Abstract
3-Hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) is associated with monitoring cholesterol levels. The presence of the single nucleotide polymorphism rs3846662 introduces alternative splicing at exon 13; the exclusion of this exon leads to a reduction in total cholesterol levels. Lower cholesterol levels are linked to a reduction in Alzheimer’s disease (AD) risk. The major allele of rs3846662, which encourages the splicing of exon 13, has recently been shown to act as a preventative allele for AD, especially in women. The purpose of our research was to replicate and confirm this finding. Using logistic regressions and survival curves, we found a significant association between AD and rs3846662, with a stronger association in individuals that carry the APOE e4 allele, supporting previously published work. The effect of rs3846662 on women is insignificant in our cohort. We confirmed that rs3846662 is associated with reduced risk for AD without gender differences; however, we failed to detect association between rs3846662 and delayed mild cognitive impairment conversion to AD for either of the APOE e4 allelic groups.
Keywords: HMGCR, Alzheimer’s disease, APOE, cholesterol synthesis, genetics, association
1. Introduction
Alzheimer’s disease (AD) is a geriatric neurodegenerative disorder characterized by extracellular senile plaques and intracellular neurofibrillary tangles (Armstrong, 2011; Newell et al., 1999; Ridge et al., 2013). The disease is thought to be caused by the malfunctioning of systems which transport, synthesize, and break down the proteins that constitute the plaques and tangles (Adlard and Cummings, 2004; Hardy and Higgins, 1992; Ridge et al., 2013; Swerdlow and Khan, 2004). Several variants associated with AD risk are in genes involved in these mechanisms, including CD33, CLU, PICALM and MS4A6A1 (Bertram et al., 2007; Lambert et al., 2013). The apolipoprotein E (APOE) gene in particular, which has deleterious (APOE e4) and protective (APOE e2) alleles, is significantly associated with AD (Corder et al., 1993). This gene regulates cholesterol metabolism in the central nervous system (Ridge et al., 2013).
Recent data suggests that the 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) gene may be another region associated with AD (Leduc et al., 2015). HMGCR is the rate-limiting step in cholesterol synthesis, and as such, is the target for low-density lipoprotein cholesterol lowering drugs known as statins (Burkhardt et al., 2008; Cano-Corres et al., 2018; Kathiresan et al., 2008; Krauss et al., 2008; Leduc et al., 2016; Medina et al., 2008). It also interacts with ABCA1 to increase AD risk (Rodriguez-Rodriguez et al., 2009). HMGCR undergoes alternative splicing at exon 13 with the presence of the intronic single-nucleotide polymorphism (SNP) rs3846662 (Burkhardt et al., 2008; Medina et al., 2008). The exclusion of the 53 amino acids in exon 13 results in a catalytically inactive protein called Δexon13 (Burkhardt et al., 2008). When compared to the full-length isoform, cells with high levels of Δexon13 have a poor response to statin therapy (Medina, 2010; Medina et al., 2008; Medina and Krauss, 2009), leading to increased concentrations of cholesterol. Since functional HMGCR is a tetramer composed of two dimers (Istvan et al., 2000), Medina and Krauss (2009) hypothesized that the combination of Δexon13 with functional proteins could be among the factors that reduces its statin sensitivity. Additional research proposed that different combinations of Δexon13 in the tetramer could lead to different levels of enzymatic activity and statin sensitivity (Leduc et al., 2016; Medina, 2010).
The frequency of Δexon13 and the resulting dysfunctional protein levels are associated with the genotype of rs3846662 (Burkhardt et al., 2008; Medina, 2010; Medina et al., 2008). The major allele at this SNP for Caucasian populations is AA; the minor allele is GG (Burkhardt et al., 2008). The major A allele promotes the skipping of exon 13 and increases the amount of circulating Δexon13, while the minor G allele retains exon 13 at a much higher rate (Burkhardt et al., 2008; Medina and Krauss, 2009; Simmons et al., 2011). The difference in abundance of Δexon13 between these two alleles is around 16 to 20 percent (Burkhardt et al., 2008; Medina et al., 2008). Heterozygotic (GA) expression of Δexon13 clearly falls between that of homozygotic AA and homozygotic GG (Burkhardt et al., 2008).
