Abstract
The results on associations of tumor necrosis factor (TNF)-receptor associated factor 1/complement component 5 (TRAF1/C5) rs10818488 and rs3761847 polymorphisms with rheumatoid arthritis (RA) are controversial, thus this study was performed to examine whether the aforementioned polymorphisms were associated with RA in a Chinese population. Furthermore, an updated meta-analysis was conducted. The polymorphisms were genotyped in 328 Chinese RA patients and 449 healthy controls. Studies examining the association of TRAF1/C5 rs10818488 and/or rs3761847 polymorphism with RA were exhaustively searched. No significant difference in either genotype or allele distribution between RA patients and controls was found. The updated meta-analysis was conducted based on 19 articles including the present study. A significant association of RA with TRAF1/C5 rs10818488 polymorphism G allele in Europeans (OR = 0.843, 95% CI = 0.730-0.975, p = 0.021) and in Asians (OR = 1.070, 95% CI = 1.009-1.136, p = 0.024) was found. Additionally, a significant association of RA with TRAF1/C5 rs10818488 polymorphism G allele under the recessive model in Asians (OR = 1.129, 95% CI = 1.023-1.246, p = 0.016) and in Africans (OR = 0.657, 95% CI = 0.507-0.851, p = 0.001) was found. Only a borderline significant association of RA with TRAF1/C5 rs3761847 polymorphism A allele was found in Europeans. Non-significant associations of RA with TRAF1/C5 rs10818488 and rs3761847 polymorphisms were found in our study. The updated meta-analysis results demonstrate that TRAF1/C5 rs10818488 polymorphism is associated with RA in Europeans, Asians and Africans, and TRAF1/C5 rs3761847 polymorphism is associated with RA in Europeans with borderline significant evidence.
Keywords: rheumatoid arthritis, TRAF1, C5, polymorphism, meta-analysis
Introduction
Rheumatoid arthritis (RA) is one of the most frequent chronic inflammatory diseases worldwide, predominantly affecting peripheral joints. Auto-antibodies such as the rheumatoid factor (RF) and anti-citrullinated protein antibody (ACPA) are commonly present in the serum of patients with RA [1]. If the optimal therapeutic opportunity of this disease is missed, cartilage and bone damage as well as disability would ensue, resulting in a huge socioeconomic burden due to disability and high medication costs [2]. Women are more affected than men, and the sex ratio of RA has been estimated to be about 1 : 3. There is a striking reduction in the incidence of RA from north to south (in the northern hemisphere) and from urban to rural areas [1]. The highest prevalence of RA has been reported in Native American populations [3] and a very low prevalence in some areas of Africa [4]. During the past decades, rapid and enormous achievements have been made in the field of genetic epidemiology of RA, and more than one hundred genetic loci/genes underpinning RA have been detected [5], and this is undoubtedly owing to the advancement in high-throughput genotyping technology.
In 2007, two single-nucleotide polymorphisms (SNPs) within the tumor necrosis factor (TNF)-receptor associated factor 1/complement component 5 (TRAF1/C5) loci were identified to be significantly associated with RA in Europeans from one large-scale candidate gene study (rs10818488) [6] and one GWAS (rs3761847) [7]. The TRAF1/C5 region located on 9q33-34 contains TRAF1 gene as well as C5 gene, both of which might be implicated in the development of RA based on their biological functions. TRAF1 is a member of the TRAF protein family, and could transduce the signal through binding multiple protein kinases and adaptor proteins for multiple receptors of TNF superfamily (TNFSF) members including TNF-α [8], which have been suggested to be therapeutic targets of rheumatic diseases including RA [9]. Indeed, TNF-α inhibitors have been proved as effective treatment drugs for RA [10]. Additionally, TRAF1 has also been involved in the activation and proliferation of T cells [11]. TRAF1-deficient KRN/I-Ag7 mice spontaneously developed severe and progressive arthritis, and the production of anti-GPI autoantibody was markedly impaired by TRAF1 deficiency [12]. The serum levels of TRAF1 were significantly higher in RA patients, and the expression of TRAF1 in serum was found to be correlated with both disease activity and the titers of GPI, RF antibodies [13]. Significantly increased levels of complement activation fragments have been found in the blood and synovial fluid of patients with RA [14-17]. The C5-deficient mice were refractory to collagen-induced arthritis [18], and anti-C5 antibody was found to ameliorate disease in animal models of arthritis [19, 20]. As a result, both TRAF1 and C5 are considered to play an important role in the pathogenesis of RA.
Based on the consideration of genetic heterogeneity among different ethnic populations and possible false positive results, multiple studies have been subsequently performed to test whether the TRAF1/C5 rs10818488 and rs3761847 polymorphisms are associated with RA in different ethnic populations [21-39]. Nevertheless, the results were inconsistent, and this discrepancy might be due to different sample sizes, genetic backgrounds, clinical heterogeneity and publication bias, etc. Meta-analysis is a useful tool to combine the results on the same topic to get a pooled estimation with an increased statistical power [40]. Indeed, several meta-analyses on the associations of TRAF1/C5 rs10818488 and rs3761847 polymorphisms with RA have been reported [41-44]. However, some additional studies on the relationships between TRAF1/C5 rs10818488 and rs3761847 polymorphisms and RA have been reported in recent years [21-24]. Thus, the aims of this study included: 1) examining the associations of TRAF1/C5 rs10818488 and rs3761847 polymorphisms with RA in a Chinese population and 2) performing an updated meta-analysis to re-summarize the effect sizes of the TRAF1/C5 rs10818488 and rs3761847 polymorphisms associated with RA.
Material and methods
Case-control study
Three hundred and twenty-eight patients with RA were recruited from the Department of Rheumatology, Ningbo First Hospital. All patients with RA were diagnosed based on the American College of Rheumatology 1987 revised criteria for the classification of RA [45] or the 2010 American College of Rheumatology/European League Against Rheumatism (EULAR) criteria for RA [46]. Patients’ information such as disease duration, anti-CCP status and RF status were collected through reviewing medical records or by questionnaire. A total of 449 healthy control subjects without any signs or symptoms of autoimmune diseases were collected from the physical examination center of Ningbo First Hospital. This study was reviewed and approved by the ethics committee of the Ningbo University, and informed consent was obtained from all participants.
DNA extraction and SNP genotyping
EDTA anti-coagulated venous blood samples were obtained from all participants. Genomic DNA was extracted from peripheral blood lymphocytes according to the standard procedures. The genotypes of TRAF1/C5 rs10818488 and rs3761847 polymorphisms were determined by Shanghai Biowing Applied Biotechnology Co., Ltd. (www.biowing.com.cn) applying the Ligase Detection ReactionPolymerase Chain Reaction (LDR-PCR) technology.
Statistical analysis
The χ2 goodness-of-fit test was applied to test whether the genotype distributions of the TRAF1/C5 rs10818488 and rs3761847 polymorphisms among the healthy control group conformed to Hardy-Weinberg equilibrium (HWE). The difference in allele frequencies between RA patients and controls was tested by chi-square test. The difference in genotype frequencies between RA patients and controls and the associations of RA with TRAF1/C5 rs10818488 and rs3761847 polymorphisms under different genetic models (dominant model and recessive model) were evaluated by the logistic regression model with the adjustment of sex and age. All corresponding odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. A two-tailed p-value less than 0.05 was considered statistically significant. All above-mentioned statistical analysis was performed by PASW Statistics 18.0 software (SPSS, Inc., Somers, NY, USA).
