Abstract
Somatic mutations have long been recognized as an important feature of cancer. However, analysis of somatic mutations, to date, has focused almost entirely on the protein coding regions of the genome. The potential roles of somatic mutations in human long noncoding RNAs (lncRNAs) are therefore largely unknown, particularly their functional significance across different cancer types. In this study, we characterized some lncRNAs whose expression was affected by somatic mutations (defined as MutLncs) and constructed global MutLnc landscapes across 17 cancer types by systematically integrating multiple levels of data. MutLncs were commonly downregulated and carried low mutation frequencies and non-silent mutations in most cancer types. Co-occurrence analysis in pan-cancer highlighted combined patterns of specific MutLncs, suggesting that a number of MutLncs influence diverse cancer types through combination effects. Several conserved and cancer-specific functions of MutLncs were determined. We further explored the somatic mutations affecting lncRNA expression via mixed and unmixed effects, which led to specific functions in pan-cancer. Survival analysis indicated that MutLncs and co-occurrence pairs can potentially serve as cancer biomarkers. Clarification of the specific roles of MutLncs in human cancers could be beneficial for understanding the molecular pathogenesis of different cancer types and developing the appropriate treatments.
Keywords: long noncoding RNAs, somatic mutation, pan-cancer, co-occurrence, mixed and unmixed effects, microRNA, methylation, gene
Introduction
Long noncoding RNAs (lncRNAs) are a large and diverse class of RNAs that do not code for proteins and are pervasively transcribed in the human genome.1 Accumulating evidence has demonstrated essential functions of lncRNAs in several biological processes, such as post-transcriptional regulation, cell differentiation, and chromatin modification.2, 3, 4, 5 Notably, lncRNAs have been implicated in the development and progression of numerous human diseases, including cancer.6, 7 Despite significant advancements in understanding lncRNA expression patterns and functions, knowledge of their involvement in the molecular mechanisms underlying cancer remains limited—in particular, the potential pathogenic mechanisms triggered by lncRNA-related somatic mutations.8, 9
Somatic mutations, considered genomic variation phenomena, directly or indirectly alter gene expression, protein activities, and signaling pathways.10 Previous studies have reported that somatic mutations affect cancer-related protein coding genes through diverse molecular mechanisms and ultimately contribute to cancer progression.11 However, little is known about the roles of lncRNAs affected by somatic mutations (designated MutLncs) in cancer. MutLncs may represent a novel type of functional molecule with potential utility as biomarkers for cancer diagnostics and treatment. Recent studies have revealed a comprehensive landscape of somatic mutations that affect the expression patterns of various genes to trigger different human cancers (pan-cancer).12, 13, 14, 15 Pan-cancer provides a comparative analysis of the genomic and cellular alterations across diverse tumor types and may be effectively applied to investigate MutLncs in cancer.16
Recent technical advances in large-scale sequencing and genomics methods have provided opportunities to understand tumor-associated somatic mutations and their complexity across the major cancer types. For example, The Cancer Genome Atlas (TCGA) project has generated genomic and transcriptomic data from multiple cancer types, facilitating systematic characterization of somatic mutations.17 The Atlas of Noncoding RNAs in Cancer (TANRIC) characterizes the expression profiles of lncRNAs in large patient cohorts for 20 cancer types, including TCGA and independent datasets (>8,000 samples overall), providing large-scale lncRNA expression data in pan-cancer.14, 18 Integrating these large-scale datasets can provide opportunities to explore the associations between somatic mutations and lncRNA expression. To date, no large-scale or systematic analyses have focused on identifying MutLncs and their specific roles in human pan-cancer.
In this study, we systematically characterized the associations between somatic mutations and lncRNA expression across 17 cancer types by integrating TCGA somatic mutation and TANRIC lncRNA expression data. Common characteristics of MutLncs across different cancer types were observed. Mutational co-occurrence analyses disclosed a combination effect of mutations in pan-cancer. Investigation of pan-cancer MuLncs revealed several conserved and cancer-specific functions. We further explored the means by which somatic mutations affect lncRNA expression (i.e., via mixed and unmixed effect processes) in pan-cancer by integrating TCGA gene expression, microRNA (miRNA) expression, methylation, transcription factor (TF)-lncRNA interaction, and miRNA-lncRNA interaction data. Assessment of the correlations between individual or co-occurring pairs of MutLncs and survival supported the potential utility of MutLncs as cancer-specific biomarkers. Thus, comprehensive evaluation of MutLncs could provide novel insights into the roles of lncRNAs in diverse cancers. An online resource to store and retrieve all MutLnc data from different cancer types (available at http://bio-bigdata.hrbmu.edu.cn/MutLncDR/) provides additional useful information that should facilitate analyses of MutLnc functions.
Results
Global MutLnc Landscapes in Human Cancers
We generated an integrative pipeline to identity MutLncs and their effect process across various cancer types. First, the associations between somatic mutations and lncRNA expression were systematically analyzed in 17 cancer types by integrating TCGA somatic mutation and TANRIC lncRNA expression data (see Materials and Methods). Consequently, we identified a minimum of 5 and a maximum of 581 MutLncs in diverse cancers (Figure S1A; Table S1). Next, we considered whether methylations, genes, TFs, and miRNAs participate in the mechanisms by which somatic mutations affect lncRNA expression. MutLncs were classified into mixed and unmixed effect groups. Mixed MutLncs included methylation-related, gene-related, TF-related, and miRNA-related MutLncs. Notably, the majority of MutLncs were categorized as unmixed (82%) in most of the cancer types examined. However, significantly more mixed effects were observed for some cancer types. For example, methylation-related MutLncs constituted a high proportion of MutLncs in glioblastoma multiforme (GBM) and TF-related MutLncs in uterine corpus endometrial carcinoma (UCEC) (Figure S1B).
