Abstract
Fifteen different genotypes of greater yam (Dioscorea alata) NGY-1, NGY-2, NGY-3, NGY-4, NGY-5, NGY-6, NGY-7, NGY-8, NGY-9, NGY-10, NGY-11, NGY-12, NGY-13, NGY-14 and Da-199 were procured from different places of south Gujarat, like Valsad, Navsari, Surat and Anand. Among the biochemical parameters total carbohydrate ranged between 51.87 and 87.85% from different genotypes, starch ranged from 47 to 80.67%, crude fat ranged from 0.6 to 2.32%, crude fibre ranged between 1.10 and 4.09%, anthocyanin content of genotypes ranged from 1.01 to 3.25 mg/g, beta-carotene content ranged between 0.97 and 1.88 µg/g. The antinutrients namely Diosgenin and Tannin ranged from 0.28 to 0.93% and 0.923 mg/100 g to 2.447 mg/100 g, respectively. RAPD analysis was also done by the help of 18 RAPD markers: OPA1, OPA2, OPA3, OPA13, OPB1, OPB6, OPB7, OPM1, OPM2, OPM4, OPM7, OPM11, OPM12, OPM13, OPM15, OPM16, OPM17 and OPM19 from which an average of 9.7 loci were detected with an average of 4.72 polymorphic loci which is 48.65% polymorphism per loci and 48.70% average polymorphism. From the overall study, it can be conferred that the study revealed highest carbohydrate content in NGY-3 (87.85%), fat (2.32%) and crude fiber content (4.09%) in NGY-11, β-carotene in NGY-7 (1.88 µg/g), anthocyanin in NGY-4(3.25 mg/g). Lowest tannin (0.923 mg/100 g) and diosgenin (0.28%) was found in NGY-6, and NGY-7 respectively. For each of the biochemical parameters, the varieties with the optimum values may be cultivated. As the molecular studies revealed NGY-2 and NGY-3 have 96% similarities, they may be the duplicate of the same genotypes which can be studied further for better germplasm conservation.
Keywords: Greater yam, Biochemical parameters, Antinutrients, RAPD
Introduction
Tuber crops are a major component of diet in Africa, America and South-East Asia. Among the different tuber crops, Dioscorea alata, commonly called greater yam or water yam, is a tropical plant originated from South East Asia, probably in Burma (Martin 1974). In India, it is extensively cultivated in Madhya Pradesh, North-eastern states, West Bengal, Bihar, Odisha, Uttar Pradesh, Kerala, Tamil Nadu and Maharashtra as a commercial crop. It is also cultivated in the districts of south Gujarat however; the present area under yam cultivation is small in Gujarat. Though the soil in Navsari district is montmorillonite clay type and suffers under water stagnant condition during kharif season, most of south Gujarat has fertile loamy and well drained soil and the sowing time of greater yam i.e.in the month of May remains between 35 °C (Max) and 25 °C (Min), an appropriate temperature for yam growing. Assured water availability, better rail and road connectivity to distant markets have further increased its scope of cultivation in south Gujarat.
Yams are primarily consumed for its carbohydrates and secondarily for protein, vitamins and other minerals (Baah et al. 2009). Behera et al. (2009) had reported highest 82.51% starch in greater yam in their study. Bhandari et al. (2003) evaluated nutritional compounds of wild yam and found that dry matter ranged from 19.8 to 30.5% on fresh weight basis and crude protein, ash, crude fat and crude fibre contents ranged from 1.6–3.1, 0.5–1.2, 0.2–0.3 and 0.6–1.5% of fresh weight, respectively. The study of antinutritional factors revealed that the water yam contains tannins (Ezeocha and Ojimelukwe 2012) and alkaloids like diosgenin have been identified as well (Contreras-Pacheco et al. 2013). Besides these high nutritive value, it has comparative advantage for sustainable production due to its better agronomic characteristics such as ease of propagation and yields, and longer storage of the fresh tubers. It is a sun loving plant and commercially propagated vegetatively by tubers.