Leduc et al. (2015) found that the AA allele of rs3846662 acts as a protective variant and delays the onset of AD (p-value = 0.017). Homozygosity for the A allele is associated with a decrease in HMGCR activity (Krauss et al., 2008; Leduc et al., 2015; Medina et al., 2008), and as such, a corresponding decrease in cholesterol levels (Aulchenko et al., 2008). Reduction in cholesterol levels has been shown to inhibit the generation of amyloid plaques (Simons et al., 1998). Leduc et al. (2015) reported that this effect was more significant in women; however, their initial results conflicted between cohorts (Quebec founder population [QFP] cohort p-value = 0.003; Alzheimer’s Disease Cooperative Study [ADCS] cohort p-value = 0.342). Leduc et al. (2015) additionally reported that the lack of the G allele had a significant effect in APOE e2 non-carriers (QFP p-value = 0.05) and APOE e4 carriers (ADCS p-value = 0.041). In this study we have evaluated Leduc et al.’s (2015) findings in samples from the Cache County Study on Memory Health and Aging. This sample is a true population-based sample of 5092 individuals. This population is representative of the general European American population (Sharp et al., 2014). Here, we test the associations between AD and HMGCR that were reported by Leduc et al. (2015).
2. Materials and Methods
2.1. Samples
The Cache County Study on Memory Health and Aging began in 1994. It is a population-based study, which recruited everyone in Cache County, Utah that was age 65 or older. Over 95% of the population, 5092 subjects, enrolled in the study. AD status was determined using a variety of assessments administered periodically over twelve years. There were no cases of early-onset AD. Additional information about this dataset, such as diagnostic and screening criteria, has been previously reported (Breitner et al., 1999; Tschanz et al., 2002). The general demographics of this sample are presented in Table 1.
Table 1.
General demographics for the Cache County Study population
| General | Cases | Controls | |
|---|---|---|---|
| Male | 1438 | 160 | 1278 |
| Female | 2035 | 330 | 1705 |
| Mean Age of AD onset ± SDa | 80.2 ± 6.45 | 82.5 ± 6.93 | 79.8 ± 6.29 |
| Mean Age of Death ± SDa | 85.5 ± 7.13 | 89.2 ± 6.22 | 84.6 ± 7.04 |
| Mean Years of Education ± SDa | 13.3 ± 2.88 | 13.1 ± 2.96 | 13.3 ± 2.87 |
| APOE e2/e2 genotype frequency | 27 (0.78%) | 1 (0.2%) | 26 (0.87%) |
| APOE e2/e3 genotype frequency | 438 (12.61%) | 33 (6.74%) | 405 (13.58%) |
| APOE e2/e4 genotype frequency | 105 (3.02%) | 26 (5.31%) | 79 (2.65%) |
| APOE e3/e3 genotype frequency | 1939 (55.83%) | 188 (38.37%) | 1751 (58.7%) |
| APOE e3/e4 genotype frequency | 878 (25.28%) | 204 (41.63%) | 674 (22.59%) |
| APOE e4/e4 genotype frequency | 86 (2.48%) | 38 (7.75%) | 48 (1.61%) |
| HMGCR GG genotype frequency | 697 (20.07%) | 105 (21.43%) | 592 (19.84%) |
| HMGCR GA genotype frequency | 1708 (49.18%) | 250 (51.02%) | 1458 (48.88%) |
| HMGCR AA genotype frequency | 1068 (30.75%) | 105 (27.55%) | 933 (31.28%) |
SD stands for standard deviation.
DNA was available for genotyping of 3473 samples, including 490 AD cases (14.1%) and 2983 controls (85.9%). Of these 1438 are male (41.4%), and 2035 are female (58.6%). The HMGCR allele status of these samples had 697 samples with the GG genotype (20.07%), 1708 with the GA genotype (49.18%), and 1068 with the AA genotype (30.75%). 1069 were APOE e4 carriers (30.78%) and 2404 were not carriers (69.22%). There are 570 APOE e2 carriers (16.41%), with 2903 non-carriers (83.59%). See Table 1 for a summary of genotype frequencies.