Meta-analysis
Identification of eligible studies and data extraction
An exhaustive search on studies examining the association of TRAF1/C5 rs10818488 and/or rs3761847 polymorphism with genetic susceptibility to RA was conducted. Databases including PubMed, China National Knowledge Infrastructure (CNKI) database and Wanfang database were searched to identify relevant articles, using the following medical subject heading (MeSH) terms and/or text words: “tumor necrosis factor-receptor associated factor 1”, “TRAF1”, “complement component 5”, “C5”, “TRAF1/C5”, “polymorphism”, “polymorphisms”, “variant”, “variants”, “rheumatoid arthritis” and “RA”. No language restrictions were applied. Additional studies were found through reviewing the references in relevant articles. The inclusion criteria were as follows: 1) being published before August 2017; 2) applying the case-control study design; 3) allele or genotype frequencies among RA patients and controls available, providing enough data to calculate the odds ratio (OR); and 4) the genotype distribution among the control group should conform to HWE, since deviation from HWE among controls could imply some potential bias in control selection or genotyping errors. When two or more studies published by the same research group included overlapped data, only the data from the study with the largest sample size were extracted. When a study contained the results in different subgroups, we treated them independently. Studies in which family members had been studied were excluded because its analysis was based on linkage consideration. The following information from each study was extracted: first author’s name, year of publication, country, ethnicity, number of cases and controls, and allele/genotype distribution in cases and controls. Two authors (Si-Chao Huang and Dong-Jin Hua) independently extracted these data, and discrepancy was addressed by discussion.
Evaluation of the statistical association
Allele frequencies of the TRAF1/C5 rs10818488 and rs3761847 polymorphisms from each study were determined by the allele counting method. Chi-square test was employed to evaluate whether the observed genotype frequencies in the control group conformed to HWE. We performed meta-analysis on A) allelic contrast, B) dominant model, and C) recessive model. The heterogeneity between studies was assessed by Cochran’s Q statistic, as well as I2 statistic, which was used to quantify the effect of heterogeneity (I2 = 100% × (Q − df) / Q), measuring the proportion of total variation in study estimates due to heterogeneity [47]. Finally, the pooled estimate of risk was obtained by a random effects (Der Simonian-Laird) or a fixed effects model (Mantel-Haenszel) according to whether there was (p ≤ 0.1 or I2 > 50%) or not (p > 0.1 and I2 ≤ 50%) heterogeneity, respectively. Subgroup meta-analysis stratified by ethnicity was performed, due to the genetic heterogeneity among different ethnicities. Statistical analysis for this meta-analysis was performed by Stata version 10.0 (Stata Corporation, College Station, TX, USA).
Evaluation of publication bias
Potential publication bias was assessed by the funnel plot, and the Egger’s linear regression test was utilized to evaluate the funnel plot asymmetry, which is a measure to detect funnel plot asymmetry on the natural logarithm scale of the OR [48]. The significance of the intercept was assessed by the t-test suggested by Egger, and the p-value less than 0.05 was considered significant publication bias. If asymmetry was found, the “trim and fill” method was applied to adjust summary estimates for observed bias [49].
Results
The mean age of RA patients was 53.77 years, and 84.45% of patients were female. The mean disease duration of RA patients was 8.55 years, according to 191 RA patients with disease duration information. Two hundred and sixty-seven and 302 patients with RA had data on anti-CCP and RF status, and the proportion of anti-CCP-positive and RF-positive RA patients were 80.15% and 65.56%, respectively. The mean age of healthy controls was 51.10 years, and 37.42% were female. There was a significant difference in age distribution (p < 0.01) and sex composition (p < 0.01) between RA patients and healthy controls, so association analysis was conducted by the multivariate logistic regression model to adjust age and sex except for the comparison of allele frequencies between RA patients and healthy controls.
Association of TRAF1/C5 rs10818488 polymorphism with genetic susceptibility to rheumatoid arthritis
The genotype and allele frequencies of the TRAF1/C5 rs10818488 polymorphism are shown in Table 1. No deviation from Hardy-Weinberg equilibrium was observed for TRAF1/C5 rs10818488 polymorphism in the control group (p = 0.57). No significant difference was found in either genotype (p = 0.23) or allele (G vs. A, p = 0.44, OR = 1.08, 95% CI = 0.88-1.32) distribution of the TRAF1/C5 rs10818488 polymorphism between patients with RA and healthy controls. The associations between the major allele G of TRAF1/C5 rs10818488 polymorphism and the risk of RA under dominant and recessive models were assessed, whereas non-significant evidence was found (G/G + A/G vs. A/A, p = 0.92, OR = 0.98, 95% CI = 0.67-1.43; G/G vs. A/G + A/A, p = 0.12, OR = 1.33, 95% CI = 0.93-1.92).
Table 1.
Genotype and allele frequencies distribution of TRAF1 rs10818488 polymorphism among rheumatoid arthritis (RA) patients and controls
| TRAF1 rs10818488 | RA patients (n = 327) n (%) | Controls (n = 446) n (%) | p | OR (95% CI) |
|---|---|---|---|---|
| Genotype | 0.23 | |||
| G/G | 100 (30.6) | 118 (26.5) | ||
| A/G | 145 (44.3) | 217 (48.7) | ||
| A/A | 82 (25.1) | 111 (24.9) | ||
| Allele | 0.44 | 1.08 (0.88-1.32) | ||
| G | 345 (52.8) | 453 (50.8) | ||
| A | 309 (47.2) | 439 (49.2) | ||
| Dominant model | 0.92 | 0.98 (0.67-1.43) | ||
| G/G + A/G | 245 (74.9) | 335 (75.1) | ||
| A/A | 82 (25.1) | 111 (24.9) | ||
| Recessive model | 0.12 | 1.33 (0.93-1.92) | ||
| G/G | 100 (30.6) | 118 (26.5) | ||
| A/G + A/A | 227 (69.4) | 328 (73.5) | ||
Regarding the genotype comparison and associations under the genetic models (dominant model and recessive model), the p values and corresponding ORs (95% CIs) were calculated with the adjustment for age and sex, and the p values were not corrected for multiple comparison
The patients with RA were classified as seropositive and seronegative groups based on their anti-CCP and RF status, then the associations between the TRAF1/C5 rs10818488 polymorphism with genetic susceptibility to different serotypes of RA were determined. However, no significant association was found (Supplementary Tables 1-4).
Association of TRAF1/C5 rs3761847 polymorphism with genetic susceptibility to rheumatoid arthritis
The genotype distribution of TRAF1/C5 rs3761847 polymorphism among the control group did not deviate from Hardy-Weinberg equilibrium (p = 0.61). As shown in Table 2, non-significant evidence was found in neither genotype (p = 0.77) nor allele (A vs. G, p = 0.69, OR = 1.04, 95% CI = 0.85-1.28) distribution of TRAF1/C5 rs3761847 polymorphism between RA patients and healthy controls. In addition, we did not find any significant association of the major allele A of TRAF1/C5 rs3761847 polymorphism with genetic susceptibility to RA under either dominant model or recessive model (A/G + A/G vs. G/G, p = 0.90, OR = 0.98, 95% CI = 0.65-1.46; A/A vs. A/G + G/G, p = 0.53, OR = 1.12, 95% CI = 0.78-1.61).
Table 2.
