Abstract
MicroRNAs bind to the 3′ untranslated regions of mRNAs, affecting translation, tumorigenesis, and apoptosis. This study evaluated the role of TYMS (rs1059394, C > T, and rs2847153, G > A), RYR3 (rs1044129, G > A), KIAA0423 (rs1053667, T > C), and GOLGA7 (rs11337, G > T) polymorphisms for assessment of glioma risk and prognosis among the Chinese Han population. Five single-nucleotide polymorphisms were assessed in 605 glioma patients and 1,300 controls. We found a significant correlation between rs1059394 and glioma susceptibility in the homozygote and dominant genetic models (TT versus CC, odds ratio [OR] = 0.71, 95% confidence interval [CI] = 0.52–0.97, p = 0.03; CT+TT versus CC, OR = 0.74, 95% CI = 0.55–0.99, p = 0.04). The results of the Kaplan-Meier and log rank tests revealed that the rs11337 GG genotype correlated with better overall survival of glioma patients (p = 0.017) than the GT genotype. Multivariate Cox regression analysis results also showed that the rs11337 GT genotype correlated with worse overall survival (p = 0.017, hazard ratio [HR] = 1.25, 95% CI = 1.04–1.5) than the GG genotype. These results suggest that GOLGA7 (rs11337) polymorphism may play a role in the prognosis of glioma patients and that TYMS (rs1059394) is associated with glioma risk.
Keywords: GOLGA7, rs11337, polymorphism, cancer risk, prognosis, microRNA-binding site
Introduction
Glioma is the most common form of brain tumor in the world that accounts for about 80% of all malignant brain tumors and has a fatal prognosis.1, 2 Previous study showed a median overall survival of 14.6 months and a 2-year survival rate of 30% in glioma patients.3 In China, 101,600 people were diagnosed with brain and CNS cancer, and 61,000 people died of brain and CNS cancer in 2015.4 Recent research suggested that age, sex, histological type and grade, extent of resection, tumor location, radiotherapy, and chemotherapy might influence survival rate in glioma patients.5 So far, there has been little progress in improving the survival rate of glioma patients and, therefore, it is necessary to find new ways to achieve this.
MicroRNAs (miRNAs) are small RNA molecules, about 22 nucleotides in length, which can regulate the expression of mRNAs by base pairing with their 3′ UTRs and thus prevent their translation.6, 7 Many previous studies have shown that the binding of miRNAs to the 3′ UTRs of mRNAs are essential for many biological processes, including translation, proliferation, tumorigenesis, and apoptosis.6, 7, 8 The genetic polymorphisms in these regions of miRNA target genes may be associated with cancer risk and prognosis. Many studies have confirmed that the genetic polymorphisms found in the 3′ UTRs play an important role in cancer risk and prognosis.9, 10, 11 Previous studies have shown that thymidine synthase (TYMS) polymorphism may be associated with gastric cancer risk and prognosis and RYR3 polymorphism may be associated with colonic cancer prognosis.12, 13 A study also found the role of KIAA0423 polymorphism in esophageal cancer survival.14 Nevertheless, GOLGA7 polymorphism has not been found to be associated with the gastric cancer and non-Hodgkin’s lymphoma risk and prognosis.8, 15 The role of these genetic polymorphisms in glioma risk and prognosis has not been explored.
In this context, our study aims at evaluating the role of polymorphisms in the 3′ UTRs of TYMS (rs1059394 and rs2847153), RYR3 (rs1044129), KIAA0423 (rs1053667), and GOLGA7 (rs11337) mRNAs in glioma risk and prognosis among the Chinese Han population.
Results
Characteristics of the Study Population
A total of 605 glioma patients (335 males and 270 females) were included in this study, with a mean age of 40.71 ± 18.28 years old. The median survival time for glioma patients is 11 months (range, 2–44 months), and overall survival is 32.16% in the first year, which reduces to 8.62% by the third year. According to the World Health Organization (WHO) classification, glioma patients were divided into two groups: 382 patients with WHO I–II and 223 patients with WHO III–IV. All patients underwent surgery: 189 patients underwent subtotal resection (STR) or near-total resection (NTR), and 416 patients underwent gross total resection (GTR). Among all the patients, 545 patients received radiotherapy (162 patients underwent conformal radiotherapy and 383 patients underwent gamma knife), and 60 patients did not receive radiotherapy. In total, 545 patients received chemotherapy (124 patients received platinum, 52 patients received temozolomide, and 74 patients received nimustine), and 355 patients did not receive any chemotherapy. The distributions of the demographic characteristics are shown in Table 1.
Table 1.
The Characteristics of Glioma Cases and Cancer-free Controls
| Characteristics | Cases | Control | p Valuea |
|---|---|---|---|
| Number | 605 | 1,300 | |
| Age (Mean ± SD) | 40.71 ± 18.28 | 41.68 ± 13.54 | 0.195 |
| <40 years | 267 | 561 | 0.688 |
| ≥40 years | 338 | 739 | |
| Sex | |||
| Male | 335 | 700 | 0.534 |
| Female | 270 | 600 | |
| WHO Grade | |||
| I + II | 382 | ||
| III + IV | 223 | ||
| Surgery | |||
| STR and NTR | 189 | ||
| GTR | 416 | ||
| Radiotherapy | |||
| No | 60 | ||
| Conformal radiotherapy | 162 | ||
| Gamma knife | 383 | ||
| Chemotherapy | |||
| No | 355 | ||
| Platinum | 124 | ||
| Temozolomide | 52 | ||
| Nimustine | 74 | ||
STR, subtotal resection; NTR, near-total resection; GTR, gross total resection.
t test or two-sided χ2 test.
Glioma Risk Assessment
We observed a significant association between rs1059394 and glioma susceptibility in the homozygote and dominant genetic models (TT versus CC, odds ratio [OR] = 0.71, 95% confidence interval [CI] = 0.52–0.97, p = 0.03; CT+TT versus CC, OR = 0.74, 95% CI = 0.55–0.99, p = 0.04). On the other hand, the association between rs1044129 and glioma risk in the six inheritance models, as well as rs2847153, rs11337, and rs1053667, was not significant, as shown in Table 2.
Table 2.
