracellular delivery systems for nucleic acids, including antisense oligonucleotides (ASOs) and plasmid DNA (pDNA), has been studied for several decades101,102. For instance, cationic polymers are able to condense nucleic acids into polyplexes via electrostatic interactions, enhancing cellular uptake and endosome escape. Notably, the physiochemical properties of polymers can be carefully adjusted through bottom-up chemical synthesis. The eventual (nano)structure can also be dictated through chemical synthesis, covering a range of assemblies including block and star shaped copolymers, micelles, dendrimers, solid nanoparticles, polyplexes, polymer-siRNA conjugates, and more. A few polymer-based siRNA delivery systems have shown therapeutic potential in clinical trials16. Examples of major classes of polymer-based delivery systems are discussed below.
6.1. siRNA-polymer bioconjugates
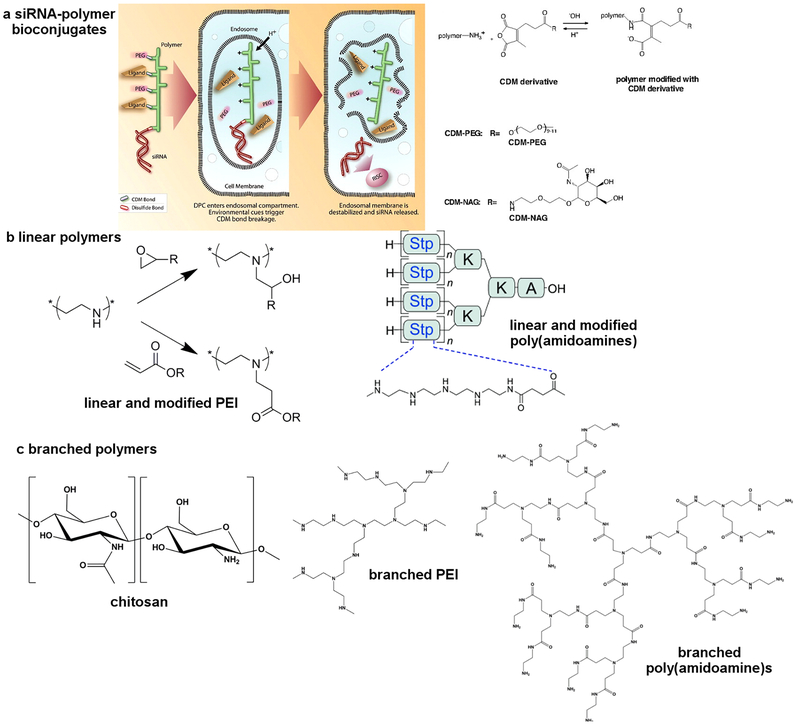
Direct conjugation of siRNA to polymers offers an attractive avenue to improve stability, pharmacokinetics, cellular uptake, and delivery. Polymers with a history of medical use, such as poly(ethylene glycol) (PEG) and poly(lactic-co-glycolic acid) (PLGA) have been conjugated to siRNA to facilitate delivery103,104. PEG conjugated to siRNA via an acid-labile linker was reported to facilitate gene silencing in hepatoma cells in vitro103 and in a tumor model104. Kataoka and co-workers pioneered the use of polyion complexes (PICs) for drug delivery, and used this system to graft siRNA through disulfide linkages to a polymer to improve the physicochemical properties and transfection efficacy105. The hydrophobic nature of PLGA within siRNA-PLGA conjugates was employed to form self-assembled micelles106. PLGA has also been used for local delivery of siRNA via the depot approach. Local drug eluter (LODER) has been developed by Silenseed107 as a potential approach to treat pancreatic cancer targeting mutant G12D KRAS. Following injection into an inoperable tumor, PLGA degrades and mediates sustained release of siRNA. This system is currently the approach in a recruiting Phase 2 clinical trial. Saltzman and coworkers reported using PLGA-spermidine nanoparticles gene silencing in the vaginal lumen and uterine horns of mice following topical delivery108. The development of Controlled Radical Polymerization (CRP) has made a major impact on polymer science, and these techniques have also been used to form bioconjugates109, star polymers, nanogels110, and other delivery materials for siRNA111. Lastly, in situ radical polymerization of siRNA enabled formation of siRNA nanoparticles and intracellular delivery112.