Genotype and allele frequencies distribution of TRAF1 rs3761847 polymorphism among rheumatoid arthritis (RA) patients and controls
| TRAF1 rs3761847 | RA patients (n = 320) n (%) | Controls (n = 445) n (%) | p | OR (95% CI) |
|---|---|---|---|---|
| Genotype | 0.77 | |||
| A/A | 98 (30.6) | 130 (29.2) | ||
| A/G | 153 (47.8) | 216 (48.5) | ||
| G/G | 69 (21.6) | 99 (22.2) | ||
| Allele | 0.69 | 1.04 (0.85-1.28) | ||
| A | 349 (54.5) | 476 (53.5) | ||
| G | 291 (45.5) | 414 (46.5) | ||
| Dominant model | 0.90 | 0.98 (0.65-1.46) | ||
| A/A + A/G | 251 (78.4) | 346 (77.8) | ||
| G/G | 69 (21.6) | 99 (22.2) | ||
| Recessive model | 0.53 | 1.12 (0.78-1.61) | ||
| A/A | 98 (30.6) | 130 (29.2) | ||
| A/G + G/G | 222 (69.4) | 315 (70.8) | ||
Regarding the genotype comparison and associations under the genetic models (dominant model and recessive model), the p values and corresponding ORs (95% CIs) were calculated with the adjustment for age and sex, and the p values were not corrected for multiple comparison
Similar to the TRAF1/C5 rs10818488 polymorphism, non-significant evidence for association of the TRAF1/C5 rs3761847 polymorphism with genetic susceptibility to different serotypes of RA was found (Supplementary Tables 5-8).
Studies included in the meta-analysis
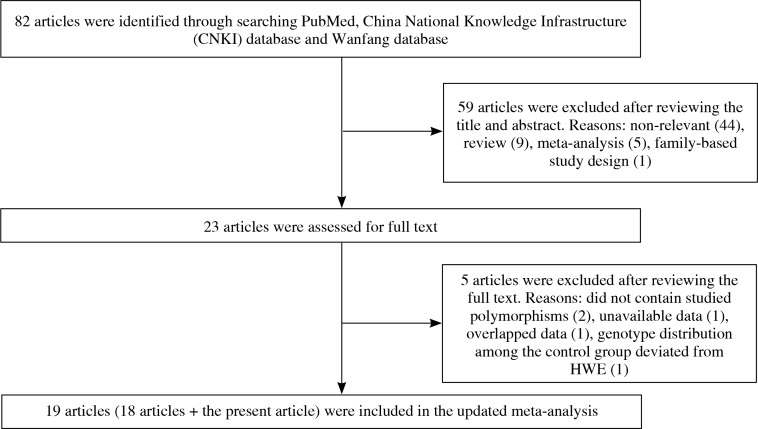
As shown in Figure 1, a total of 23 relevant articles examining the association of TRAF1/C5 rs10818488 and/or rs3761847 polymorphism with genetic susceptibility to RA were identified through PubMed, CNKI database, Wanfang database search and a review of the references [6, 7, 21-39, 50, 51], and 5 articles were excluded [37-39, 50, 51]. One article was excluded since the detailed genotype and allele information is unavailable [37]. Two articles were published by the same research group, so only the article with the largest sample size was included [31] and the other one was excluded [38]. One article in which the genotype distributions of TRAF1/C5 rs10818488 and rs3761847 polymorphisms among the control group did not conform to HWE was excluded [39]. Two articles were excluded, in which the TRAF1/C5 rs10818488 and/or rs3761847 polymorphism was not included [50, 51]. Thus, the updated meta-analysis was conducted based on a total of 19 articles including the present study.
Fig. 1.
The flowchart for the study selection process
Among the 19 articles, there are 8 articles in which only TRAF1/C5 rs10818488 polymorphism has been investigated [6, 21, 22, 24-26, 33, 35], 4 articles in which only TRAF1/C5 rs3761847 polymorphism has been investigated [7, 27, 29, 31], and the other 7 articles in which both TRAF1/C5 rs10818488 and rs3761847 polymorphisms have been studied [present study, 23, 28, 30, 32, 34, 36]. Three article contained data on four different subgroups [6, 7, 30], and we treated them independently. Finally, a total of 19 articles with 21 case-control studies for TRAF1/C5 rs3761847 polymorphism and 17 case-control studies for TRAF1/C5 rs3761847 polymorphism were eligible for this updated meta-analysis. The characteristics of the selected studies are summarized in Table 3 (the genotype and allele frequencies in selected studies are shown in Supplementary Tables 9 and 10).
Table 3.
Characteristics of individual studies included in the present meta-analysis
| Author | Year | Country | Ethnicity | Sample size | Polymorphisms studied | HWE (p-value) | |
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| The present study | 2017 | China | Asian | 328 | 449 | rs10818488; rs3761847 | 0.57; 0.61 |
| Moaaz et al. [21] | 2016 | Egypt | African | 105 | 75 | rs10818488 | 0.23 |
| Fodil et al. [22] | 2015 | Western Algeria | African | 110 | 196 | rs10818488 | 0.56 |
| Liang et al. [23] | 2015 | China | Asian | 300 | 300 | rs10818488; rs3761847 | 0.73; 0.84 |
