Abstract
Protein translocation by the bacterial type VI secretion system (T6SS) is driven by a rapid contraction of a sheath assembled around a tube with associated effectors. Here, we show that TssA‐like or TagA‐like proteins with a conserved N‐terminal domain and varying C‐terminal domains can be grouped into at least three distinct classes based on their role in sheath assembly. The proteins of the first class increase speed and frequency of sheath assembly and form a stable dodecamer at the distal end of a polymerizing sheath. The proteins of the second class localize to the cell membrane and block sheath polymerization upon extension across the cell. This prevents excessive sheath polymerization and bending, which may result in sheath destabilization and detachment from its membrane anchor and thus result in failed secretion. The third class of these proteins localizes to the baseplate and is required for initiation of sheath assembly. Our work shows that while various proteins share a conserved N‐terminal domain, their roles in T6SS biogenesis are fundamentally different.
Keywords: ImpA_N domain proteins, protein localization, protein–protein interactions, sheath assembly, type VI secretion system
Subject Categories: Microbiology, Virology & Host Pathogen Interaction
Introduction
The type VI secretion system (T6SS) is a membrane‐anchored contractile nanomachine used by many Gram‐negative bacteria to deliver proteins from the cytosol directly to an extracellular space or across a target cell membrane. The nanomachine structurally and functionally resembles the contractile tail of bacteriophages and R‐type pyocins (Brackmann et al, 2017). T6SS biogenesis proceeds in a strictly hierarchical order (Wang et al, 2019). First, a protein complex spanning both membranes forms (Aschtgen et al, 2010; Felisberto‐Rodrigues et al, 2011; Durand et al, 2015). On this membrane complex, the baseplate forms and subsequently the sheath‐tube complex polymerizes (Brunet et al, 2015; Nguyen et al, 2017; Wang et al, 2017; Nazarov et al, 2018). Once fully extended, the sheath rapidly contracts, propelling the inner tube and associated effector proteins out of the cell (Basler et al, 2012; Cianfanelli et al, 2016). The contracted sheath is then disassembled by an ATP‐dependent unfoldase, and the components are recycled for another round of sheath extension and contraction (Bönemann et al, 2009; Basler & Mekalanos, 2012; Kapitein et al, 2013).
Based on phylogenetic analyses, T6SSs have been classified into three different types (i, ii, and iii) and type i can be further split into subclasses i1, i2, i3, i4a, i4b, and i5 (Barret et al, 2013; Russell et al, 2014; Li et al, 2015). The current model of T6SSi includes a minimal set of 13 proteins to assemble a functional T6SS (Mougous et al, 2006; Pukatzki et al, 2006). TssJ, TssL, and TssM form the membrane complex (Brunet et al, 2015; Durand et al, 2015); VgrG, TssE, TssF, TssG, and TssK form the baseplate (Nguyen et al, 2017; Nazarov et al, 2018); and TssB/VipA, TssC/VipB, and Hcp form the long sheath‐tube polymer (Wang et al, 2017; Szwedziak & Pilhofer, 2019). The AAA(+) ATPase ClpV disassembles contracted sheath, and its subunits can be reused to build another T6SS sheath (Bönemann et al, 2009; Basler & Mekalanos, 2012; Basler et al, 2012; Kapitein et al, 2013; Förster et al, 2014; Douzi et al, 2016). T6SSii is exclusively populated by the Francisella pathogenicity island and contains a set of 17 core components (Bröms et al, 2010; de Bruin et al, 2011), while the type iii system is found only in Bacteroidetes and contains 12 core components (De Maayer et al, 2011; Russell et al, 2014). Recently, a fourth type (T6SSiv) that is closely related to extracellular injection machineries such as R‐type pyocins and antifeeding prophages has been described (Böck et al, 2017). This particular system does not contain a canonical transmembrane anchor and also lacks ClpV unfoldase (Böck et al, 2017). However, similarly to Francisella, contracted sheaths may be refolded by a related ATPase (Brodmann et al, 2017).
TssA proteins have been initially shown to play an essential role in assembly of baseplate and the sheath‐tube in Pseudomonas aeruginosa (TssA1PA) and Escherichia coli (TssAEC) (Planamente et al, 2016; Zoued et al, 2016). Interestingly, TssAs can be categorized into different classes harboring distinct protein domain architectures. The specific architecture presumably affects their function during biogenesis of the T6SS. All TssA‐like proteins harbor a conserved ImpA_N domain (PF06812) located at the N‐terminal end, while the C‐terminal part differs in its composition. Recent analyses showed that domains can be further segregated into ImpA containing domain (Nt1), middle domain (Nt2), and C‐terminal domain (CTD) (Dix et al, 2018). A high‐resolution crystal structure of TssAEC C‐terminus harboring a VasJ domain (PF16989) was recently obtained, and it was shown that this part of the protein forms two stacked hexameric rings (Zoued et al, 2016). Dynamic rearrangement of wedges connecting the six helices supposedly leads to a ~90 Å opening of the structure. TssAEC interacts with membrane complex and baseplate components, but also with sheath component TssC/VipB. Thus, it was proposed that TssAEC might coordinate sheath‐tube assembly and guarantee its stability in the extended state (Zoued et al, 2016). However, another recent study showed that the C‐terminal domain (CTD, G388‐L472, helices α8–α12) of a closely related TssA from Aeromonas hydrophila forms a high‐order oligomer with D5 symmetry (Dix et al, 2018). The CTD is connected to the middle N‐terminal domain (Nt2, R232‐L374, helices α1–α7) through a ~21 residues flexible linker. Neighboring Nt2 domains form dimers, which do not follow D5 symmetry of the CTD oligomer (Dix et al, 2018).
TssA1PA was suggested to contain partial secondary structure homologies to the phage baseplate component gp6 in its C‐terminal part (Planamente et al, 2016). It forms a dodecameric ring with dimensions that are similar to sheath‐tube ring and has a central hole that could accommodate Hcp. TssA1PA was further shown to interact with baseplate components TssK, TssF, and VgrG1a, sheath‐tube and ClpV, but in contrast to TssAEC not with components of the membrane complex. Due to these properties, it was proposed that TssA1PA might be a baseplate component (Planamente et al, 2016).
Lastly, some TssA‐like proteins harbor a transmembrane region and a C‐terminal VasL domain of unknown function (PF12486). These TssAs were suggested to play an accessory role and corresponding genes thus referred to as tagA (type VI secretion accessory gene with ImpA domain) (Zoued et al, 2017). Recently, a TagA protein from E. coli (TagAEC) was shown to interact with TssAEC, localize at the distal end of sheath once it was fully extended and to stabilize the extended structure. Deletion of tagA EC caused excessive sheath polymerization, bending, and breaking of sheath structures and thus reduced efficiency of killing target cells (Santin et al, 2018). In addition, TagAEC was shown to be required for contraction of a part or the full‐length sheath toward the distal end. While it is unclear if these non‐canonical sheath contraction events result in protein secretion, these contractions constitute up to one‐third of observed contractions in E. coli (Szwedziak & Pilhofer, 2019).
Importantly, certain TssA proteins seem to be indispensable for proper T6SS assembly. Hcp secretion was not detectable in a ΔtssA EC strain, and no sheath structures were observed in a ΔtssA EC TssB‐mCherry strain (Zoued et al, 2016). Similarly, secretion of Hcp, VgrG1a, and Tse3 was not detectable in a tssA1 PA knockout strain and sheath formation in a TssB1‐sfGFP ΔtssA1 PA strain was severely decreased (Planamente et al, 2016).
Here, we investigated the role of several distinct proteins sharing the ImpA_N domain and we show that their functions differ significantly. The Vibrio cholerae and P. aeruginosa TssA proteins TssAVC and TssA2PA with C‐terminal VasJ domain (Class A) facilitate sheath assembly initiation and polymerization by forming a stable dodecamer at the end of the sheath that is distal from the membrane anchor. The TagA protein of V. cholerae (TagAVC) with the ImpA_N domain followed by a hydrophobic domain (Class B) localizes to cell membrane and prevents sheath assembly likely by competing with TssAVC. Finally, we show that a third class of ImpA_N domain containing proteins (Class C), represented by TssA1 in P. aeruginosa, localizes to the site of sheath assembly initiation. Our data show that ImpA_N domain containing proteins have diverse functions in the biogenesis of T6SS and that their role in sheath assembly is likely dictated by the structure and function of their C‐terminal domains.
Results
TssAVC and TssA2PA facilitate sheath assembly initiation and polymerization
Proteins that have ImpA_N domain followed by C‐terminal VasJ domain form Class A of TssA‐like proteins (Fig EV1A). This class is represented by E. coli TssAEC (EC042_4540) and A. hydrophila TssAAH (AHA1844) (Zoued et al, 2016; Dix et al, 2018), as well as V. cholerae TssAVC (VCA0119). TssAVC and TssAEC share 19.9% sequence identity, while TssAVC and TssAAH share 32.8% sequence identity (Appendix Table S1). To investigate the role of these proteins, we first imaged VipA‐mCherry2 sheath assembly in V. cholerae in the presence or absence of tssA VC (Appendix Table S2). We found that the parental strain usually forms five T6SS structures at any given time during logarithmic growth (Fig 1A, Movie EV1). This is in agreement with number of sheaths detected in the strain expressing VipA‐msfGFP (Vettiger & Basler, 2016). Image analysis showed that the tssA VC‐negative strain formed mostly dynamic sheath spots and only few structures that fully extended and contracted (Fig 1A, Movie EV1). In a bacterial competition assay, tssA VC knockout strain was able to kill E. coli prey cells only at a reduced rate (Fig EV1B). This resembled the reduced prey cell killing by a strain lacking the baseplate component tssE, which forms about 1000 times less structures compared to the parental strain (Vettiger & Basler, 2016). In addition, speed of sheath polymerization dropped from 23 nm per second measured in the parental strain to 3 nm per second in the strain lacking tssA VC (Fig EV1C).
Figure EV1. TssA protein domain organization and mutant phenotypes.
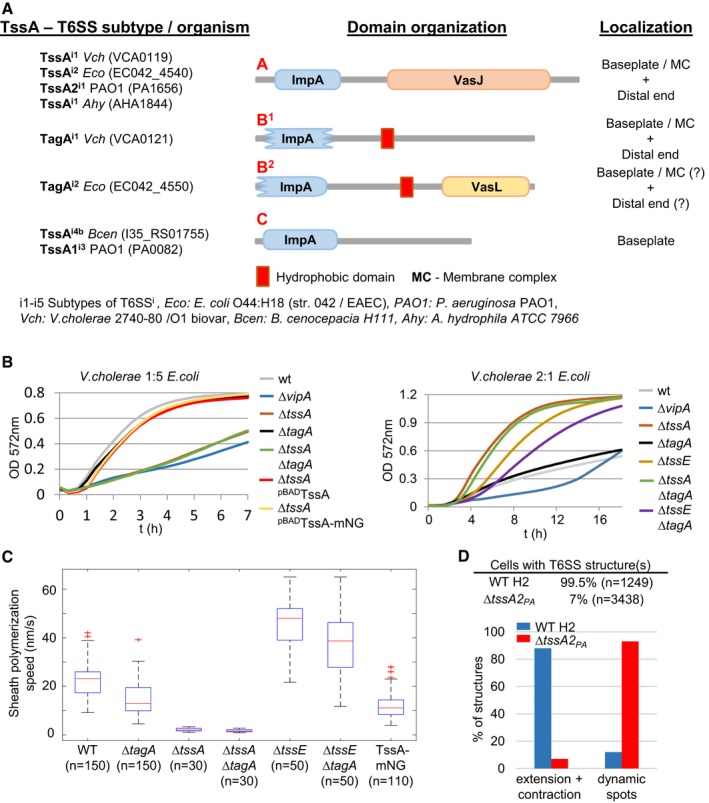
- Protein domain organization and proposed subcellular localization of V. cholerae 2,740–80/O1, P. aeruginosa PAO1, E. coli O44:H18 (str. 042/EAEC), A. hydrophila ATCC7966, and B. cenocepacia H111 TssA proteins. TssA proteins display three specific protein domain architectures A, B1/B2, and C. Most common architecture of each class is shown.