One example of a polymeric siRNA delivery system in clinical trials is the Dynamic PolyConjugates system, which enabled >60% silencing of apoB in hepatocytes in vivo at a dose of about 2.5 mg/kg (50 μg siRNA and 800 μg polymer per mouse)113 Key components of this design included an amphipathic poly(vinyl ether) with acid-labile maleamate bonds that cleave inside of the endosome, PEG chains for shielding, and N-acetylgalactosamine ligands to target hepatocytes. Similarly, a poly(butyl amino vinyl ether) (PBAVE) polymer was designed with side chains of alkyl and amino groups to mediate cellular uptake and endosomal release, and GalNac groups for hepatocyte targeting. This polymer was conjugated to siRNA, providing a single-component system with multiple functionalities for distinct purposes. The Dynamic PolyConjugate design has shown promising results or the treatment of Hepatitis B114. Newer generation PolyConjugates, that improve on the initial design have been reported to fully silence liver genes (0.2 mg/kg in non-human primates), producing a 7 week effect.115 A co-injection strategy using PBAVE and cholesterol-siRNA is a clinical candidate for the treatment of hepatitis B (HBV).116 Furthermore, the original PBAVE polymer has been alternatively modified with a melittin-like peptide to endow similar reversibly masked endosomolytic properties and entered a Phase I clinical trial.117,118
Merck & Co. have reported a similar design that utilizes a cationic, amphilphilic poly(vinyl ether) copolymer backbone with pendant carboxy dimethylmaleic (CDM) pH masking groups, liver targeting N-acetylgalactosamine (NAG), and immunostealth (PEG) groups119. siRNA against ApoB was covalently attached, and the polymer conjugates showed effective single digit mg/kg silencing in the liver. A biodegradable design based on polypeptide120 and poly(amido amine)121 conjugates further expanded the in vivo activity and tolerability of the siRNA conjugates.
6.2. Polymeric complexes
One of the initial requirements of polymeric carriers is the ability to complex siRNA. This has most frequently been achieved using electrostatic interactions between positively charged groups on polymer chains and the negatively charged phosphates in siRNA molecules. Tertiary amines are particularly well suited for siRNA delivery because they can be charged at low pH during self-assembly with siRNA, neutral at extracellular pH, and positively charged after endocytosis to enable endosomal release. This process may involve charge mixing with endosomal lipids, pH buffering / proton sponge effects, among other mechanisms to facilitate escape of siRNA into the cytoplasm122–124. Although efforts have been made to use other interaction parameters, such as intercalation and physical entrapment, these efforts have yielded less efficacious carriers125. In addition to functional groups that mediate siRNA binding, a second fundamental feature of efficacious polymers for siRNA delivery is hydrophobicity. It is understood that hydrophobic interactions can further stabilize siRNA-polymer nanoparticles above the nanoparticle pKa. This can be accomplished by inclusion of hydrophobic molecules (e.g. cholesterol) or by chemical modification of polymers with hydrophobic domains (e.g. alkyl chains). Thus, an optimal balance between pKa (6.0 – 6.5) and hydrophobicity is implicated in effective materials for siRNA delivery126–131.
(Linear (LPEI) and branched (BPEI) PEIs have been extensively investigated for pDNA and siRNA delivery. In general, BPEI provides greater complexation ability with siRNA due to the flexible structure, an increased number of charges per volume, and more folding options.132 Modification of PEI with neutral or anionic moieties has been shown to reduce cytotoxic effects, sometimes without loss of the endosomal rupture abilities. In particular, hydrophobic modifications have been shown to improve siRNA delivery133,134. For example, lipid derived BPEI can efficiently deliver siRNA to endothelial cells and silence multiple endothelial genes in mice135,136. Moreover, the formulation showed selective silencing in endothelial cells. In addition, branched polyamidoamine dendrimers (PAMAM) have been investigated for siRNA delivery137–140. Interestingly, by tuning the formulation, PAMAM-based delivery systems can deliver siRNA to endothelial cells, hepatocytes, or tumor cells.141 In addition, formulated with small molecule amines or lipids, non-cationic polymers such as PLGA are capable of delivery siRNA108,142. PAMAM-RNA complexes have also been incorporated into degradable polymer scaffolds to mediate controlled local release and sustained gene silencing to increase survival of mice bearing aggressive triple-negative cancer143.
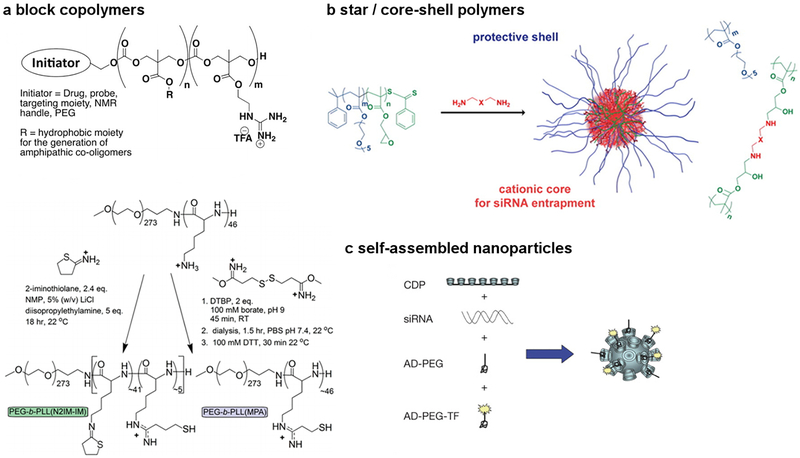
Block copolymers of PEG-b-PLA and PEG-b-PLGA have been used to create core-shell hybrid nanoparticles, often with inclusion of cationic lipids to enable siRNA encapsulation and release. For example, Yang et al.144,145 utilized PEG-b-PLA/BHEM-Chol nanoparticles to silence polo-like kinase 1 (Plk1) to inhibit tumor growth in mice. PLGA-based block copolymer systems have also been developed by Farokhzad and coworkers146,147 for systemic delivery of siRNA against various targets, including demonstration of efficacy in a prostate cancer model. Linear poly(β-amino esters) (PBAEs) have also been extensively explored for the in vivo delivery of nucleic acids, including siRNA148–153.