| Ahmadlou et al. [24] | 2014 | Iran | Asian | 362 | 422 | rs10818488 | 0.58 |
| Fakhfakh et al. [25] | 2012 | Tunisia | African | 108 | 161 | rs10818488 | 0.42 |
| Mohamed et al. [26] | 2012 | Egypt | African | 172 | 160 | rs10818488 | 0.10 |
| Danoy et al. [27] | 2011 | China | Asian | 1035 | 1702 | rs3761847 | NA |
| El-Gabalawy et al. [28] | 2011 | Canada | North American Native | 333 | 490 | rs10818488; rs3761847 | 0.66; 0.66 |
| Zhu et al. [29] | 2011 | China | Asian | 576 | 689 | rs3761847 | 0.06 |
| Nishimoto et al. (1) [30] | 2010 | Japan | Asian | 1504 | 752 | rs10818488; rs3761847 | 0.13; 0.18 |
| Nishimoto et al. (2) [30] | 2010 | Japan | Asian | 830 | 658 | rs10818488; rs3761847 | 0.80; 0.90 |
| Nishimoto et al. (3) [30] | 2010 | Japan | Asian | 1113 | 940 | rs10818488; rs3761847 | 0.49; 0.60 |
| Nishimoto et al. (4) [30] | 2010 | Japan | Asian | 950 | 507 | rs10818488; rs3761847 | 0.09; 0.10 |
| Han et al. [31] | 2009 | Korea | Asian | 1301 | 996 | rs3761847 | 0.48 |
| Stark et al. [32] | 2009 | Slovakia | European | 520 | 303 | rs10818488; rs3761847 | 0.29; 0.28 |
| Zervou et al. [33] | 2008 | Greece | European | 311 | 344 | rs10818488 | 0.16 |
| Barton et al. [34] | 2008 | UK | European | 3418 | 3337 | rs10818488; rs3761847 | 0.35; 0.33 |
| Palomino-Morales et al. [35] | 2008 | Colombia | Colombian | 274 | 421 | rs10818488 | 0.44 |
| Kurreeman et al. (1) [6] | 2007 | Netherlands | European | 251 | 241 | rs10818488 | NA |
| Kurreeman et al. (2) [6] | 2007 | Netherlands | European | 406 | 262 | rs10818488 | NA |
| Kurreeman et al. (3) [6] | 2007 | Sweden | European | 1500 | 912 | rs10818488 | NA |
| Kurreeman et al. (4) [6] | 2007 | USA | European | 470 | 469 | rs10818488 | NA |
| Plenge et al. (1) [7] | 2007 | USA | European | 863 | 1182 | rs3761847 | NA |
| Plenge et al. (2) [7] | 2007 | Sweden | European | 632 | 653 | rs3761847 | NA |
| Plenge et al. (3) [7] | 2007 | USA | European | 485 | 1282 | rs3761847 | NA |
| Plenge et al. (4) [7] | 2007 | Sweden | European | 568 | 516 | rs3761847 | NA |
| WTCCC [36] | 2007 | UK | European | 1858 | 2935 | rs10818488; rs3761847 | 0.26; 0.25 |
HWE – Hardy-Weinberg equilibrium, NA – not available, WTCCC – Wellcome Trust Case Control Consortium
Meta-analysis of the TRAF1/C5 rs10818488 polymorphism with genetic susceptibility to rheumatoid arthritis
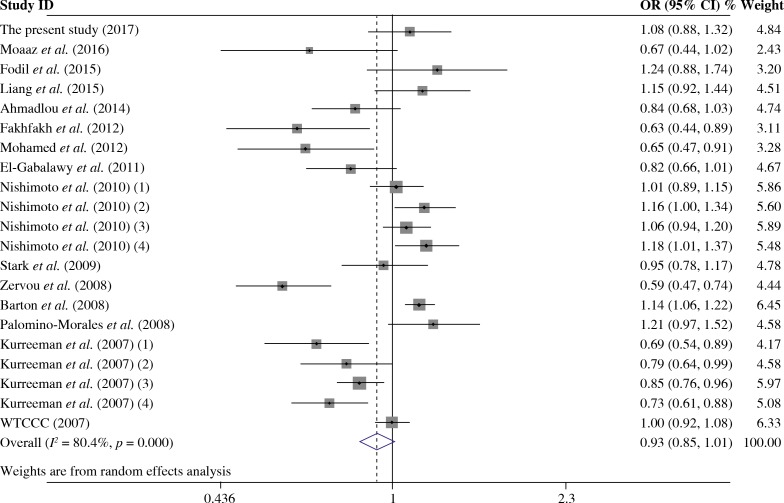
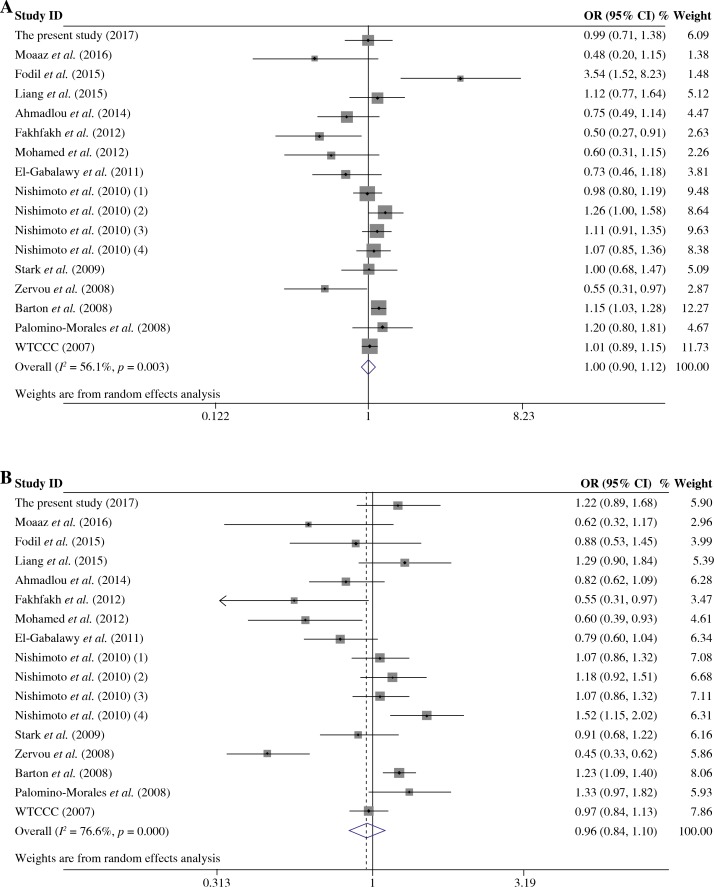
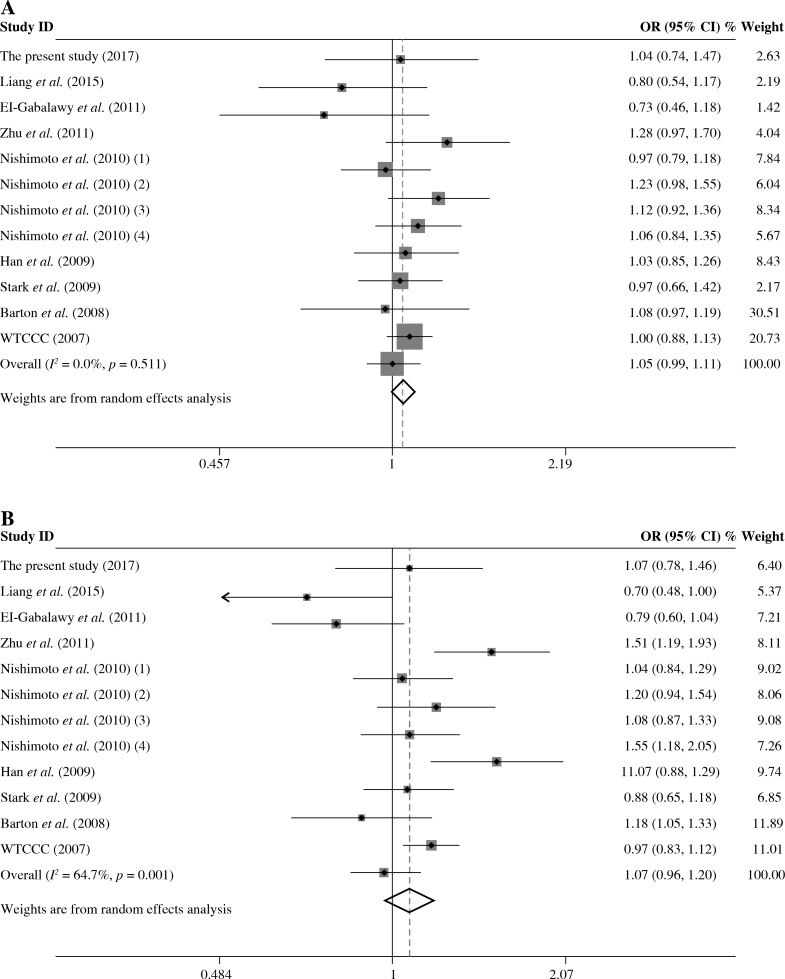
The summary of meta-analysis for association of the TRAF1/C5 rs10818488 polymorphism with genetic susceptibility to RA is shown in Table 4, and a total of 15171 cases and 13998 controls in 21 case-control studies were eligible for this meta-analysis. Among these eligible studies, eight studies were from Europeans, seven from Asians, four from Africans, one from North American Native and one from Colombian. Non-significant evidence for the association of TRAF1/C5 rs10818488 polymorphism with RA was detected in the overall meta-analysis (G vs. A, OR = 0.925, 95% CI = 0.851-1.006, p = 0.069; G/G + A/G vs. A/A, OR = 1.002, 95% CI = 0.899-1.117, p = 0.966; G/G vs. A/G + A/A, OR = 0.961, 95% CI = 0.838-1.102, p = 0.569) (Figs. 2 and 3). When we pooled the results stratified by ethnicity, significant evidence was detected for the association of RA with the TRAF1/C5 rs10818488 polymorphism G allele in Europeans (G vs. A, OR = 0.843, 95% CI = 0.730-0.975, p = 0.021) and in Asians (G vs. A, OR = 1.070, 95% CI = 1.009-1.136, p = 0.024). In addition, a significant association was found for the association of RA with TRAF1/C5 rs10818488 polymorphism G allele under the recessive model in Asians (G/G vs. A/G + A/A, OR = 1.129, 95% CI = 1.023-1.246, p = 0.016) and in Africans (G/G vs. A/G + A/A, OR = 0.657, 95% CI = 0.507-0.851, p = 0.001).
Table 4.