Genotype Frequencies of TYMS, GOLGA7, RYR3, and KIAA0423 Polymorphism in Cases and Controls
| Model | Genotype | Control (%) | Case (%) | OR (95% CI) | p Value |
|---|---|---|---|---|---|
| rs11337 HWE: p = 0.51 | |||||
| Co-dominant | GG | 779 (59.9) | 384 (63.5) | 1.00 (reference) | |
| Heterozygote | GT | 460 (35.4) | 194 (32.0) | 0.86 (0.70–1.05) | 0.14 |
| Homozygote | TT | 61 (4.7) | 27 (4.5) | 0.90 (0.56–1.43) | 0.65 |
| Dominant | GG | 779 (59.9) | 384 (63.5) | 1.00 (reference) | 0.14 |
| GT+TT | 521 (40.1) | 221 (36.5) | 0.86 (0.71–1.05) | ||
| Recessive | GG+GT | 1,239 (95.3) | 578 (95.5) | 1.00 (reference) | 0.82 |
| TT | 61 (4.7) | 27 (4.5) | 0.95 (0.60–1.51) | ||
| Overdominant | GG+TT | 840 (64.6) | 411 (68) | 1.00 (reference) | 0.16 |
| GT | 460 (35.4) | 194 (32) | 0.86 (0.7–1.06) | ||
| Allele |
G | 2,018 (77.6) | 962 (79.5) | 1.00 (reference) | 0.19 |
| T | 582 (22.4) | 248 (20.5) | 0.89 (0.76–1.06) | ||
| rs1044129 HWE: p = 0.86 | |||||
| Co-dominant | GG | 259 (19.9) | 106 (17.5) | 1.00 (reference) | |
| Heterozygote | GA | 639 (49.2) | 315 (52.1) | 1.20 (0.93–1.57) | 0.17 |
| Homozygote | AA | 402 (30.9) | 184 (30.4) | 1.12 (0.84–1.49) | 0.44 |
| Dominant | GG | 259 (19.9) | 106 (17.5) | 1.00 (reference) | 0.22 |
| GA+AA | 1,041 (80.1) | 499 (82.5) | 1.17 (0.91–1.50) | ||
| Recessive | GG+GA | 898 (69.1) | 421 (69.6) | 1.00 (reference) | 0.82 |
| AA | 402 (30.9) | 184 (30.4) | 0.98 (0.79–1.20) | ||
| Overdominant | GG+AA | 661 (50.8) | 290 (47.9) | 1.00 (reference) | 0.24 |
| GA | 639 (49.2) | 315 (52.1) | 1.12 (0.93–1.36) | ||
| Allele |
G | 1,157 (44.5) | 527 (43.6) | 1.00 (reference) | 0.58 |
| A | 1,443 (55.5) | 683 (56.4) | 1.04 (0.91–1.19) | ||
| rs1053667 HWE: p = 0.96 | |||||
| Co-dominant | TT | 969 (74.5) | 465 (76.9) | 1.00 (reference) | |
| Heterozygote | CT | 307 (23.6) | 135 (22.3) | 0.92 (0.73–1.15) | 0.46 |
| Homozygote | CC | 24 (1.9) | 5 (0.8) | 0.43 (0.16–1.14) | 0.08 |
| Dominant | TT | 969 (74.5) | 465 (76.9) | 1.00 (reference) | 0.27 |
| CT+CC | 331 (25.5) | 140 (23.1) | 0.88 (0.70–1.10) | ||
| Recessive | TT+CT | 1,276 (98.1) | 600 (99.2) | 1.00 (reference) | 0.09 |
| CC | 24 (1.9) | 5 (0.8) | 0.44 (0.17–1.17) | ||
| Overdominant | TT+CC | 993 (76.4) | 470 (77.7) | 1.00 (reference) | 0.53 |
| CT | 307 (23.6) | 135 (22.3) | 0.93 (0.74–1.17) | ||
| Allele |
T | 2,245 (86.3) | 1,065 (88) | 1.00 (reference) | 0.16 |
| C | 355 (23.7) | 145 (12) | 0.86 (0.70–1.06) | ||
| rs1059394 HWE: p = 0.53 | |||||
| Co-dominant | CC | 131 (10.1) | 80 (13.2) | 1.00 (reference) | |
| Heterozygote | CT | 548 (42.1) | 255 (42.2) | 0.76 (0.56–1.04) | 0.09 |
| Homozygote | TT | 621 (47.8) | 270 (44.6) | 0.71 (0.52–0.97) | 0.03* |
| Dominant | CC | 131 (10.1) | 80 (13.2) | 1.00 (reference) | 0.04* |
| CT+TT | 1,169 (89.9) | 525 (86.8) | 0.74 (0.55–0.99) | ||
| Recessive | CC+CT | 679 (52.2) | 335 (55.4) | 1.00 (reference) | 0.20 |
| TT | 621 (47.8) | 270 (44.6) | 0.88 (0.73–1.07) | ||
| Overdominant | CC+TT | 752 (51.9) | 350 (57.8) | 1.00 (reference) | 1.00 |
| CT | 548 (42.1) | 255 (42.2) | 1.00 (0.82–1.22) | ||
| Allele |
C | 810 (31.2) | 415 (34.5) | 1.00 (reference) | 0.05 |
| T | 1,790 (68.8) | 795 (65.5) | 0.87 (0.75–1.00) | ||
| rs2847153 HWE: p = 0.47 | |||||
| Co-dominant | GG | 534 (41.1) | 223 (36.9) | 1.00 (reference) | |
| Heterozygote | GA | 589 (45.3) | 295 (48.9) | 1.20 (0.97–1.48) | 0.09 |
| Homozygote | AA | 177 (13.6) | 86 (14.2) | 1.16 (0.86–1.57) | 0.32 |
| Dominant | GG | 534 (41.1) | 223 (36.9) | 1.00 (reference) | 0.09 |
| GA+AA | 766 (58.9) | 381 (63.1) | 1.19 (0.98–1.45) | ||
| Recessive | GG+GA | 1,123 (86.4) | 518 (85.8) | 1.00 (reference) | 0.71 |
| AA | 177 (13.6) | 86 (14.2) | 1.05 (0.80–1.39) | ||
| Overdominant | GG+AA | 711 (44.7) | 309 (51.2) | 1.00 (reference) | 0.15 |
| GA | 589 (45.3) | 295 (48.8) | 1.15 (0.95–1.39) | ||
| Allele | G | 1,657 (63.7) | 741 (61.3) | 1.00 (reference) | 0.16 |
| A | 943 (36.3) | 467 (38.7) | 1.11 (0.96–1.28) | ||
HWE, Hardy-Weinberg equilibrium; STR, subtotal resection; NTR, near-total resection; GTR, gross total resection; reference, OR = 1 is the reference compared with other genotypes. *p ≤ 0.05 indicates statistical significance.