The use of well-defined polymer architecture and precise cross-linking chemistry are also useful strategies for siRNA delivery. Wagner and co-workers showed that the incorporation of two or three cysteine cross-links into poly(amido amines) was not necessary for pDNA delivery, but was absolutely critical to achieve siRNA delivery154,155. It has also been shown that delivery via block156 and core-shell127 type architectures helps to increase electrostatic interactions by physically concentrating the cationic charges. Dimethylamine and piperazine groups, cross-linked into the cores of well-defined polymers with PEG shells, provided enhanced complexation and delivery as compared to more than 100 other amine-based cross-linkers screened in a 1,536-member combinatorial library127. Triblock copolymers of poly(LPEI-b-(propylene glycol)-b-LPEI) nicely elucidated the effect of polymer composition and architecture on delivery. Whereas LPEI50-b-PPG36-b-LPEI50 showed poor efficacy, decreasing the LPEI block length and increasing the hydrophobic block to LPEI14-b-PPG-b-LPEI14 greatly improved delivery157. The use of endosome destabilizing agents is an additional way to enhance block copolymer-mediated delivery158. Overall, tertiary amines and alkyl chains have been identified as key functional groups for effective siRNA delivery across a variety of siRNA delivery materials including lipids and polymers.93,126–130,159
6.3. Biopolymers
Natural materials have also been used for siRNA delivery. Of note, Davis and co-workers demonstrated that cyclodextrin-based self-assembling polymeric nanoparticles can facilitate siRNA delivery, including in humans160,161. For instance, a cyclodextrin-containing polymer (CDP), a PEG stabilization agent, and human transferrin have been used in self-assembled nanoparticles where transferrin acts as a targeting ligand for transferrin receptors that are frequently overexpressed on cancer cells. The self-assembled four component formulation was delivered IV to patients with solid cancers in a Phase 1b clinical trial. Following siRRM2 nanoparticle treatment, mRNA levels (M2 subunit of ribonucleotide reductase (RRM2)) and the protein (RRM2) was reduced. Significant silencing has been measured in humans at doses of 18–30 mg m−2 and 2.5–5.0 mg/kg in mice160,161. Chitosan is another natural cationic polymer that has been used for siRNA delivery. It has been shown that higher MW chitosan provides better complexation and stability, whereas lower MW chitosan with specific degrees of deacetylation offer better intracellular release162,163. Huang and co-workers showed that a liposomeprotamine-hyaluronic acid (LPH) nanoparticle formulation could enable silencing of CD47 in tumor tissues after IV administration164. This siRNA-mediated silencing of CD47 inhibited the growth of tumors in multiple models, including melanoma and lung metastasis.
Proteins have also been utilized for siRNA delivery. Lieberman, Song, and co-corkers designed a protamine-HIV-1 envelope antibody fusion protein that could deliver siRNA selectively to cells expressing the HIV-1 envelope165. This targeting approach was subsequently applied Her2+ breast cancer models using siRNAs complexed with a Her2-ScFv-protamine peptide fusion protein166. Additionally, Liu and coworkers demonstrated that supercharged green fluorescent protein (GFP) with a net theoretical charge of +36 could form a siRNA-protein complex and enable delivery to a variety of cell lines167. Synthetic amino acid-based polymers are also attractive for delivery. A derivative of polyglutamic acid that incorporated a cell-penetrating helical structure was used for siRNA delivery and shown to cause pore formation in cell membranes, thereby enhancing delivery168.
7. Outlook
In the past two decades, significant advances have been made in the development of siRNA therapeutics for treating diverse diseases. Chemists are capable of synthesizing siRNA with modified nucleotides to achieve high efficacy, high stability, and high specificity. However, in order to maximize the advantages of siRNA therapeutics in humans, there are still formidable challenges for the delivery systems. To address the issues associated with potency, selectivity, and safety, many strategies have been applied to develop new delivery materials.
Chemists often design delivery systems considering elements, bonds, and functional groups. The key chemical properties of successful carriers are largely consistent between lipid and polymer-based systems. These include hydrophobic modifications, tertiary amines, and the ability to interact with short siRNA strands via multiple types of bonding interactions. Currently, lipid- and lipidoid-based siRNA materials are highly effective delivery systems. The key components of lipids and lipidoids are cationic/ionizable groups, functional linkers, and lipid tails. Extensive SAR studies have provided design criteria, as described above. Moreover, formulation methods are also very important to the efficacy in vivo. Direct conjugation of small molecule ligands or polymers to siRNA offers the advantage of a being single component delivery system with defined composition. For example, GalNAc-siRNA conjugates not only provide an approach for ligand based cell internalization without the need of cationic materials, but also target hepatocytes specifically. In addition, synthetic and natural materials also offer ways to tune the degradation and responsiveness of the delivery system. For efficient siRNA delivery, cationic materials that include additional stabilizing interactions (e.g. hydrophobic modifications) and utilize architecture (e.g. well defined polymer morphology, cross-linking) are the most promising. Outside of the classical drug delivery systems, there are also new strategies to develop siRNA therapeutics. For instance, DNA origami, RNA nanoparticles, SNA were utilized to deliver siRNA to tumor cells and other forms of RNA can activate RNAi. Overall, the major challenges for siRNA therapeutics are to increase the efficacy, enhance cell-tissue specificity, and improve the safety.