The experiments contained in this publication was conducted at Navsari Agricultural University, situated in south Gujarat. A total of 15 genotypes used in this experiment were collected from different districts of south Gujarat like Navsari, Surat, Dang and Valsad and middle Gujarat i.e.; Anand. These genotypes were named as Navsari Greater Yam (NGY). Da-199 (a better adopting variety for this zone) was collected from Central Tuber Crops Research Institute (CTCRI), Thiruvananthapuram, India, and as check variety in this experiment. This collection was established over the last 7 years. Screening for quality parameters (nutritional and antinutritional factors) for establishment as variety of the genotype/s maintained by selection method was the prime objective of this project, as this is a clonally propagated crop and cross pollination is problematic. The different genotypes collected from different areas of south Gujarat may be similar due to its extensive clonal propagation and may had been introduced under different local name in different places. Hence, the objective of present study was to chararcterize biochemical composition of starch, total soluble sugar (sweeter test after cooking), crude fiber (better digestibility), crude oil (smooth food texture), total carbohydrate (if lower then antidiabetic), anthocyanin content, β-carotene (antioxidant) and antinutrients content (tannin, diosgenin) of 15 different genotypes of Dioscorea alata. This data set was then used for the hierarchical classification of the genotypes based on biochemical as well as molecular parameters to screen the best suited genotype in this region and to identify the duplication of the clonal propagated material under different local names.
Materials and methods
Experimental site
The present study was conducted at the Department of Soil Science and Agricultural Chemistry, N. M. College of Agriculture, Navsari Agricultural University, Navsari. The fresh tuber samples for experiment were collected from Horticultural farm, Navsari Agricultural University. Navsari, annual rainfall averages between 1000 and 1500 mm. Deep black and coastal alluvial soil is predominant in this region. Navsari is located at 20.95°N 72.93°E and it has an average elevation of 9 m (29′) above sea level.
Experimental materials
The genotypes of D. Alata cultivated by Horticultural Farm, Navsari Agricultural University Campus collected from different districts of south Gujarat region like Valsad, Navsari, Dang, Surat and the middle Gujarat region Anand are as follows: NGY-1, NGY-2, NGY-3, NGY-4, NGY-5, NGY-6, NGY-7, NGY-8, NGY-9, NGY-10, NGY-11, NGY-12, NGY-13, NGY-14 and Da-199. Afterwards these genotypes were procured by the Department of Soil Science and Agricultural Chemistry, NAU, Navsari for further investigation. The uninfected, healthy tubers were cured by the method of Eshel (2011). The selected tubers were cleaned with tissue paper without washing and the soils were removed followed by storing at 35 °C temperature with 90% relative humidity for 4 days. The tubers after curing were kept under the shade in open air and all the biochemical analyses were completed within 7 days after harvest.
Biochemical parameters
Total carbohydrates
Total carbohydrates were analyzed by anthrone method as described by Hedge and Hofreiter (1962). Dried tuber samples of 0.5 gm were mixed with 5 ml of 2.5 N HCl in a test tube and boiled over water bath at 85–90 °C for 3 h followed by extraction for 10 min. The extract was reacted with 4 ml of 0.2% anthrone reagent and heated for 8 min in boiling water bath. The intensity of the green color was read at 630 nm using Spectrophotometer. The amount of total carbohydrate was determined from the standard curve of glucose and expressed as gm/100 gm (%) dry weight.
Total soluble sugar
The soluble sugars were determined by anthrone method as described by Timpa et al. (1985). Dried tuber samples of 0.5 gm were mixed with 15 ml of 80% ethanol in a test tube and boiled over water bath at 85–90 °C for 30 min followed by extraction. Supernatant were collected and evaporated in a water bath at 85–90 °C, resulting residues were used to determine the soluble sugar content. For reaction mixture, 0.5 ml aliquot was taken in a test tube and volume was made up to 1 ml with distilled water and mixed with 4 ml of 0.2% anthrone followed by 8 min in boiling and reading the colour intensity at 630 nm in spectrophotometer. The amount of soluble sugar was determined from the standard curve of glucose and expressed as gm/100 gm (%) dry weight.