Prognostic Values of Various Factors in the Overall Survival (OS) of Glioma Patients
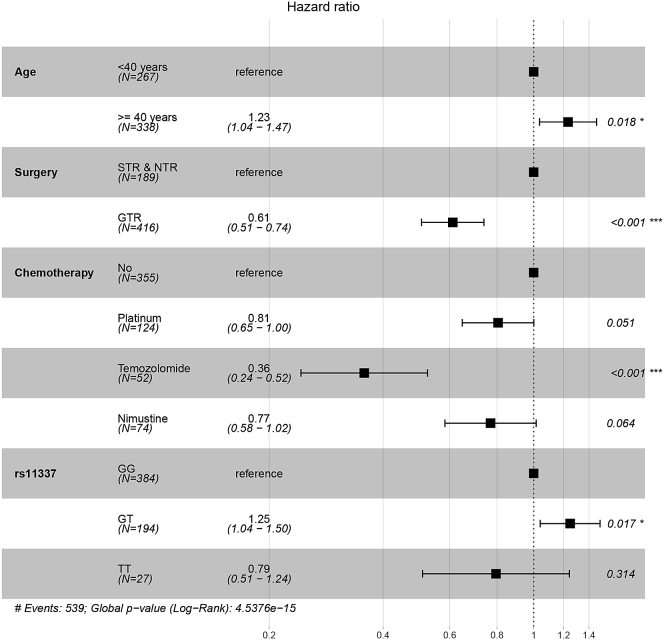
We conducted a univariate analysis and found that age, extent of resection, chemotherapy, and rs11337 were significant factors and hence included these factors into the multivariate Cox regression for analysis. The results of the Cox proportional hazards regression model showed that age, extent of resection, chemotherapy, and rs11337 were independent risk factors for OS. Patients aged ≥ 40 years had a worse OS (p = 0.018, hazard ratio [HR] = 1.23, 95% CI = 1.04–1.47), compared to patients aged < 40 years. The patients who underwent GTR had a better OS (p < 0.001, HR = 0.61, 95% CI = 0.51–0.74), compared to the patients who underwent STR or NTR. Similarly, patients who underwent temozolomide treatment had a better OS (p < 0.001, HR = 0.36, 95% CI = 0.24–0.52), compared to the patients who did not receive chemotherapy. We also saw that the GT genotype was associated with worse OS (p = 0.017, HR = 1.25, 95% CI = 1.04–1.50) as compared to the rs11337 GG genotype, whereas patients with TT genotype had a non-significant HR of 0.79 (95% CI = 0.51–1.24; p = 0.314). The details were shown in Table 3 and Figure 1.
Table 3.
Analysis of Polymorphism and Clinical Features in Glioma Patient Overall Survival
| Characteristic | Patients (n) | Events (n) | Rate (%) | Univariate Analysis |
Multivariable Analysis |
||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | ||||
| Age | |||||||
| <40 years | 267 | 229 | 85.77 | ref | ref | ref | ref |
| ≥40 years | 338 | 310 | 91.72 | 1.20 (1.01–1.42) | 0.0386* | 1.23 (1.04–1.47) | 0.018* |
| WHO Grade | |||||||
| I–II | 382 | 336 | 87.96 | ref | ref | ||
| III–IV | 223 | 206 | 92.38 | 1.18 (0.99–1.40) | 0.0626 | ||
| Surgery | |||||||
| STR and NTR | 189 | 186 | 98.41 | ref | ref | ref | ref |
| GTR | 416 | 353 | 84.86 | 0.58 (0.49–0.70) | <0.001* | 0.61 (0.51–0.74) | <0.001* |
| Chemotherapy | |||||||
| No | 355 | 333 | 93.80 | ref | ref | ref | ref |
| Platinum | 124 | 112 | 90.32 | 0.84 (0.68–1.04) | 0.116 | 0.81 (0.65–1.00) | 0.0513 |
| Temozolomide | 52 | 30 | 57.69 | 0.33 (0.22–0.48) | <0.001* | 0.36 (0.24–0.52) | <0.001* |
| Nimustine | 74 | 64 | 86.49 | 0.64 (0.49–0.84) | 0.0013* | 0.77 (0.58–1.02) | 0.0639 |
| rs11337 | |||||||
| GG | 384 | 338 | 88.02 | ref | ref | ref | ref |
| GT | 194 | 180 | 92.78 | 1.24 (1.04–1.49) | 0.019* | 1.25 (1.04–1.50) | 0.0169* |
| TT | 27 | 21 | 77.78 | 0.85 (0.55–1.32) | 0.467 | 0.79 (0.51–1.24) | 0.314 |
| Sex | |||||||
| Male | 335 | 297 | 88.66 | ref | ref | ||
| Female | 270 | 242 | 89.63 | 1.08 (0.91–1.28) | 0.355 | ||
| Radiotherapy | |||||||
| No | 60 | 49 | 81.67 | ref | ref | ||
| Conformal radiotherapy | 162 | 133 | 82.10 | 1.08 (0.78–1.51) | 0.622 | ||
| Gamma knife | 383 | 357 | 93.21 | 1.17 (0.87–1.58) | 0.303 | ||
| rs1044129 | |||||||
| GG | 106 | 97 | 91.51 | ref | ref | ||
| GA | 315 | 277 | 87.94 | 0.93 (0.74–1.18) | 0.557 | ||
| AA | 184 | 165 | 89.67 | 0.97 (0.76–1.25) | 0.836 | ||
| rs1053667 | |||||||
| TT | 465 | 416 | 89.46 | ref | ref | ||
| TC | 135 | 120 | 88.89 | 0.94 (0.77–1.51) | 0.549 | ||
| CC | 5 | 3 | 60.00 | 0.60 (0.19–1.86) | 0.371 | ||
| rs1059394 | |||||||
| CC | 80 | 70 | 87.50 | ref | ref | ||
| CT | 255 | 224 | 87.84 | 0.90 (0.69–1.18) | 0.451 | ||
| TT | 270 | 245 | 90.74 | 1.00 (0.77–1.30) | 0.992 | ||
| rs2847153 | |||||||
| GG | 223 | 204 | 91.48 | ref | ref | ||
| GA | 295 | 258 | 87.46 | 0.93 (0.77–1.12) | 0.434 | ||
| AA | 86 | 76 | 88.37 | 0.91 (0.70–1.19) | 0.509 | ||
STR, subtotal resection; NTR, near-total resection; GTR, gross total resection; ref, reference compared with other indicators. *p ≤ 0.05 indicates statistical significance.