Future efforts may include 1) delivery using new targeting ligands and chemical probes that specifically bind to surface markers on diseased cell populations, 2) increasing efficacy (particularly with regard to understanding how much siRNA enters the cytoplasm and how to better facilitate that process169,170), 3) widening of therapeutic window using materials with low toxicity, 4) design of materials with defined degradation products that can be metabolized (important when siRNA-based drugs will be repeatedly dosed), 5) simplification of the formulation procedure, and 6) delivery to organs other than the liver. Chemists have the ability to build materials on the molecular level and are well suited to meet these challenges to make siRNA therapeutics successful in the clinic for a broad range of diseases. We hope that ONPATTRO is the first of many future RNAi therapeutics to effectively treat human disease.
Figure 7.
Selected examples of conjugate, linear, and branched type architectures for siRNA delivery.
Figure 8.
Selected examples of block copolymer, star / core-shell, and self-assembled designs for siRNA delivery.
Acknowledgements
The authors thank Dr. Christopher Alabi for providing critical feedback and useful suggestions. Y.D. acknowledges the support from the Maximizing Investigators’ Research Award R35GM119679 from the National Institute of General Medical Sciences as well as the start-up fund from the College of Pharmacy at The Ohio State University. D.J.S. acknowledges financial support from the Cancer Prevention and Research Institute of Texas (CPRIT grant R1212) and the Welch Foundation (I-1855).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Garber K Alnylam launches era of RNAi drugs. Nat Biotechnol 36, 777–778 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Robb GB & Rana TM RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell 26, 523–537 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R & Anderson DG Knocking down barriers: advances in siRNA delivery. Nature Reviews Drug Discovery 8, 129–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaynor JW, Campbell BJ & Cosstick R RNA interference: a chemist’s perspective. Chem. Soc. Rev 39, 4169–4184 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Watts JK, Deleavey GF & Damha MJ Chemically modified siRNA: tools and applications. Drug Discov Today 13, 842–855 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Lima WF, et al. Single-stranded siRNAs activate RNAi in animals. Cell 150, 883–894 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Prakash TP, et al. Identification of metabolically stable 5’-phosphate analogs that support single-stranded siRNA activity. Nucleic Acids Res 43, 2993–3011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juliano R, Bauman J, Kang H & Ming X Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol. Pharmaceut 6, 686–695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castanotto D & Rossi JJ The promises and pitfalls of RNA-interference-based therapeutics. Nature 457, 426–433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JB & Siegwart DJ Design of synthetic materials for intracellular delivery of RNAs: From siRNA-mediated gene silencing to CRISPR/Cas gene editing. Nano Res 11, 5310–5337 (2018). [Google Scholar]
- 12.Lu Y, Aimetti AA, Langer R & Gu Z Bioresponsive materials. Nat Rev Mater 2(2017). [Google Scholar]
- 13.Whitehead KA, Langer R & Anderson DG Knocking down barriers: advances in siRNA delivery. Nature Reviews Drug Discovery 8, 129–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnett JC, Rossi JJ & Tiemann K Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnology Journal 6, 1130–1146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Fougerolles A, Vornlocher HP, Maraganore J & Lieberman J Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discovery 6, 443–453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanasty R, Dorkin JR, Vegas A & Anderson D Delivery materials for siRNA therapeutics. Nat Mater 12, 967–977 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Kaczmarek JC, Kowalski PS & Anderson DG Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med 9, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuckerman JE & Davis ME Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat Rev Drug Discov 14, 843–856 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Eckstein F Nucleoside phosphorothioates. Annual Review of Biochemistry 54, 367–402 (1985). [DOI] [PubMed] [Google Scholar]
- 20.Braasch DA, et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42, 7967–7975 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Chiu YL & Rana TM siRNA function in RNAi: a chemical modification analysis. RNA 9, 1034–1048 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JD & Dunn DB An additional sugar component of ribonucleic acids. Biochimica et Biophysica Acta 31, 573–575 (1959). [DOI] [PubMed] [Google Scholar]
- 23.Broom AD & Robins RK The Direct Preparation of 2’-O-Methyladenosine from Adenosine. J. Am. Chem. Soc 87, 1145–1146 (1965). [DOI] [PubMed] [Google Scholar]
- 24.Obika S, et al. Synthesis of 2’-O,4’-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3’-endo sugar puckering. Tetrahedron Letters 38, 8735–8738 (1997). [Google Scholar]
- 25.Koshkin AA, et al. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerization, and unprecedented nucleic acid recognition. Tetrahedron 54, 3607–3630 (1998). [Google Scholar]