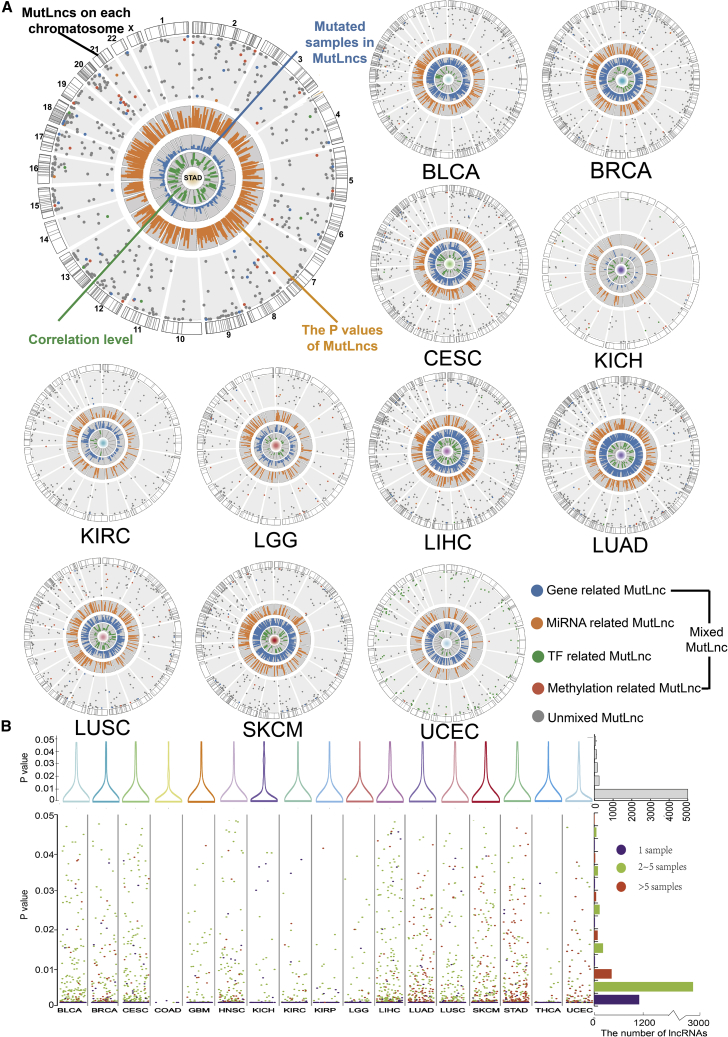
We constructed circular maps to obtain a global overview of the basic information, effect process, and underlying mechanisms for each MutLnc across cancer types (Figure 1A). Other information, including the p values of MutLncs, samples with mutations, and correlation levels, was included in the map. We further provided an outline of p values and mutated sample number distributions. The number of MutLncs was reduced with higher p values. The majority of p values were concentrated between 0 and 0.01, indicating that the associations identified were reliable (Figure 1B). For all MutLncs, 2 to 5 samples with the mutations were predominantly detected. For example, in the region of 0 to 0.01, 1,503 MutLncs were identified in a single sample, 2,887 MutLncs in 2–5 samples, and >606 MutLncs in over 5 samples across the 17 cancer types. Low-frequency mutations also showed significant p values, indicating no intrinsic bias of our method to infer mutation-correlated lncRNA expression from mutational frequency data.
Figure 1.
The Global Landscape of MutLncs in Human Cancers
(A) Global map of MutLncs affected by mixed and unmixed effected processes across cancer types (magnified image of the map for STAD). The gray bands in the outer circle of the map represent MutLncs on each chromosome. The dots in the map represent MutLncs with positions of −log10 (p values), and different colors of dots signify different effect processes. Bar plots in the inner circles of the map represent the distribution of p values (dark orange), numbers of mutated samples (dark blue), and correlation level (dark green). (B) The t test p value distribution of MutLncs across cancer types is shown at the top using vioplots. MutLncs mutated in 1 sample, 2–5 samples, and more than 5 samples are designated in purple, green, and red, respectively. The summary of data for all lncRNAs is indicated on the right.
Common Characteristics of MutLncs across Cancer Types
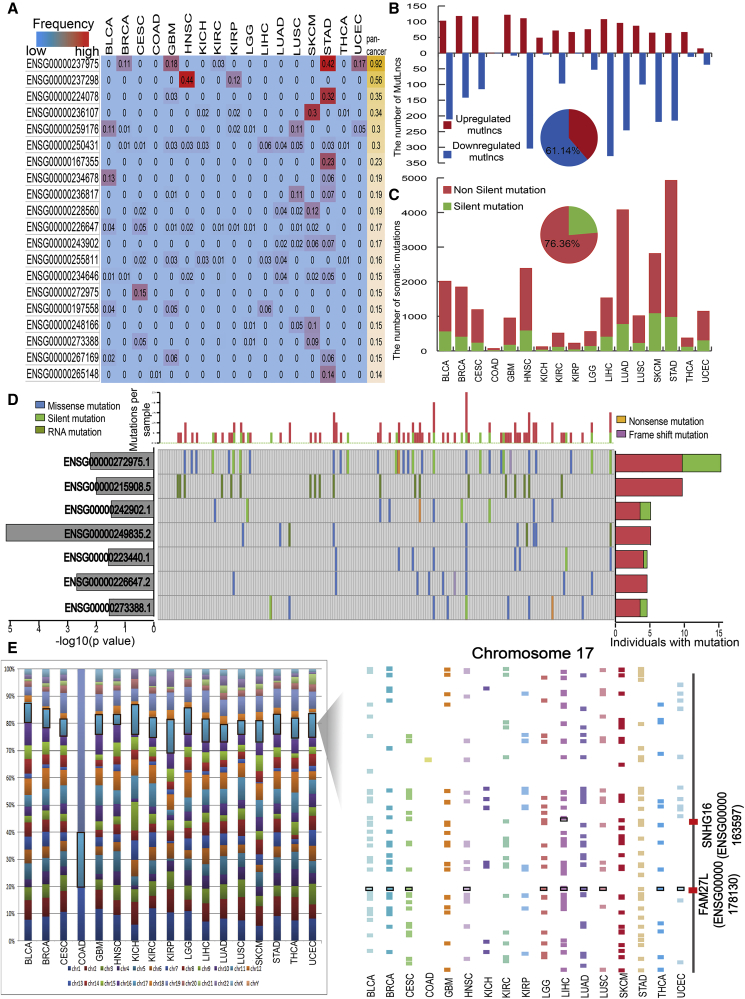
We characterized MutLnc features from multiple perspectives across cancer types. First, we analyzed the percentages of mutated samples in each tumor type (Figure 2A). Mutational frequencies tended to be low for the majority of MutLncs in each cancer type. These findings are concordant with previous studies showing that somatic mutation is a low-frequency alteration type.19 Mutational frequencies of MutLncs were cancer type-specific to a certain extent. For instance, the MutLnc FLG-AS1 (Ensembl: ENSG00000237975) was mutated in 42% samples in stomach adenocarcinoma (STAD) and 18% samples in GBM, but no samples for some other cancer types. Another example, TTN-AS1 (Ensembl: ENSG00000237298), displayed 44% mutational frequency in head-neck squamous cell carcinoma (HNSC), which was the highest for all MutLncs across all the cancer types examined. Notably, however, TTN-AS1 was not mutated in other cancer types, except kidney renal papillary cell carcinoma (KIRP) (mutational frequency = 0.12). A previous study reported dysregulation of TTN-AS1 in nasopharyngeal nonkeratinizing carcinoma but did not discuss the underlying reasons. Results from our analysis provide an explanation at the genome alteration level.20 TTN-AS1, a lncRNA located on the antisense strand of the TTN gene, has been widely studied in muscle contractile machinery, chromosomes, and oncogenes.21, 22 Although TTN is considered a cancer gene based on mathematical predictions, no direct biological evidence has been obtained to explain its role in cancer. Characterization of MutLnc TTN-AS1 may therefore provide novel insights into the specific roles of TTN in cancer.
Figure 2.
Common Characteristics of MutLncs across Cancer Types
(A) Mutational frequency of part MutLncs in individual cancer types and pan-cancer are shown. MutLncs with higher mutation frequencies are colored red, while those with lower mutation frequencies are blue. (B) Bar chart showing the number of upregulated (red) and downregulated (blue) MutLncs across cancer types. (C) Bar chart showing the number of non-silent (red) and silent (green) MutLncs across cancer types. (D) MutLncs are listed vertically by mutation frequency in CESC. The colored bars in the map represent different mutations of lncRNAs occurring in a specific sample. Percentages of samples with mutations are specified on the right. Silent and non-silent mutations are colored green and red, respectively. The p values of MutLncs are shown on the left in a bar plot. Samples are presented as columns with the overall number of mutations plotted at the top, and the mutations classified as silent and non-silent clusters. (E) Distribution of MutLncs on different chromosomes for each cancer type (schematic of the locations of MutLncs within chromosome 17 across diverse cancer types). Two examples of MutLncs are shown on the right-hand column, denoted by a red line.