Figure 1.
Forest Plots of Multivariate Cox Regression Analysis for OS
STR, subtotal resection; NTR, near-total resection; GTR, gross total resection.
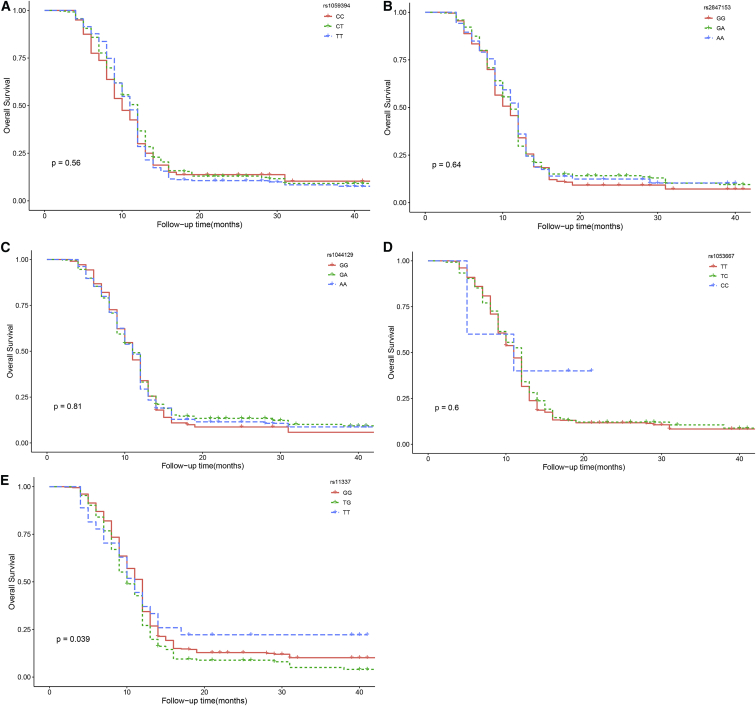
The results of the Kaplan-Meier and log rank tests showed the difference among three rs11337 genotypes (p = 0.039; Figure 2) and further analysis revealed that rs11337 GG genotype was associated with better OS in glioma patients (p = 0.017), compared to the GT genotype. As for rs1059394 (p = 0.56) and rs2847153 (p = 0.64) in TYMS, rs1044129 (p = 0.81) in RYR3, and rs1053667 in KIAA0423 (p = 0.6), we observed no significant difference among the different genotypes. This was in accordance with the results of Cox regression.
Figure 2.
Kaplan-Meier Analysis of Overall Survival Are Shown for Different Genotypes
(A) rs1059394 in TYMS; (B) rs2847153 in TYMS; (C) rs1044129 in RYR3; (D) rs1053667 in KIAA0423; (E) rs11337 in GOLGA7.
Prognostic Values of Various Factors in the Progression-free Survival (PFS) of Glioma Patients
We conducted a univariate analysis and found that age, extent of resection, and chemotherapy were significant factors and then included these factors into the multivariate Cox regression for analysis. As shown in Table 4, the results of the Cox proportional hazards regression model showed that age, extent of resection, and chemotherapy were independent risk factors for PFS. Patients aged ≥ 40 years compared with those aged < 40 years had worse PFS (p = 0.0191, HR = 1.22, 95% CI = 1.03–1.45). The patients who underwent GTR had a better PFS (p < 0.001, HR = 0.61, 95% CI = 0.51–0.74), compared to the patients who underwent STR or NTR. The patients who underwent temozolomide treatment also had a better PFS (p < 0.001, HR = 0.37, 95% CI = 0.26–0.54), compared to the patients who did not receive chemotherapy.
Table 4.