- 26.Hanessian S, et al. Structure-based design of a highly constrained nucleic acid analogue: improved duplex stabilization by restricting sugar pucker and torsion angle gamma. Angew. Chem. Int. Ed 51, 11242–11245 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat. Chem. Bio 3, 709–710 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Wu SY, et al. 2’-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat. Commun 5(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen PE, Egholm M, Berg RH & Buchardt O Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254, 1497–1500 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Herbert B, et al. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. U.S.A 96, 14276–14281 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, Sergueeva ZA, Dobrikov M & Shaw BR Nucleoside and oligonucleoside boranophosphates: chemistry and properties. Chem. Rev 107, 4746–4796 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Hall AH, Wan J, Shaughnessy EE, Ramsay Shaw B & Alexander KA RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 32, 5991–6000 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stirchak EP, Summerton JE & Weller DD Uncharged stereoregular nucleic acid analogs: 2. Morpholino nucleoside oligomers with carbamate internucleoside linkages. Nucleic Acids Res. 17, 6129–6141 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alter J, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat. Med 12, 175–177 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Langkjaer N, Pasternak A & Wengel J UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorganic and Medicinal Chemistry 17, 5420–5425 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Laursen MB, et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol. Biosyst 6, 862–870 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Fisher M, et al. Biological effects of hexitol and altritol-modified siRNAs targeting B-Raf. European Journal of Pharmacology 606, 38–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bramsen JB, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 37, 2867–2881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salinas JC, Yu J, Ostergaard M, Seth PP & Hanessian S Conception and Synthesis of Oxabicyclic Nucleoside Phosphonates as Internucleotidic Phosphate Surrogates in Antisense Oligonucleotide Constructs. Org Lett 20, 5296–5299 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Sipa K, et al. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA 13, 1301–1316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia J, et al. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chemical Biology 1, 176–183 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Suter SR, et al. Controlling miRNA-like off-target effects of an siRNA with nucleobase modifications. Org Biomol Chem 15, 10029–10036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner AF & Khorana HG Studies on polynucleotides. VI. Experiments on the chemical polymerization of mononucleotides. Oligonucleotides derived from thymidine-3’-phosphate. Journal of the American Chemical Society 81, 4651–4656 (1959). [Google Scholar]
- 44.Zamecnik PC & Stephenson ML Inhibition of Rous-Sarcoma Virus-Replication and Cell Transformation by a Specific Oligodeoxynucleotide. Proc. Natl. Acad. Sci. U.S.A 75, 280–284 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephenson ML & Zamecnik PC Inhibition of Rous-Sarcoma Viral-Rna Translation by a Specific Oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. U.S.A 75, 285–288 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raal FJ, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet 375, 998–1006 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Bennett CF Therapeutic Antisense Oligonucleotides Are Coming of Age. Annu Rev Med 70, 307–321 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Watts JK & Corey DR Silencing disease genes in the laboratory and the clinic. J Pathol 226, 365–379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Micura R Small interfering RNAs and their chemical synthesis. Angew. Chem. Int. Ed 41, 2265–2269 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Marshall WS & Kaiser RJ Recent advances in the high-speed solid phase synthesis of RNA. Current Opinion in Chemical Biology 8, 222–229 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Jeong JH, Mok H, Oh YK & Park TG siRNA conjugate delivery systems. Bioconjugate Chem. 20, 5–14 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Turanov AA, et al. RNAi modulation of placental sFLT1 for the treatment of preeclampsia. Nature Biotechnology 36, 1164–+ (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J, et al. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics 8, 1575–1590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kruspe S & Giangrande PH Aptamer-siRNA Chimeras: Discovery, Progress, and Future Prospects. Biomedicines 5(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dassie JP, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol 27, 839–849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sehgal A, et al. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat Med 21, 492–497 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Muratovska A & Eccles MR Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Letters 558, 63–68 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Chiu YL, Ali A, Chu CY, Cao H & Rana TM Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chemistry & Biology 11, 1165–1175 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Cesarone G, Edupuganti OP, Chen CP & Wickstrom E Insulin receptor substrate 1 knockdown in human MCF7 ER+ breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analogue of insulin-like growth factor 1. Bioconjugate Chem. 18, 1831–1840 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Letsinger RL, Zhang GR, Sun DK, Ikeuchi T & Sarin PS Cholesteryl-conjugated oligonucleotides: synthesis, properties, and activity as inhibitors of replication of human immunodeficiency virus in cell culture. Proc. Natl. Acad. Sci. U.S.A 86, 6553–6556 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol 25, 1149–1157 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Rajeev KG, Zimmermann T, Manoharan M, Maier M & Fitzgerald K Inhibitory RNA interference agents modified with saccharide ligands. (Alnylam Pharmaceuticals, Inc., USA, WO 2012/037254, 2012). [Google Scholar]
- 64.http://investors.alnylam.com/news-releases/news-release-details/alnylam-provides-rd-updates-and-announces-2019-product-and. (2018).