2.2. Statistical Analyses
We ran our analysis on R version 3.3.2 (Sincere Pumpkin Patch) (R Core Team, 2016). To conform with the analyses conducted by Leduc et al. (2015) we used a dominant model with respect to allele “G” for coding the genotypes of rs3846662: (a) G carriers, which accounts for both the minor allele homozygote, GG, and the heterozygotic GA genotype; and (b) G non-carriers, which is the AA genotype. We used logistic regression models to assess the association of AD with HMGCR status in order to allow for the inclusion of age, the number of APOE e4 alleles, and the number of APOE e2 alleles as covariates. All p-values reported from our cohort are one-tailed, in contrast to Leduc et al.’s (2015) two-tailed p-values, as we are restricted to the hypothesis that the G non-carrier status is protective. A significant outcome in the other direction is viewed as a failure to replicate, and as such, this analysis is a classic case for a one-tailed test (Bland and Altman, 1994; Kimmel, 1957; Ruxton and Neuhäuser, 2010). By restricting our analyses to a one-tailed model and the specific genetic models used by Leduc et al. (2015), statistical power to validate their findings is maximized. We first replicated the analyses of Leduc et al. (2015) and evaluated differential effects related to gender (see Tables 2 and 3).
Table 2.
Odds ratios and one-sided p-values from the seven replication logistic regressions (LR)
| Analysis | Cases / Controls | Intercept | HMGCRa | APOE e4 | APOE e2 | Gender | Power | |
|---|---|---|---|---|---|---|---|---|
| General | LR p-value | 490 / 2983 | < 1e-16 | 0.049* | < 1e-16* | 0.086b | 1.685e-05* | 100% |
| Odds ratio | 0.055 | 0.832 | 3.231 | 0.814 | 1.549 | |||
| General Male | LR p-value | 160 / 1278 | <1e-16 | 0.111 | 4.87e-10* | 0.285 | NA | 98% |
| Odds ratio | 0.091 | 0.791 | 2.862 | 0.865 | NA | |||
| General Female | LR p-value | 330 / 1705 | <1e-16 | 0.129 | <1e-16* | 0.101 | NA | 100% |
| Odds ratio | 0.127 | 0.856 | 3.449 | 0.789 | NA | |||
| APOE e4− | LR p-value | 222 / 2182 | <1e-16 | 0.414 | NA | NA | NA | 99% |
| Odds ratio | 0.103 | 0.967 | NA | NA | NA | |||
| APOE e4+ | LR p-value | 268 / 801 | <1e-16 | 0.016* | NA | NA | NA | 99% |
| Odds ratio | 0.369 | 0.712 | NA | NA | NA | |||
| APOE e2− | LR p-value | 430 / 2473 | <1e-16 | 0.029* | NA | NA | NA | 100% |
| Odds ratio | 0.186 | 0.802 | NA | NA | NA | |||
| APOE e2+ | LR p-value | 60 / 510 | <1e-16 | 0.395 | NA | NA | NA | 83% |
| Odds ratio | 0.115 | 1.081 | NA | NA | NA | |||
indicates significance
refers to rs3846662. Odds ratios in this column are relative to the G-negative genotype
indicates a trend
+ and − indicate carrier and non-carrier respectively
Table 3.
Odds ratios and one-sided p-values from the eight additional logistic regressions (LR) that examined smaller gender subgroupings
| Analysis | Cases / Controls | Intercept | HMGCRa | Power | |
|---|---|---|---|---|---|
| Female APOE e4− | LR p-value | 145 / 1252 | <1e-16 | 0.337 | 96% |
| Odds ratio | 0.113 | 1.082 | |||
| Female APOE e4+ | LR p-value | 185 / 453 | 5.8e-15 | 0.022* | 96% |
| Odds ratio | 0.456 | 0.669 | |||
| Male APOE e4− | LR p-value | 77 / 930 | <1e-16 | 0.154 | 85% |
| Odds ratio | 0.089 | 0.757 | |||
| Male APOE e4+ | LR p-value | 83 / 348 | <1e-16 | 0.022* | 81% |
| Odds ratio | 0.253 | 0.824 | |||
| Female APOE e2− | LR p-value | 390 / 1407 | <1e-16 | 0.057b | 99% |
| Odds ratio | 0.113 | 1.082 | |||
| Female APOE e2+ | LR p-value | 40 / 298 | <1e-16 | 0.334 | 79% |
| Odds ratio | 0.456 | 0.669 | |||
| Male APOE e2− | LR p-value | 140 / 1066 | <1e-16 | 0.137 | 95% |
| Odds ratio | 0.139 | 0.801 | |||
| Male APOE e2+ | LR p-value | 20 / 212 | <1e-16 | 0.459 | 75% |
| Odds ratio | 0.096 | 0.948 | |||
indicates significance
refers to rs3846662. Odds ratios in this column are relative to the G-negative genotype
indicates a trend
+ and − indicate carrier and non-carrier respectively
We additionally conducted a survival analysis, to replicate and extend the results of Leduc et al. (2015) with respect to AD-free survival. Survivor curves by HMGCR status and gender were generated using Kaplan-Meier estimators in R. We formally compared differences in survival between allelic variants using Cox Proportional Hazards Regression Models. We applied the same model to determine if rs3846662 is associated with a delay in the conversion time from normal and mild cognitive impairment (MCI) to AD.