Analysis of Polymorphism and Clinical Features in Glioma Patient Progression-free Survival
| Characteristic | Patients (n) | Events (n) | Rate (%) | Univariate Analysis |
Multivariable Analysis |
||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | ||||
| Age | |||||||
| <40 years | 267 | 239 | 89.51 | ref | ref | ref | ref |
| ≥40 years | 338 | 324 | 95.86 | 1.19 (1.00–1.40) | 0.047* | 1.22 (1.03–1.45) | 0.0191* |
| WHO Grade | |||||||
| I–II | 382 | 353 | 92.41 | ref | ref | ||
| III–IV | 223 | 210 | 94.17 | 1.15 (0.97–1.36) | 0.116 | ||
| Surgery | |||||||
| STR and NTR | 189 | 183 | 96.83 | ref | ref | ref | ref |
| GTR | 416 | 380 | 91.35 | 0.58 (0.48–0.69) | <0.001* | 0.61 (0.51–0.74) | <0.001* |
| Chemotherapy | |||||||
| No | 335 | 351 | 104.78 | ref | ref | ref | ref |
| Platinum | 124 | 116 | 93.55 | 0.99 (0.80–1.22) | 0.916 | 0.97 (0.80–1.20) | 0.759 |
| Temozolomide | 52 | 32 | 61.54 | 0.35 (0.24–0.50) | <0.001* | 0.37 (0.26–0.54) | <0.001* |
| Nimustine | 74 | 64 | 86.49 | 0.73 (0.56–0.96) | 0.022* | 0.88 (0.67–1.16) | 0.377 |
| rs11337 | |||||||
| GG | 384 | 355 | 92.45 | ref | ref | ref | ref |
| GT | 194 | 187 | 96.39 | 1.21 (1.01–1.44) | 0.0367* | 1.19 (0.99–1.43) | 0.0519 |
| TT | 27 | 21 | 77.78 | 0.75 (0.49–1.17) | 0.209 | 0.88 (0.67–1.16) | 0.113 |
| Sex | |||||||
| Male | 335 | 310 | 92.54 | ref | ref | ||
| Female | 270 | 253 | 93.70 | 1.10 (0.93–1.30) | 0.263 | ||
| Radiotherapy | |||||||
| No | 60 | 55 | 91.67 | ref | ref | ||
| Conformal radiotherapy | 162 | 137 | 84.57 | 1.13 (0.83–1.56) | 0.436 | ||
| Gamma knife | 383 | 371 | 96.87 | 1.21 (0.91–1.60) | 0.199 | ||
| rs1044129 | |||||||
| GG | 105 | 102 | 97.14 | ref | ref | ||
| GA | 312 | 290 | 92.95 | 0.92 (0.73–1.15) | 0.453 | ||
| AA | 183 | 171 | 93.44 | 0.95 (0.75–1.22) | 0.698 | ||
| rs1053667 | |||||||
| TT | 465 | 423 | 90.97 | ref | ref | ||
| TC | 135 | 126 | 93.33 | 0.95 (0.78–1.16) | 0.604 | ||
| CC | 5 | 4 | 80.00 | 0.66 (0.25–1.77) | 0.411 | ||
| rs1059394 | |||||||
| CC | 80 | 76 | 95.00 | ||||
| CT | 225 | 233 | 103.56 | 0.89 (0.69–1.15) | 0.368 | ||
| TT | 270 | 254 | 94.07 | 0.96 (0.74–1.24) | 0.762 | ||
| rs2847153 | |||||||
| GG | 223 | 211 | 94.62 | ||||
| GA | 295 | 272 | 92.20 | 0.91 (0.76–1.09) | 0.299 | ||
| AA | 86 | 79 | 91.86 | 0.91 (0.70–1.17) | 0.456 | ||
STR, subtotal resection; NTR, near-total resection; GTR, gross total resection; ref, reference compared with other indicators. *p ≤ 0.05 indicates statistical significance.
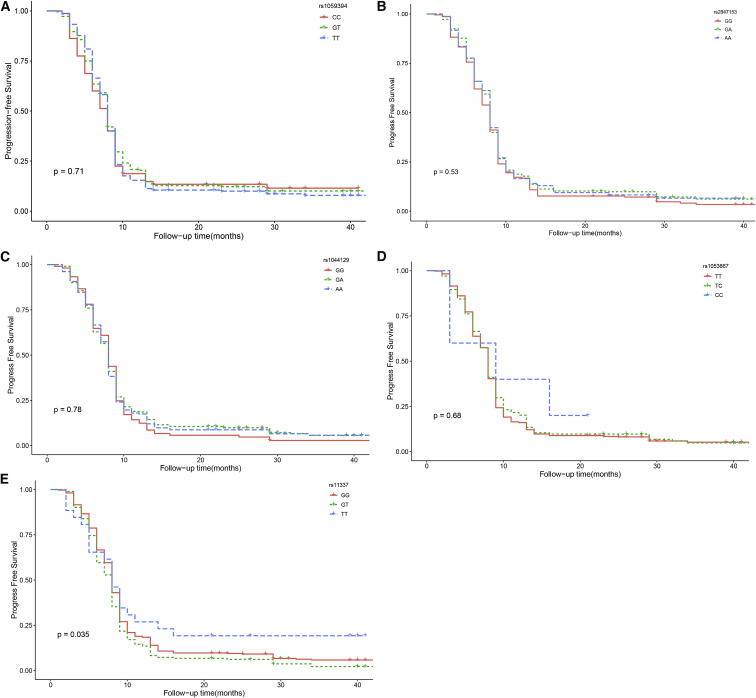
The results of the Kaplan-Meier and log rank tests showed that the rs11337 GG genotype had a better PFS in glioma patients (p = 0.035; Figure 3). After taking into account factors such as age, extent of resection, and chemotherapy, patients with the TT genotype (p = 0.113, HR = 0.88, 95% CI = 0.67–1.16) or TG genotype (p = 0.0519, HR = 1.19, 95% CI = 0.99–1.43) had a non-significant HR compared with patients with the GG genotype. As for rs1059394 (p = 0.71) and rs2847153 (p = 0.53) in TYMS, rs1044129 (p = 0.78) in RYR3, and rs1053667 in KIAA0423 (p = 0.68), we observed no significant difference among different genotypes. This was in accordance with the results of Cox regression analysis.
Figure 3.
Kaplan-Meier Analysis of Progression-free Survival Are Shown for Different Genotypes
(A) rs1059394 in TYMS; (B) rs2847153 in TYMS; (C) rs1044129 in RYR3; (D) rs1053667 in KIAA0423; (E) rs11337 in GOLGA7.
Expression Quantitative Trait Loci
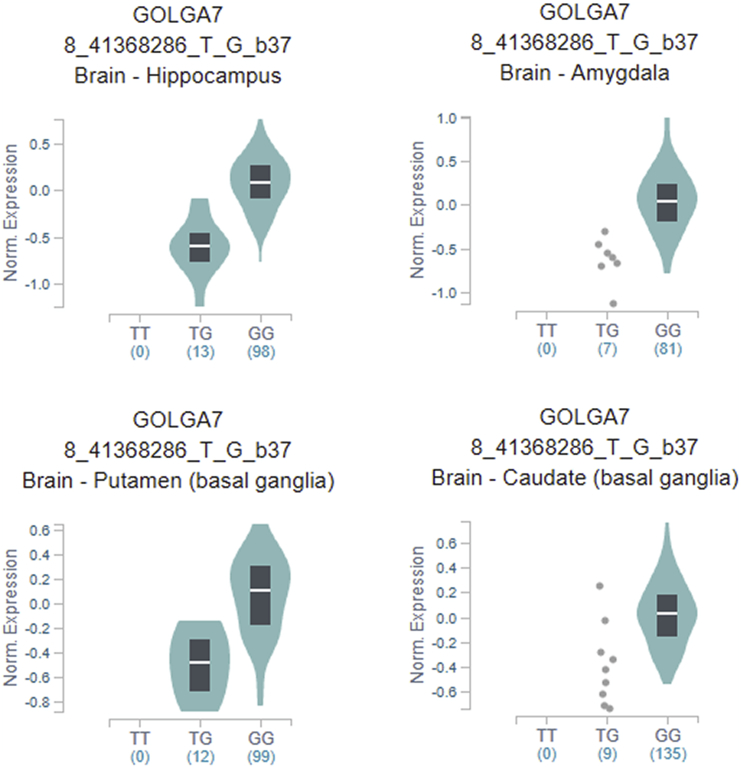
We further used the GTEx portal to explore biological effects of rs11337 in the GOLGA7 gene expression. The results showed that genotypes of rs11337 was significantly associated with GOLGA7 gene expression in four brain tissues (Figure 4; all p values were less than 0.001).