- 65.Matsuda S, et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem Biol 10, 1181–1187 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Parmar RG, et al. Facile Synthesis, Geometry, and 2’-Substituent-Dependent in Vivo Activity of 5’-(E)- and 5’-(Z)-Vinylphosphonate-Modified siRNA Conjugates. J Med Chem 61, 734–744 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Seeman NC Nanomaterials based on DNA. Annual Review of Biochemistry 79, 65–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol 7, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cutler JI, Auyeung E & Mirkin CA Spherical nucleic acids. J Am Chem Soc 134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Jensen SA, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med 5, 209ra152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jasinski D, Haque F, Binzel DW & Guo P Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 11, 1142–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo P The emerging field of RNA nanotechnology. Nat Nanotechnol 5, 833–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shu D, Shu Y, Haque F, Abdelmawla S & Guo P Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6, 658–667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shu Y, et al. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliver Rev 66, 74–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JB, Hong J, Bonner DK, Poon Z & Hammond PT Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater 11, 316–322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allen TM & Cullis PR Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliver. Rev (2012). [DOI] [PubMed] [Google Scholar]
- 77.Schroeder A, Levins CG, Cortez C, Langer R & Anderson DG Lipid-based nanotherapeutics for siRNA delivery. Journal of Internal Medicine 267, 9–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heyes J, Palmer L, Bremner K & MacLachlan I Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Controlled Release 107, 276–287 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angewandte Chemie International Edition (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Allen TM & Cullis PR Drug delivery systems: entering the mainstream. Science 303, 1818–1822 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nature Biotechnology 28, 172–176 (2010). [DOI] [PubMed] [Google Scholar]
- 82.He W, et al. Discovery of siRNA Lipid Nanoparticles to Transfect Suspension Leukemia Cells and Provide In Vivo Delivery Capability. Mol Ther 22, 359–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jayaraman M, et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew Chem Int Ed Engl (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alabi CA, et al. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc Natl Acad Sci U S A 110, 12881–12886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abrams MT, et al. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol Ther 18, 171–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maier MA, et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther 21, 1570–1578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coelho T, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med 369, 819–829 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Zhi D, et al. The headgroup evolution of cationic lipids for gene delivery. Bioconjug Chem 24, 487–519 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Desigaux L, et al. Self-assembled lamellar complexes of siRNA with lipidic aminoglycoside derivatives promote efficient siRNA delivery and interference. Proc. Natl. Acad. Sci. U.S.A 104, 16534–16539 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santel A, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 13, 1222–1234 (2006). [DOI] [PubMed] [Google Scholar]
- 91.Adami RC, et al. An amino acid-based amphoteric liposomal delivery system for systemic administration of siRNA. Mol. Ther 19, 1141–1151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abrams MT, et al. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther 18, 171–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol 26, 561–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol 26, 561–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love Kevin T, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proceedings of the National Academy of Sciences of the United States of America 107, 1864–1869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novobrantseva TI, et al. Systemic RNAi-mediated gene silencing in nonhuman primate and rodent myeloid cells. Mol. Ther. - Nucl. Acids 1, e4/1–e4/13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alabi CA, Sahay G, Langer R & Anderson DG Development of siRNA-probes for studying intracellular trafficking of siRNA nanoparticles. Integr Biol (Camb) 5, 224–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong Y, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Whitehead KA, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun 5, 4277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou K, et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc Natl Acad Sci U S A 113, 520–525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pack DW, Hoffman AS, Pun S & Stayton PS Design and development of polymers for gene delivery. Nat. Rev. Drug Discovery 4, 581–593 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Mintzer MA & Simanek EE Nonviral Vectors for Gene Delivery. Chem. Rev 109, 259–302 (2009). [DOI] [PubMed] [Google Scholar]
- 103.Oishi M, Nagasaki Y, Itaka K, Nishiyama N & Kataoka K Lactosylated poly(ethylene glycol)-siRNA conjugate through acid-labile ss-thiopropionate linkage to construct pH-sensitive polyion complex micelles achieving enhanced gene silencing in hepatoma cells. J. Am. Chem. Soc 127, 1624–1625 (2005). [DOI] [PubMed] [Google Scholar]
- 104.Kim SH, Jeong JH, Lee SH, Kim SW & Park TG Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J. Controlled Release 129, 107–116 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Takemoto H, et al. Polyion complex stability and gene silencing efficiency with a siRNA-grafted polymer delivery system. Biomaterials 31, 8097–8105 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Lee SH, Mok H, Lee Y & Park TG Self-assembled siRNA-PLGA conjugate micelles for gene silencing. J. Controlled Release 152, 152–158 (2011). [DOI] [PubMed] [Google Scholar]
- 107.Zorde Khvalevsky E, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A 110, 20723–20728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woodrow KA, et al. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat. Mater 8, 526–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith D, Holley & McCormick CL RAFT-synthesized copolymers and conjugates designed for therapeutic delivery of siRNA. Polym. Chem 2, 1428–1441 (2011). [Google Scholar]
- 110.Averick SE, et al. Preparation of cationic nanogels for nucleic acid delivery. Biomacromolecules 13, 3445–3449 (2012). [DOI] [PubMed] [Google Scholar]
- 111.Siegwart DJ, Oh JK & Matyjaszewski K ATRP in the design of functional materials for biomedical applications. Prog. Polym. Sci 37, 18–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yan M, et al. Single siRNA nanocapsules for enhanced RNAi delivery. J. Am. Chem. Soc 134, 13542–13545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rozema DB, et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A 104, 12982–12987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wooddell CI, et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol Ther 21, 973–985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lewis D Dynamic polyconjugates (DPC) technology: an elegant solution to the siRNA delivery problem. (2006).