Next, we classified MutLncs into upregulated and downregulated groups based on fold change values (fold change values >2 and <0.05, respectively) (Figure 2B; Table S2). We identified more downregulated (61.14%) than upregulated MutLncs, but with distinct patterns across different cancer types. For example, >75% and 73% MutLncs were downregulated in liver hepatocellular carcinoma (LIHC) and HNSC, while >94% MutLncs were upregulated in GBM. Thus, the expression patterns of MutLncs appear dependent on the cancer type (Figure 2B).
We further considered whether the somatic mutation frequency and type lead to different features of MutLnc expression (Figures 2C and 2D). The numbers of non-silent mutations were greater than silent mutations for each cancer type, with around 76% MutLncs being non-silent (Figure 2C). We have provided a list of significant MutLncs with different somatic mutational frequencies and types in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), including CROCCP2 (Ensembl: ENSG00000215908), RP11-309L24.2 (Ensembl: ENSG00000242902), and VCAN-AS1 (Ensembl: ENSG00000249835). Expression patterns of these lncRNAs were affected by mutations (Figure 2D). MYHAS (Ensembl: ENSG00000272975.1) was mutated in 15.3% samples and VCAN-AS1 only in 5.1% samples. However, expression of the latter MutLnc was more significantly correlated to mutations, as confirmed in other examples (Figure S2A). Moreover, a higher number of samples carried non-silent mutations than silent mutations for 7 MutLncs. We propose that the non-silent mutation may exert a more significant effect on lncRNA expression. For example, over half of the observed mutations of Ensembl: ENSG00000272975.1 were non-silent, and expression of non-silent MutLncs in samples was lower than that in samples with silent mutations. This finding is consistent with the theory that non-silent mutations induce higher-level genomic alterations (Figure S2B).
Finally, we observed that MutLncs in different cancer types show similar chromosome distribution (Figure 2E) and are dispersed throughout multiple chromosomes. Overall, chromosomes 17 and 1 were more highly enriched in MutLncs. Previous studies have reported a correlation between chromosome 17 and cancer.23 Accordingly, we further analyzed the genomic distribution of MutLncs on chromosome 17 (Figure 2E). MutLncs were dispersed along the chromosome 17 region in different cancer types, with some located on the centromere. Mutations in the chromosome 17 centromere region have been linked to cancer.24 For example, FAM27L, located near the chromosome 17 centromere, has been identified as a MutLnc in 10 cancer types including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), CESC, HNSC, brain lower grade glioma (LGG), LIHC, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), thyroid carcinoma (THCA), and UCEC. FAM27L may play a role in the malignant transformation and/or metastasis of collateral tumors.25 We hypothesize that the somatic mutation affects FAM27L expression, in turn, leading to functional changes that trigger cancer development.
Co-occurrence Analysis Highlights Particular MutLnc Combined Patterns in Pan-Cancer
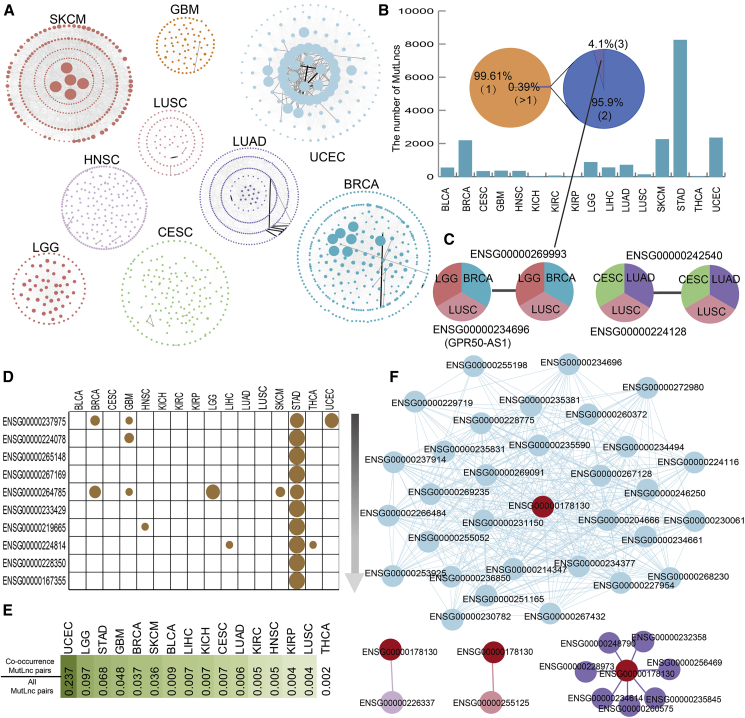
We constructed a co-occurrence combined effect network across cancer types to further determine the ways in which MutLncs contribute to cancer (Figure 3A). Topological analysis indicated that the degree of networks follows a scale-free distribution in most cancer types (Figure S3A).
Figure 3.
Co-occurrence Patterns of MutLncs in Different Cancer Types
(A) Co-occurrence network of MutLncs across the cancers examined. The node in the network represents a MutLnc, and the edge signifies two MutLncs with significant co-occurrence. Different colors represent different cancer types. The node size indicates the degree of the node in the network, and the thickness of the edge represents the p value of the MutLnc pairs. (B) Number of co-occurring MutLnc pairs across cancer types. A pie chart shows the proportion of MutLnc pairs co-occurring in different cancers. (C) Two co-occurrence MutLnc pairs appearing in three cancer types are shown. (D) The highest degree nodes in the co-occurrence network. (E) Ratio between co-occurrence MutLncs and all MutLnc pairs. (F) The MutLnc FAM27L and co-occurrence pairs in LUSC, LUAD, HNSC, and UCEC.