Starch content
For starch analysis (Hedge and Hofreiter 1962), the dried tuber samples of 0.2 gm was added with hot 80% alcohol followed by centrifuging at 3000 rpm for 10 min. The supernatant was then decanted. And the process was repeated 5–6 times to remove sugar contents. The residue left behind was then cooled by placing it in ice water followed by addition of 6.5 ml of 52% perchloric acid with constant stirring using a glass rod, centrifuged and the supernatant was collected. The step was repeated 4–5 times and the supernatants were then pooled and the volume made up to 100 ml with water. After that 0.5 ml aliquots were reacted with 0.2% 10 ml of freshly prepared anthrone reagent and placed in boiling water bath for 7.5 min. After cooling, the absorbance of the mixture was noted at 630 nm in a spectrophotometer. Standard curve was prepared using 0–100 μg of glucose and starch content was expressed in % (gm/100 gm).
Crude fat
Total fat content of the sample was determined by the procedure of AOAC (1984). Dried powdered tuber sample of 5 gm was taken in cellulose thimble, held in extraction tube for continuous extraction with petroleum ether (boiling point 40 °C–60 °C) till solvent in the extractor became colourless. After removal of ether by distillation, the flask containing the residue was kept in an oven at 80 °C for 10 min, cooled in desiccators and weighed. The crude fat content was expressed as % dry weight.
Crude fibre
Crude fibre estimation was carried out by Maynard (1970). Two gm of sample was extracted with ether or petroleum ether to remove fat followed by drying and then was boiled with 200 ml of H2SO4 for 30 min and then filtered through muslin cloth and washed with boiling water until washing are free of acid. The residue was boiled with 200 ml of NaOH for 30 min followed by filtering through muslin cloth again and washed with 25 ml of boiling H2SO4, three 50 ml portion of water and 25 ml alcohol. The difference in the weight of the residue before and after of ashing at 600 ± 150 °C was recorded and fibre content in the tubers was expressed as % dry matter.
Anthocyanin content
Anthocyanin content of the sample was determined using a pH differential method described by Hosseinian et al. (2008). Raw tissues (10 gm) were homogenized with 15 ml of 80% methanol and centrifuged at 2000 rpm for 20 min. The supernatant was collected and final volume made up to 100 ml with distilled water. For analysis, two separate solutions of each samples were prepared, one for pH 1.0 using 0.025 M potassium chloride buffer, with hydrochloric acid (HCl) slowly added to the mixture to adjust the pH to 1.0. The other for pH 4.5 using 0.4 M sodium acetate buffer, using acetic acid to adjust the pH of the mixture to 4.5. The pH of the mixture was read using a calibrated pH meter and 0.5 ml of the sample was added to 1.5 ml of each buffer solution followed by adjustment to the suitable pH. The absorbance of each mixture was measured at both 520 and 700 nm against distilled water as a blank. Anthocyanin pigment concentration was calculated using molar extinction coefficient (ε = 26 900) and expressed as cyanidine-3-glucoside equivalents and the units used was mg/gm.
Beta (β) carotene
Total beta carotene content was determined by a slight modification of the method of Karnjanawipagul et al. (2010). Standard β-carotene for identification was prepared in hexane to obtain 4 μg/mL. Five grams of fresh tubers was blended with a pinch of sodium carbonate and mixed with a mechanical blender. Then the sample mixture was transferred into a centrifuge tube, added with 10 ml Tetrahydrofuran and mixed for 2 min under cold water. The mixture was centrifuged at 5000 g for 5 min and the supernatant was collected. Extraction was performed by adding 15 mL dichloromethane and 15 mL of 10% w/v NaCl into the supernatant and shaken for 2 min. The extraction was repeated twice, organic layer was collected and evaporated under nitrogen steam and the residue dissolved in 25 ml hexane followed by spectrophotometric observation at 450 nm and comparing with the standard curve of beta carotene for different concentrations and was expressed as (µg/gm).