- Bacterial competition assay comparing T6SS‐dependent killing efficiency in different strain backgrounds. Fast killing kinetics via T6SS can be best observed by mixing strains in a ratio of 1:5 (V. cholerae : E. coli) (left panel), while less efficient or slow killing kinetics can be observed by mixing strains in a ratio of 2:1 (V. cholerae: E. coli) (right panel). ∆tssA mutant kills prey E. coli cells at a very slow rate, comparable to a ∆tssE mutant strain (right panel). ∆tagA mutant shows similar killing kinetics to WT (left and right panel). ∆tagA ∆tssE double mutant shows a shift in killing kinetics compared to ∆tagA mutant.
- T6SS sheath polymerization speed in WT and different mutant backgrounds. Polymerization speed in the ∆tagA mutant is reduced (13 nm/s) compared to WT speed (23 nm/s) and drastically reduced in the ∆tssA mutant (3 nm/s). Polymerization speed in the ∆tssE mutant is 48 nm/s and decreases to 38 nm/s in the ∆tagA ∆tssE double mutant. The strain harboring TssAVC‐mNeonGreen fusion displays decreased polymerization speed (11 nm/s) compared to parental strain without a tag on TssA (23 nm/s).
- Quantification of structures per cell and percentage of dynamic sheath spots versus extending and contracting structures in H2‐T6SS WT and ΔtssA2 PA strain. ΔtssA2 PA strain displays very few structures and mostly dynamic spots.
Figure 1. TssAVC and TssA2PA influence T6SS sheath assembly dynamics.
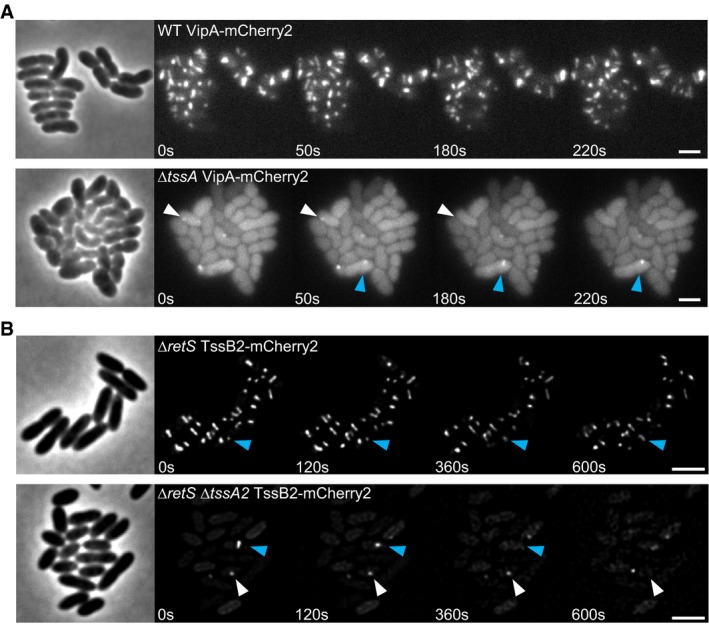
- Time lapse images of T6SS activity in tagged parental strain (VipA‐mCherry2) and ∆tssA mutant background. The parental strain forms multiple dynamic structures per cell. ∆tssA mutant predominantly forms dynamic sheath spots (white arrow) and few WT‐like sheath structures (blue arrow). Scale bars: 2 μm.
- H2‐T6SS dynamics in tagged parental strain (∆retS TssB2‐mCherry2, referred to as WT H2) and ΔtssA2 PA strain. Blue arrow indicates extending and contracting T6SS sheath structures, white arrow points to dynamic sheath spots. The WT H2 strain harbors multiple dynamic sheath structures per cell, while the ΔtssA2 PA strain displays very few dynamic spots or extended and contracting structures. Scale bars: 2 μm.
Similarly, we analyzed a second member of Class A, the TssA2 from H2‐T6SS of P. aeruginosa (PA1656) (Sana et al, 2012; Allsopp et al, 2017). TssA2PA shares 21,8% sequence identity with TssAVC and 25.2% sequence identity with TssAEC (Appendix Table S1). We show that multiple structures of TssB2‐mCherry2 sheath reside in single cells (Fig 1B, Movie EV2). Dynamics of H2‐T6SS sheaths are, however, significantly slower than V. cholerae sheaths. Full extension of one sheath can take up to 10 min, and sheath structures stay in extended state for at least 5 min (Fig 1B). Deletion of tssA2 PA severely decreased number of T6SS sheaths and mostly dynamic spots were visible; however, few fully extending and contracting sheath structures could be observed (Figs 1B and EV1D, Movie EV2).
TssAVC and TssA2PA localize to the distal end of an assembling sheath
Previous work of Zoued et al demonstrated that another member of TssA Class A, TssAEC of E. coli, first localizes to membrane complex and then coordinates sheath‐tube assembly at the distal end, presumably by incorporating new tube and sheath components (Zoued et al, 2016). Since TssA of V. cholerae is closely related to TssAEC (Appendix Table S1), we wondered if the two proteins share similar role in T6SS biogenesis. We fused mNeonGreen to TssAVC and observed its localization using fluorescence microscopy in a strain background with mCherry2‐tagged sheath (VipA‐mCherry2). While sheath assembly was about two times slower (Fig EV1C), the T6SS in TssAVC‐mNeonGreen/VipA‐mCherry2 remained fully functional (Fig EV1B). We found that, in most cases, TssAVC first localized to T6SS assembly initiation site before sheath signal appeared and then colocalized with a distal end of a polymerizing sheath (Fig 2A, Movie EV3).
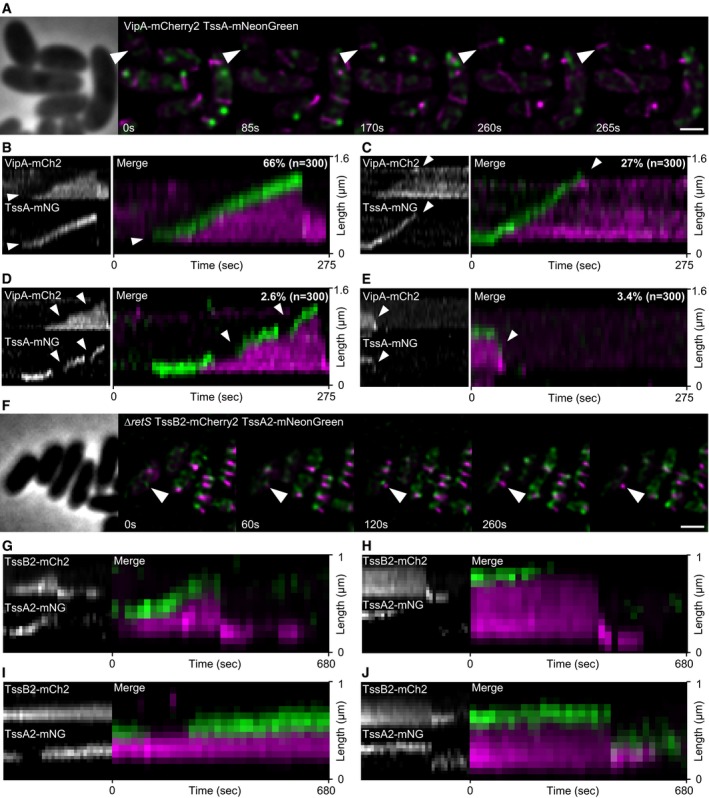
Figure 2. Localization and dynamics of TssAVC and TssA2PA .
-
ATime lapse images of T6SS activity in tagged strain (VipA‐mCherry2 TssAVC‐mNeonGreen). White arrow indicates path of TssAVC localization during one cycle of T6SS dynamics. Scale bar: 1 μm.
-
B–EKymographs of dynamics observed for TssAVC. Frequency of specific dynamics is indicated in the upper right corner of the merge image. (B) Sheath and TssAVC dynamics from A. TssAVC localizes to T6SS initiation site and to distal end of growing sheath (white arrow). (C) Once sheath reaches cell periphery, TssAVC dissociates from distal end (white arrow) and sheath stays extended a prolonged period of time. (D) TssAVC dissociates from growing sheath structure and re‐associates at a later time point (white arrows) to continue catalysis of polymerization. (E) TssAVC stays attached to sheath distal end after contraction (white arrow).
-
FFluorescence microscopy of T6SS dynamics in a double‐tagged strain (∆retS TssB2‐mCherry2 TssA2PA‐mNeonGreen). White arrow indicates full cycle of extending and contracting T6SS sheath. Scale bar: 1 μm.
-
G–JKymographs of TssB2 and TssA2PA dynamics. TssA2PA localizes at sheath initiation site and on distal end during polymerization (G), dissociates before contraction (H), dissociates and re‐associated during sheath extension (I), or stays attached to distal end after contraction (J).
For analysis of sheath dynamics in time lapse movies, we generally used kymographs generated with Fiji (Schindelin et al, 2012). A straight line was drawn along an assembling sheath, and the signals of the underlying pixels were replotted in a new XY coordinate system where the pixels along the Y‐axis represent the pixels along the line drawn over an assembling sheath and the individual time points are shown along the X‐axis. Such representation allows simple visualization of sheath assembly, measurement of sheath length, and speed of assembly as well as detection of colocalization of two proteins. Kymographs of TssAVC‐mNeonGreen and VipA‐mCherry2 movies of 300 cells revealed that in about two‐thirds of the analyzed cases TssAVC stayed attached to assembling sheaths until their contraction shortly (< 5 s) after full assembly (Fig 2B). In about 27% of the cases, TssAVC dissociated from the sheath after its extension to the opposite side of the cell, which was followed by prolonged period of stable extended sheath (Fig 2C). However, in few cases (about 3%) TssA also dissociated during sheath assembly causing a delay or stalling of the sheath polymerization while TssA re‐association resumed polymerization (Fig 2D). In about 3% of the cases, TssAVC stayed attached to the sheath distal end even after its contraction (Fig 2E).
To test whether TssA forms a stable complex on the sheath end, we photobleached cytosolic TssAVC‐mNeonGreen subunits during sheath polymerization. This was achieved by incubating the cells in the presence of ampicillin, which leads to formation of large viable spheroplasts with functional T6SS (Vettiger et al, 2017). Large cells allow controlled photobleaching of a relatively small section of the cell. We found that TssAVC‐mNeonGreen signal of the complex at the distal end of the polymerizing sheath remained constant after photobleaching of the cytosol, suggesting that the TssAVC subunits are not exchanged with the cytosolic TssAVC pool during sheath polymerization (Appendix Fig S1, Movie EV4).
Since TssA2PA is in the same TssA class as TssAVC and TssAEC (Class A), we hypothesized that all TssA proteins from this class might play the same role in T6SS biogenesis. Consequently, we used fluorescence microscopy to observe TssA2PA‐mNeonGreen dynamics in TssB2‐mCherry2 strain (Fig 2F–J, Movie EV5). We found that TssA2PA displays almost identical dynamics to TssAVC. TssA2PA localized to T6SS assembly initiation sites before sheath polymerization and then localized to the distal end of the polymerizing sheath (Fig 2G). Further, we often detected TssA2PA dissociating from sheath distal end after full extension (Fig 2H) or during extension followed by its re‐association to the polymerizing sheath (Fig 2I), but also residing on contracted sheath structures (Fig 2J). This suggests that all TssAs within the Class A play the same role in T6SS assembly.
TssAVC forms a dodecamer in vivo
Recent in vitro studies have shown that TssAs from E. coli, A. hydrophila, and Burkholderia cenocepacia form rings with 6‐fold, 5‐fold, and 16‐fold symmetry (Zoued et al, 2016; Dix et al, 2018). We aimed to estimate the oligomeric state of the TssAVC complex in vivo. We used the LacI–lacO system where two LacI repressor molecules, lacking tetramerization domain, bind to one lacO operator sequence (Belmont & Straight, 1998; Dong et al, 1999). First, we generated strains harboring 3, 6, or 12 copies of the lacO integrated into V. cholerae chromosome at the site of the disrupted lacZ gene (Fig EV2A). Expressing the lacI repressor fused to mNeonGreen (LacI‐mNeonGreen) in these strains yielded fluorescent LacI‐mNeonGreen spots with 6, 12, or 24 copies of mNeonGreen, respectively (Fig EV2B). The expression of LacI‐mNeonGreen in the parental strain lacking lacO sequences yielded no detectable foci (Fig EV2B). Similarly, no foci were detected when LacI‐mNeonGreen was expressed in a strain harboring 12 copies of lacO but supplemented with IPTG to disrupt binding of LacI to lacO. We quantified the signal emitted from LacI‐mNeonGreen spots and TssAVC‐mNeonGreen signal using ImageJ (Fig EV2C and D). Signals observed for TssAVC‐mNeonGreen fusions were most similar to the signal produced by LacI‐mNeonGreen molecules binding to 6 copies of lacO. This suggests that TssAVC forms a dodecamer in vivo.