We identified MutLnc co-occurrence pairs in which two MutLncs are significantly mutated in the same samples via Fisher’s exact test (p < 0.01). Pairwise co-occurrence analysis for all cancer types (except colon adenocarcinoma [COAD] because of quantitative restriction) disclosed 18,803 co-occurring MutLnc pairs, from 2 in kidney chromophobe (KICH) to 8,240 in STAD (Figure 3B). Further evaluation of the specific and common features of these co-occurrence pairs showed that only 0.39% of pairs appeared over one cancer type, which were classified as “common.” Overall, 95.9% and 4.1% common co-occurrence MutLnc pairs appeared in two and three cancer types, respectively (Figure 3B). Frequency analysis of MutLnc in co-occurring MutLnc pairs showed a range of frequencies, from 3 in KICH to 461 in STAD. A relatively low number of MutLncs displayed co-occurrence, and most MutLncs in co-occurring pairs were dysregulated in multiple cancer types (Figure S3B). We also detected two co-occurrence pairs in three cancer types. One is GPR50, which has been identified as a melatonin-related receptor related to cancer26 (Figure 3C). LUSC and LUAD, two similar cancer types, also shared a common co-occurrence pair. Co-occurrence networks in some cancer types, such as STAD and UCEC, involved a large degree of MutLncs and appeared to have a compact structure. In contrast, networks of other cancers, such as CESC, contained a low degree of MutLncs (Figure 3D), signifying differences in the MutLnc combined modes in diverse cancers. Next, we calculated the ratio between a specific co-occurring MutLnc pair and all MutLnc pairs, representing the co-occurrence pattern level in each cancer type. The highest ratio was obtained for UCEC, indicating high complexity of the MutLnc network in this cancer type (Figure 3E). In particular, FAM27L, which is dysregulated in ten cancer types, showed co-occurrence patterns in LUSC, LUAD, HNSC, and UCEC (Figure 3F). However, the partners of FAM27L in diverse cancer types were distinct and type-specific. It indicated that the same MutLnc can play its role by combining with different MutLncs in diverse cancer type. We further observed that most MutLncs only exert a combined effect with particular MutLncs in different cancer types. For example, MutLnc AC108025.2 (Ensembl: ENSG00000230090.1) displayed co-occurrence with AC005550.5 (Ensembl: ENSG00000225974.1) in kidney renal clear cell carcinoma (KIRC), but occurred alone in CESC. MutLncs in other cancer types, such as BRCA, LGG, and GBM, showed similar patterns (Figure S3C), supporting the theory that cancer is a heterogeneous disease.
Pan-Cancer Investigation of MutLncs Reveals Several Conserved and Cancer-Specific Functions
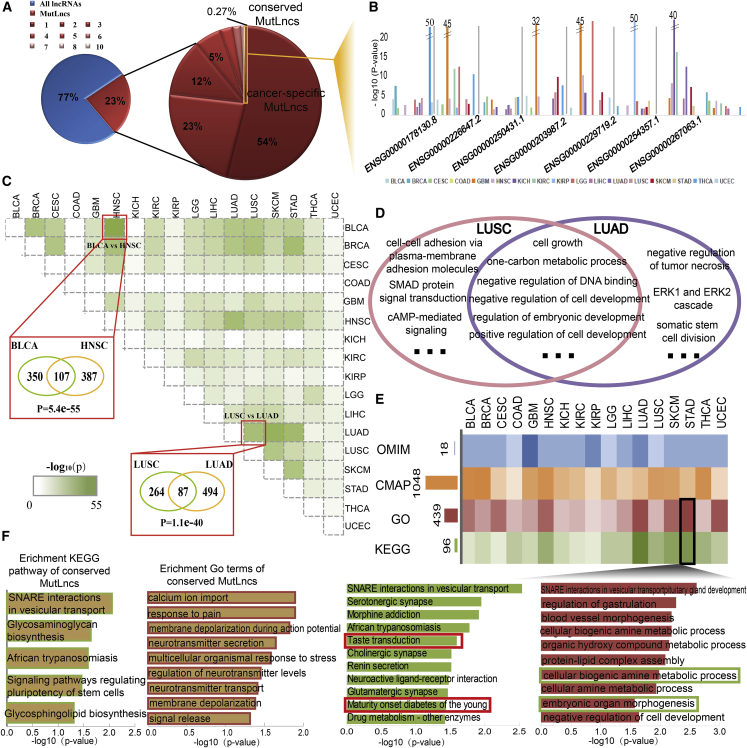
Although MutLncs shared several common characteristics, analysis of the MutLncs across cancer types highlighted both common and specific features among cancers. Among all of the lncRNAs examined, 23% were identified as MutLncs. We found that ∼54% of MutLncs occurred only in one cancer and only 0.27% of MutLncs were dysregulated in more than eight cancers (Figure 4A). This group of MutLncs, categorized as “common”, displayed differential dysregulation in multiple cancer types (Figure 4B). Although most MutLncs were cancer-specific, subtypes with similar tissue-of-origin shared common MutLncs. MutLncs in each cancer type varied from other cancer MutLncs, and our results revealed both known and new relationships among these cancers (Figure 4C). Among the two subtypes of lung cancers, LUAD and LUSC, the similarity of MutLncs was higher. Approximately 33% of MutLncs in LUSC also occurred in LUAD, which showed significance with Fisher’s test (p < 1.1e−40). However, BLCA and HNSC, two cancers with no obvious tissue-of-origin relationship, shared the most common MutLncs. We further tested the MutLncs in these two cancers for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment to explain this phenomenon. The MutLncs examined were highly enriched for basic cellular processes related to multiple cancer types (Figure S4A). We further examined the similarities between LUSC and LUAD via functional annotation of the respective MutLncs. The results indicate that the two cancer types share common functions, such as cell growth and positive regulation of cell development. Some particular functions were also cancer subtype-specific. For example, MutLncs in LUAD were enriched in the ERK1 and ERK2 cascades, which may show different mechanisms between cancer subtypes (Figure 4D).
Figure 4.
Conserved and Cancer-Specific Functions of MutLncs in Pan-Cancer Analysis
(A) The pie chart shows the proportion of MutLncs in different cancer types. The majority of MutLncs are cancer-specific. (B) Distribution of MutLncs identified in more than 7 cancer types. The bar plot represents the p value of MutLncs, and each cancer type is shown as a different color. (C) The matrix shows similarities between pairs of cancer types. The darker green color represents greater similarity between two cancers. Cancer pairs with the same origin and highest similarity are shown. (D) GO terms of MutLncs in LUSC and LUAD. (E) Heatmap showing all enriched KEGG pathways, GO terms, CMAP drugs, and OMIM diseases of MutLncs across cancer types. The darker color represents greater enrichment of MutLncs in these functions. (F) KEGG pathways and GO terms enriched for conserved MutLncs in all cancer types ranked by −log10(P) are presented as bar plots. GO terms and KEGG pathways enriched for MutLncs in STAD ranked by −log10(P) are presented as bar plots.
We further explored both common and specific features of MutLncs across the cancer types by performing functional enrichment analysis. Additionally, co-expression between MutLncs and their neighbor coding genes was analyzed to confirm the exactitude of the functional annotation, which showed strong correlations for the majority of the relationships (Figure S4B). Most MutLncs were identified in relation to known Gene Ontology (GO) terms, KEGG pathways, Connectivity Map (CMAP) drugs, and Online Mendelian Inheritance in Man (OMIM) diseases across cancer types (Figure 4E). All conserved MutLncs in the 17 cancer types were enriched in a number of pathways and GO terms related to cancer-like signaling regulating the pluripotency of stem cells (Figure 4F). However, distinct biological processes were captured for different cancer types. For example, MutLncs in STAD were enriched in specific pathways and GO terms related to stomach function, such as “taste transduction” and “maturity onset diabetes of the young” pathways. We additionally detected common functions of MutLncs in different cancer types, such as synaptic transmission, shown to be related to cancer-associated pain and metastasis27, 28 (Figure S4C).
Somatic Mutations Exert Mixed or Unmixed Effects on lncRNAs in Each Cancer Type
Somatic mutations are usually considered the initiator of cancer by altering genetic and epigenetic mechanisms, which in turn influence lncRNA expression through various effects.29, 30 Here we considered two major processes through which somatic mutations impact lncRNA expression: (1) unmixed, in which the somatic mutation affects lncRNA expression without the participation of other confounding factors; and (2) mixed, in which the mutations have an impact on lncRNA expression and other factors, including methylation, gene expression, miRNA expression, and TF expression simultaneously. The somatic mutations may first affect methylation, gene expression, miRNA expression, and TF expression, then these factors further impact lncRNA expression (see Materials and Methods).