Anti-nutritional factors
Diosgenin
The quantity of Diosgenin in samples was determined as described by Baccou et al. (1977) and Umetsu et al. (2000). The dried flakes of tuber were oven dried at 70 °C for 72 h., 1 g of the pulverized material was placed in a plastic tube (50 ml capacity, centrifugable grade) and 30 ml methanol was added to the tubes and left on the shaker overnight. The extract was centrifuged at 3500 rpm for 18 min and the supernatant collected. Two subsequent extractions were carried out and the supernatants collected. The final volume was adjusted to 100 ml. For analysis, two colour developing reagent solutions were prepared. Solution A consisted of 0.5 ml p-anisaldehyde in 99.5 ml ethyl acetate. Solution B was made by mixing 50 ml concentrated sulphuric acid with 50 ml ethyl acetate. 100 μl of the methanol extract was taken in a tube and the methanol was evaporated under reduced pressure. The residue left behind was dissolved in 2 ml of ethyl acetate and 1 ml each of reagents A and reagent B and stirred well. The test tube was placed in a water bath maintained at 60 °C for 10 min to develop colour. It was then allowed to cool for 10 min in 25 °C water bath. The absorbance of the coloured solution was measured in a spectrophotometer at 430 nm. As a reagent blank, 2 ml ethyl acetate was placed in tube and assayed in similar manner. For calibration curve, 2–70 μg standard diosgenin in 2 ml ethyl acetate was used. Each sample was repeated thrice and the average was taken. The diosgenin content was expressed as % in dry matter.
Tannin content
Tannin content was determined by slight modification of the method of Trupti P. Durgawale et al. (2016) by HPLC. Dry powdered sample (2 gm) was homogenized in 20 ml of distilled water and then was boiled in water bath (1000 C) for 30 min. After that, the sample was centrifuged in 2000 rpm for 20 min and the supernatant was collected in a volumetric flask and made the volume up to 100 ml with distilled water. The resulting extract was subjected for HPLC analysis.
HPLC condition
Standard of tannin (1gm enclosed in vial) was obtained from Sigma. Stock solution of tannin was prepared by taking 2 mg in 100 ml milipore water. The concentration of stock solution was equal to 20 ppm. The standard solution was injected into Knauer (Germany) HPLC system and chromatographic condition was set with LC-1000 pump (Isocratic), having C18 column and connected with LC PDA detector. Peak identification and quantification was made by “Clarity chrome software” for HPLC system. HPLC was calibrated by running mobile phase (Methanol and water by the ratio of 50:50, respectively) at the rate of 1 ml per minute. Wave length was fixed at 270 nm. The pressure of the column was kept at 1800–2000 PSI. Tannic acid standard solution of 20 ppm was injected when the injector was in load mode. The standard tannin peak was achieved at the retention time of 2.350 min (Rt = 2.350). Sample chromatogram was compared to pure standard of tannin (20 ppm) obtained from Sigma. Tannin content was expressed as mg/100gm.
Molecular studies
DNA isolation
DNA were isolated from younger leaves of the plants by CTAB method proposed by Doyle and Doyle (1990) with slight modification. DNA was isolated from 250 mg of young leaves by using DNeasy Plant Mini Kit (QIAGEN). The RAPD primer amplification was done by the protocol of Williams et al. (1990).
PCR amplification
Amplification was done in 25 µl PCR tube containing template DNA (25–30 ng), 2.5 U Taq polymerase (Qiagen, USA), 0.4 µM each dNTP (Qiagen, USA), 2.5 mM MgCl2 (Qiagen, USA), 1X Taq buffer containing 15 mM MgCl2 (Qiagen, USA) and 0.4 µM decamer primer. The PCR reaction was done by using applied biosystemsVeriti Thermal Cycler and the cycle followed for the same was as follows: (a) premelting for 5 min in 94 °C, (b). 35 cycles of 1 min denaturation at 94 °C, (c). 1.20 min annealing at 38 °C followed by 10 min extension at 72 °C and a final extension of 10 min at 72 °C. Amplified product was run under agarose gel (0.8%) and gel picture was captured by gel documentation unit.