Figure 4.
Analysis of the rs11337 G > T Polymorphisms in the GOLGA7 Gene in Four Brain Tissues
Brain-hippocampus, p = 7.6e−13; brain-amygdala, p = 3.2e−8; brain-putamen (basal ganglia), 7.6e−11; brain-caudate (basal ganglia), p = 1.3e−8
Discussion
To our knowledge, this is the first study to explore the role of TYMS (rs1059394 and rs2847153), RYR3 (rs1044129), KIAA0423 (rs1053667), and GOLGA7 (rs11337) 3′ UTR polymorphisms in glioma risk and prognosis among the Chinese Han population. We observed that rs1059394 and glioma risk were significantly correlated. In addition, we found that glioma patients with rs11337 GT genotype had a worse OS compared with patients with the GG genotype. We also observed that the association between the other four gene variants (rs1059394 and rs2847153 in TYMS, rs1044129 in RYR3, and rs1053667 in KIAA0423) and glioma patient prognosis was not significant.
Chemotherapy and surgery are usually the first choice of treatment for glioma patients. A study conducted by Ma et al.16 on 205 glioma patients suggested that age, preoperative Karnofsky’s performance status score, tumor location, radiotherapy, radical surgery, and chemotherapy were independent factors of prognosis among the Han population of China. The conclusions of this study were roughly consistent with ours. In our study, we found that age, the extent of surgical resection, and chemotherapy were independent prognostic factors for glioma survival. Glioma patients with age < 40 years old have a better prognosis. We also saw that GTR might be associated with a better prognosis than STR or NTR. In addition, we found that temozolomide therapy showed the best curative effect among all the four chemotherapy agents used. However, there was no significant association between radiotherapy and prognosis of glioma patients. This was inconsistent with the conclusion of the abovementioned study, which stated that radiotherapy was the strongest predictor of prognosis. We believe that our conclusions are more convincing as we have used a larger sample size, and studies on a large scale are required to confirm the role of radiotherapy in the prognosis of glioma patients.
miRNAs seem to be critical in many treatments, especially cancer treatment.8 As most of the miRNAs bind to the target sequence located in the 3′ UTR of the mRNA, the genetic polymorphism of target genes in these regions may act as a tumor suppressor or oncogene and alter the interactions between miRNA and mRNA, leading to carcinogenesis or progression.17, 18, 19 Recently, more and more single-nucleotide polymorphisms (SNPs) in the 3′ UTR of mRNA have been reported to be associated with cancer risk and prognosis.8, 20, 21 Previous studies have reported the role of these five gene variants in hepatocellular carcinoma, lymphoma, gastric carcinoma, breast cancer, and esophageal cancers, but not in glioma.8, 14, 15, 19, 22
TYMS participates in folate metabolism and provides nucleotides needed for DNA synthesis and repair.23 The damage of TYMS enzymes is related to chromosome damage and increased induction of fragile sites, which may lead to the development of cancer.24, 25 Therefore, functional genetic variants in TYMS may be associated with cancer risk and prognosis. GOLGA7 is a member of the Golgi family, anchored in the middle of the Golgi membrane molecules.26 The results of expression quantitative trait loci (eQTL) analysis for rs11337 revealed that rs11337 GT genotype was associated with less expression of GOLGA7 as compared to the rs11337 GG genotype, and expression of GOLGA7 influences protein transport from Golgi apparatus to cell surface, which may affect cancer prognosis.26
However, there were also some limitations to our study. First, as a single-center study, selection bias was unavoidable. Second, the sample size in this study was relatively small because gliomas were relatively rare compared to other tumors. Therefore, these findings need to be validated using studies on larger sample sizes. Third, the number of SNPs analyzed was limited. Finally, the conclusions drawn from this study cannot be directly extrapolated to other races, because all the patients were from the Han population of China.
In summary, our findings indicate that rs11337 (G > T) in GOLGA7 may play a role in survival of glioma patients and rs1059394 in TYMS is associated with glioma risk. While these findings may contribute to personalized treatment in the future, they need to be further validated using larger sample sizes.
Materials and Methods
Ethics Statement
The protocol used in this study was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University Shaanxi Province (Xi’an, China). All participants signed the informed consent.
Study Population
A total of 605 glioma patients and 1300 healthy controls were included in this study. Selected controls were matched to cases based on age (p = 0.195) and sex (p = 0.534). All patients were consecutively recruited between September 2010 and May 2014 at Tangdu Hospital, which was affiliated with the Fourth Military Medical University in China. At least two senior neuropathologists confirmed the histopathological diagnosis. All healthy controls underwent a checkup at the same hospital during the same period of time. The clinical information was collected and updated regularly by follow-up and hospital records, including ethnicity, age, sex, WHO grade, radiotherapy record, surgery record, chemotherapy record, and the condition of the patient at the last follow-up.
Genotyping Assay
SNPs in 3′ UTRs were selected after conducting a literature review. DNA samples were extracted from whole blood using the universal genomic DNA extraction kit (TaKaRa, Kyoto, Japan).27, 28 DNA concentrations were assessed through spectrophotometry (DU530 UV/VIS spectrophotometer, Beckman Instruments, Fullerton, CA, USA). The multiplexed SNP mass EXTEND assay was designed using Sequenom mass ARRAY assay design (version3.0, Agena Bioscience, San Diego, CA, USA).29, 30, 31 SNP genotyping was performed using Sequenom mass ARRAY RS1000.32 The Sequenom Typer 4.0 software was used to analyze the data.32, 33 Primers for each SNP were shown in Table 5. In total, five SNPs (rs1059394 and rs2847153 in TYMS, rs1044129 in RYR3, rs1053667 in KIAA0423, and rs11337 in GOLGA7) were successfully genotyped.