Figure EV2. Fluorescence quantification.
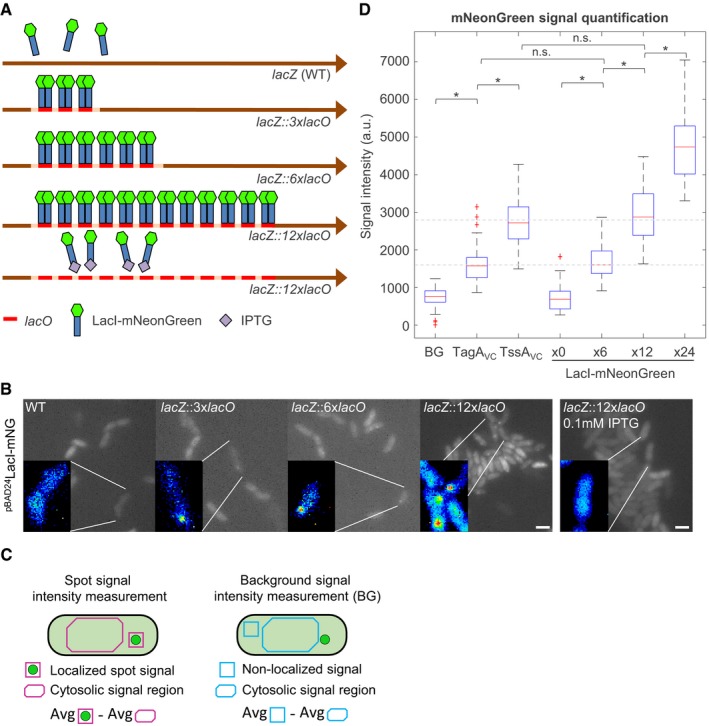
- Scheme of lacO array integration into V. cholerae chromosome at the lacZ locus. 3, 6, or 12 copies of the lacO operator accommodate 6, 12, or 24 molecules of LacI‐mNeonGreen, respectively. IPTG (isopropyl‐β‐D‐thiogalactopyranoside) binds to the lac repressor LacI and disrupts its ability to bind DNA.
- Fluorescence microscopy of strains harboring different lacO arrays. LacI‐mNeonGreen was expressed from pBAD24 plasmid by induction with 0.002% l‐arabinose. Images depicted for each strain are summed stacks of a short time lapse series. Inlays show magnified regions of interest, false colored with rainbow LUT using ImageJ. Cells contained one or two discrete foci of LacI‐mNeonGreen fluorescence. Scale bars: 2 μm.
- Scheme of cells harboring a mNeonGreen protein fusion and quantification procedure. Raw mNeonGreen signal was collected using ImageJ. Spot fluorescence signal (either localized signal of a complex or non‐localized signal) of three consecutive frames (red or blue square, approximately 5 × 5 pixels) in a cell was averaged. An average signal of the cell cytosol (red or blue corner rectangle) of the corresponding three frames was then subtracted from the spot signal. This resulted in fluorescence signal intensity of a spot (red square) or background signal intensity (blue square).
- Quantification of mNeonGreen fluorescence signals. Distributions of signal intensities measured in raw data for each strain are represented in the graph. 0× LacI‐mNeonGreen corresponds to the parental strain (WT), which does not harbor a lacO array. Signal distributions of 6×, 12×, and 24× LacI‐mNeonGreen correspond to strains harboring 3xlacO, 6xlacO, and 12xlacO array, respectively. Distribution of signal intensities for TagAVC corresponds to the strain harboring TagAVC‐mNeonGreen chromosomal fusion. Distribution of signal intensities for TssAVC corresponds to the strain harboring TssAVC‐mNeonGreen chromosomal fusion. BG corresponds to cytosolic mNeonGreen background signal in the strain harboring TssAVC‐mNeonGreen chromosomal fusion. BG (n = 41), TagAVC (n = 111), TssAVC (n = 76), x0 (n = 44), x3 (n = 76), x6 (n = 62), x12 (n = 88). n.s.: Distributions are not significantly different at alpha = 0.005 (two‐sample Kolmogorov–Smirnov test). * distributions are significantly different from each other, P < 0.001 (P‐values Bonferroni corrected).
High‐resolution structure of TssAVC
To further analyze the TssA of V. cholerae, we purified the protein and used cryo‐electron microscopy (cryo‐EM) to solve its structure (Fig 3). Choice of symmetry during refinement was dictated by 2D class averages (Fig 3, Appendix Fig S2). To test the symmetry, an initial 3D reference was built and refined without imposing any symmetry (C1), which resulted in clear six‐pointed star reconstruction. Further refinements have been done utilizing C6 symmetry. The outer and lumenal diameters are 132 and 53 Å, and the height of the assembly is 38 Å. Molecular weight estimation from the size‐exclusion chromatography (SEC) profile is ~600 kDa, which corresponds to twelve copies of TssAVC in one oligomer (634 kDa from sequence) (Appendix Fig S3).
Figure 3. Cryo‐EM of TssAVC .
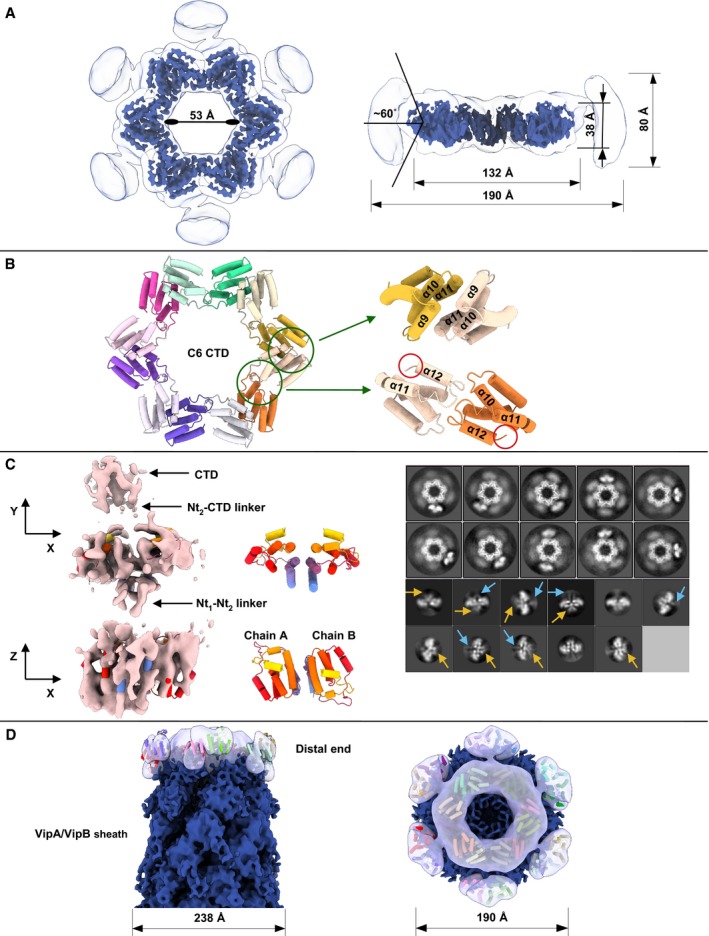
- Top view and side cutaway view of the TssAVC cryo‐EM reconstruction, shown low‐pass filtered at a lower (white, transparent) and higher (royal blue, non‐transparent) threshold. Possible (−60°; 60°) range of motion of Nt2 domain relative to the CTD ring plane is shown.
- Top view of the ribbon diagram of TssAVC CTD model. Two interfaces are highlighted with green circles. Enlarged side view of interface between helices α9–α11 of two neighboring subunits (top right), enlarged side view of interface between α11–α12 linker of one subunit and helix α10 from the neighboring subunit based on conserved WEP motif (bottom right). Part of Nt2‐CTD linkers are highlighted with red circles.
- Top view (XY‐plane) and side view (XZ‐plane) of the TssAVC Nt2 domain cryo‐EM reconstruction (pink, non‐transparent) shown with fitted Nt2‐dimer model (left). Partial Nt2‐CTD and Nt1‐Nt2 linker densities are highlighted with black arrows. Top and side views of ribbon diagram of Nt2 dimer (middle). Representative 2D class averages of TssAVC particles with visible connections between Nt2 dimer and CTD (top right), representative 2D class averages of Nt2‐dimer particles, Nt2‐CTD linker and Nt1‐Nt2 linker densities are highlighted with blue and yellow arrows (bottom right).
- Side and top views of putative model of TssAVC Nt2‐CTD ring fitted into cryo‐EM reconstruction (royal blue, non‐transparent) of the distal end of TssAVC T6SS (EMD‐3878). TssAVC Nt2‐CTD symmetrized reconstruction shown (white, transparent), fitted Nt2‐CTD model shown as a ribbon diagram.
The model built in cryo‐EM density of TssAVC is composed of twelve C‐terminal subunits (G376‐T466, helices α8–α12) tightly packed as two interpenetrating rings, similar to A. hydrophila TssA (TssAAH), but different from head‐to‐head stacked hexameric rings of the TssAEC assembly (Fig 3A and B, Appendix Fig S2) (Zoued et al, 2016; Dix et al, 2018). Each of the C‐terminal subunits forms two interfaces with its neighbors. The first 688 Å2 PISA‐predicted interface is based on three conserved residues (W454, E455, and P456) from α11–α12 linker of one subunit and helix α10 from the neighboring subunit (Fig 3B). Phylogenetic analyses have shown that the WEP motif is only present in TssA proteins of Class A (Dix et al, 2018). Second 672 Å2 interface forms a sixfold axis between helices α9–α11 of two neighboring subunits (Fig 3B). Well‐resolved TssAVC star‐like CTD is surrounded by disordered density (Fig 3A–D), which most likely represents the middle N‐terminal domain (Nt2). 2D class averages show the presence of dimers (most likely Nt2 dimers) connected to the CTD ring (Fig 3C). Seven helices α1–α7 constituting each Nt2 subunit (D218‐S363) are visible in 2D class averages, as well as two ~27 residues Nt2‐CTD interdomain linkers (Fig 3C). Due to linker mobility and length, Nt2 dimers are not dominated by sixfold symmetry of the CTD assembly. Instead, they show moderate degree of motion relative to the sixfold axis and degree of rotation around linker axis (Fig 3A and C). Similar behavior of Nt2 dimers was observed for crystal packaging of Nt2‐CTD TssAAH (Dix et al, 2018).
To improve the resolution of the density corresponding to the Nt2 dimer, symmetry of TssAVC reconstruction was relaxed from C6 to C1 and focused classification and refinement were performed. Resulting 3D reconstruction was used for picking template creation. Picked particles after standard processing pipeline were reconstructed to 6.6 Å resolution (Appendix Fig S2F and G). A homology model of TssAVC Nt2 domain was fitted and refined into cryo‐EM reconstruction using molecular dynamics flexible fitting (MDFF) (Fig 3C). Cryo‐EM density outlines all helices α1–α7 of both Nt2 subunits, assembled into a dimer with 591 Å2 interface between helices α1–α3. In addition, partial densities of Nt2‐CTD and Nt1‐Nt2 linkers are present in the reconstruction (Fig 3C). Based on this, we predict that the Nt1 domain is on the periphery of the CTD‐Nt2 assembly, however, likely connected through flexible linkers and thus not resolved in our cryo‐EM structure. Our reconstruction of TssAVC CTD fits as a rigid body with CC = 0.87 into the distal end of VipA‐N3 sheath mutant reconstruction (Fig 3D).