Details of RAPD markers are as follows
| S. no. | RAPD primer | Primer sequence | Melting temperature (°C) |
|---|---|---|---|
| 1 | OPA1 | CAGGCCCTTC | 38 |
| 2 | OPA2 | TGCCGAGCTG | 38 |
| 3 | OPA3 | AGTCAGCCAC | 38 |
| 4 | OPA13 | CAGCACCCAC | 38 |
| 5 | OPB1 | GTTTCGCTCC | 38 |
| 6 | OPB6 | TGCTCTGCCC | 38 |
| 7 | OPB7 | GGTGACGCAG | 38 |
| 8 | OPM1 | GTTGGTGGCT | 38 |
| 9 | OPM2 | ACAACGCCTC | 38 |
| 10 | OPM4 | GGCGGTTGTC | 38 |
| 11 | OPM7 | CCGTGACTCA | 38 |
| 12 | OPM11 | GTCCACTGTG | 38 |
| 13 | OPM12 | GGGACGTTGG | 38 |
| 14 | OPM13 | GGTGGTCAAG | 38 |
| 15 | OPM15 | GACCTACCAC | 38 |
| 16 | OPM16 | GTAACCAGCC | 38 |
| 17 | OPM17 | TCAGTCCGGG | 38 |
| 18 | OPM19 | CCTTCAGGCA | 38 |
Statistical analysis
Statistical analysis was carried out by using Statistical Software R 3.5.3. Hierarchical cluster analysis (de Amorim 2015) using Ward’s minimum variance method and square Euclidian distance was performed to prepare the dendogram. In the gel picture, each individual band was considered as different alleles of the particular RAPD markers used. After recording the allele frequency, data was exported in the binary format using the AlphaEaseFC software where presence of the allele was recorded as 1 and absent as 0 respectively. The dendrogram was raised form Jaccards coefficient similarity matrix by the experimental data collected from RAPD analysis done by NTsysPC version 2.11 software using SHAN function following the UPGMA method to estimate similarity indices and genetic relatedness among and within the different yam genotypes.
Results
The highest total carbohydrate content (Table 1) was found in NGY-3 (87.85%), followed by NGY-2 (83.61%) which is at par with Da-199 (85.34%) while lowest total carbohydrate content had been found in NGY-9 (51.87%) and NGY-6 (53.75%).
Table 1.
Different nutritional (carbohydrate, starch, total soluble sugar, crude fat, crude fiber), antioxidant (anthocyanin, beta carotene) and antinutritional factors (diosgenin and tannin content) of fifteen genotypes of greater yam
| Varieties | Carbohydrate (%) |
Starch (%) |
Total soluble sugar (%) |
Crude fat (%) |
Crude fiber (%) |
Anthocyanin (mg/g) |
Beta carotene (µg/g) |
Diosgenin (powder) (%) |
Tanin (mg/100 gm) |
|---|---|---|---|---|---|---|---|---|---|
| NGY-1 | 75.10 (f) | 67.33(ef) | 7.52 (a) | 1.45 (c) | 3.96(ab) | 1.98 (de) | 1.02 (h) | 0.91 (ab) | 1.470 (ef) |
| NGY-2 | 83.61 (b) | 75.00(bc) | 6.90 (c) | 0.83 (i) | 2.27 (g) | 2.46 (c) | 1.02 (h) | 0.57 (e) | 0.933 (j) |