In different cancer types, MutLncs displayed similar patterns, the majority of which were unmixed. However, some cancers, such as UCEC, had different patterns, with the TFs group exerting a major effect. Overall, 82% MutLncs were classified as unmixed and 18% as mixed (including 8% methylation-, 1% miRNA-, 5% gene-, and 4% TF-related MutLncs). We additionally validated the correlation intensity distribution between MutLnc expression and methylation, miRNA, gene, or TF expression to evaluate the accuracy of the process, which revealed strong correlations (Figures S5A–S5D).
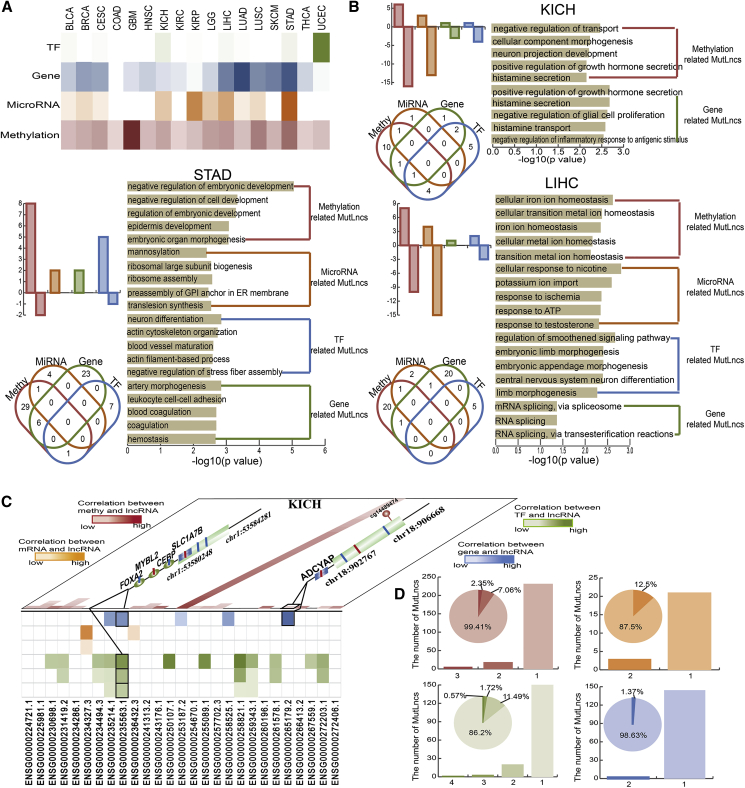
Some cancer types, such as STAD, contained numerous mixed MutLncs, indicative of the complexity at the MutLnc level (Figure 5A). Three major cancer types, including LIHC, KICH, and STAD, with more than two MutLncs in the mixed effect group were analyzed. We found no obvious tendencies or fixed patterns of upregulation or downregulation of different effect groups of MutLncs. For example, 16 downregulated and 6 upregulated methylation-related MutLncs were identified in STAD, while 2 downregulated and 8 upregulated methylation-related MutLncs were observed in KICH (Figure 5B). Moreover, some MutLncs were associated with two mixed effects and showed more complex effect mechanisms in three cancer types. We additionally performed gene ontology analysis on all MutLncs across the cancer types to explore the distinct roles of TF-, gene-, methylation-, and miRNA-related MutLncs (Figure S5E). In some cancers, such as STAD, most methylation-related MutLncs were associated with embryonic development and miRNA-related MutLncs with the ribosome, as reported in previous studies31, 32 (Figure 5B).
Figure 5.
Somatic Mutations with Mixed and Unmixed Effects on lncRNA Expression in Pan-Cancer
(A) Heatmap of all unmixed type MutLncs in each cancer type. The darker color indicates a greater number of MutLncs in this group. (B) Bar plots represent the number of upregulated and downregulated MutLncs in KICH, LIHC, and STAD. The Venn diagrams show the MutLncs shared among the methylation, gene, TF, and microRNA effect process. GO terms enriched for different types of MutLncs ranked by −log10(P) are shown as bar plots. Methylation-, microRNA-, TF-, and gene-related MutLncs are presented in red, orange, blue, and green, respectively. (C) Two MutLncs simultaneously affected in different mixed effect groups in KICH. (D) Bar plots showing the number of cancer types containing MutLncs. Red, orange, green, and blue represent methylation, microRNA, TF, and gene-related mixed MutLncs, respectively.
Furthermore, simultaneous mixed effects on the same MutLnc were observed, and 38% of MutLncs were affected via a mixed effect mechanism, with methylation-related MutLncs having a major status in KICH (Figure S5F). The MutLnc RP11-334A14.8 (Ensembl: ENSG00000235563.1) was affected by three TFs (FOXA2, MYBL2, and CEBP) and one gene (SLC1A). MutLnc RP11-672L10.2 (Ensembl: ENSG00000265179) was linked to one gene, ADCYAP1B, and one methylation event, cg14489474 (Figure 5C). All of the above TFs, genes, and methylation expression were affected by mutations, which further affected lncRNA expression. Overall, these results reflect the interdependency of multiple layers of variation and complex biological processes in determining lncRNA expression levels. Although the processes through which somatic mutations exert an impact on lncRNA expression are complex, we can consider the mutation the anchor that plays a leading role in MutLnc-associated processes. Our data further showed that most methylation-, miRNA-, gene-, and TF-related MutLncs are cancer-specific and that a number of TF-related MutLncs are dysregulated in four cancer types (Figure 5D).
Cancer-Specific MutLncs Contribute to Prognosis in Human Cancers
We further examined whether the MutLncs or co-occurrence pairs are correlated with cancer survival and, further, whether the mutation status could aid in distinguishing the two groups of patients (see Materials and Methods). It would be helpful to ascertain the potential of MutLncs as prognostic biomarkers with clinical implications.