Table 5.
Primers Used in This Study
| SNP_ID | 1st PCRP (5′–3′) | 2nd PCRP (5′–3′) | UEP_SEQ (5′–3′) |
|---|---|---|---|
| rs1044129 | ACGTTGGATGACCCTGGAGGTATTGGTACG | ACGTTGGATGAGTGGAGCTGCTCTGTTTAG | TAGGTGAATCTCCTCAAATACA |
| rs11337 | ACGTTGGATGCGAAATCCAGTATTAGCACC | ACGTTGGATGTTGAGAGCGCTGTATTTGGG | CATTAAAAGTTTCACTGTCAGA |
| rs1053667 | ACGTTGGATGGGGCAACAAATTGTAGTTTC | ACGTTGGATGAATCTGAGTCACATGGGATG | gtttgGAGAAAAGTCCTGCTCA |
| rs1059394 | ACGTTGGATGGTATCGACAGGATCATACTC | ACGTTGGATGCGACCTGTTGTAATTGCTCC | cATTGCTCCTCATGTCC |
| rs2847153 | ACGTTGGATGTCTTTAAGTAGGCTGGTCCC | ACGTTGGATGAGAAAAGATCTGGGAGGGTG | gCAAAGAAGGGATCAGACT |
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) was calculated using the χ2 test.34 The population gene diversity was considered genetically balanced when the p value was > 0.05. Six inheritance models were created to assess cancer risk. ORs and 95% CIs were calculated to assess the association between the polymorphisms in these five variants and glioma susceptibility.35 OS and PFS were calculated to evaluate the prognosis. The Kaplan-Meier and log rank tests were performed to assess the effect of the genotype on the prognosis of patients. Univariate and multivariate Cox regression analysis were conducted to analyze the prognostic factors in glioma patients. We further used the GTEx (http://www.gtexportal.org/home/index.html) portal to explore biological effects of rs11337 in the GOLGA7 gene expression in four brain tissues.36 All statistical tests were two-sided, with statistical significance evaluated at the 0.05 α-level. All calculations were performed using the R software (version 3.5.1).
Author Contributions
L.Z. performed experiments, analyzed data, and wrote the paper, performed some experiments, and analyzed data; Z.D. initiated the study and designed experiments. S.D., Y.D., P.Y., Y.Z., L.Y., M.Z., S.Y., Y.W., Z.Z., N.L., and H.K. read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 81471670), the International Cooperative Project of Shaanxi Province, China (no. 2016KW-008), and the Key Research and Development Plan, Shaanxi province, China (2017ZDXM-SF-066).
Contributor Information
Huafeng Kang, Email: kanghuafeng1973@126.com.
Zhijun Dai, Email: dzj0911@126.com.
References
- 1.Rasmussen B.K., Hansen S., Laursen R.J., Kosteljanetz M., Schultz H., Nørgård B.M., Guldberg R., Gradel K.O. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish Neuro-Oncology Registry. J. Neurooncol. 2017;135:571–579. doi: 10.1007/s11060-017-2607-5. [DOI] [PubMed] [Google Scholar]
- 2.Barnholtz-Sloan J.S., Ostrom Q.T., Cote D. Epidemiology of Brain Tumors. Neurol. Clin. 2018;36:395–419. doi: 10.1016/j.ncl.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Grossman S.A., Ye X., Piantadosi S., Desideri S., Nabors L.B., Rosenfeld M., Fisher J., NABTT CNS Consortium Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin. Cancer Res. 2010;16:2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Wrensch M., Wiencke J.K., Wiemels J., Miike R., Patoka J., Moghadassi M., McMillan A., Kelsey K.T., Aldape K., Lamborn K.R. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006;66:4531–4541. doi: 10.1158/0008-5472.CAN-05-4032. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang C., Zhao Y., Ming Y., Zhao S., Guo Z. A polymorphism at the microRNA binding site in the 3′-untranslated region of C14orf101 is associated with the risk of gastric cancer development. Exp. Ther. Med. 2016;12:1867–1872. doi: 10.3892/etm.2016.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin T.B., Du S., Zhu X.K., Li G., Ouyang Y., He N., Zhang Z., Zhang Y., Kang L., Yuan D. Polymorphism in the IL4R gene and clinical features are associated with glioma prognosis: Analyses of case-cohort studies. Medicine (Baltimore) 2016;95:e4231. doi: 10.1097/MD.0000000000004231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin T., Wang Y., Li G., Du S., Yang H., Geng T., Hou P., Gong Y. Analysis of difference of association between polymorphisms in the XRCC5, RPA3 and RTEL1 genes and glioma, astrocytoma and glioblastoma. Am. J. Cancer Res. 2015;5:2294–2300. [PMC free article] [PubMed] [Google Scholar]
- 11.Mistry A.M., Vnencak-Jones C.L., Mobley B.C. Clinical prognostic value of the isocitrate dehydrogenase 1 single-nucleotide polymorphism rs11554137 in glioblastoma. J. Neurooncol. 2018;138:307–313. doi: 10.1007/s11060-018-2796-6. [DOI] [PubMed] [Google Scholar]
- 12.Shen R., Liu H., Wen J., Liu Z., Wang L.E., Wang Q., Tan D., Ajani J.A., Wei Q. Genetic polymorphisms in the microRNA binding-sites of the thymidylate synthase gene predict risk and survival in gastric cancer. Mol. Carcinog. 2015;54:880–888. doi: 10.1002/mc.22160. [DOI] [PubMed] [Google Scholar]
- 13.Chae Y.S., Kim J.G., Kang B.W., Lee S.J., Lee Y.J., Park J.S., Choi G.S., Lee W.K., Jeon H.S. Functional polymorphism in the MicroRNA-367 binding site as a prognostic factor for colonic cancer. Anticancer Res. 2013;33:513–519. [PubMed] [Google Scholar]