We conducted a post-hoc power analysis to determine the statistical power to observe an effect of HMGCR on AD status in our cohort. We used the number of cases and controls with the frequency of the HMGCR allele to calculate the probability that we could detect the odds ratio reported by Leduc et al. (2015) at the 0.05 significance level. We used the calculator provided by Skol et al. (2006) to calculate these probabilities.
3. Results
Our logistic regression analysis indicated that rs3846662 was significantly correlated with AD status (p-value = 0.049). This effect was more evident in all carriers of APOE e4 (p-value = 0.016), in both male (p-value = 0.022) and female (p-value = 0.022) carriers. Additionally, significance was observed in non-carriers of APOE e2 (p-value = 0.029). See Table 4 for a summary of the regressions that revealed significant results.
Table 4.
Summary of all five logistic regressions that generated statistically significant results
| General | APOE e4+ | Female APOE e4+ | Male APOE e4+ | APOE e2− | |
|---|---|---|---|---|---|
| Cases / Controls | 490 / 2983 | 268 / 801 | 185 / 453 | 83 / 348 | 430 / 2473 |
| HMGCRa Sig (One-tailed) | 0.049* | 0.016* | 0.022* | 0.022* | 0.029* |
| HMGCRa Odds Ratiob | 0.832 | 0.711 | 0.669 | 0.824 | 0.802 |
| Power | 100% | 99% | 96% | 81% | 100% |
indicates significance.
refers to rs3846662
relative to the G-negative genotype
+ and − indicate carrier and non-carrier respectively.
We were unable to replicate Leduc et al.’s (2015) finding that rs3846662 was significantly correlated with female AD status in the general female group (p-value = 0.129). We were able to replicate the finding that the lack of the G allele was associated with protection from AD (p-value = 0.049). See Table 5 for a direct comparison of these three regressions to the findings of Leduc et al. (2015).
Table 5.
Direct comparison of results with Leduc et al. (2015) findings
| Cohort | Cases / Controls | HMGCRa | APOE e4+ | APOE e2+ | Power | |
|---|---|---|---|---|---|---|
| Cache Countyb | Overall Effect | 490 / 2983 | 0.049* | <1e-16* | 0.086 | 100% |
| Females | 330 / 1705 | 0.129 | <1e-16* | 0.101 | 100% | |
| Males | 160 / 1278 | 0.111 | 4.87e-10* | 0.285 | 98% | |
| QFPc | Overall Effect | 574 / 250 | 0.024* | 0.001* | 0.001* | NA |
| Females | 334 / 250 | 0.003* | 0.001* | 0.001* | NA | |
| Males | 240 / 250 | 0.686 | 0.001* | 0.293 | NA | |
| ADCSc | Overall Effect | 409 / 409 | 0.129 | 0.029* | 0.118 | NA |
| Females | 164 / 409 | 0.342 | 0.017* | 0.209 | NA | |
| Males | 245 / 409 | 0.145 | 0.285 | 0.296 | NA | |
indicates significance
refers to rs3846662
indicates Cache County cohorts with a one-tailed significance test
indicates cohorts from Leduc et al. (2015) that are two-tailed significance values
indicates carrier.
Key: QFP = Quebec Founder Population; ADCS = Alzheimer’s Disease Cooperative Study
We also examined the effect of rs3846662 in APOE allele subgroups reported by Leduc et al. (2015). We found that APOE e4 carriers without the G allele experience a protective effect (p-value = 0.016), as do APOE e2 non-carriers (p-value = 0.029), which is in concordance with the findings in Leduc et al. (2015). See Table 6 for a direct comparison of these models to the findings of Leduc et al. (2015).
Table 6.