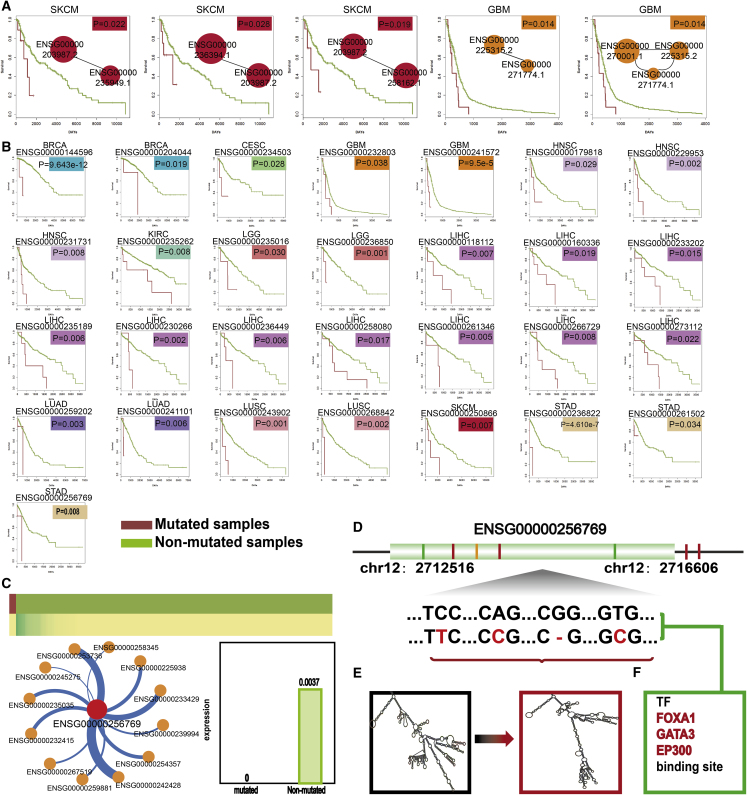
First, we identified some co-occurrence pairs in SKCM and GBM that were related with survival. These findings support the importance of MutLnc combined events in tumorigenesis and their prognostic value in clinical practice (Figure 6A). Moreover, 29 MutLncs were correlated with survival, and mutated samples were significantly associated with decreased survival in 14 types of cancer (Figure 6B). For example, the MutLnc CACNA1C-AS3 (Ensembl: ENSG00000256769.1), with 7 mutated samples, was significantly related to survival (p = 0.00796), with shorter survival in mutated samples in STAD (Figure 6B). Mutational features of the lncRNA CACNA1C-AS3 were additionally analyzed. As a result, we identified 71% non-silent mutations and 42.9% variant sites with a C to T conversion (Figures S6A and S6B). We further hypothesized that the mutation affects lncRNA expression via three major mechanisms. Take CACNA1C-AS3, for instance. First, the MutLnc CACNA1C-AS3 co-occurred with 12 MutLncs, such as HOTAIRM1 (Ensembl: ENSG00000233429), which is located between HOXA1 and HOXA2 genes (Figure 6C). Second, prediction of the minimum free energy (MFE) changes caused by mutations in MutLnc CACNA1C-AS3 revealed significant effects on predicted lncRNA secondary structures, which, in turn, impacted lncRNA expression33 (Figures 6D and 6E). Third, the mutations located on the TF FOXA1, GATA3, and EP300 binding sites may affect the binding of TF to alter lncRNA expression (Figure 6F). Previous studies have consistently demonstrated that mutations in TF binding sites lead to lncRNA expression.34, 35
Figure 6.
MutLncs as Specific Biomarkers of Cancer
(A) Survival analysis of co-occurring MutLnc pairs: Kaplan-Meier survival analysis of two groups of patients with mutation (red line) and without mutation (green line). Survival days are shown along the x axis. Overall survival rates are shown along the y axis. (B) Kaplan-Meier survival analysis of a single MutLnc. (C) MutLnc CACNA1C-AS3 (Ensembl: ENSG00000256769) as an example. Expression profiles of CACNA1C-AS3 in STAD are shown in the right panel. High expression values are depicted in green and low expression in yellow. Co-occurrence status and expression bar plots of CACNA1C-AS3 are shown. (D) Mutation sites and types of CACNA1C-AS3. (E) Secondary structure alterations induced by the mutations. (F) Changes in TF binding sites induced by the mutations.
Discussion
In this study, we systematically examined the associations between somatic mutations and lncRNA expression across 17 cancer types. Global MutLnc landscapes were constructed to determine the features and roles of MutLncs in multiple cancer types. Co-occurrence analysis highlights particular MutLnc combined patterns in pan-cancer. One MutLnc can combine with different MutLncs in diverse cancers. Comparative analyses showed that only a small proportion of MutLncs are conserved and that they vary greatly among diverse cancer types. In addition, cancers with similar tissues of origin displayed higher MutLnc similarities. Somatic mutations exert mixed or unmixed effects on lncRNAs in each cancer type. The strong correlations between MutLncs and survival support their potential as specific cancer biomarkers. Moreover, MutLnc co-occurrence pairs might be effectively applied as new possible prognostic biomarkers for particular cancers.
Cancers are clonal proliferation disorders that arise owing to mutations that confer a selective growth advantage to cells. A number of studies have confirmed the somatic mutations were an essentiality of carcinogenesis and cancer development. The majority of work to date has focused on the mutations impacting gene expression, with limited documentation of indications that mutations in the non-coding region are also important.36 In addition, accumulating studies have demonstrated the significance of lncRNAs in cancer, including roles as drivers of tumor-suppressive and oncogenic functions, microRNA competitors, and diagnostic biomarkers.37, 38, 39, 40 In the current study, we comprehensively evaluated the properties of MutLncs from different cancer types in an attempt to obtain novel insights into genome alterations at the lncRNA level in cancer.
We successfully determined that somatic mutations impact lncRNA expression profiles in a pan-cancer analysis. Notably, our method could be used to identify MutLncs with both high and low mutation frequencies. The identified associations were accurate and unbiased, since most lncRNAs were tested regardless of prior association with cancer, previously curated pathways, or interaction databases. Our data may facilitate the elucidation of novel correlations that add to the emerging blueprint of cancer in general. Strict distance limits (5 kb from lncRNAs), large sample numbers, and permutation tests were additionally employed to ensure the reliability of associations. Multidimensional genomics data provide more extensive insights into the mutations affecting lncRNA functions and related pathways. Cancer primarily develops due to somatic alterations in the genome. Thus, investigation and interpretation of lncRNA mutations should provide novel and useful insights into the mechanisms underlying the functions of these molecules in cancer. Further work could focus on these MutLncs and their complex regulatory patterns and reveal novel mechanism underlying carcinogenesis and cancer treatment.
Our study identified some MutLncs in diverse cancer types, and we inferred the possible mechanism of MutLncs in cancer development and drug resistance. We found that numerous somatic mutations were located on lncRNA TF binding site (TFBS) regions in multiple cancer types (Figure S7A). These mutations accounted for a large proportion of all of the MutLncs (Figure S7B). MutLncs such as ESR1, TRPS1, ERG, and RUNX1 might interfere with some important functions of TFs (Figure S7C). These TFs all played essential roles in cancer development and progression. For example, we found that some somatic mutations were located on lncRNA SNHG16, and these mutations caused the function loss of TFBSs for TF ESR1 in breast cancer. Somatic mutation in the lncRNA SNHG16 lost binding sites for TF ESR1, which promoted low expression of lncRNA SNHG16. Low expression of lncRNA SNHG16 promoted the overexpression of oncogene E2F5, resulting in breast cancer development and progression (Figure S8A). Similarly, somatic mutation in the lncRNA HOTAIR lost binding sites for TF CTCF, thus promoting low expression of lncRNA HOTAIR. Low expression of lncRNA HOTAIR promoted the overexpression of oncogene Akt and resulted in the drug resistance of Calycosin and genistein in breast cancer41 (Figure S8B). This finding could provide a novel insight for exploring the role of MutLncs in drug resistance for cancer.
In summary, the MutLnc profiles provide a global overview of the dysregulated lncRNAs affected by somatic mutations across different cancer types. Our findings expand the existing knowledge about lncRNA characteristics in relation to cancer. Integrating mutational and lncRNA expression data from tumor samples enhances the interpretation capacity of the mutations identified, which may facilitate optimal selectivity of targets for functional studies and the development of novel cancer therapeutics.