- 116.Wong SC, et al. Co-Injection of a Targeted, Reversibly Masked Endosomolytic Polymer Dramatically Improves the Efficacy of Cholesterol-Conjugated Small Interfering RNAs In Vivo. Nucleic Acid Ther 22, 380–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Macron D Arrowhead presents preclinical data on HBV candidate, subcutaneous delivery tech. (2012).
- 118.Rozema DB, Ekena K, Lewis DL, Loomis AG & Wolff JA Endosomolysis by masking of a membrane-active agent (EMMA) for cytoplasmic release of macromolecules. Bioconjugate Chem. 14, 51–57 (2003). [DOI] [PubMed] [Google Scholar]
- 119.Guidry EN, et al. Improving the In Vivo Therapeutic Index of siRNA Polymer Conjugates through Increasing pH Responsiveness. Bioconjugate Chem. 25, 296–307 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Barrett SE, et al. Development of a liver-targeted siRNA delivery platform with a broad therapeutic window utilizing biodegradable polypeptide-based polymer conjugates. J. Controlled Release 183, 124–137 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Parmar RG, et al. Novel Endosomolytic Poly(amido amine) Polymer Conjugates for Systemic Delivery of siRNA to Hepatocytes in Rodents and Nonhuman Primates. Bioconjugate Chem. 25, 896–906 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Sahay G, Alakhova DY & Kabanov AV Endocytosis of nanomedicines. J. Controlled Release 145, 182–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zelphati O & Szoka FC Jr. Mechanism of oligonucleotide release from cationic liposomes. Proc. Natl. Acad. Sci. U.S.A 93, 11493–11498 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hafez IM, Maurer N & Cullis PR On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 8, 1188–1196 (2001). [DOI] [PubMed] [Google Scholar]
- 125.Zhou K, et al. Intercalation-mediated nucleic acid nanoparticles for siRNA delivery. Chem. Commun 52, 12155–12158 (2016). [DOI] [PubMed] [Google Scholar]
- 126.Kanasty R, Dorkin JR, Vegas A & Anderson D Delivery materials for siRNA therapeutics. Nat. Mater 12, 967–977 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Siegwart DJ, et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proceedings of the National Academy of Sciences of the United States of America 108, 12996–13001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed 51, 8529–8533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scholz C & Wagner E Therapeutic plasmid DNA versus siRNA delivery: Common and different tasks for synthetic carriers. J. Controlled Release 161, 554–565 (2012). [DOI] [PubMed] [Google Scholar]
- 130.Nelson CE, et al. Balancing Cationic and Hydrophobic Content of PEGylated siRNA Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano 7, 8870–8880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hao J, et al. Rapid synthesis of a lipocationic polyester library via ring-opening polymerization of functional valerolactones for efficacious siRNA delivery. J. Am. Chem. Soc 137, 9206–9209 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Kwok A & Hart SL Comparative structural and functional studies of nanoparticle formulations for DNA and siRNA delivery. Nanomedicine: Nanotechnology, Biology and Medicine 7, 210–219 (2011). [DOI] [PubMed] [Google Scholar]
- 133.Philipp A, Zhao XB, Tarcha P, Wagner E & Zintchenko A Hydrophobically modified oligoethylenimines as highly efficient transfection agents for siRNA delivery. Bioconjugate Chem. 20, 2055–2061 (2009). [DOI] [PubMed] [Google Scholar]
- 134.Schroeder A, et al. Alkane-modified short polyethyleneimine for siRNA delivery. J. Controlled Release 160, 172–176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dahlman JE, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nature Nanotech. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xue W, et al. Small RNA combination therapy for lung cancer. Proc. Natl. Acad. Sci. U.S.A 111, E3553–E3561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee JH, et al. Polyplexes assembled with internally quaternized PAMAM-OH dendrimer and plasmid DNA have a neutral surface and gene delivery potency. Bioconjugate Chem. 14, 1214–1221 (2003). [DOI] [PubMed] [Google Scholar]
- 138.Patil ML, et al. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjugate Chem. 19, 1396–1403 (2008). [DOI] [PubMed] [Google Scholar]
- 139.Zhou JH, et al. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem. Commun, 2362–2364 (2006). [DOI] [PubMed] [Google Scholar]
- 140.Liu XX, et al. PAMAM dendrimers mediate siRNA delivery to target Hsp27 and produce potent antiproliferative effects on prostate cancer cells. Chemmedchem 4, 1302–1310 (2009). [DOI] [PubMed] [Google Scholar]
- 141.Khan OF, et al. Ionizable Amphiphilic Dendrimer-Based Nanomaterials with Alkyl-Chain-Substituted Amines for Tunable siRNA Delivery to the Liver Endothelium In Vivo. Angew Chem Int Edit 53, 14397–14401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hasan W, et al. Delivery of multiple siRNAs using lipid-coated PLGA nanoparticles for treatment of prostate cancer. Nano Lett. 12, 287–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Conde J, Oliva N, Atilano M, Song HS & Artzi N Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat Mater 15, 353–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yang X-Z, et al. Systemic delivery of siRNA with cationic lipid assisted PEG-PLA nanoparticles for cancer therapy. J. Controlled Release 156, 203–211 (2011). [DOI] [PubMed] [Google Scholar]