Direct comparison of results with Leduc et al. (2015) findings
| Cohort | Cases / Controls | HMGCRa | Power | |
|---|---|---|---|---|
| Cache Countyb | APOE e4− | 222 / 2182 | 0.414 | 99% |
| APOE e4+ | 268 / 801 | 0.016* | 99% | |
| APOE e2− | 430 / 2473 | 0.029* | 100% | |
| APOE e2+ | 60 / 510 | 0.395 | 83% | |
| QFPc | APOE e4− | 308 / 250 | 0.634 | NA |
| APOE e4+ | 262 / 250 | 0.183 | NA | |
| APOE e2− | 469 / 250 | 0.05* | NA | |
| APOE e2+ | 101 / 250 | 0.304 | NA | |
| ADCSc | APOE e4− | 140 / 409 | 0.476 | NA |
| APOE e4+ | 268 / 409 | 0.041* | NA | |
| APOE e2− | 392 / 409 | 0.156 | NA | |
| APOE e2+ | 17 / 409 | 0.579 | NA | |
indicates significance
refers to rs3846662
indicates Cache County cohorts with a one-tailed significance test
indicates cohorts from Leduc et al. (2015) that are two-tailed significance values
+ and − indicate carrier and non-carrier respectively.
Key: QFP = Quebec Founder Population; ADCS = Alzheimer’s Disease Cooperative Study
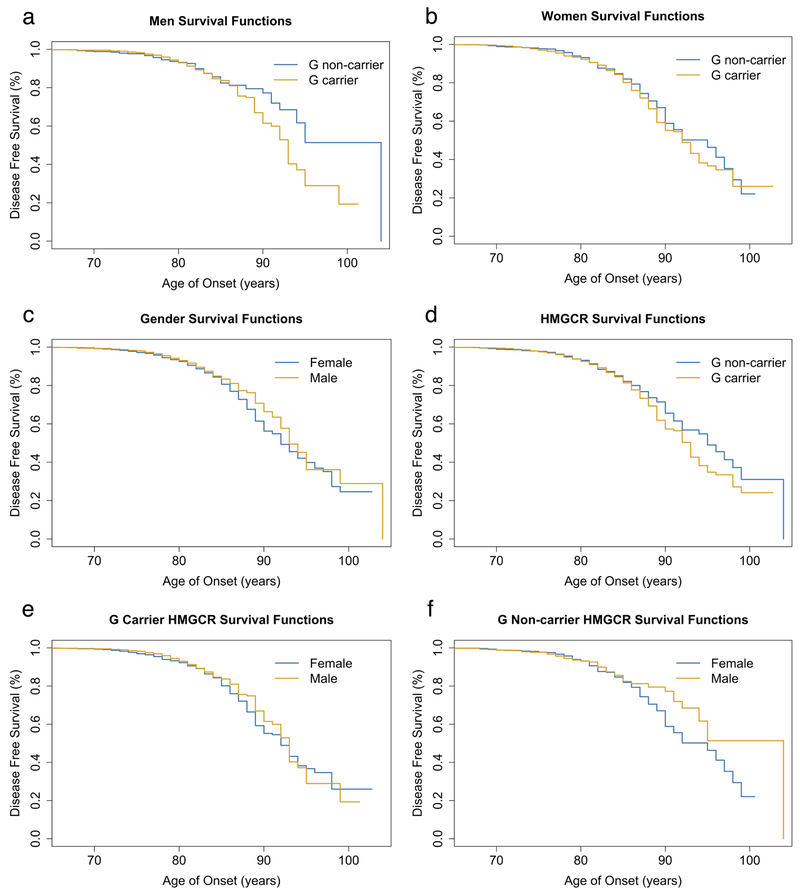
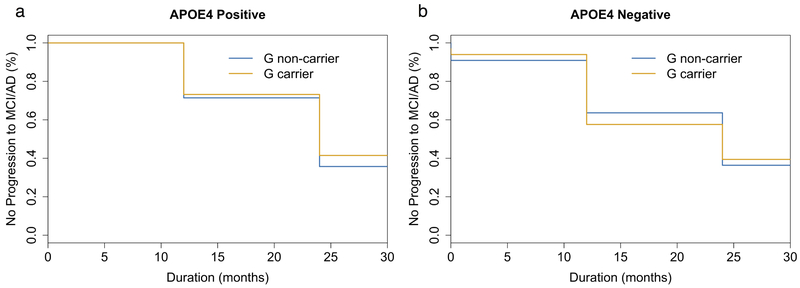
The survival curves compared the onset of AD between males and their rs3846662 allele status (p-value = 0.161; Figure 1a) and the onset of AD between females and their rs3846662 allele status (p-value = 0.327; Figure 1b). We then created survival curves for four more additional comparisons: in Figure 1c, a comparison between genders (p-value = 0.059); in Figure 1d, a comparison between the allele status of rs3846662 (p-value = 0.102); in Figure 1e, a comparison between genders for G carrier rs3846662 allele status (p-value = 0.209); and in Figure 1f, a comparison between genders for G non-carrier rs3846662 allele status (p-value = 0.134). We found no difference in conversion time from MCI to AD between G non-carrier and G carrier patients with the APOE e4 allele (p-value = 0.663; Figure 2a) and without the APOE e4 allele (p-value = 0.671; Figure 2b).