Materials and Methods
Sources and Scope of Cancer Data
We obtained lncRNA expression data on 17 cancer types from TANRIC.18 Somatic mutation, methylation, gene expression and miRNA expression data on these 17 cancers were acquired from the TCGA Pan-Cancer project. Cancer samples with clinical follow-up information were retained for further analysis. The cancer name abbreviations of TCGA and number of cancer samples are listed in Table S3. The platforms of somatic mutations across the different cancer types are listed in Table S4.
Human Gene and lncRNA Annotation Data
GENCODE (Release 19) annotation files, including comprehensive gene and lncRNA annotations in a GTF format, were used for mapping the mutations upstream and downstream of specific lncRNAs and genes. Annotation information on methylation and miRNAs in TCGA was used for mapping upstream and downstream mutations.
TF-lncRNA and miRNA-lncRNA Interaction Data
To ensure high network interaction quality, we obtained high-throughput experimentally verified TF-lncRNA interaction data by collecting lncRNAs from five databases (Ensemble, NONCODEv4, LNCipedia, LncRBase, and GENCODE) and identified their verified TFBSs via chromatin immunoprecipitation sequencing (ChIP-seq) from the University of California, Santa Cruz (UCSC) Genome Browser.42, 43, 44, 45, 46, 47 We defined the sequence 5 kb upstream to 1 kb downstream of the transcription start site (TSS) of each lncRNA as its promoter region based on a previous study.48 Our miRNA-lncRNA interaction data were downloaded from starBase v2.0, which provides comprehensive crosslinking immunoprecipitation sequencing (CLIP-seq) experimentally supported miRNA-lncRNA interactions.49
Comprehensive Mutation Profiles of lncRNAs
Mutation profiles for lncRNAs across the cancer types were constructed. First, the somatic mutations, which are annotated as confirmed single nucleotide polymorphisms (SNPs), would be screened out. Then, with the BEDtools, we mapped all somatic mutations between 5 kb upstream of the lncRNA TSSs and 5 kb downstream of lncRNA transcription termination sites (TTS). Next, we constructed mutation profiles for genes, methylations, TFs, and miRNAs following the above benchmark method. We further constructed methylation and gene profiles for lncRNAs by mapping methylation sites and genes to ±10 kb from the lncRNA TSSs. Finally, TF and miRNA profiles for lncRNAs were generated using experimentally verified TF-lncRNA and miRNA-lncRNA interactions. Thus, for individual MutLncs, we obtained information on the corresponding mutations, methylation sites, gene, TF, and miRNA profiles for use in subsequent analyses.
Identification of MutLncs in Pan-Cancer Based on Somatic Mutations and lncRNA Expression Patterns
An integrative pipeline to detect MutLncs in different cancer types was developed by integrating somatic mutation and lncRNA expression data (Figure S9). To this end, we first built two matrices (expression and mutation) presented as lncRNA (row) by sample (column). The elements in the expression matrix were the true values of lncRNA expression, and the mutation matrix was binary: 1 (true) if mutation occurs in a particular lncRNA in a particular sample or 0 (false). The t test was used to identify lncRNAs that were differentially expressed between samples with and without somatic mutations. The one-sample t test was employed to identify MutLncs with only one mutation. Finally, 1,000 random permutations of the samples with mutations were generated, and MutLncs were obtained based on permutation p values (p < 0.05). The t test p values were retained for subsequent analyses.
Mixed and Unmixed Effects of Somatic Mutations on lncRNA Expression in Pan-Cancer
As reported widely, cancer samples frequently acquire genetic and epigenetic alterations that influence lncRNA expression through diverse mechanisms.50 Accordingly, we assessed the involvement of other potential confounding factors, including methylation, specific genes, TFs, and miRNAs (Figure S10A). MutLncs were defined as mixed if somatic mutations simultaneously influenced methylation, gene, TF, miRNA, and lncRNA expression patterns. In the mixed effects group, the somatic mutation first affected methylation, gene, TF, or miRNA expression patterns, which further influenced lncRNA expression. For identification of such MutLncs, we initially determined their interactions with each methylation site, gene, TF, and miRNA from the above profiles. Next, we identified the differential patterns of each methylation site, gene, TF, and miRNA via t test between the samples with and without mutations. Significantly different patterns were selected as candidate interactions. Third, we calculated Pearson correlation coefficients (PCCs) for each candidate interaction pair to ensure concordant changes in methylation, gene, TF, miRNA, and lncRNA expression levels. The process was defined as mixed based on the following criteria: corr (lncRNA, methylation) < −0.3; corr (lncRNA, gene) > 0.75; corr (lncRNA, TF) > 0.75 and corr (lncRNA, miRNA) < −0.25, where (lncRNA, methylation), (lncRNA, gene), corr (lncRNA, TF) and corr (lncRNA, miRNA) represent the Pearson correlation coefficients of lncRNA-methylation, lncRNA-gene, lncRNA-TF, and lncRNA-miRNA interactions, respectively, based on expression values. The process was considered unmixed when mutations affected lncRNA expression alone, independently of the other factors, which did not meet the above criteria.
Identification of Co-occurring MutLnc Pairs
Two-by-two contingency tables were constructed for every pairwise MutLncs vector to determine significant (p < 0.01, Fisher’s exact test) mutational co-occurrence. Coincident pairwise mutation in at least three samples was additionally required for classification as significant mutational co-occurrence. For each pair of MutLncs, we quantified the number of samples with (1) common mutations, (2) the first MutLnc mutation only, (3) the second MutLnc mutation only, and (4) mutations other than the pair examined. These four values were used to calculate the odds ratio using Fisher’s exact test (Figure S10B).
Pan-Cancer Analysis of MutLncs
We additionally employed Fisher’s exact test to analyze specific and common features of MutLncs in pan-cancer as well as cancer similarities of MutLncs. For each cancer pair, we quantified the number of MutLncs (1) in both, (2) only dysregulated in the first cancer, (3) only dysregulated in the second cancer, and (4) dysregulated in other cancers. These four values were used to calculate the odds ratio using Fisher’s exact test (Figure S10C).
Functional Enrichment Analysis
Functional enrichment was performed for MutLncs across cancer types with the Enrichr tool online web server using default parameters.51 We obtained enriched GO terms (p < 0.01), KEGG pathways (p < 0.05), OMIM disease (p < 0.05), and CMAP drugs (p < 0.01).
Survival Analysis
For each MutLnc or co-occurrence pairs across cancers, we classified samples into “mutation” and “non-mutation” groups. Kaplan-Meier survival analysis was performed for the two clustered groups, and statistical significance assessed using the log-rank test. All analyses were performed within the R 2.15.3 framework.
Additional Files
MutLnc data of the 17 cancer types can be accessed from the resource http://bio-bigdata.hrbmu.edu.cn/MutLncDR/.
Author Contributions
Xia Li, S.C., and S.N. conceived and designed the experiments. Y.G., Xin Li, H.Z., and Y.Z. analyzed data. P.W. and Y.W. collected the data. S.S., Y.F., and W.S. validated the method and data. Y.G. and S.N. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31601080 and 61603116) and the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2017071 and UNPYSCT-2017060). The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.08.004.