| NGY-3 | 87.85 (a) | 80.67 (a) | 5.81 (e) | 2.00 (b) | 3.14(de) | 2.05 (de) | 1.25 (d) | 0.42 (g) | 2.447 (a) |
| NGY-4 | 77.71 (e) | 75.00(bc) | 2.16 (l) | 0.80 (i) | 3.91 (b) | 3.25 (a) | 0.97 (i) | 0.36 (h) | 1.550 (d) |
| NGY-5 | 80.37 (cd) | 73.33(cd) | 6.93 (c) | 0.97(gh) | 4.08 (a) | 2.82 (b) | 1.16 (ef) | 0.52 (f) | 1.350 (h) |
| NGY-6 | 53.75 (i) | 50.33 (h) | 3.14 (j) | 1.46 (c) | 1.88 (h) | 1.33 (gh) | 1.87 (a) | 0.33 (h) | 0.923 (j) |
| NGY-7 | 60.55 (h) | 55.00 (g) | 4.29 (g) | 0.93 (h) | 3.20 (d) | 2.07 (d) | 1.88 (a) | 0.28 (i) | 2.277 (b) |
| NGY-8 | 74.83 (f) | 70.33(de) | 3.82 (h) | 2.05 (b) | 1.10 (h) | 1.69 (f) | 1.17 (e) | 0.89 (b) | 1.417 (fg) |
| NGY-9 | 51.87 (i) | 47.00 (h) | 3.53 (i) | 1.03 (fg) | 2.14 (g) | 1.63 (f) | 1.07 (g) | 0.91 (ab) | 1.117 (i) |
| NGY-10 | 67.61 (g) | 64.67 (f) | 2.59 (k) | 1.20 (d) | 3.08 (de) | 1.38 (g) | 1.12 (f) | 0.78 (d) | 1.457 (f) |
| NGY-11 | 78.42 (de) | 71.67(cd) | 5.92 (e) | 2.32 (a) | 4.09 (a) | 1.71 (f) | 1.78 (b) | 0.50 (f) | 1.527 (de) |
| NGY-12 | 74.75 (f) | 68.00(ef) | 6.12 (d) | 0.61 (j) | 2.90 (f) | 1.25 (h) | 1.52 (c) | 0.49 (f) | 1.363 (gh) |
| NGY-13 | 75.46 (f) | 67.33(ef) | 7.11 (b) | 1.14 (e) | 3.04 (ef) | 1.32 (gh) | 1.51 (c) | 0.59 (e) | 1.333 (h) |
| NGY-14 | 81.42 (c) | 77.33(ab) | 3.15 (j) | 1.08 (ef) | 3.75 (c) | 1.96 (e) | 1.81 (b) | 0.81 (c) | 1.457 (d) |
| Da-199 | 85.34 (b) | 79.67 (a) | 5.11 (f) | 1.50 (c) | 3.05 (de) | 1.01 (i) | 1.78 (b) | 0.93 (a) | 1.617 (c) |
| SEM | 0.676 | 1.176 | 0.052 | 0.022 | 0.050 | 0.036 | 0.015 | 0.010 | 0.020 |
| CD @ 5% | 1.953 | 3.398 | 0.151 | 0.063 | 0.144 | 0.103 | 0.043 | 0.029 | 0.057 |
| CV % | 1.58 | 2.99 | 1.84 | 2.93 | 2.79 | 3.35 | 1.84 | 2.84 | 2.297 |
Starch content was highest in NGY-3 (80.67%) which was significantly at par with Da-199 (79.67%), while lowest starch content was found in NGY-9 (47%), which was significantly at par with NGY-6 (50.33%) (Table 1).
Total soluble sugar content was highest in NGY-1 (7.52%) followed by NGY-13 (7.11%), while lowest total soluble Sugar content was found in NGY-4 (2.16%) (Table 1).
Crude fat content (Table 1) was highest in NGY-11 (2.32%), followed by NGY-8 (2.05%) and NGY-3 (2%) while lowest crude fat content was found in NGY-12 (0.61%).
The crude fibre content in different genotypes of greater yam had varied significantly (1.10 to 4.09%) in tubers (Table 1). Crude fibre content was highest in NGY-11 (4.09%) which was significantly at par with NGY-5 (4.08%), while lowest crude fibre content was found in NGY-8 (1.10%) at par with NGY-6 (1.88%).