Figure 1.
Survival curves measuring effect of HMGCR rs3846662 intron 13 on Alzheimer’s Disease (AD) free survival. (a) Male age of onset of AD in rs3846662 G non-carriers vs. G carriers (p-value = 0.161). (b) Female age of onset of AD in rs3846662 G non-carriers vs. G carriers (p-value = 0.327). (c) Age of onset of AD in males vs females (p-value = 0.059). (d) Age of onset of AD in rs3846662 G non-carriers vs. G carriers (p-value = 0.102). (e) Age of onset of AD in rs3846662 G carrier males vs. females (p-value =0.209). (f) Age of onset of AD in rs3846662 G non-carrier males vs. females (p-value = 0.134)
Figure 2.
Survival curves measuring the effect of HMGCR rs3846662 intron 13 on mild cognitive impairment (MCI) conversion to Alzheimer’s Disease (AD). (a) APOE e4 carriers comparing rs3846662 G non-carriers vs. G carriers (p-value = 0.663). (b) APOE e4 non-carriers comparing rs3846662 G non-carriers vs. G carriers (p-value = 0.671)
Power analyses demonstrated that each experiment had adequate statistical power (Tables 2, 3, 5, and 6). The lowest power observed in the seven replication experiments was found in the male APOE e2 carrier group at 75 percent.
4. Discussion
We have conducted a well-powered validation study of previous reports of a protective role in AD for rs3846662. Our findings provide support for several of Leduc et al.’s (2015) reported associations, including an overall protective effect and associations within APOE e2 non-carriers and APOE e4 carriers. These replication findings are in concordance with other recent studies. Chang et al. (2016) reported that they were able to confirm that the rs3846662 G non-carrier allele was significant (p-value = 0.02) and acted as a protective variant for AD in a northern Han Chinese population.
Since the majority of statins target HMGCR, several studies have analyzed the effect differing levels of Δexon13 may have on treatment efficacy (Cano-Corres et al., 2018; Leduc et al., 2016; Medina, 2010; Medina et al., 2008; Medina and Krauss, 2009; Simmons et al., 2011). Since rs3846662 modulates levels of Δexon13, the genotype of this variant could be used to predict the effectiveness of treatment. However, results have been inconclusive. A 2015 study found that only women with higher levels of Δexon13 had a worse response to statin therapy (Leduc et al., 2016). Other studies have suggested that individuals with an abundance of Δexon13 have poor response to statin treatment (Medina et al., 2008), suggesting that statin therapy might be a viable option in individuals that are G carriers, as G non-carriers produce higher amounts of Δexon13 (Simmons et al., 2011). However, a recent study by Cano-Corres et al. (2018) found that statin therapy was ineffective in G carrier patients despite the lower levels of the inactive protein. Discrepancy in statin response experiments may possibly be influenced by a SNP in linkage disequilibrium with rs3846662 instead, prompting the need for further analysis.
We were unable to confirm the larger effect in women that was reported previously. We also failed to detect significant association in our survival analyses. Differences in our findings and those of Leduc et al. (2015) could be due to differences in sample sizes: our sample size was over 3000 individuals with only 490 cases from a population-based cohort, while the population of Leduc et al. (2015) had 334 cases from a total of 584 individuals in a clinical case/control cohort. There were also differences in the age at onset and age at death of AD subjects in the Cache County Study. The mean age of onset for our cases was 82.5 years; the Leduc et al. (2015) population had a mean onset age of 71.7 years. The mean age of death for our cases was 89.2 years, which is ten years higher than the population of Leduc et al. (2015) at 79.2 years. The difference between the especially long-lived Cache participants and the populations reported in Leduc et al. (2015) may contribute to the divergence in our findings.