Contributor Information
Shangwei Ning, Email: ningsw@ems.hrbmu.edu.cn.
Steven Xi Chen, Email: steven.chen@miami.edu.
Xia Li, Email: lixia@hrbmu.edu.cn.
Supplemental Information
References
- 1.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthès P., Kokkinaki M., Nef S., Gnirke A. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 3.Bardeesy N., DePinho R.A. Pancreatic cancer biology and genetics. Nat. Rev. Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 4.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin R., Maeda S., Liu C., Karin M., Edgington T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson I.R., Takahashi K., Futreal P.A., Chin L. Emerging patterns of somatic mutations in cancer. Nat. Rev. Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akavia U.D., Litvin O., Kim J., Sanchez-Garcia F., Kotliar D., Causton H.C., Pochanard P., Mozes E., Garraway L.A., Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jörnsten R., Abenius T., Kling T., Schmidt L., Johansson E., Nordling T.E., Nordlander B., Sander C., Gennemark P., Funa K. Network modeling of the transcriptional effects of copy number aberrations in glioblastoma. Mol. Syst. Biol. 2011;7:486. doi: 10.1038/msb.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding J., McConechy M.K., Horlings H.M., Ha G., Chun Chan F., Funnell T., Mullaly S.C., Reimand J., Bashashati A., Bader G.D. Systematic analysis of somatic mutations impacting gene expression in 12 tumour types. Nat. Commun. 2015;6:8554. doi: 10.1038/ncomms9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masica D.L., Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 2011;71:4550–4561. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B., Wang D., Wu J., Tang J., Chen W., Chen X., Zhang D., Deng Y., Guo M., Wang Y. Expression profiling and functional prediction of long noncoding RNAs in nasopharyngeal nonkeratinizing carcinoma. Discov. Med. 2016;21:239–250. [PubMed] [Google Scholar]
- 21.Machado C., Andrew D.J. D-Titin: a giant protein with dual roles in chromosomes and muscles. J. Cell Biol. 2000;151:639–652. doi: 10.1083/jcb.151.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zastrow M.S., Flaherty D.B., Benian G.M., Wilson K.L. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci. 2006;119:239–249. doi: 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- 23.Wertheim I., Tangir J., Muto M.G., Welch W.R., Berkowitz R.S., Chen W.Y., Mok S.C. Loss of heterozygosity of chromosome 17 in human borderline and invasive epithelial ovarian tumors. Oncogene. 1996;12:2147–2153. [PubMed] [Google Scholar]
- 24.Marchiò C., Lambros M.B., Gugliotta P., Di Cantogno L.V., Botta C., Pasini B., Tan D.S., Mackay A., Fenwick K., Tamber N. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J. Pathol. 2009;219:16–24. doi: 10.1002/path.2574. [DOI] [PubMed] [Google Scholar]
- 25.Kamińska K., Szczylik C., Bielecka Z.F., Bartnik E., Porta C., Lian F., Czarnecka A.M. The role of the cell-cell interactions in cancer progression. J. Cell. Mol. Med. 2015;19:283–296. doi: 10.1111/jcmm.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill S.M., Belancio V.P., Dauchy R.T., Xiang S., Brimer S., Mao L., Hauch A., Lundberg P.W., Summers W., Yuan L. Melatonin: an inhibitor of breast cancer. Endocr. Relat. Cancer. 2015;22:R183–R204. doi: 10.1530/ERC-15-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takasu K., Ogawa K., Nakamura A., Kanbara T., Ono H., Tomii T., Morioka Y., Hasegawa M., Shibasaki M., Mori T. Enhanced GABAergic synaptic transmission at VLPAG neurons and potent modulation by oxycodone in a bone cancer pain model. Br. J. Pharmacol. 2015;172:2148–2164. doi: 10.1111/bph.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai A.C., Xia Y.F., Suzuki E., Kessler M., Civelli O., Nothacker H.P. Cancer metastasis-suppressing peptide metastin upregulates excitatory synaptic transmission in hippocampal dentate granule cells. J. Neurophysiol. 2005;94:3648–3652. doi: 10.1152/jn.00590.2005. [DOI] [PubMed] [Google Scholar]
- 29.Cheng E.C., Lin H. Repressing the repressor: a lincRNA as a MicroRNA sponge in embryonic stem cell self-renewal. Dev. Cell. 2013;25:1–2. doi: 10.1016/j.devcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi H., Ning S., Li X., Li Y., Wu W., Li X. A novel reannotation strategy for dissecting DNA methylation patterns of human long intergenic non-coding RNAs in cancers. Nucleic Acids Res. 2014;42:8258–8270. doi: 10.1093/nar/gku575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Meyenn F., Iurlaro M., Habibi E., Liu N.Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z. Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Mol. Cell. 2016;62:983. doi: 10.1016/j.molcel.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berindan-Neagoe I., Monroig Pdel.C., Pasculli B., Calin G.A. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian E., Børset M., Sawyer J.R., Brede G., Våtsveen T.K., Hov H., Waage A., Barlogie B., Shaughnessy J.D., Jr., Epstein J., Sundan A. Allelic mutations in noncoding genomic sequences construct novel transcription factor binding sites that promote gene overexpression. Genes Chromosomes Cancer. 2015;54:692–701. doi: 10.1002/gcc.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinhold N., Jacobsen A., Schultz N., Sander C., Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredriksson N.J., Ny L., Nilsson J.A., Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik R., Patel L., Prensner J.R., Shi Y., Iyer M.K., Subramaniyan S., Carley A., Niknafs Y.S., Sahu A., Han S. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol. Cancer Res. 2014;12:1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P., Ning S., Zhang Y., Li R., Ye J., Zhao Z., Zhi H., Wang T., Guo Z., Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W.T., Sun Y.M., Huang W., He B., Zhao Y.N., Chen Y.Q. Genome-wide Long Non-coding RNA Analysis Identified Circulating LncRNAs as Novel Non-invasive Diagnostic Biomarkers for Gynecological Disease. Sci. Rep. 2016;6:23343. doi: 10.1038/srep23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J., Lin C., Yong W., Ye Y., Huang Z. Calycosin and genistein induce apoptosis by inactivation of HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells. Cell. Physiol. Biochem. 2015;35:722–728. doi: 10.1159/000369732. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., Chen R. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44(D1):D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakraborty S., Deb A., Maji R.K., Saha S., Ghosh Z. LncRBase: an enriched resource for lncRNA information. PLoS ONE. 2014;9:e108010. doi: 10.1371/journal.pone.0108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44(D1):D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kersey P.J., Lawson D., Birney E., Derwent P.S., Haimel M., Herrero J., Keenan S., Kerhornou A., Koscielny G., Kähäri A. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ning S., Yue M., Wang P., Liu Y., Zhi H., Zhang Y., Zhang J., Gao Y., Guo M., Zhou D. LincSNP 2.0: an updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017;45(D1):D74–D78. doi: 10.1093/nar/gkw945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popadin K., Gutierrez-Arcelus M., Dermitzakis E.T., Antonarakis S.E. Genetic and epigenetic regulation of human lincRNA gene expression. Am. J. Hum. Genet. 2013;93:1015–1026. doi: 10.1016/j.ajhg.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.