TagAVC stabilizes extended sheath and prevents its excessive polymerization
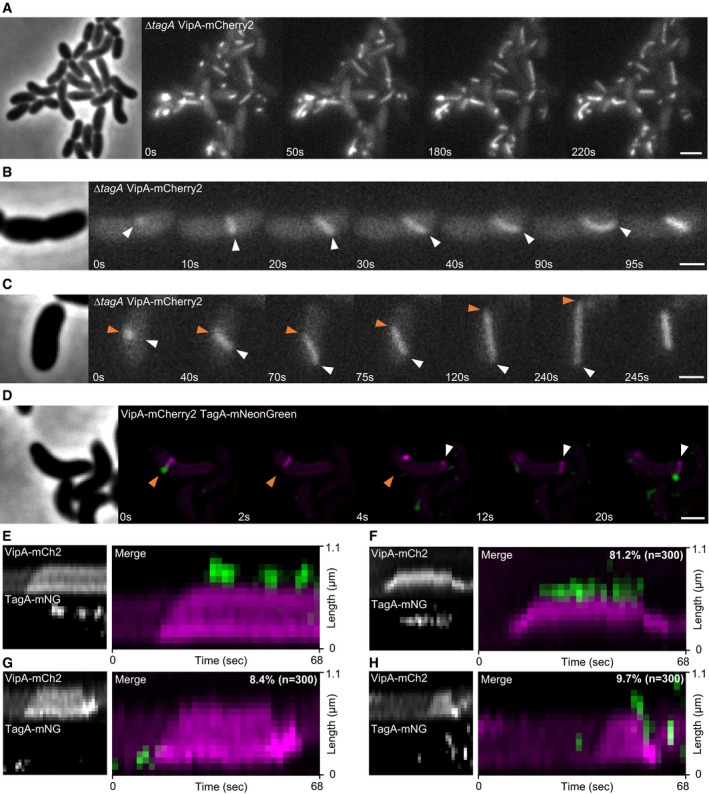
Class B of TssA‐like proteins is represented by V. cholerae TagA (VCA0121) and the recently characterized E. coli TagA (Santin et al, 2018). TagAVC shares 19.3% sequence identity with TagAEC (Appendix Table S1). To test the role of TagAVC on sheath dynamics, we first deleted tagA VC and imaged sheath assembly. Manual inspection of about 250 cells imaged for 5 min showed that the deletion strain had fivefold decreased number of sheath structures per cell. Overall sheath contractions per cell per minute decreased by threefold when compared to the parental strain (Figs 1A and 4A, Movie EV6 and Fig EV3D). This suggests that sheaths in ΔtagA VC strain are less stable overall, in particular that the sheaths contract soon after full assembly, and thus, fewer fully assembled sheaths are detected in these cells (Fig EV3A and B). Indeed, in a bacterial competition assay the ΔtagA VC mutant showed killing kinetics similar to the parental strain (Fig EV1B left). It is likely that the very high overall T6SS activity in V. cholerae compensates threefold reduction in contractions per minute in the ΔtagA VC strain, and thus, no difference in killing of prey cells is observed using this assay. Therefore, we compared the ΔtssE mutant, a strain that is significantly less efficient in killing E. coli cells, to the strain lacking both tssE and tagA VC in bacterial competition assay performed with higher ratio of V. cholerae to E. coli (Fig EV1B right). We observed slower killing kinetics in the double mutant showing that TagAVC contributes to T6SS function. In both the parental strain and the ΔtssE background strain, the presence of TagA slightly increases speed of sheath polymerization (Fig EV1C).
Figure 4. TagAVC localization and influence on sheath stability.
-
AThe ∆tagA mutant displays long and bent but still dynamic structures. Scale bar: 2 μm.
-
B, CFrames from time lapse movies showing sheath dynamics in the ∆tagA mutant. Sheath polymerization does not stop when the distal end reaches opposing membrane (white arrow). Continued polymerization causes the sheath to tilt and distal end to move toward the bacterial cell pole, ultimately effecting bending of the whole structure. (C) Sheath detaches from its membrane anchor (orange arrow), keeps extending to maximum cell length, and then contracts. Scale bars: 1 μm.
-
DTime lapse series of T6SS activity in tagged strain (VipA‐mCherry2 TagAVC‐mNeonGreen). Orange arrow indicates sheath distal end, and white arrow indicates initiation site of another T6SS sheath. Scale bar: 1 μm.
-
E–HKymographs of dynamics observed for TagAVC. (E) TagAVC localizes at the distal end when sheath reaches the cell periphery. Signal of TagAVC‐mNeonGreen disappeared and reappeared on this site repeatedly. (F) TagAVC localizes at the distal end when sheath reaches the cell periphery and stays associated with this site until contraction. (G) TagAVC briefly localizes at T6SS initiation site. (H) TagAVC briefly localizes at T6SS initiation site and at distal end, where it stays attached after contraction.
Figure EV3. TagAVC effect on sheath and TssAVC dynamics.
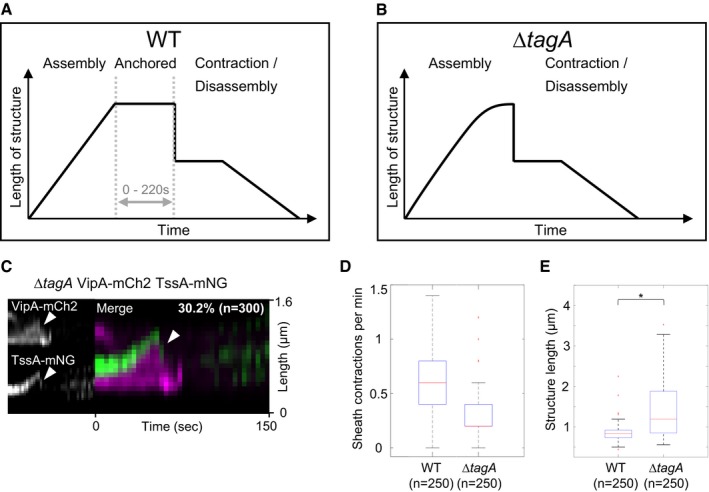
- Representative scheme of T6SS sheath dynamics in wild type. T6SS sheath stays extended or presumably anchored to the membrane via TagA for up to 220 s before contraction (average anchored time: 50 s, n = 150).
- Representative scheme of T6SS sheath dynamics in the tagA mutant. Sheath does not stay extended before contraction.
- Representative kymograph of VipA‐mCherry2 and TssA‐mNeonGreen dynamics in a tagA mutant strain background. 30% of all sheath structures show TssA‐mNeonGreen signal at distal end (white arrows) after contraction.
- Quantification of number of sheath contractions. 0.6 sheath contractions per minute per cell were measured in WT and 0.2 contractions per minute per cell in the ∆tagA mutant.
- Sheath length significantly increases in the ∆tagA mutant. * Samples are significantly different from each other (two‐sample t‐test, P < 0.001).
Importantly, we noticed that the lengths of T6SS sheaths are significantly increased in ΔtagA VC mutant (Fig EV3E). In the parental strain, sheath assembly is initiated at the membrane and continues until the opposing side of the cell. On the other hand, sheaths in the ΔtagA VC strain often continue polymerizing even after reaching the opposite side of the cell. This leads to extension along the longitudinal axis of the cell until reaching the cell pole (Fig 4A–C). This continued sheath polymerization results in various amount of sheath bending. Such bent sheath structures are not readily observed in the parental strain, suggesting that TagAVC could interact with the extended sheath and prevent further polymerization upon reaching membrane opposite to the site of assembly initiation.
Interestingly, while most sheath structures fully extend and contract normally despite significant bending, we also observed that during polymerization sheaths in ΔtagA VC mutant cells may apparently detach from their membrane anchor (Fig 4C). Surprisingly, this did not result in immediate sheath contraction and the sheaths further extended to the whole cell length before subsequent contraction (Fig 4C), suggesting that those structures still harbored an intact baseplate. Contraction of such a detached sheath structure most likely failed to secrete proteins unless it reattached to another trans‐envelope complex. Altogether, the absence of tagA VC resulted in sheath bending as well as detaching from its membrane anchor, which are likely the main reasons for lower overall T6SS activity. The observed phenotypes are consistent with the observations made recently for TagAEC (Santin et al, 2018).
TagAVC localizes to membrane and prevents sheath assembly upon overexpression
Analysis of TagAVC sequence suggested that the protein may localize to the membrane (Fig EV1A) similarly to TagAEC (Santin et al, 2018). Since the overall signal of chromosomally expressed TagA‐mNeonGreen was low, we decided to first overexpress the TagAVC protein. We tested if TagAVC overexpression changes T6SS activity and found that overexpression of TagAVC led to complete inhibition of T6SS‐dependent killing of E. coli, while a strain overexpressing TssAVC was still able to kill like the parental strain (Fig EV4A). Interestingly, fluorescence microscopy revealed that no sheaths were assembled when TagAVC was overexpressed (Fig EV4B). In addition, overexpressed TagAVC‐mNeonGreen localized to the membrane and inhibited sheath assembly, suggesting that TagAVC‐mNeonGreen has the same properties as untagged protein (Fig EV4B). In contrast, overexpression of TssAVC‐mNeonGreen had no influence on sheath assembly (Fig EV4C). When we removed the inducer after prolonged overexpression, we observed that during the next 2 h the cells that overproduced TssAVC‐mNeonGreen displayed regular T6SS activity, while the cells that overproduced TagAVC‐mNeonGreen only slowly regained T6SS activity (Fig EV4C). Interestingly, when TagAVC was overexpressed in the VipA‐mCherry2 TssAVC‐mNeonGreen strain, the TssAVC‐mNeonGreen spots remained closely associated with the membrane and colocalized with sheath spots (Fig EV4D). High concentration of TagAVC thus results in retention of TssAVC at sheath assembly initiation sites and inhibition of sheath assembly.
Figure EV4. Overproduction of TagA inhibits T6SS in V. cholerae .
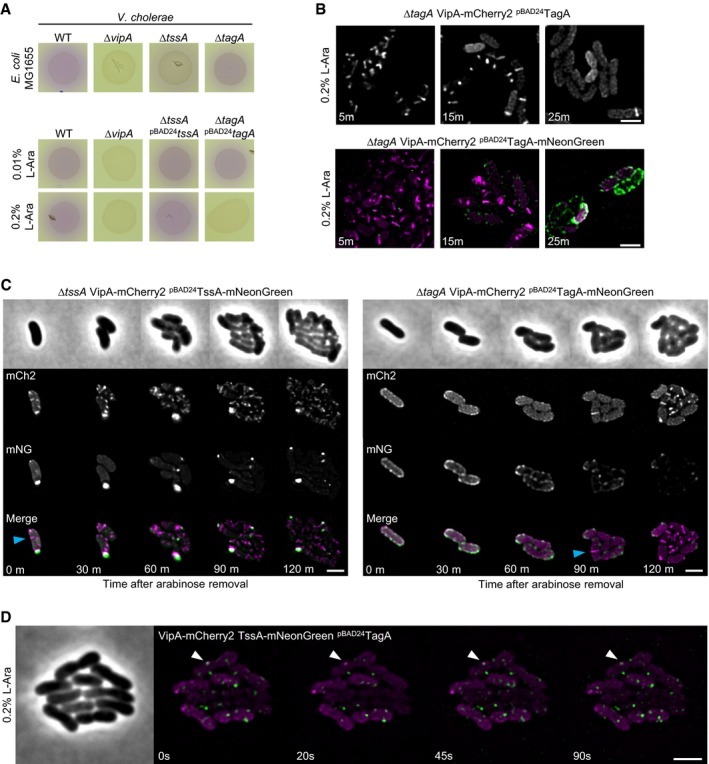
- Bacterial competition assay. Killing of E. coli (MG1655, lacZ+) by V. cholerae T6SS can be observed as CPRG substrate is converted by released β‐galactosidase over time (red color). ΔtssA VC strain shows significantly decreased killing efficiency, while tagA VC knockout is indistinguishable from WT killing efficiency. ΔvipA strain was used as T6SS‐negative control (upper panel). Overproduction of TagAVC but not TssAVC leads to inhibition of T6SS‐mediated killing (lower panel).
- Overproduction of TagAVC and TagAVC‐mNeonGreen leads to inhibition of T6SS sheath formation. Overproduced TagAVC‐mNeonGreen localized predominantly at the cell periphery in discrete foci. Scale bars: 2 μm.
- Indicated strains were pre‐induced with 0.2% l‐arabinose for 1 h, then washed, and observed using fluorescence microscopy. Dynamic structures (blue arrow) can be detected directly after washing in the TssAVC‐mNeonGreen overproducing strain (left panel). The strain overproducing TagAVC‐mNeonGreen was only able to form T6SS structures after 90‐min recovery period (right panel). Scale bar: 2 μm.
- Time lapse series of a VipA‐mCherry2 TssAVC‐mNeonGreen double‐tagged strain overproducing TagAVC from plasmid. TagAVC expression was induced for 1 h with 0.2% l‐arabinose prior to imaging. TssAVC‐mNeonGreen displays non‐dynamic behavior and co‐localizes with sheath foci in the cell periphery (white arrow). Scale bar: 2 μm.