Meta-analysis of the TRAF1 rs10818488 polymorphism with genetic susceptibility to rheumatoid arthritis (RA)
| Polymorphism | Comparison | Population | Sample size | No. of studies | Test of association | Model | Test of heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR | 95% CI | p | Q | p | I2 (%) | |||||
| TRAF1 rs10818488 | G vs. A | Overall | 15171 | 13998 | 21 | 0.925 | 0.851-1.006 | 0.069 | R | 102.10 | < 0.001 | 80.4 |
| European | 8716 | 8496 | 8 | 0.843 | 0.730-0.975 | 0.021 | R | 61.48 | < 0.001 | 88.6 | ||
| Asian | 5353 | 3999 | 7 | 1.070 | 1.009-1.136 | 0.024 | F | 9.12 | 0.167 | 34.2 | ||
| African | 495 | 592 | 4 | 0.768 | 0.552-1.069 | 0.117 | R | 10.22 | 0.017 | 70.7 | ||
| NAN | 333 | 490 | 1 | 0.816 | 0.658-1.011 | 0.062 | NA | NA | NA | NA | ||
| Colombian | 274 | 421 | 1 | 1.215 | 0.974-1.515 | 0.084 | NA | NA | NA | NA | ||
| G/G + A/G vs. A/A | Overall | 12544 | 12114 | 17 | 1.002 | 0.899-1.117 | 0.966 | R | 36.45 | 0.003 | 56.1 | |
| European | 6089 | 6612 | 4 | 1.018 | 0.864-1.201 | 0.828 | R | 7.87 | 0.049 | 61.9 | ||
| Asian | 5353 | 3999 | 7 | 1.065 | 0.968-1.170 | 0.196 | F | 5.95 | 0.429 | 0 | ||
| African | 495 | 592 | 4 | 0.825 | 0.350-1.947 | 0.661 | R | 16.45 | 0.001 | 81.8 | ||
| NAN | 333 | 490 | 1 | 0.733 | 0.457-1.177 | 0.199 | NA | NA | NA | NA | ||
| Colombian | 274 | 421 | 1 | 1.202 | 0.799-1.810 | 0.377 | NA | NA | NA | NA | ||
| G/G vs. A/G + A/A | Overall | 12544 | 12114 | 17 | 0.961 | 0.838-1.102 | 0.569 | R | 68.38 | < 0.001 | 76.6 | |
| European | 6089 | 6612 | 4 | 0.860 | 0.618-1.197 | 0.370 | R | 35.31 | < 0.001 | 91.5 | ||
| Asian | 5353 | 3999 | 7 | 1.129 | 1.023-1.246 | 0.016 | F | 10.50 | 0.105 | 42.8 | ||
| African | 495 | 592 | 4 | 0.657 | 0.507-0.851 | 0.001 | F | 1.88 | 0.598 | 0 | ||
| NAN | 333 | 490 | 1 | 0.789 | 0.597-1.043 | 0.097 | NA | NA | NA | NA | ||
| Colombian | 274 | 421 | 1 | 1.328 | 0.971-1.816 | 0.075 | NA | NA | NA | NA | ||
OR – odds ratio, CI – confidence interval, F – fixed effects model, R – random effects model, NAN – North American native, NA – not applicable, WTCCC – Wellcome Trust Case Control Consortium
Fig. 2.
Forest plot for the meta-analysis of the association between TRAF1/C5 rs10818488 polymorphism G allele and rheumatoid arthritis
Fig. 3.
Forest plot for the meta-analysis of the association between TRAF1/C5 rs10818488 polymorphism G allele and rheumatoid arthritis (A) under the dominant model, (B) under the recessive model
Meta-analysis of the TRAF1/C5 rs3761847 polymorphism with genetic susceptibility to rheumatoid arthritis
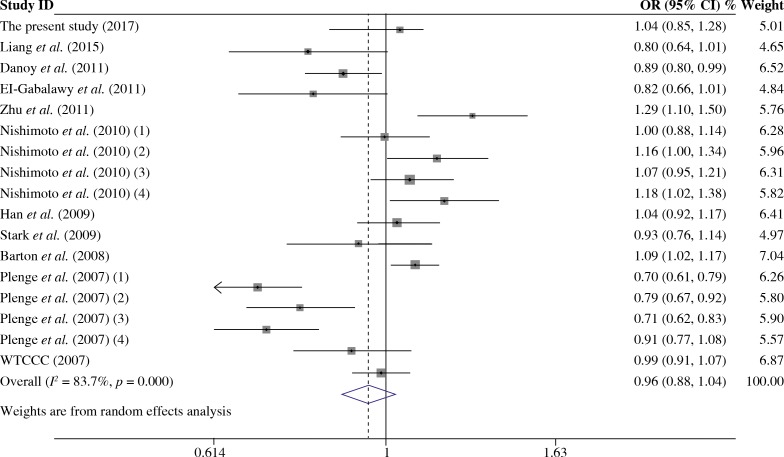
As presented in Table 5, a total of 17 studies including 16553 cases and 17659 were included for the meta-analysis on the association between the TRAF1/C5 rs3761847 polymorphism with genetic susceptibility to RA. Among these eligible studies, seven studies were from Europeans, nine from Asians and one from North American Native. Overall meta-analysis revealed that the TRAF1/C5 rs3761847 polymorphism was not significantly associated with genetic susceptibility to RA (A vs. G, OR = 0.955, 95% CI = 0.882-1.035, p = 0.264; A/A + A/G vs. G/G, OR = 1.051, 95% CI = 0.993-1.112, p = 0.083; A/A vs. A/G + G/G, OR = 1.074, 95% CI = 0.964-1.197, p = 0.193) (Figs. 4 and 5). When the results were combined and stratified by ethnicity, only borderline significant evidence was detected for the association of RA with TRAF1/C5 rs3761847 polymorphism A allele in Europeans (A vs. G, OR = 0.867, 95% CI = 0.751-1.000, p = 0.050). However, non-significant association between the TRAF1/C5 rs3761847 polymorphism and RA was found in Asians (A vs. G, OR = 1.048, 95% CI = 0.962-1.142, p = 0.286; A/A + A/G vs. G/G, OR = 1.074, 95% CI = 0.988-1.168, p = 0.095; A/A vs. A/G + G/G, OR = 1.138, 95% CI = 0.968-1.313, p = 0.077).
Table 5.
Meta-analysis of the TRAF1 rs3761847 polymorphism with genetic susceptibility to rheumatoid arthritis (RA)
| Polymor-phism | Comparison | Population | Sample size | No. of studies | Test of association | Model | Test of heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR | 95% CI | p | Q | p | I2 (%) | |||||
| TRAF1 rs3761847 | A vs. G | Overall | 16553 | 17659 | 17 | 0.955 | 0.882-1.035 | 0.264 | R | 98.24 | < 0.001 | 83.7 |
| European | 8340 | 10204 | 7 | 0.867 | 0.751-1.000 | 0.050 | R | 59.49 | < 0.001 | 89.9 | ||
| Asian | 7880 | 6965 | 9 | 1.048 | 0.962-1.142 | 0.286 | R | 25.57 | 0.001 | 68.7 | ||
| NAN | 333 | 490 | 1 | 0.816 | 0.658-1.011 | 0.062 | NA | NA | NA | NA | ||
| A/A + A/G vs. GG | Overall | 12970 | 12324 | 12 | 1.051 | 0.993-1.112 | 0.083 | F | 10.21 | 0.511 | 0 | |
| European | 5792 | 6571 | 3 | 1.042 | 0.965-1.125 | 0.297 | F | 1.00 | 0.606 | 0 | ||
| Asian | 6845 | 5263 | 8 | 1.074 | 0.988-1.168 | 0.095 | F | 6.69 | 0.462 | 0 | ||
| NAN | 333 | 490 | 1 | 0.733 | 0.457-1.177 | 0.199 | NA | NA | NA | NA | ||
| A/A vs. A/G + GG | Overall | 12970 | 12324 | 12 | 1.074 | 0.964-1.197 | 0.193 | R | 31.17 | 0.001 | 64.7 | |
| European | 5792 | 6571 | 3 | 1.029 | 0.866-1.222 | 0.748 | R | 6.05 | 0.049 | 66.9 | ||
| Asian | 6845 | 5263 | 8 | 1.138 | 0.968-1.313 | 0.077 | R | 18.68 | 0.009 | 62.5 | ||
| NAN | 333 | 490 | 1 | 0.789 | 0.597-1.043 | 0.097 | NA | NA | NA | NA | ||
OR – odds ratio, CI – confidence interval, F – fixed effects model, R – random effects model, NAN – North American native, NA – not applicable, WTCCC – Wellcome Trust Case Control Consortium
Fig. 4.