The anthocyanin content was highest in NGY-4 (3.25 mg/g) followed by NGY-5 (2.82 mg/g) while lowest anthocyanin content was found in Da-199 (1.01 mg/g) (Table 1).
The highest Beta carotene content (Table 1) was observed in NGY-7(1.88 µg/g), which was significantly at par with one other genotype such as NGY-6 (1.77 µg/g) and lowest Beta carotene content was found in the NGY-4 (0.97 µg/g) genotype of greater yam.
The diosgenin content of greater yam shown in Table 1 was highest in Da-199 (0.93%) which was significantly at par with NGY-1 (0.91%) and lowest in NGY-7 (0.28%).
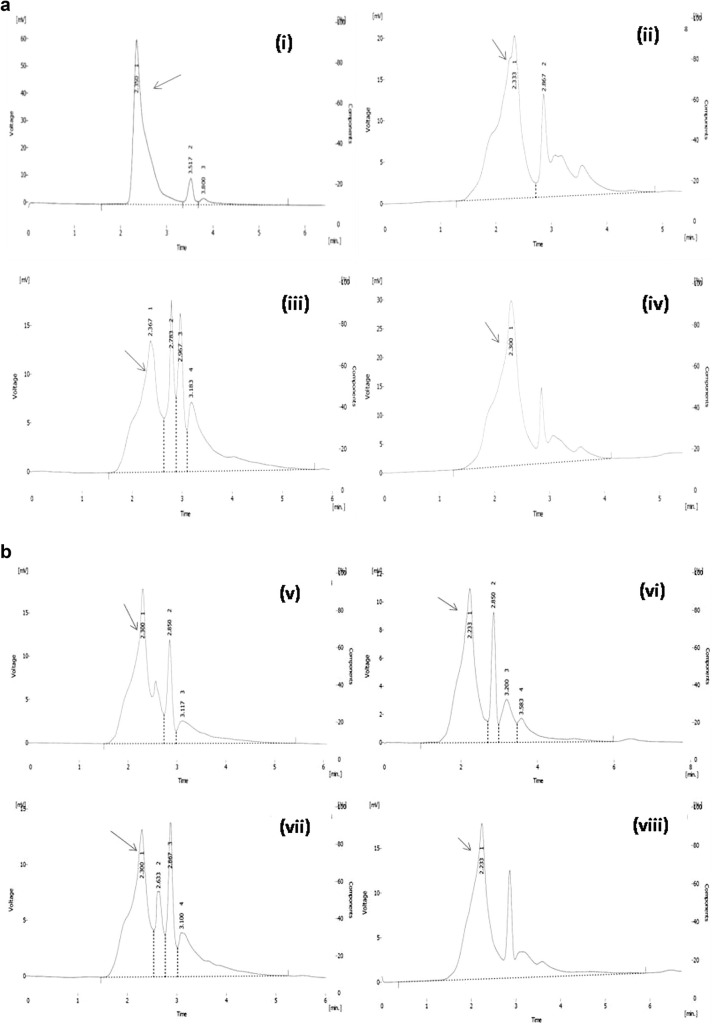
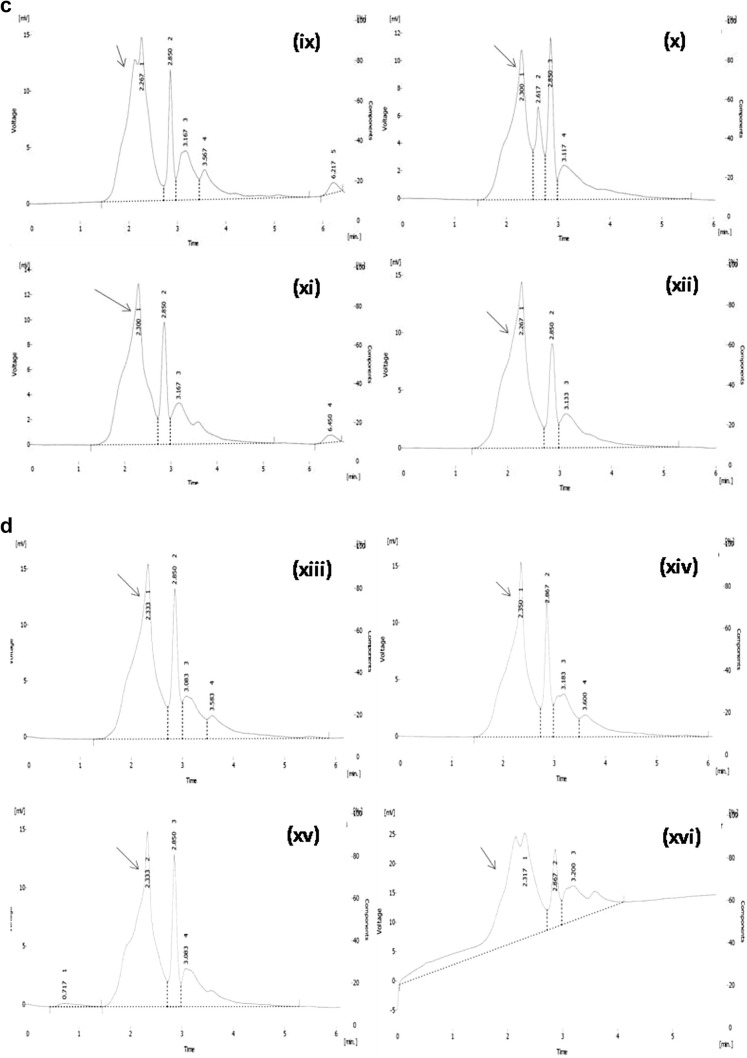
Tannin content was measured in 15 genotypes of greater yam by High Pressure Liquid Chromatography (HPLC) (Table 1). The desirable peak for tannic acid in all the chromatograms had