TagAVC forms a hexamer at the distal end of an assembled sheath
To further understand the function of TagAVC protein, we imaged localization of chromosomally expressed TagAVC‐mNeonGreen during sheath assembly. Localized TagAVC‐mNeonGreen signal, stable for at least two subsequent frames (2s apart), was occasionally detected near a sheath structure. In over 80% of these cases, TagAVC‐mNeonGreen localized at the site where the sheath distal end reached the cell periphery after full extension (Fig 4D–H, Movie EV7). Further analysis of time lapse movies revealed several distinct dynamics of TagAVC localization. In most cases, TagAVC‐mNeonGreen signal disappeared very briefly before sheath contraction (< 2 s) (Fig 4D). We also observed that TagAVC displayed a rather dynamic localization at the distal end of the sheath by appearing and disappearing periodically (Fig 4E). We cannot exclude, however, that photobleaching occurred or TagAVC spot shifted out of focus. Disappearance of TagAVC signal was not immediately followed by sheath contraction in these cases. Further, we found that in about 10% of the cases TagAVC localized to sheath assembly initiation sites but remained there only for few seconds (2–6 s) prior to sheath assembly (Fig 4G). In about 10% of the cases, we detected TagAVC on the distal end of contracted structures (Fig 4H). The fact that many sheaths contract soon after assembly (Fig 2B) likely explains the overall low frequency of formation of stable TagAVC‐mNeonGreen spots. In general, the observed dynamics of TagAVC is consistent with dynamics and localization of TagAEC (Santin et al, 2018).
To estimate the oligomerization status of TagAVC, we used the same approach as described above and compared brightness of TagAVC‐mNeonGreen spots to the brightness of LacI‐mNeonGreen spots bound to lacO arrays of different length (Fig EV2C and D). TagAVC‐mNeonGreen spots were as bright as LacI‐mNeonGreen spots on 3× lacO array, which was near the detection limit of our approach. This suggests that TagAVC forms a hexamer (Fig EV2D).
Interaction partners of TssA and TagA in V. cholerae
We noticed that the fraction of sheaths with TssAVC‐mNeonGreen spots still attached to sheath distal end after contraction increased from 3% in the parental strain to 30% in the ΔtagA VC mutant (Figs 2E and EV3C). Together with the sheath assembly phenotype of the ΔtagA VC strain and the TagAVC localization data, this suggested a possible interaction between TagAVC and TssAVC as also shown for these proteins in E. coli (Santin et al, 2018). To identify interaction partners of TssAVC and TagAVC, we first performed a pulldown experiment using hemagglutinin (HA)‐tagged proteins as baits, and second, to confirm the identified interaction partners, we used bacterial two‐hybrid system (Karimova et al, 2017). We identified sheath component VipB, TagAVC, and ClpV as interacting proteins of TssAVC in both pulldown and bacterial two‐hybrid system (Appendix Fig S4A and B). This is consistent with observation for TssAEC; however, unlike TssAEC, which was shown to directly interact with Hcp and several baseplate components (Zoued et al, 2016), we found no such interaction for TssAVC using pulldown or bacterial two‐hybrid assays (Appendix Fig S4A and B). Using TagAVC‐HA as bait revealed interaction with ClpV, sheath components VipA and VipB as well as baseplate component TssK (Appendix Fig S4A). Bacterial two‐hybrid assay confirmed TssK interaction; however, interactions with sheath subunits were not confirmed (Appendix Fig S4B).
Baseplate localization of TssA1 in P. aeruginosa
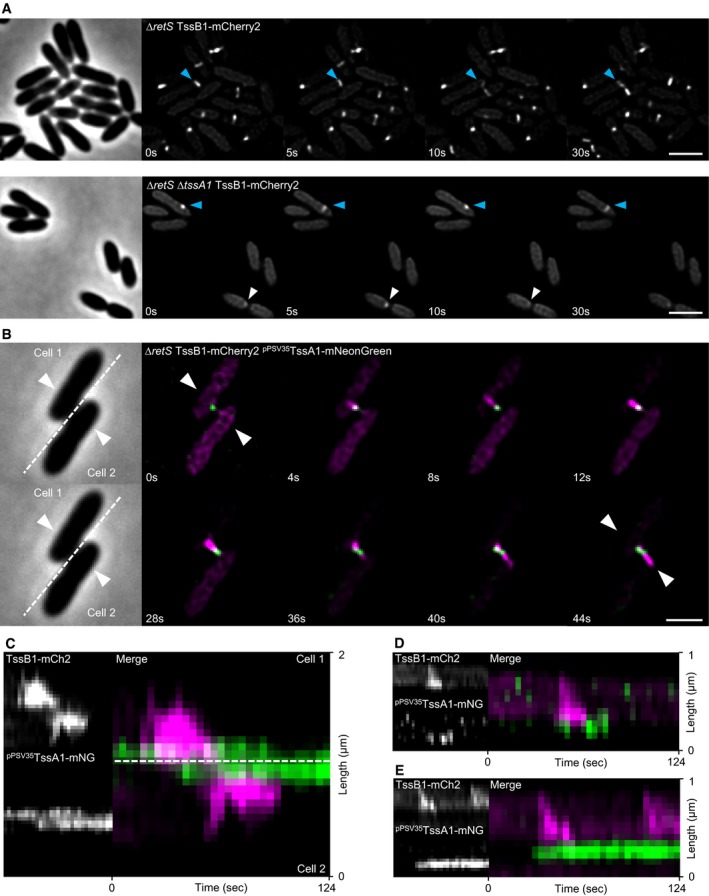
To investigate the role of Class C TssA‐like proteins, we turned to TssA1 in H1‐T6SS of P. aeruginosa. It was reported earlier that deletion of tssA1 in P. aeruginosa PAK led to severe decrease in T6SS activity (Planamente et al, 2016). However, T6SS activity was not completely abolished, and thus, we decided to investigate the role of TssA1 in the closely related strain P. aeruginosa PAO1 using fluorescence microscopy. We used a ΔretS mutant strain background, since RetS negatively affects H1‐T6SS expression (Mougous et al, 2006). In the ΔretS TssB1‐mCherry2 strain, H1‐T6SS shows very fast sheath dynamics as one cycle of T6SS extension and contraction usually takes only 5–20 s. Extending and contracting structures can be observed in up to 50% of cells (Fig 5A, Movie EV8, Fig EV5C). Deletion of tssA1 PA decreased the amount of detected sheath structures, and the majority of observed H1‐T6SS activity consisted of dynamic sheath spots, but extending and contracting sheath structures were also detectable (Fig 5A, Movie EV8, Fig EV5C). Importantly, deletion of tssA1 PA had little to no influence on the speed of sheath assembly (Figs 5A and EV5D).
Figure 5. TssA1PA localizes at T6SS initiation sites in P. aeruginosa PAO1.
-
AH1‐T6SS dynamics in tagged parental strain (∆retS TssB1‐mCherry2, referred to as WT H1) and ΔtssA1 PA strain. Blue arrow indicates extending and contracting T6SS sheath structures, and white arrow points to dynamic sheath spots. Scale bars: 2 μm.
-
BFluorescence microscopy of T6SS dynamics in ∆retS TssB1‐mCherry2 strain harboring pPSV35 plasmid with TssA1PA‐mNeonGreen fusion. Scale bar: 2 μm.
-
C–EKymographs of TssB1 and TssA1PA dynamics. TssA1PA localizes exclusively at sheath initiation site (B–E), disappears after contraction (C,D), or remains in place for longer timeframe (C,E). TssA1PA signal appears in neighboring cell shortly after T6SS attack (B,C).
Figure EV5. TssA1PA complementation and quantification of dynamic sheaths.
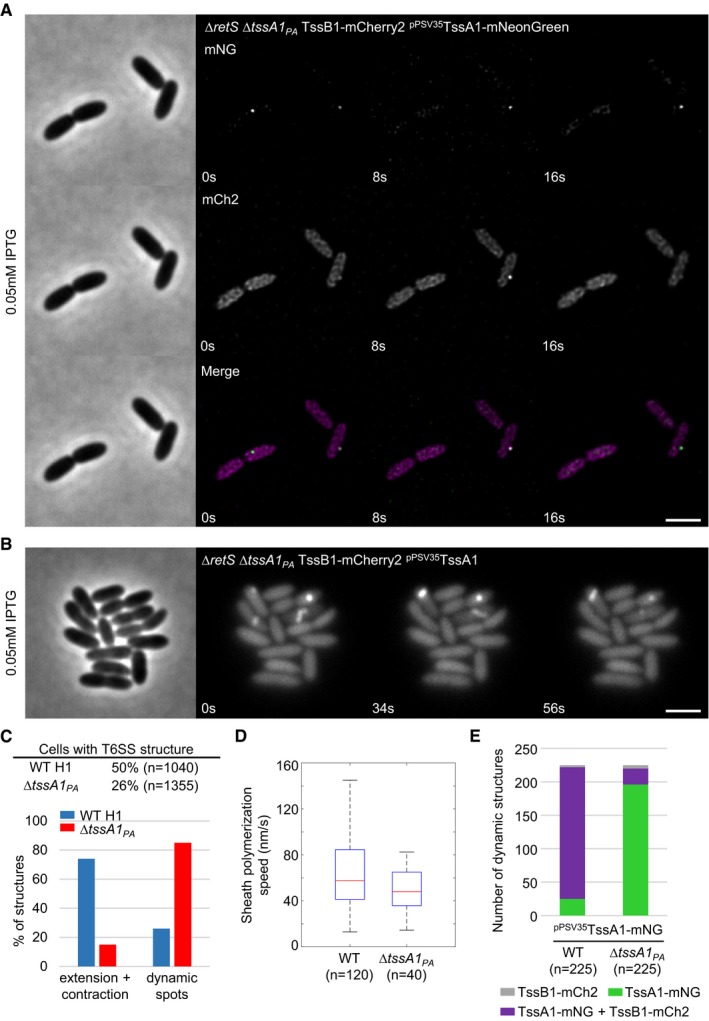
- Time lapse fluorescence microscopy of ∆retS ∆tssA1 PA TssB1‐mCherry2 strain harboring pPSV35 plasmid with TssA1PA‐mNeonGreen fusion. No dynamic T6SS sheath structures can be observed. Scale bar: 2 μm.
- Time lapse series of ∆retS ∆tssA1 TssB1‐mCherry2 strain harboring pPSV35 plasmid with TssA1PA. ∆tssA1 mutation can be complemented with native copy of TssA1 expressed from pPSV35 plasmid. Several dynamic sheath structures can be observed. Scale bar: 2 μm.
- Quantification of structures per cell and percentage of dynamic sheath spots versus extending and contracting structures. ΔtssA1 PA strain displays less structures and mostly dynamic spots.
- Polymerization speed of T6SS sheath in ∆retS TssB1‐mCherry2 (WT) and ∆retS ∆tssA1 PA TssB1‐mCherry2 strains. Polymerization speed is 58 nm/s on average in WT and 48 nm/s on average in the ∆tssA1 PA mutant.
- Colocalization of TssA1PA‐mNeonGreen with TssB1 sheath during T6SS dynamics. WT, ∆retS TssB1‐mCherry2 strain harboring pPSV35 plasmid with TssA1PA‐mNeonGreen fusion. ∆tssA1 PA, ∆retS ∆tssA1 PA TssB1‐mCherry2 strain harboring pPSV35 plasmid with TssA1PA‐mNeonGreen fusion. Proportion of dynamic structures consisting of TssA1PA‐mNeonGreen spots that colocalize with sheath structures and spots (WT) or spots (∆tssA1 PA) is labeled in magenta. Fraction of dynamic TssA1PA‐mNeonGreen spots without sheath signal in close proximity is labeled in green. Percentage of sheath signals (spots or structures) without TssA1PA‐mNeonGreen spot in proximity is labeled in gray.
So far, only indirect evidence pointed to localization of TssA1PA at the baseplate (Planamente et al, 2016). Consequently, we attempted to complement the ΔtssA1 PA mutant strain with mNeonGreen‐tagged TssA1PA to observe its localization and dynamics. Unfortunately, full complementation of T6SS dynamics in the strain harboring TssB1‐mCherry2 fusion was only possible with untagged version of TssA1PA (Fig EV5A, B and E). Additionally, in the absence of the native copy of TssA1PA, most detected dynamic structures (87%) contained only TssA1PA‐mNeonGreen (Fig EV5E), suggesting that the complex formed solely from tagged TssA1PA‐mNeonGreen fails to recruit TssB1‐mCherry2.