- 14.Faluyi O.O., Eng L., Qiu X., Che J., Zhang Q., Cheng D., Ying N., Tse A., Kuang Q., Dodbiba L. Validation of microRNA pathway polymorphisms in esophageal adenocarcinoma survival. Cancer Med. 2017;6:361–373. doi: 10.1002/cam4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang B., Liu C., Diao L., Wang C., Guo Z. A polymorphism at the microRNA binding site in the 3′ untranslated region of C14orf101 is associated with non-Hodgkin lymphoma overall survival. Cancer Genet. 2014;207:141–146. doi: 10.1016/j.cancergen.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Ma X., Lv Y., Liu J., Wang D., Huang Q., Wang X., Li G., Xu S., Li X. Survival analysis of 205 patients with glioblastoma multiforme: clinical characteristics, treatment and prognosis in China. J. Clin. Neurosci. 2009;16:1595–1598. doi: 10.1016/j.jocn.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Song F., Zheng H., Liu B., Wei S., Dai H., Zhang L., Calin G.A., Hao X., Wei Q., Zhang W., Chen K. An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin. Cancer Res. 2009;15:6292–6300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 18.Chin L.J., Ratner E., Leng S., Zhai R., Nallur S., Babar I., Muller R.U., Straka E., Su L., Burki E.A. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Liu Y., Song F., Zheng H., Hu L., Lu H., Liu P., Hao X., Zhang W., Chen K. Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. USA. 2011;108:13653–13658. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vymetalkova V., Pardini B., Rosa F., Jiraskova K., Di Gaetano C., Bendova P., Levy M., Veskrnova V., Buchler T., Vodickova L. Polymorphisms in microRNA binding sites of mucin genes as predictors of clinical outcome in colorectal cancer patients. Carcinogenesis. 2017;38:28–39. doi: 10.1093/carcin/bgw114. [DOI] [PubMed] [Google Scholar]
- 21.Schneiderova M., Naccarati A., Pardini B., Rosa F., Gaetano C.D., Jiraskova K., Opattova A., Levy M., Veskrna K., Veskrnova V. MicroRNA-binding site polymorphisms in genes involved in colorectal cancer etiopathogenesis and their impact on disease prognosis. Mutagenesis. 2017;32:533–542. doi: 10.1093/mutage/gex026. [DOI] [PubMed] [Google Scholar]
- 22.Peng C., Guo Z., Wu X., Zhang X.L. A polymorphism at the microRNA binding site in the 3′ untranslated region of RYR3 is associated with outcome in hepatocellular carcinoma. OncoTargets Ther. 2015;8:2075–2079. doi: 10.2147/OTT.S85856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan X., Liu H., Ju J., Li Y., Li P., Wang L.E., Brewster A.M., Buchholz T.A., Arun B.K., Wei Q., Liu Z. Genetic variant rs16430 6bp > 0bp at the microRNA-binding site in TYMS and risk of sporadic breast cancer risk in non-Hispanic white women aged ≤ 55 years. Mol. Carcinog. 2015;54:281–290. doi: 10.1002/mc.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hori T., Ayusawa D., Shimizu K., Koyama H., Seno T. Chromosome breakage induced by thymidylate stress in thymidylate synthase-negative mutants of mouse FM3A cells. Cancer Res. 1984;44:703–709. [PubMed] [Google Scholar]
- 25.Hori T., Ayusawa D., Glover T.W., Seno T. Expression of fragile site on the human X chromosome in somatic cell hybrids between human fragile X cells and thymidylate synthase-negative mouse mutant cells. Jpn. J. Cancer Res. 1985;76:977–983. [PubMed] [Google Scholar]
- 26.Ohta E., Misumi Y., Sohda M., Fujiwara T., Yano A., Ikehara Y. Identification and characterization of GCP16, a novel acylated Golgi protein that interacts with GCP170. J. Biol. Chem. 2003;278:51957–51967. doi: 10.1074/jbc.M310014200. [DOI] [PubMed] [Google Scholar]
- 27.Dai Z., Tian T., Wang M., Yang T., Li H., Lin S., Hao Q., Xu P., Deng Y., Zhou L. Genetic polymorphisms of estrogen receptor genes are associated with breast cancer susceptibility in Chinese women. Cancer Cell Int. 2019;19:11. doi: 10.1186/s12935-019-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M., Liu X., Lin S., Tian T., Guan F., Guo Y., Li X., Deng Y., Zheng Y., Xu P. FABP1 Polymorphisms Contribute to Hepatocellular Carcinoma Susceptibility in Chinese Population with Liver Cirrhosis: A Case-Control Study. J. Cancer. 2018;9:4294–4300. doi: 10.7150/jca.27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong Y., Hsieh C.H., Alonso L.C. ANRIL: A lncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front. Endocrinol. (Lausanne) 2018;9:405. doi: 10.3389/fendo.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z., Yu X., Shen J. ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol. 2016;37:5657–5661. doi: 10.1007/s13277-016-4808-5. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Pan S., Liu L., Zhai X., Liu J., Wen J., Zhang Y., Chen J., Shen H., Hu Z. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS ONE. 2012;7:e35145. doi: 10.1371/journal.pone.0035145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabriel S., Ziaugra L., Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2009;2 doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 33.Thomas R.K., Baker A.C., Debiasi R.M., Winckler W., Laframboise T., Lin W.M., Wang M., Feng W., Zander T., MacConaill L. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 34.Zhu J., Fu W., Jia W., Xia H., Liu G.C., He J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol. Ther. Nucleic Acids. 2018;12:854–860. doi: 10.1016/j.omtn.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J., Zou Y., Liu X., Zhu J., Zhang J., Zhang R., Yang T., Xia H. Association of Common Genetic Variants in Pre-microRNAs and Neuroblastoma Susceptibility: A Two-Center Study in Chinese Children. Mol. Ther. Nucleic Acids. 2018;11:1–8. doi: 10.1016/j.omtn.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuo Z.J., Liu W., Zhang J., Zhu J., Zhang R., Tang J., Yang T., Zou Y., He J., Xia H. Functional Polymorphisms at ERCC1/XPF Genes Confer Neuroblastoma Risk in Chinese Children. EBioMedicine. 2018;30:113–119. doi: 10.1016/j.ebiom.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]