While our findings do not definitely characterize the relationship between rs3846662 and AD, they do provide support for a protective effect for non-carriers of the “G” allele, which is pronounced in APOE e4 carriers and APOE e2 non-carriers. These findings suggest that further study of the role of rs3846662 in AD risk and conversion from MCI to AD is warranted.
Highlights.
rs3846662 is associated with reduced risk for Alzheimer’s disease
Individuals with APOE e4 experience a greater protective effect from rs3846662
Failed to detect gender-specific association of rs3846662, contrasting prior reports
Failed to detect association between rs3846662 and delayed MCI conversion to AD
Acknowledgements
The authors thank the participants and staff of the Dementia Progression Study and the Cache County Study on Memory Health and Aging for their important contributions to this work.
Funding
This work was supported by the National Institutes of Health (R01AG11380, R01AG21136, R01AG31272, R01AG042611, RF1AG054052). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- AD
Alzheimer’s disease
- QFP
Quebec Founder Population
- ADCS
Alzheimer’s Disease Cooperative Study
- MCI
mild cognitive impairment
Footnotes
Disclosure Statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adlard PA, Cummings BJ, 2004. Alzheimer’s disease - A sum greater than its parts? Neurobiol. Aging 25, 725–733. 10.1016/j.neurobiolaging.2003.12.016 [DOI] [PubMed] [Google Scholar]
- Armstrong RA, 2011. The pathogenesis of alzheimer’s disease: A reevaluation of the “amyloid cascade hypothesis.” Int. J. Alzheimers. Dis 630865 10.4061/2011/630865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BWJH, Janssens ACJW, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin M-R, Gyllensten U, Campbell H, Rudan I, Johansson Å, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga J-J, de Geus EJC, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJG, Uitterlinden AG, Witteman JCM, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Döring A, Wichmann H-E, Smit JH, McCarthy MI, van Duijn CM, Peltonen L, 2008. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet 41, 47–55. 10.1038/ng.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE, 2007. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet 39, 17–23. 10.1038/ng1934 [DOI] [PubMed] [Google Scholar]
- Bland MJ, Altman DG, 1994. One and two sided tests of significance. BMJ 309, 248 10.1136/bmj.309.6949.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitner JCS, Wyse BW, Anthony JC, Welsh-Bohmer KA, Steffens DC, Norton MC, Tschanz JT, Plassman BL, Meyer MR, Skoog I, Khachaturian A, 1999. APOE-ε4 count predicts age when prevalence of AD increases, then declines. Neurology 53, 321–331. 10.1212/WNL.3.2.321 [DOI] [PubMed] [Google Scholar]
- Burkhardt R, Kenny EE, Lowe JK, Birkeland A, Josowitz R, Noel M, Salit J, Maller JB, Pe’er I, Daly MJ, Altshuler D, Stoffel M, Friedman JM, Breslow JL, 2008. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler. Thromb. Vasc. Biol 28, 2078–2084. 10.1161/ATVBAHA.108.172288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Corres R, Candás-Estébanez B, Padró-Miquel A, Fanlo-Maresma M, Pintó X, Alía-Ramos P, 2018. Influence of 6 genetic variants on the efficacy of statins in patients with dyslipidemia. J. Clin. Lab. Anal e22566 10.1002/jcla.22566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Tan L, Tan M, Wang H, 2016. Association of HMGCR polymorphism with late-onset Alzheimer 's disease in Han Chinese. Oncotarget 7, 22746–22751. 10.18632/oncotarget.8176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA, 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science (80-. ). 261, 921–923. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA, 1992. Alzheimer’s disease: the amyloid cascade hypothesis. Science (80-. ). 256, 184–185. 10.1126/science.1566067 [DOI] [PubMed] [Google Scholar]
- Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J, 2000. Crystal structure of the catalytic portion of human HMG-CoA reductase: Insights into regulation of activity and catalysis. EMBO J. 19, 819–830. 10.1093/emboj/19.5.819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M, 2008. Polymorphisms Associated with Cholesterol and Risk of Cardiovascular Events. N. Engl. J. Med 358, 1240–1249. 10.1056/NEJMoa0706728 [DOI] [PubMed] [Google Scholar]
- Kimmel HD, 1957. Three criteria for the use of one-tailed tests. Psychol. Bull 10.1037/h0046737 [DOI] [PubMed] [Google Scholar]
- Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI, 2008. Variation in the 3-Hydroxyl-3-Methylglutaryl Coenzyme A Reductase Gene Is Associated With Racial Differences in Low-Density Lipoprotein Cholesterol Response to Simvastatin Treatment. Circulation 117, 1537–1544. 10.1161/CIRCULATIONAHA.107.708388 [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin C-F, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau M-T, Choi S-H, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MCD, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, (EADI), E.A.D.I., (GERAD), G. and E.R. in A.D., (ADGC), A.D.G.C., (CHARGE), C. for H. and A.R. in G.E., Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RFAG, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JSK, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang L-S, Dartigues J-F, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc V, Bourque L, Poirier J, Dufour R, 2016. Role of rs3846662 and HMGCR alternative splicing in statin efficacy and baseline lipid levels in familial hypercholesterolemia. Pharmacogenet. Genomics 26, 1–11. 10.1097/FPC.0000000000000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc V, De Beaumont L, Théroux L, Dea D, Aisen P, Petersen RC, Initiative, the A.D.N., Dufour R, Poirier J, 2015. HMGCR is a genetic modifier for risk, age of onset and MCI conversion to Alzheimer’s disease in a three cohorts study. Mol. Psychiatry 20, 867–873. 10.1038/mp.2014.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MW, 2010. The relationship between HMGCR genetic variation, alternative splicing, and statin efficacy. Discov. Med 9, 495–499. [PubMed] [Google Scholar]
- Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM, 2008. Alternative Splicing of 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Is Associated With Plasma Low-Density Lipoprotein Cholesterol Response to Simvastatin. Circulation 118, 355–362. 10.1161/CIRCULATIONAHA.108.773267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MW, Krauss RM, 2009. The Role of HMGCR Alternative Splicing in Statin Efficacy. Trends Cardiovasc. Med 19, 173–177. 10.1016/j.tcm.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell KL, Hyman BT, Growdon JH, Hedley-Whyte ET, 1999. Application of the National Institute on Aging (NIA)-Reagan Institute criteria for the neuropathological diagnosis of Alzheimer disease. J. Neuropathol. Exp. Neurol 58, 1147–1155. [DOI] [PubMed] [Google Scholar]
- Ridge PG, Ebbert MTW, Kauwe JSK, 2013. Genetics of Alzheimer’s disease. Biomed Res. Int 2013, e254954 10.1155/2013/254954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez E, Mateo I, Infante J, Llorca J, Garcia-Gorostiaga I, Vazquez-Higuera JL, Sanchez-Juan P, Berciano J, Combarros O, 2009. Interaction between HMGCR and ABCA1 cholesterol-related genes modulates Alzheimer’s disease risk. Brain Res. 1280, 166–171. 10.1016/j.brainres.2009.05.019 [DOI] [PubMed] [Google Scholar]
- Ruxton GD, Neuhäuser M, 2010. When should we use one-tailed hypothesis testing? Methods Ecol. Evol 1, 114–117. 10.1111/j.2041-210X.2010.00014.x [DOI] [Google Scholar]
- Sharp AR, Ridge PG, Bailey MH, Boehme KL, Norton MC, Tschanz JT, Munger RG, Corcoran CD, Kauwe JSK, Initiative ADN, 2014. Population substructure in Cache County, Utah: the Cache County study. BMC Bioinformatics 15, S8 10.1186/1471-2105-15-S7-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CR, Zou F, Younkin SG, Estus S, 2011. Evaluation of the global association between cholesterol-associated polymorphisms and Alzheimer’s disease suggests a role for rs3846662 and HMGCR splicing in disease risk. Mol. Neurodegener 6, 62 10.1186/1750-1326-6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K, 1998. Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc. Natl. Acad. Sci 95, 6460–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M, 2006. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet 38, 209–213. 10.1038/ng1706 [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Khan SM, 2004. A “mitochondrial cascade hypothesis” for sporadic Alzheimer’s disease. Med. Hypotheses 63, 8–20. 10.1016/j.mehy.2003.12.045 [DOI] [PubMed] [Google Scholar]
- Team, R.C., 2016. R: A language and environment for statistical computing.
- Tschanz JT, Welsh-Bohmer KA, Plassman BL, Norton MC, Wyse BW, Breitner JCS, 2002. An adaptation of the modified mini-mental state examination: analysis of demographic influences and normative data: the cache county study. Neuropsychiatry. Neuropsychol. Behav. Neurol 15, 28–38. [PubMed] [Google Scholar]