We next expressed TssA1PA‐mNeonGreen in the presence of the native copy of TssA1PA in the strain harboring TssB1‐mCherry2 fusion. Colocalization analysis showed that the majority (197 out of 225) of dynamic structures formed in such strain contained both TssA1PA‐mNeonGreen and TssB1‐mCherry2 (Fig EV5E). In 163 cases, a TssA1PA‐mNeonGreen spot colocalized with a site from which an extended sheath assembled (as shown in Fig 5B). In 34 cases, the TssA1PA‐mNeonGreen spot colocalized with a dynamic sheath spot. Similar ratio between formation of dynamic sheath spots and contractile sheaths was observed for the cells expressing only the native copy of TssA1PA (Fig EV5C).
In the presence of native TssA1PA, the tagged protein TssA1PA‐mNeonGreen formed a spot at the membrane before sheath signal appeared (Fig 5B–E, Movie EV9), which was followed by sheath extension away from this spot and later sheath contraction. Interestingly, after sheath contraction, TssA1PA signal disappeared (Fig 5D) or stayed in place for another round of sheath assembly and contraction from the same site (Fig 5E). During the dueling behavior of P. aeruginosa H1‐T6SS, a new spot of TssA1PA‐mNeonGreen appeared in a cell within few seconds after a T6SS attack from the neighboring cell and before a new sheath assembled to carry out the counterattack (Fig 5B and C). Overall, these results suggest that TssA1PA is localized at the baseplate and increases the rate of sheath assembly initiation but likely plays no role in sheath polymerization. However, since the tagged TssA1PA is only functional in the presence of the untagged native copy of TssA1PA, we cannot rule out a dual role of TssA1PA protein.
Discussion
Here, we show that proteins with a conserved N‐terminal domain PF06812 cluster into three main classes and mainly differ in their C‐terminal sequence. These proteins are usually named either TssA or TagA or ImpA_N containing proteins (Mintz & Fives‐Taylor, 2000; Zoued et al, 2017). One class of TssA proteins contains recently characterized TssA from E. coli (Zoued et al, 2016). This protein forms a dodecamer in vitro, interacts with TssK, TssE, VgrG, Hcp, TssB, and TssC, and localizes to the distal end of an assembling sheath (Zoued et al, 2016). Interestingly, Zoued et al showed that the N‐terminal part of TssAEC containing ImpA_N domain might be responsible for interaction with sheath and to a lesser extent with baseplate components, while the C‐terminus preferably interacted with Hcp. The TssA protein of V. cholerae is from the same class, and we show here that it plays the same role in T6SS biogenesis. Similarly to TssAEC, most of the time TssAVC localizes to the distal end of assembling sheaths (Fig 2B). In addition to our high‐resolution dodecameric cryo‐EM structure, we show that intensity of one TssAVC‐mNeonGreen spot likely represented 12 molecules of mNeonGreen, suggesting that TssAVC forms a dodecamer during sheath assembly (Fig EV2D). Our photobleaching experiment suggests that once this dodecamer forms, it is stable during the whole sheath assembly process (Appendix Fig S1). However, TssAVC is not strictly required for sheath assembly as tssA VC‐negative strain still kills prey cells, assembles short dynamic sheaths and occasionally also full‐length sheath with significantly reduced assembly speed (Fig EV1). The H2‐T6SS cluster in P. aeruginosa also encodes a TssA protein of the same class. Live‐cell imaging of the sheath assembly and TssA2PA localization confirmed that also this protein localizes to the distal end of the assembling sheath (Fig 2). Taking these observations together, it is very likely that all TssA proteins of the Class A (Fig EV1) perform the same function in T6SS biogenesis and are required for fast and efficient polymerization of T6SS sheath. Interestingly, elongation of sheath of bacteriophage T4 does not require the action of chaperones (Arisaka et al, 1979). Therefore, while potentially not being strictly essential, Class A TssAs act as chaperones assisting sheath‐tube copolymerization, which likely requires assistance in proper formation of the sheath hand‐shake domain (Ge et al, 2015; Kudryashev et al, 2015; Wang et al, 2017). We thus propose to rename this class of TssA proteins to TsaC (T6SS sheath assembly chaperone).
TagA protein encoded by V. cholerae is a representative of the ImpA_N domain containing class B proteins (Fig EV1) that were initially proposed to have only accessory role in T6SS biogenesis (Zoued et al, 2017). A recent study provided evidence that a TagA protein from E. coli localizes to the membrane at the distal end of the sheath and holds it in place (Santin et al, 2018). Similarly, we show that TagAVC localizes to the cell periphery and most of the times forms hexameric spots at the distal ends of fully assembled sheaths (Fig 4). Deletion of tagA VC resulted in about fivefold decrease in number of sheath assemblies, but only about threefold decrease in sheath contractions, and thus, the influence on the overall ability of V. cholerae to kill prey cells was negligible (Fig EV1B). Consistently with observations made in E. coli (Santin et al, 2018; Szwedziak & Pilhofer, 2019), the sheaths in tagA VC‐negative strain were mostly very long, stretching from one pole of a cell to another. In addition, the assembling sheaths were often bent and occasionally consequently detached from their membrane anchor (Fig 4). This suggests that TagAVC prevents extensive sheath polymerization, which may result in bending and sheath instability. Furthermore, sheaths in tagA VC knockout mutant did not stay extended or anchored to the membrane as observed in the parental strain (Fig EV3A and B), which was also observed in E. coli (Santin et al, 2018). Interestingly, Szwedziak and Pilhofer showed recently that TagAEC is also required for fast sheath assembly as the tagA EC‐negative cells assembled the sheaths about 7‐fold slower than wild type. In addition, electron cryotomography showed that the sheaths in E. coli are attached at both ends to the cell membrane, one end is connected to a baseplate and the second end to an uncharacterized structure likely composed of TagAEC and potentially other proteins (Szwedziak & Pilhofer, 2019). Moreover, the TagAEC‐mediated membrane attachment was also required for non‐canonical sheath contractions observed in one‐third of the contraction events where the full‐length sheath or its part contracted away from the baseplate toward the distal end (Szwedziak & Pilhofer, 2019). However, whether such non‐canonical contractions lead to protein secretion and delivery to target cells remains to be shown.
We show that TagAVC interacts with TssAVC, TssK, ClpV, VipA, and VipB. Interestingly, we observed that TssAVC spots disappeared from the fully extended sheaths (Fig 2C). Additionally, in the tagA VC mutant strain harboring VipA‐mCherry2 and TssAVC‐mNeonGreen fusions, TssAVC often remained attached to the distal end of sheath even after contraction (Fig EV3C). This indicates that TagAVC blocks function of TssAVC potentially by removing TssAVC from the fully assembled sheath. Removal of TssAVC stops or slows down further sheath polymerization and thus stabilizes the extended sheath (Fig EV3A and B and summarized in Fig 6A). Moreover, when we overexpressed TagAVC, sheath assembly was completely blocked and only spots of sheath signal colocalizing with TssAVC spots were detectable in the cells (Fig EV4D). This suggests that TagAVC may inhibit TssAVC function both during initiation of sheath assembly and after its extension. This could be mediated by TssA C‐terminus since deletion of TagAEC and deletion of C‐terminal part of TssAEC had similar effect on sheath dynamics (Szwedziak & Pilhofer, 2019). We propose to rename the class of TagA proteins (Fig EV1) to TsmA (T6SS sheath membrane anchor).
Figure 6. Role of TsaC, TsmA, and TsaB in T6SS dynamics.
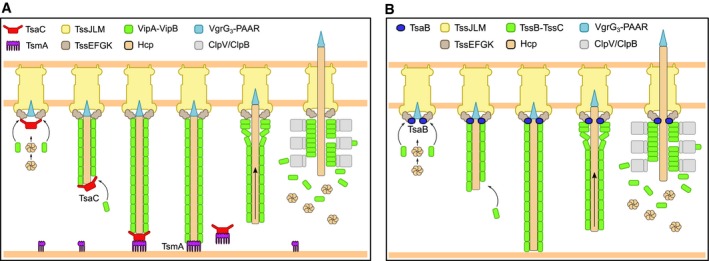
- Model of T6SS dynamics. After membrane complex has formed, TsaC assists in recruitment of baseplate components and sheath‐tube copolymerization. Once extending sheath‐tube has reached opposing cell membrane, TsmA replaces TsaC at the distal end and holds it in place. Shortly after TsmA disappearance, sheath contracts and releases spike tube to the environment. Sheath components are then recycled by the ClpV ATPase.
- TsaB localizes at the baseplate and assists in the initiation of T6SS biogenesis.
Interestingly, many T6SS clusters lack genes encoding TagA‐like proteins, notably the H1‐T6SS of P. aeruginosa. The sheath dynamics of this cluster is, however, very different from dynamics of sheath assembly of TagA‐containing E. coli or V. cholerae clusters. Even though a model excluding sheath contraction was proposed for H1‐T6SS (Corbitt et al, 2018), we show here that H1‐T6SS sheaths assemble very quickly within 5‐20s and immediately contract (Fig 5), which makes it technically challenging to detect the whole sheath assembly, contraction, and disassembly cycle. We also often observed dynamic sheath spots for H1‐T6SS, which could be functional contractile sheath assemblies shorter than the diffraction limit of the used fluorescence microscopy technique. It was shown previously that short sheath structures formed upon limitation of Hcp protein availability can still deliver effectors into target cells (Vettiger & Basler, 2016). This is also consistent with the observation that the tssA VC‐negative strain mostly forms small dynamic sheath spots but is still capable of prey killing, albeit with much lower efficiency than the parental strain. It is therefore possible that certain sheath‐tube assemblies evolved to be more prone to contraction and contract either before or right after reaching the opposite side of the cell similarly to what was observed in Acinetobacter baylyi (Ringel et al, 2017). This could be achieved by evolution of less stable baseplate or membrane complex that may trigger sheath contraction before extending excessively and potentially bending and detaching from the cell envelope. On the other hand, the H2‐T6SS cluster apparently lacks a homolog of TagA protein while the sheaths are very stable in the extended form. Those sheaths do not bend or extend from bacterial pole to pole as in the case of tagA‐negative V. cholerae T6SS. Therefore, there might be yet another TagA‐independent mechanism that stabilizes properly extended sheaths or this could be a consequence of a very slow polymerization speed of H2‐T6SS sheaths (Fig 2F).
TssA1 protein of P. aeruginosa H1‐T6SS cluster (Fig EV1, Class C) was shown to bind TssK1, TssF1, ClpV1, Hcp1, and TssB1, form ring‐like structures, and localized to ends of contracted sheaths and thus was suggested to be a baseplate component (Planamente et al, 2016). Here, we show that TssA1PA‐mNeonGreen indeed localized to sheath assembly initiation sites (summarized in Fig 6B). When we expressed TssA1PA‐mNeonGreen in the absence of the native copy of tssA1 PA, the majority of TssA1PA‐mNeonGreen spots localized to the membrane and disappeared without subsequent sheath formation (Fig EV5E). However, in the strain harboring the native copy of tssA1 PA, localization of TssA1PA‐mNeonGreen at the membrane was generally followed by sheath assembly (Fig EV5E). Because both native and tagged TssA1PA were present in the cells, the oligomeric complex forming the TssA1PA‐mNeonGreen spots was likely composed of both proteins and such complex was thus able to initiate sheath assembly. It is important to note that nearly all dynamic sheath structures detected in the strain expressing both native and tagged TssA1PA proteins colocalized with TssA1PA‐mNeonGreen spots. Therefore, it is unlikely that TssA1PA‐mNeonGreen spot formation and sheath assembly initiation are two separate events or that the TssA1PA spot formation is an artifact of tagging by mNeonGreen. When we deleted tssA1 PA and analyzed the sheath assembly dynamics, we observed dramatic decrease in the number of detectable sheath assemblies (Fig 5). However, unlike deletion of tssA VC, removing tssA1 PA had only subtle influence on sheath assembly speed (Figs 5 and EV5D). This suggests that TssA1PA is required only for sheath assembly initiation and raises the question whether there are other proteins encoded by H1‐T6SS that may chaperone sheath‐tube assembly. We propose renaming the class of proteins similar to TssA1PA to TsaB (T6SS sheath assembly baseplate).
In summary, we show here that sheath‐tube assembly is a highly coordinated process, which requires various proteins sharing a conserved N‐terminal domain. The precise understanding of the roles of these proteins will require high‐resolution structures of their complex with the sheath or baseplate as well as structure‐based mutagenesis followed by detailed analysis of sheath dynamics.