Forest plot for the meta-analysis of the association between TRAF1/C5 rs3761847 polymorphism A allele and rheumatoid arthritis
Fig. 5.
Forest plot for the meta-analysis of the association between TRAF1/C5 rs3761847 polymorphism A allele and rheumatoid arthritis (A) under the dominant model, (B) under the recessive model
Evaluation of heterogeneity and publication bias
Heterogeneity was detected for the association of RA with TRAF1/C5 rs10818488 polymorphism in the overall meta-analysis. When we pooled results according to the ethnicity of study populations, the heterogeneity disappeared in the subgroup analysis of Asians and in the subgroup analysis of the association between RA and the TRAF1/C5 rs10818488 polymorphism G allele under the recessive model in Africans, but remained significant in other subgroup analysis. In addition, heterogeneity was found for the association of RA with TRAF1/C5 rs3761847 polymorphism A allele under allelic contrast and the recessive model in the overall meta-analysis. When we pooled results according to the ethnicity of study populations, the heterogeneity remained significant in the subgroup analysis of Europeans and Asians.
As shown in Table 6, publication bias analysis on the associations between the TRAF1/C5 rs10818488 and rs3761847 polymorphisms and genetic susceptibility to RA was performed. The only significant evidence for publication bias was found for the overall meta-analysis of the association between RA and the TRAF1/C5 rs10818488 polymorphism under allelic contrast (t = 2.71, p = 0.014). Therefore, the “trim and fill” method was used to adjust publication bias. However, the adjusted OR calculated remained the same and non-significant.
Table 6.
Publication bias tested by Egger’s test in the overall population
| Polymorphism | Comparisons | t | p-value |
|---|---|---|---|
| TRAF1 rs10818488 | G vs. A | 2.71 | 0.014 |
| G/G + A/G vs. A/A | 1.80 | 0.091 | |
| G/G vs. A/G + A/A | 1.96 | 0.069 | |
| TRAF1 rs3761847 | A vs. G | 1.19 | 0.251 |
| A/A + A/G vs. GG | 0.80 | 0.445 | |
| A/A vs. A/G + GG | 0.64 | 0.536 |
Discussion
Since the discoveries that the TRAF1/C5 rs10818488 and rs3761847 polymorphisms were associated with genetic predisposition to RA reaching a genome-wide significance level have been reported [6, 7], multiple studies have been performed to examine whether these two genetic polymorphisms are involved in the genetic background of RA among different ethnic populations. In the present study, we first determined whether the TRAF1/C5 rs10818488 and rs3761847 polymorphisms were associated with RA in a Chinese population, then conducted an updated meta-analysis to combine the results from different studies to re-summarize the effect sizes. However, non-significant evidence for the associations of TRAF1/C5 rs10818488 and rs3761847 polymorphisms with RA was detected in our study population. Emerging evidence suggests that RA is a heterogeneous disease with a significant discrepancy in the phenotype as demonstrated by differences in autoantibody positivity, and it has been accepted that the anti-CCP/RF-positive RA patients and anti-CCP/RF-negative RA patients have different genetic architectures [52]. Thus, the relationships between TRAF1/C5 rs10818488 and rs3761847 polymorphisms and different serotypes of RA patients were also evaluated, whereas non-significant evidence was found.
In 2007, in view of the functions of C5 and TRAF1, which are encoded by immune-related genes and adjacent to each other on chromosome 9q33-34, Kurreeman et al. selected the TRAF1/C5 genes as candidate genes for RA and performed a multitiered case-control study, and the peak evidence for association was detected from the rs10818488 polymorphism surpassing the genome-wide significance level, which is located in the intergenic region between C5 and TRAF1 [6]. In order to investigate the potential functional effect of the rs10818488 polymorphism, they scanned for transcription factor binding sites, and this polymorphism was found to be a potential binding site for EP300, a histone acetyltransferase that regulates transcription via chromatin remodeling. Thus, the rs10818488 polymorphism may be involved in the regulation of the neighboring TRAF1 and/or C5 gene [6]. Afterwards, another study found a significant difference between risk allele carriers and non-carriers of rs10818488 for the expression level of TRAF1 in phorbol myristate acetate-stimulated lymphoblastoid cell lines, indicating this polymorphism might be involved in the pathogenesis of RA through regulating the expression of TRAF1 under certain stimulation signals [30]. Subsequently, many studies have been performed to evaluate the association between TRAF1/C5 rs10818488 polymorphism and RA in different ethnic populations, whereas the results were inconsistent. In the current study, no significant evidence was detected for difference in either genotype or allele distribution between RA patients and healthy controls, and our results were consistent with Liang et al. [23], another study performed in Chinese population.
In addition to the rs10818488 polymorphism, the rs3761847 polymorphism, also located in the intergenic region between C5 and TRAF1, was found to be associated with RA reaching a genome-wide level of significance from a GWAS [7]. Notably, a recent study has revealed a possible mechanism by which the TRAF1/C5 rs3761847 polymorphism contributed to RA [53]. In that study, the monocytes from healthy human subjects with the homozygotes of risk allele of rs3761847 was found to express less TRAF1 protein but greater amounts of inflammatory cytokines in response to lipopolysaccharide (LPS). Further experiments showed that the TRAF1 MATH domain could bind directly to the linear ubiquitination (LUBAC) complex, and interfere with the recruitment and linear ubiquitination of NEMO, resulting in decreased NF-κB activation and cytokine production. In addition, TRAF1-/- mice showed increased susceptibility to LPS-induced septic shock. All these findings provided a mechanistic explanation for the increased inflammation caused by RA-associated rs3761847 polymorphism [53]. Similar to the rs10818488 polymorphism, the rs3761847 polymorphism has also attracted attention and many studies have been performed to test the relationship between the rs3761847 polymorphism and genetic susceptibility to RA. In our study, we also did not find significant evidence for the association between this polymorphism and RA, and our results were consistent with Danoy et al. [27], another study performed in Chinese population. However, a significant association was found in another study performed in Chinese population by Zhu et al. [29], and the estimated effect size for allelic comparison was moderate (OR = 1.28).
Although we did not find significant associations of RA with the TRAF1/C5 rs10818488 and rs3761847 polymorphisms, it should be noted that the non-significant results might be owing to the relatively lower power caused by a small sample size or the “winner’s curse” [54] – an overestimation of the true effect in the positive report accounting for the lack of power.