- 145.Yang X-Z, et al. Single-Step Assembly of Cationic Lipid–Polymer Hybrid Nanoparticles for Systemic Delivery of siRNA. ACS Nano 6, 4955–4965 (2012). [DOI] [PubMed] [Google Scholar]
- 146.Shi J, Xiao Z, Votruba Alexander R, Vilos C & Farokhzad Omid C Differentially Charged Hollow Core/Shell Lipid–Polymer–Lipid Hybrid Nanoparticles for Small Interfering RNA Delivery. Angew. Chem. Int. Ed 50, 7027–7031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu X, et al. Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc. Natl. Acad. Sci. U.S.A 110, 18638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kozielski KL, Tzeng SY & Green JJ A bioreducible linear poly(β-amino ester) for siRNA delivery. Chem. Commun 49, 5319–5321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kozielski KL, Tzeng SY, Hurtado De Mendoza BA & Green JJ Bioreducible Cationic Polymer-Based Nanoparticles for Efficient and Environmentally Triggered Cytoplasmic siRNA Delivery to Primary Human Brain Cancer Cells. ACS Nano 8, 3232–3241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lynn DM, Anderson DG, Putnam D & Langer R Accelerated Discovery of Synthetic Transfection Vectors: Parallel Synthesis and Screening of a Degradable Polymer Library. J. Am. Chem. Soc 123, 8155–8156 (2001). [DOI] [PubMed] [Google Scholar]
- 151.Green JJ, Langer R & Anderson DG A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Acc. Chem. Res 41, 749–759 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kaczmarek JC, et al. Polymer-lipid nanoparticles for systemic delivery of mRNA to the lungs. Angew. Chem. Int. Ed 55, 13808–13812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Su X, Fricke J, Kavanagh DG & Irvine DJ In Vitro and in Vivo mRNA Delivery Using Lipid-Enveloped pH-Responsive Polymer Nanoparticles. Mol. Pharmaceut 8, 774–787 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schaffert D, et al. Solid-phase synthesis of sequence-defined T-, i-, and U-shape polymers for pDNA and siRNA delivery. Angew. Chem. Int. Ed 50, 8986–8989 (2011). [DOI] [PubMed] [Google Scholar]
- 155.Schaffert D, Badgujar N & Wagner E Novel fmoc-polyamino acids for solid-phase synthesis of defined polyamidoamines. Org. Lett 13, 1586–1589 (2011). [DOI] [PubMed] [Google Scholar]
- 156.Matsumoto S, et al. Environment-responsive block copolymer micelles with a disulfide cross-linked core for enhanced siRNA delivery. Biomacromolecules 10, 119–127 (2009). [DOI] [PubMed] [Google Scholar]
- 157.Brissault B, Leborgne C, Scherman D, Guis C & Kichler A Synthesis of poly(propylene glycol)-block-polyethylenimine triblock copolymers for the delivery of nucleic acids. Macromol Biosci 11, 652–661 (2011). [DOI] [PubMed] [Google Scholar]
- 158.Yu HJ, et al. Overcoming endosomal barrier by amphotericin B-loaded dual pH-responsive PDMA-b-PDPA micelleplexes for siRNA delivery. ACS Nano 5, 9246–9255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Love K, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A 107, 1864–1869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Davis ME The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: From concept to clinic. Mol. Pharmaceut 6, 659–668 (2009). [DOI] [PubMed] [Google Scholar]
- 161.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–U1140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Howard KA, et al. RNA interference in vitro and in vivo using a chitosan/siRNA nanoparticle system. Mol. Ther 14, 476–484 (2006). [DOI] [PubMed] [Google Scholar]
- 163.Lavertu M, Methot S, Tran-Khanh N & Buschmann MD High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 27, 4815–4824 (2006). [DOI] [PubMed] [Google Scholar]
- 164.Wang Y, et al. Intravenous delivery of siRNA targeting CD47 effectively inhibits melanoma tumor growth and lung metastasis. Mol Ther 21, 1919–1929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Song EW, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol 23, 709–717 (2005). [DOI] [PubMed] [Google Scholar]
- 166.Yao YD, et al. Targeted Delivery of PLK1-siRNA by ScFv Suppresses Her2(+) Breast Cancer Growth and Metastasis. Sci. Transl. Med 4(2012). [DOI] [PubMed] [Google Scholar]
- 167.McNaughton BR, Cronican JJ, Thompson DB & Liu DR Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proc. Natl. Acad. Sci. U.S.A 106, 6111–6116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gabrielson NP, Lu H, Yin LC, Kim KH & Cheng JJ A cell-penetrating helical polymer for siRNA delivery to mammalian cells. Mol. Ther 20, 1599–1609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sahay G, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol 31, 653–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gilleron J, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol 31, 638–646 (2013). [DOI] [PubMed] [Google Scholar]