Materials and Methods
Strains and culture conditions
Vibrio cholerae 2,740–80 and P. aeruginosa PAO1 ∆retS fluorescent protein‐tagged strains as well as knockout strains were generated as described previously (Basler & Mekalanos, 2012). Recombinant clones were checked by colony PCR and sequence‐verified. Escherichia coli MG1655 (lacZ+) was used as prey strain in bacterial competition assays. Bacterial two‐hybrid strains were generated using the instructions from the manufacturer (Euromedex). For bacterial two‐hybrid assay, cells were incubated at 30°C for 24–48 h according to instructions. Stains used in this study are listed in Appendix Table S2. Bacteria were grown in LB broth at 37°C aerobically. Ampicillin (300–500 μg/ml), streptomycin (100 μg/ml), kanamycin (50 μg/ml), irgasan (20 μg/ml), and gentamicin (15 μg/ml) were used as supplements in growth media when necessary.
Bacterial competition assay
Vibrio cholerae can lyse or permeabilize E. coli (MG1655, lacZ+) with its active T6SS, indicated by conversion of a membrane impermeable β‐galactosidase substrate (chlorophenol red‐β‐D‐galactopyranoside, CPRG) over time. Strains were mixed in a ratio of 1:5 or 2:1 (V. cholerae: E. coli) and co‐incubated on solid LB agar supplemented with 40 μg/ml CPRG and 300 μg/ml Ampicillin when strains harboring pBAD24 plasmid were used. Killing of E. coli prey cells was recorded by monitoring OD572 increase on a 96‐well plate reader. At least three independent experiments were run on different days. Average of these experiments is shown.
Image analysis
ImageJ was used to measure frequency of contractions and length of WT and ∆tagA mutant sheath structures before contraction. Sample images were taken from at least two different days of acquisition and three different time points within a time lapse series.
Fluorescence microscopy
Fluorescence microscopy and processing of images were done as described in Brackmann et al (2017). Briefly, overnight cultures were washed with LB medium, diluted 1:100, supplemented with antibiotics, and cultivated to an optical density (OD) at 600 nm of 1. Then, cells were concentrated to OD 10, spotted on a LB 1% agarose pad, and covered with a glass coverslip. Bacteria were either directly imaged at 30°C or directly incubated on the agarose pad for 45 min prior to imaging (P. aeruginosa PAO1). We used a Nikon Ti‐E inverted motorized microscope with Perfect Focus System and Plan Apo 100× Oil Ph3 DM (NA 1.4) objective lens. The microscope was equipped with SPECTRA X light engine (Lumencor), and ET‐EGFP (Chroma #49002) and ET‐mCherry (Chroma #49008) filter sets were used to excite and filter fluorescence. The setup further contained a sCMOS camera pco.edge 4.2 (PCO, Germany) (pixel size 65 nm) and VisiView software (Visitron Systems, Germany) to record images. Temperature control was set to 30°C, and humidity was regulated to 95% by an Okolab T‐unit (Okolab). Additional image processing was carried out using Fiji (Schindelin et al, 2012) as described previously (Basler et al, 2013). For photobleaching, mNeonGreen fluorescence was reduced using a VS‐AOTF 488 nm Laser system mounted with iLas2 laser merge on the microscope, allowing simultaneous laser and LED illumination. Photobleaching experiments were carried out as described earlier (Vettiger et al, 2017). Image series of all experiments with TssA‐ or TagA‐mNeonGreen fusions as well as all P. aeruginosa image series were subjected to deconvolution. Deconvolution was carried out using Huygens Remote Manager (http://huygens-rm.org). Background estimation was set to auto, 40 iterations were run, quality change stopping criterion was 0.1, and deconvolution algorithm used was classic maximum‐likelihood estimation.
Fluorescence quantification
lacO arrays were designed to contain repeats of the operator sequence (aattgtgagcggataacaatt) with 15 random nucleotide spacers. LacI351 lacking C‐terminal tetramerization domain, which binds DNA like WT (Gregory et al, 2010) was fused to mNeonGreen and used to estimate number of mNeonGreen molecules needed to detect fluorescent spots of certain intensity in the cells. Raw mNeonGreen signal from LacI, TagAVC, and TssAVC spots was collected using ImageJ. mNeonGreen signal of three consecutive images of LacI or TssAVC/TagAVC spots (approximately 5 × 5 pixels) in a cell was averaged. Subsequently, an average signal of the cell cytosol of the corresponding frames was subtracted from the average signal of the spot and compared to random spots of similar size inside the cell. All measurements were done on unprocessed images.
Co‐IP and mass spectrometry
Co‐IP was performed using Pierce Anti‐HA Magnetic Beads. Cells were grown in LB medium to OD 600 nm of 1 with appropriate supplements, and HA‐tagged protein expression was induced with 0.002% l‐arabinose. Cells were lysed using lysozyme and CelLytic B (Sigma), and lysates were used directly for binding to Anti‐HA Magnetic Beads. Binding and washing were done according to the recommended protocol (Pierce). Proteins were eluted by boiling at 98°C for 10 min and resuspended in TN‐buffer, reduced, and alkylated. Samples were subsequently used for mass spectrometry analysis as described previously (Brackmann et al, 2017). Briefly, proteins were digested and supplemented with TFA to a final concentration of 1% overnight. Peptides were cleaned with PreOmics Cartridges (PreOmics, Martinsried, Germany) following the manufacturer's instructions. After drying under vacuum, peptides were resuspended in 0.1% aqueous formic acid solution at a concentration of 0.5 mg/ml. 0.5 μg of peptides of each sample was subjected to LC‐MS analysis as described earlier (Brackmann et al, 2017). MS1 and MS2 scans were recorded at a target of 1E6 ions and 10,000 ions, respectively. One microscan was acquired for each spectrum, and collision energy was 35%. All raw data acquired by DDA were converted to mgf format (version 3.0, ProteoWizard, http://proteowizard.sourceforge.net/). Resulting files were searched against a decoy (consisting of forward and reverse protein sequences) database of predicted protein sequence of V. cholerae (UniProt, Organism ID: 243277, download date 11/09/2017, containing known contaminants) using Mascot (Matrix Science, version 2.4). Search parameters were as follows: Full tryptic specificity was required (cleavage after lysine and arginine residues unless followed by proline); up to three missed cleavages allowed; carbamidomethyl (C) was set as a fixed modification; oxidation (M) and acetyl (protein N‐term) were set as variable modifications; 0.6‐Da fragment mass tolerance for CID tandem mass spectra; and 10 ppm precursor mass tolerance. After importing results to Scaffold (http://www.proteomesoftware.com, version 4), FDR rate was set to < 1% for protein identifications by the local Scaffold FDR algorithm based on the number of decoy hits. Co‐IP and mass spectrometry results are summarized in Appendix Table S3.
tssA VC gene subcloning and protein expression
tssA VC (amino acids 1–469, Gene number: VC_A0119) of V. cholerae VC2740‐80 was cloned into a modified pACEBACI (Geneva Biotech) expression vector containing a GATEWAY (Life Technologies) cassette with N‐terminal His10 tag according to the manufacturer's protocol. Bacmid and virus were generated in Sf21 cells (Expression systems) in Insect‐Xpress medium (Lonza), following the MultiBac instructions. Mycoplasma contamination was tested using MycoAlert™ Mycoplasma Detection Kit (Lonza), and no contamination was detected. Protein expression was done according to MultiBac instructions. Cells were harvested 3 days after infection by centrifugation and stored at −80°C until further use.
TssAVC protein purification
TssAVC protein was purified at 4°C. 40 g of cell pellet was resuspended in 160 mL of lysis buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 20 mM imidazole, 10% glycerol, 1× complete protease inhibitors, and 5 mM 2‐mercaptoethanol) and passed 3 times on a microfluidizer operating at 15k psi. The cell lysate was clarified by centrifugation at 95k g for 1 h. The supernatant was filtered using 0.22‐μm filters and applied to a 5 ml HisTrap HP affinity column (GE Healthcare) pre‐equilibrated in lysis buffer. The column was washed with 10 CV of NiW buffer (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 50 mM imidazole, 10% glycerol, and 5 mM 2‐mercaptoethanol) and eluted with elution buffer (25 mM Tris–HCl pH 7, 100 mM NaCl, 750 mM imidazole, 5% glycerol, and 5 mM 2‐mercaptoethanol). Wash and elution fractions were checked on an SDS gel (Appendix Fig S3A). Eluate fractions were pooled, mixed with TEV protease at 1:10 (m/m), and dialyzed overnight against gel filtration buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT) in 10 kDa MWCO membrane. TEV protease digested protein was concentrated using a Millipore 100 kDa centrifugal device and injected to a 10/300 Superose 6 increase column (GE Healthcare) equilibrated with gel filtration buffer. Peak fractions at 2 mg/ml were immediately used for cryo‐EM grid preparation. Due to the preferential orientation of the particles in the vitreous ice, sodium cholate at CMC concentration was added. Detergent selection was monitored by tilt angular distribution of reconstructed volumes.
Cryo‐EM data acquisition
An aliquot of 3 μl of TssAVC sample was applied onto non‐glow‐discharged Lacey holey carbon grids (Carbon Cu 300 mesh, Ted Pella), blotted for 3 s, and vitrified using a Leica EM GP2 (Leica Microsystems). The chamber was kept at 20°C and 90% humidity during the blotting process. Images were collected on an FEI Titan Krios TEM (ThermoFisher Scientific), operated at 300 kV, and equipped with a Gatan Quantum‐LS energy filter (zero loss slit width 20 eV; Gatan Inc.); data were recorded with a post‐GIF K2‐Summit direct electron detector (Gatan Inc.) in electron counting mode (60 e/Å2 total dose, fractionated into 40 frames), at a calibrated pixel size of 1.058 Å. A total of 2,963 movies were collected using SerialEM (Mastronarde, 2005).
Image processing and model building
TssAVC
Recorded movies were preprocessed online with FOCUS (Biyani et al, 2017). Movies were drift‐corrected and dose‐weighted using MotionCor2 (Zheng et al, 2017). The contrast transfer function of each micrograph was estimated using the CTFFIND4.1 (Rohou & Grigorieff, 2015). An initial particle set was picked using Gautomatch without templates (developed by Zhang, K., MRC Laboratory of Molecular Biology, Cambridge, UK) and then 2D classified in cryoSPARC v1 (Punjani et al, 2017). Particles selected from the best 2D classes were used for ab‐initio reconstruction and subsequent 3D refinement to 10 Å resolution without imposing symmetry. Reference‐based projections were generated, low‐pass filtered to 20 Å using EMAN2 (Tang et al, 2007), and used as templates to pick 261,412 particles with Gautomatch. After several rounds of 2D classification in RELION3 (Zivanov et al, 2018), best 93,235 particles were selected from 2D class averages showing tilted and side views in addition to preferential top or bottom views. Consensus 3D refinement iterated with two rounds of 3D classification revealed the best class with well‐resolved CTD density. A final 3D refinement with a soft‐mask that included only CTD density, followed by postprocessing resulted in a map with a global resolution of 3.9 Å (Appendix Fig S2). Global auto‐sharpening using PHENIX (Afonine et al, 2018) was applied to the map after final 3D refinement, and resulted sharpened map was used for model building. Local resolution calculation was done using the ResMap web service (Kucukelbir et al, 2014), and 3D Fourier shell correlation was calculated with 3DFSC web service (Tan et al, 2017). One dimensional histogram for a tilt angle was plotted using plot_indivEuler_histogram_fromStarFile.py script (https://github.com/leschzinerlab/).
Homology model of CTD TssAVC was generated by SWISS‐MODEL (Waterhouse et al, 2018) based on the X‐ray structure of C‐terminal domain from Nt2‐CTD domains of TssAAH from Dix et al (PDB‐6G7C). Initial rigid‐body fitting was done with COLORES (Wriggers, 2012) and further refined with PHENIX real space refinement (Adams et al, 2010). Rounds of manual adjustment in Coot (Emsley et al, 2010) were iterated with structure reliability evaluation using MolProbity web service (Appendix Table S4) (Chen et al, 2010).