Despite the associations between TRAF1/C5 rs10818488 and rs3761847 polymorphisms and RA have been assessed in different ethnic populations, the results are controversial, and this discrepancy might be attributed to many factors including different sample sizes, genetic backgrounds and clinical heterogeneity, etc. Meta-analysis is a useful tool to combine the results on the same topic to get a pooled estimation with an increased statistical power [40]. Although some meta-analyses on the relationships of TRAF1/C5 rs10818488 and rs3761847 polymorphisms with RA have been reported [41-44], some additional studies on this topic have been reported in recent years. Thus, we performed this updated meta-analysis to re-evaluate whether the TRAF1/C5 rs10818488 and rs3761847 polymorphisms are associated with the genetic predisposition to RA and to re-summarize the effect sizes of these polymorphisms associated with RA.
Our overall meta-analysis results revealed that non-significant association of the TRAF1/C5 rs10818488 polymorphism G allele with RA was detected. Since heterogeneity was found in the overall meta-analysis of the TRAF1/C5 rs10818488 polymorphism G allele with RA susceptibility, we performed stratified meta-analysis by ethnicity, and the heterogeneity disappeared in the subgroup analysis of Asians but remained significant in Europeans and Africans, and a significant association of RA and TRAF1/C5 rs10818488 polymorphism G allele was observed in Europeans and Asians. Notably, the TRAF1/C5 rs10818488 polymorphism G allele was found to be a protective allele in Europeans, but a risk allele in Asians. This might be due to the genetic heterogeneity between Europeans and Asians, and this might also be caused by chance. Thus, further studies with larger sample sizes are needed to clarify whether the TRAF1/C5 rs10818488 polymorphism G allele is associated with RA with the opposite direction in different ethnic populations. Non-significant evidence for association between RA and the TRAF1/C5 rs10818488 polymorphism G allele under the dominant model and recessive model was found in the overall metaanalysis. There was also significant heterogeneity when the overall meta-analysis of the association between RA and the TRAF1/C5 rs10818488 polymorphism G allele under different genetic models was performed, so subgroup meta-analysis stratified by ethnicity was conducted, and the results showed that a significant association was found for the association of RA with the TRAF1/C5 rs10818488 polymorphism G allele under the recessive model in Asians and Africans. Our updated meta-analysis indicated that the TRAF1/C5 rs10818488 polymorphism is involved in the genetic background of RA in Asians, Europeans and Africans.
The overall meta-analysis did not reveal significant association between RA and the TRAF1/C5 rs3761847 polymorphism A allele under allelic contrast, dominant model and recessive model. Heterogeneity was detected for the association of RA with the TRAF1/C5 rs3761847 polymorphism A allele under allelic contrast and recessive model in the overall meta-analysis. When the results were pooled according to the ethnicity, the heterogeneity remained significant in the subgroup analysis of Europeans and Asians. In the subgroup meta-analysis stratified by ethnicity, only board-line significant evidence was found for the association of RA with TRAF1/C5 rs3761847 polymorphism A allele under allelic contrast. Thus, the present updated meta-analysis showed that the TRAF1/C5 rs3761847 polymorphism might be involved in the genetic background of RA in Europeans.
Several limitations of the present study should be acknowledged. First, the sample size of our case-control study is not large enough, leading to a relatively lower power to detect associations with moderate effect sizes. Second, age and gender were imbalanced between RA patients and controls in the present study, whereas the logistic regression model with the adjustment of age and sex was employed to overcome this limitation. Third, significant heterogeneity was found among several comparisons, which might distort the meta-analysis. Finally, this meta-analysis was performed based on uncorrected estimate; if the potential confounding factors, such as age, sex, clinical characteristic, and environmental factors were available, a more precise meta-analysis could be conducted to obtain reliable results.
In conclusion, we did not find significant evidence for associations of RA with TRAF1/C5 rs10818488 and rs3761847 polymorphisms in the present study. The updated meta-analysis results demonstrate that the TRAF1/C5 rs10818488 polymorphism is involved in the genetic susceptibility of RA in Asians, Europeans and Africans, and the TRAF1/C5 rs3761847 polymorphism is associated with genetic susceptibility to RA in Europeans with borderline significant evidence.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81602921), Nature Science Foundation of Ningbo city (Grant No. 2016A610159), Medical and Health Planned Science and Technology Project of Zhejiang Province (Grant No. 2017KY582), Ningbo Scientific Innovation Team for Environmental Hazardous Factor Control and Prevention (Grant No. 2016C51001) and K.C. Wong Magna Fund in Ningbo University.
Footnotes
The authors declare no conflict of interest.
References
- 1.Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388:2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 2.Schneider M, Ostendorf B, Specker CH. Early diagnosis of rheumatoid arthritis. Z Rheumatol. 2005;64:516–523. doi: 10.1007/s00393-005-0790-9. [DOI] [PubMed] [Google Scholar]
- 3.Harvey J, Lotze M, Stevens MB, et al. Rheumatoid arthritis in a Chippewa Band. I. Pilot screening study of disease prevalence. Arthritis Rheum. 1981;24:717–721. doi: 10.1002/art.1780240515. [DOI] [PubMed] [Google Scholar]
- 4.Silman AJ, Ollier W, Holligan S, et al. Absence of rheumatoid arthritis in a rural Nigerian population. J Rheumatol. 1993;20:618–622. [PubMed] [Google Scholar]
- 5.Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurreeman FA, Padyukov L, Marques RB, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4:e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis – a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 9.Croft M, Siegel RM. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat Rev Rheumatol. 2017;13:217–233. doi: 10.1038/nrrheum.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolen JS, Landewé R, Bijlsma J, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann Rheum Dis. 2017;76:960–977. doi: 10.1136/annrheumdis-2016-210715. [DOI] [PubMed] [Google Scholar]
- 11.So T, Nagashima H, Ishii N. TNF Receptor-Associated Factor (TRAF) Signaling Network in CD4(+) T-Lymphocytes. Tohoku J Exp Med. 2015;236:139–154. doi: 10.1620/tjem.236.139. [DOI] [PubMed] [Google Scholar]
- 12.Cheng T, Choi Y, Finkel TH, et al. Tumor necrosis factor receptor-associated factor 1 influences KRN/I-Ag7 mouse arthritis autoantibody production. J Clin Immunol. 2013;33:759–766. doi: 10.1007/s10875-013-9866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng T, Sun X, Wu J, et al. Increased serum levels of tumor necrosis factor receptor-associated factor 1 (TRAF1) correlate with disease activity and autoantibodies in rheumatoid arthritis. Clin Chim Acta. 2016;462:103–106. doi: 10.1016/j.cca.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Morgan BP, Daniels RH, Williams BD. Measurement of terminal complement complexes in rheumatoid arthritis. Clin Exp Immunol. 1988;73:473–478. [PMC free article] [PubMed] [Google Scholar]
- 15.Corvetta A, Pomponio G, Rinaldi N, et al. Terminal complement complex in synovial tissue from patients affected by rheumatoid arthritis, osteoarthritis and acute joint trauma. Clin Exp Rheumatol. 1992;10:433–438. [PubMed] [Google Scholar]
- 16.Høgåsen K, Mollnes TE, Harboe M, et al. Terminal complement pathway activation and low lysis inhibitors in rheumatoid arthritis synovial fluid. J Rheumatol. 1995;22:24–28. [PubMed] [Google Scholar]