TssAVC Nt2
Symmetry of the C6 reconstruction of TssAVC was relaxed to C1, and one of the six Nt2 dimers was focused 3D refined to 10 Å resolution. The resulting reconstruction was used for Gautomatch template creation. 1,184,380 particles were picked and 2D classified several times to clean the data set from false positives. Best 33,501 particles from 2D class averages with visible alpha helices were selected and processed first with RELION3, and further refined using non‐uniform refinement in cryoSPARC v2, resulted in 6.6 Å resolution reconstruction. A homology model of Nt2 TssAVC was generated with SWISS‐MODEL based on the X‐ray structure of Nt2 domain from Nt2‐CTD domains of Ah TssA from Dix et al (PDB‐6G7C) and further refined with molecular dynamics flexible fitting (MDFF) and phenix.real_space_refine using Namdinator web service (Kidmose et al, 2019). To find relative orientations of Nt2 dimer and CTD ring, partial signal subtraction and 3D classification without alignment were used. Two masks were created, first around one of Nt2‐dimers corresponding density and second around five symmetry‐related copies. Symmetry of the C6 reconstruction of TssAVC was relaxed to C1, and five symmetry‐related copies of Nt2 dimer were subtracted from all C6‐C1 expanded particles, and 3D classified without alignment into ten classes using T‐factor of 10. 3D class with improved Nt2‐dimer density was selected, and corresponding particles were re‐extracted without subtraction, and further 3D refined with local angular searches without imposing symmetry (C1) to 10 Å resolution. Resulted reconstruction was C6 symmetrized and rigid‐body fitted into cryo‐EM reconstruction of the TssAVC T6SS distal end (EMD‐3878) (Fig 3D). Finally, previously reconstructed Nt2‐dimer domain (Fig 3C) was rigid‐body fitted into symmetrized reconstruction (Fig 3D).
Author contributions
JPS generated strains, performed experiments, and processed and analyzed data. SN and RA performed sample purification, SN performed cryo‐EM data collection, processing, and model building. ML generated strains and performed bacterial two‐hybrid experiments. PDR assisted in image data analysis. HS assisted and supported cryo‐EM data collection. MB conceived the project and analyzed the data. JPS and MB wrote the manuscript. All authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Review Process File
Acknowledgements
The work was supported by Swiss National Science Foundation Grants BSSGI0_155778 and 31003A_159525 as well as the University of Basel. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing core facility at University of Basel. We acknowledge the Biozentrum BioEM Lab for access to their microscopes, and Mohamed Chami and Lubomir Kovacik for support with cryo‐EM data collection. We also thank Dr. Anna Hagmann and Prof. Timm Maier for their help with cloning and initial trials of TssAVC protein expression.
The EMBO Journal (2019) 38: e100825
Data availability
EM maps: Electron Microscopy Data Bank EMD‐4898 (https://www.ebi.ac.uk/pdbe/emdb/).
Electron micrographs: Electron Microscopy Public Image Archive EMPIAR‐10271 (https://www.ebi.ac.uk/pdbe/emdb/empiar/).
Atomic coordinates: Research Collaboratory for Structural Bioinformatics Protein Data Bank PDB 6RIU (http://www.rcsb.org).
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, Urzhumtsev A (2018) New tools for the analysis and validation of cryo‐EM maps and atomic models. Acta Crystallogr Sect Struct Biol 74: 814–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A (2017) RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 114: 7707–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F, Tschopp J, Van Driel R, Engel J (1979) Reassembly of the bacteriophage T4 tail from the core‐baseplate and the monomeric sheath protein P18: a co‐operative association process. J Mol Biol 132: 369–386 [DOI] [PubMed] [Google Scholar]
- Aschtgen M‐S, Thomas MS, Cascales E (2010) Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence 1: 535–540 [DOI] [PubMed] [Google Scholar]
- Barret M, Egan F, O'Gara F (2013) Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environ Microbiol Rep 5: 117–126 [DOI] [PubMed] [Google Scholar]
- Basler M, Mekalanos JJ (2012) Type 6 secretion dynamics within and between bacterial cells. Science 337: 815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail‐like structure. Nature 483: 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Ho BT, Mekalanos JJ (2013) Tit‐for‐Tat: type VI secretion system counterattack during bacterial cell‐cell interactions. Cell 152: 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Straight AF (1998) In vivo visualization of chromosomes using lac operator‐repressor binding. Trends Cell Biol 8: 121–124 [DOI] [PubMed] [Google Scholar]
- Biyani N, Righetto RD, McLeod R, Caujolle‐Bert D, Castano‐Diez D, Goldie KN, Stahlberg H (2017) Focus: the interface between data collection and data processing in cryo‐EM. J Struct Biol 198: 124–133 [DOI] [PubMed] [Google Scholar]
- Böck D, Medeiros JM, Tsao H‐F, Penz T, Weiss GL, Aistleitner K, Horn M, Pilhofer M (2017) In situ architecture, function, and evolution of a contractile injection system. Science 357: 713–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A (2009) Remodelling of VipA/VipB tubules by ClpV‐mediated threading is crucial for type VI protein secretion. EMBO J 28: 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackmann M, Nazarov S, Wang J, Basler M (2017) Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol 27: 623–632 [DOI] [PubMed] [Google Scholar]
- Brodmann M, Dreier RF, Broz P, Basler M (2017) Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat Commun 8: 15853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms JE, Sjöstedt A, Lavander M (2010) The role of the francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol 1: 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE (2011) The biochemical properties of the Francisella pathogenicity island (FPI)‐encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology 157: 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E (2015) The type VI secretion TssEFGK‐VgrG phage‐like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet 11: e1005545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli FR, Monlezun L, Coulthurst SJ (2016) Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24: 51–62 [DOI] [PubMed] [Google Scholar]
- Corbitt J, Yeo JS, Davis CI, LeRoux M, Wiggins PA (2018) Type VI secretion system dynamics reveals a novel secretion mechanism in Pseudomonas aeruginosa . J Bacteriol 200: e00744–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer P, Venter SN, Kamber T, Duffy B, Coutinho TA, Smits THM (2011) Comparative genomics of the Type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genom 12: 576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix SR, Owen HJ, Sun R, Ahmad A, Shastri S, Spiewak HL, Mosby DJ, Harris MJ, Batters SL, Brooker TA et al (2018) Structural insights into the function of type VI secretion system TssA subunits. Nat Commun 9: 4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Spott S, Zimmermann O, Kisters‐Woike B, Müller‐Hill B, Barker A (1999) Dimerisation mutants of lac repressor. I. a monomeric mutant, L251A, that binds lac operator DNA as a dimer 11 Edited by M. Yaniv. J Mol Biol 290: 653–666 [DOI] [PubMed] [Google Scholar]
- Douzi B, Brunet YR, Spinelli S, Lensi V, Legrand P, Blangy S, Kumar A, Journet L, Cascales E, Cambillau C (2016) Structure and specificity of the Type VI secretion system ClpV‐TssC interaction in enteroaggregative Escherichia coli . Sci Rep 6: 34405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Nguyen VS, Zoued A, Logger L, Péhau‐Arnaudet G, Aschtgen M‐S, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A et al (2015) Biogenesis and structure of a type VI secretion membrane core complex. Nature 523: 555–560 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felisberto‐Rodrigues C, Durand E, Aschtgen M‐S, Blangy S, Ortiz‐Lombardia M, Douzi B, Cambillau C, Cascales E (2011) Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ‐TssM complex of an Escherichia coli pathovar. PLoS Pathog 7: e1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster A, Planamente S, Manoli E, Lossi NS, Freemont PS, Filloux A (2014) Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J Biol Chem 289: 33032–33043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH (2015) Atomic structures of a bactericidal contractile nanotube in its pre‐ and postcontraction states. Nat Struct Mol Biol 22: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Jung J, Tuwatananurak I, Pogliano K (2010) Transposon Assisted Gene Insertion Technology (TAGIT): a tool for generating fluorescent fusion proteins. PLoS ONE 5: e8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein N, Bönemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A (2013) ClpV recycles VipA/VipB tubules and prevents non‐productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87: 1013–1028 [DOI] [PubMed] [Google Scholar]
- Karimova G, Gauliard E, Davi M, Ouellette SP, Ladant D (2017) Protein‐protein interaction: bacterial two‐hybrid. Methods Mol Biol 1615: 159–176 [DOI] [PubMed] [Google Scholar]
- Kidmose RT, Juhl J, Nissen P, Boesen T, Karlsen JL, Pedersen BP (2019) Namdinator – automatic molecular dynamics flexible fitting of structural models into cryo‐EM and crystallography experimental maps. IUCrJ 6: 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD (2014) Quantifying the local resolution of cryo‐EM density maps. Nat Methods 11: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashev M, Wang RY‐R, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M (2015) Structure of the type VI secretion system contractile sheath. Cell 160: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yao Y, Xu HH, Hao L, Deng Z, Rajakumar K, Ou H‐Y (2015) SecReT6: a web‐based resource for type VI secretion systems found in bacteria. Environ Microbiol 17: 2196–2202 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51 [DOI] [PubMed] [Google Scholar]
- Mintz KP, Fives‐Taylor PM (2000) impA, a gene coding for an inner membrane protein, influences colonial morphology of Actinobacillus actinomycetemcomitans. Infect Immun 68: 6580–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S et al (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312: 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov S, Schneider JP, Brackmann M, Goldie KN, Stahlberg H, Basler M (2018) Cryo‐EM reconstruction of Type VI secretion system baseplate and sheath distal end. EMBO J 37: e97103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VS, Logger L, Spinelli S, Legrand P, Huyen Pham TT, Nhung Trinh TT, Cherrak Y, Zoued A, Desmyter A, Durand E et al (2017) Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor‐binding proteins and evolved to bind the membrane complex. Nat Microbiol 2: 17103 [DOI] [PubMed] [Google Scholar]
- Planamente S, Salih O, Manoli E, Albesa‐Jové D, Freemont PS, Filloux A (2016) TssA forms a gp6‐like ring attached to the type VI secretion sheath. EMBO J 35: 1613–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103: 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA (2017) cryoSPARC: algorithms for rapid unsupervised cryo‐EM structure determination. Nat Methods 14: 290–296 [DOI] [PubMed] [Google Scholar]
- Ringel PD, Hu D, Basler M (2017) The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep 21: 3927–3940 [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N (2015) CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192: 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB et al (2014) A type VI secretion‐related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S (2012) The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and fur and modulates internalization in epithelial cells. J Biol Chem 287: 27095–27105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin YG, Doan T, Lebrun R, Espinosa L, Journet L, Cascales E (2018) In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat Microbiol 3: 1304 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Pilhofer M (2019) Bidirectional contraction of a type six secretion system. Nat Commun 10: 1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, Lyumkis D (2017) Addressing preferred specimen orientation in single‐particle cryo‐EM through tilting. Nat Methods 14: 793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46 [DOI] [PubMed] [Google Scholar]
- Vettiger A, Basler M (2016) Type VI secretion system substrates are transferred and reused among sister cells. Cell 167: 99–110.e12 [DOI] [PubMed] [Google Scholar]
- Vettiger A, Winter J, Lin L, Basler M (2017) The type VI secretion system sheath assembles at the end distal from the membrane anchor. Nat Commun 8: 16088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Brackmann M, Castaño‐Díez D, Kudryashev M, Goldie KN, Maier T, Stahlberg H, Basler M (2017) Cryo‐EM structure of the extended type VI secretion system sheath–tube complex. Nat Microbiol 2: 1507–1512 [DOI] [PubMed] [Google Scholar]
- Wang J, Brodmann M, Basler M (2019) Assembly and subcellular localization of bacterial type VI secretion systems. Annu Rev Microbiol 73: 28.1–28.18 [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L et al (2018) SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46: W296–W303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W (2012) Conventions and workflows for using Situs. Acta Crystallogr D Biol Crystallogr 68: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J‐P, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. Elife 7: e42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoued A, Durand E, Brunet YR, Spinelli S, Douzi B, Guzzo M, Flaugnatti N, Legrand P, Journet L, Fronzes R et al (2016) Priming and polymerization of a bacterial contractile tail structure. Nature 531: 59–63 [DOI] [PubMed] [Google Scholar]
- Zoued A, Durand E, Santin YG, Journet L, Roussel A, Cambillau C, Cascales E (2017) TssA: the cap protein of the type VI secretion system tail. BioEssays 39: 1600262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Review Process File
Data Availability Statement
EM maps: Electron Microscopy Data Bank EMD‐4898 (https://www.ebi.ac.uk/pdbe/emdb/).
Electron micrographs: Electron Microscopy Public Image Archive EMPIAR‐10271 (https://www.ebi.ac.uk/pdbe/emdb/empiar/).
Atomic coordinates: Research Collaboratory for Structural Bioinformatics Protein Data Bank PDB 6RIU (http://www.rcsb.org).