- 17.Brodeur JP, Ruddy S, Schwartz LB, et al. Synovial fluid levels of complement SC5b-9 and fragment Bb are elevated in patients with rheumatoid arthritis. Arthritis Rheum. 1991;34:1531–1537. doi: 10.1002/art.1780341209. [DOI] [PubMed] [Google Scholar]
- 18.Hietala MA, Jonsson IM, Tarkowski A, et al. Complement deficiency ameliorates collagen-induced arthritis in mice. J Immunol. 2002;169:454–459. doi: 10.4049/jimmunol.169.1.454. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Rollins SA, Madri JA, et al. Anti-C5 monoclonal antibody therapy prevents collagen-induced arthritis and ameliorates established disease. Proc Natl Acad Sci U S A. 1995;92:8955–8959. doi: 10.1073/pnas.92.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macor P, Durigutto P, De Maso L, et al. Treatment of experimental arthritis by targeting synovial endothelium with a neutralizing recombinant antibody to C5. Arthritis Rheum. 2012;64:2559–2567. doi: 10.1002/art.34430. [DOI] [PubMed] [Google Scholar]
- 21.Moaaz M, Mohannad N. Association of the polymorphisms of TRAF1 (rs10818488) and TNFAIP3 (rs2230926) with rheumatoid arthritis and systemic lupus erythematosus and their relationship to disease activity among Egyptian patients. Cent Eur J Immunol. 2016;41:165–175. doi: 10.5114/ceji.2016.60991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fodil M, Benzaoui A, Zemani-Fodil F, et al. Association of PTPN22 (rs2476601) and STAT4 (rs7574865) polymorphisms with Rheumatoid Arthritis in the Western Algerian population. Acta Reumatol Port. 2015;40:56–62. [PubMed] [Google Scholar]
- 23.Liang AF, Fei Y. Association of the TRAF1/C5 gene polymorphism with rheumatoid arthritis in Han population of Guizhou Province. Int J Lab Med. 2015;36:310–312. [Google Scholar]
- 24.Ahmadlou S, Akhiani M, Salimzadeh A, et al. Lack of association between single nucleotide polymorphism rs10818488 in TRAF1/C5 region and rheumatoid arthritis in iranian population. Iran J Allergy Asthma Immunol. 2014;13:19–25. [PubMed] [Google Scholar]
- 25.Fakhfakh Karray E, Chalbi H, Ben Dhifallah I, et al. Association study of TRAF1-C5 polymorphism with susceptibility to rheumatoid arthritis in Tunisian population. Joint Bone Spine. 2012;79:331–332. doi: 10.1016/j.jbspin.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Mohamed RH, Pasha HF, El-Shahawy EE. Influence of TRAF1/C5 and STAT4 genes polymorphisms on susceptibility and severity of rheumatoid arthritis in Egyptian population. Cell Immunol. 2012;273:67–72. doi: 10.1016/j.cellimm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Danoy P, Wei M, Johanna H, et al. Association of variants in MMEL1 and CTLA4 with rheumatoid arthritis in the Han Chinese population. Ann Rheum Dis. 2011;70:1793–1797. doi: 10.1136/ard.2010.144576. [DOI] [PubMed] [Google Scholar]
- 28.El-Gabalawy HS, Robinson DB, Daha NA, et al. Non-HLA genes modulate the risk of rheumatoid arthritis associated with HLA-DRB1 in a susceptible North American Native population. Genes Immun. 2011;12:568–574. doi: 10.1038/gene.2011.30. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J, Zhang D, Wu F, et al. Single nucleotide polymorphisms at the TRAF1/C5 locus are associated with rheumatoid arthritis in a Han Chinese population. BMC Med Genet. 2011;12:53. doi: 10.1186/1471-2350-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimoto K, Kochi Y, Ikari K, et al. Association study of TRAF1-C5 polymorphisms with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese. Ann Rheum Dis. 2010;69:368–373. doi: 10.1136/ard.2008.104315. [DOI] [PubMed] [Google Scholar]
- 31.Han TU, Bang SY, Kang C, et al. TRAF1 polymorphisms associated with rheumatoid arthritis susceptibility in Asians and in Caucasians. Arthritis Rheum. 2009;60:2577–2584. doi: 10.1002/art.24759. [DOI] [PubMed] [Google Scholar]
- 32.Stark K, Rovenský J, Blazicková S, et al. Association of common polymorphisms in known susceptibility genes with rheumatoid arthritis in a Slovak population using osteoarthritis patients as controls. Arthritis Res Ther. 2009;11:R70. doi: 10.1186/ar2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zervou MI, Sidiropoulos P, Petraki E, et al. Association of a TRAF1 and a STAT4 gene polymorphism with increased risk for rheumatoid arthritis in a genetically homogeneous population. Hum Immunol. 2008;69:567–571. doi: 10.1016/j.humimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Barton A, Thomson W, Ke X, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17:2274–2279. doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palomino-Morales RJ, Rojas-Villarraga A, González CI, et al. STAT4 but not TRAF1/C5 variants influence the risk of developing rheumatoid arthritis and systemic lupus erythematosus in Colombians. Genes Immun. 2008;9:379–382. doi: 10.1038/gene.2008.30. [DOI] [PubMed] [Google Scholar]
- 36.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vernerova L, Spoutil F, Vlcek M, et al. A Combination of CD28 (rs1980422) and IRF5 (rs10488631) Polymorphisms Is Associated with Seropositivity in Rheumatoid Arthritis: A Case Control Study. PLoS One. 2016;11:e0153316. doi: 10.1371/journal.pone.0153316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HS, Korman BD, Le JM, et al. Genetic risk factors for rheumatoid arthritis differ in Caucasian and Korean populations. Arthritis Rheum. 2009;60:364–371. doi: 10.1002/art.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu TT, Zhao DB. Association of TRAF1 genes polymorphism with increased risk of rheumatoid arthritis in Han people. Chin J Rheumatol. 2011;15:22–25. [Google Scholar]
- 40.Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997;315:1533–1537. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Li W, Zhang X, et al. Association between polymorphism in TRAF1/C5 gene and risk of rheumatoid arthritis: a meta-analysis. Mol Biol Rep. 2014;41:317–324. doi: 10.1007/s11033-013-2864-0. [DOI] [PubMed] [Google Scholar]
- 42.Song GG, Bae SC, Kim JH, et al. Associations between TRAF1-C5 gene polymorphisms and rheumatoid arthritis: a meta-analysis. Immunol Invest. 2014;43:97–112. doi: 10.3109/08820139.2013.837917. [DOI] [PubMed] [Google Scholar]
- 43.Xu K, Peng H, Zhou M, et al. Association study of TRAF1/C5 polymorphism (rs10818488) with susceptibility to rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Gene. 2013;517:46–54. doi: 10.1016/j.gene.2012.12.092. [DOI] [PubMed] [Google Scholar]
- 44.Patsopoulos NA, Ioannidis JP. Susceptibility variants for rheumatoid arthritis in the TRAF1-C5 and 6q23 loci: a meta-analysis. Ann Rheum Dis. 2010;69:561–566. doi: 10.1136/ard.2009.109447. [DOI] [PubMed] [Google Scholar]
- 45.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 46.Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 47.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 48.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 50.Plant D, Flynn E, Mbarek H, et al. Investigation of potential non-HLA rheumatoid arthritis susceptibility loci in a European cohort increases the evidence for nine markers. Ann Rheum Dis. 2010;69:1548–1553. doi: 10.1136/ard.2009.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang M, Rowland CM, Garcia VE, et al. A large-scale rheumatoid arthritis genetic study identifies association at chromosome 9q33.2. PLoS Genet. 2008;4:e1000107. doi: 10.1371/journal.pgen.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirino Y, Remmers EF. Genetic architectures of seropositive and seronegative rheumatic diseases. Nat Rev Rheumatol. 2015;11:401–414. doi: 10.1038/nrrheum.2015.41. [DOI] [PubMed] [Google Scholar]
- 53.Abdul-Sater AA, Edilova MI, Clouthier DL, et al. The signaling adaptor TRAF1 negatively regulates Toll-like receptor signaling and this underlies its role in rheumatic disease. Nat Immunol. 2017;18:26–35. doi: 10.1038/ni.3618. [DOI] [PubMed] [Google Scholar]
- 54.Kraft P. Curses – winner’s and otherwise – in genetic epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. discussion 657-658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.