been indicated by arrow sign (Fig. 1a–d) were significantly differing in mean value of tannin (0.923 mg/100 g to 2.447 mg/100 g) in different genotypes of greater yam tubers. The highest tannin was observed in the genotype NGY-3 (2.45 mg/100 g), while the lowest amount of tannin observed in genotype NGY- 6 (0.92 mg/100 g), which was significantly at par with other one genotype NGY-2 (0.93 mg/100 g). The other two genotypes like NGY-4 and NGY-14 contained 1.55 mg/100 g and 1.46 mg/100 g tannin respectively which were at par.
Fig. 1.
Chromatogram of tannin for fifteen different genotypes for greater yam. i Tannin standard 20 ppm, ii NGY-1, iii NGY-2, iv NGY-3, v NGY-4, vi NGY-5, vii NGY-6, viii NGY-7, ix NGY-8, x NGY-9, xi NGY-10, xii NGY-11, xiii NGY-12, xiv NGY-13, xv NGY-14, xvi Da-199
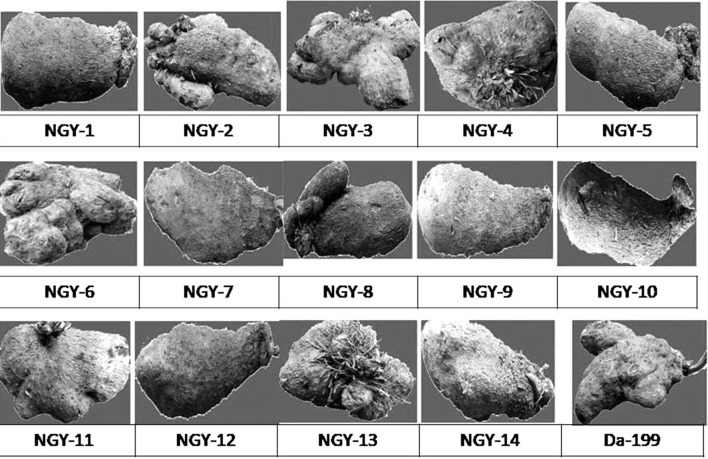
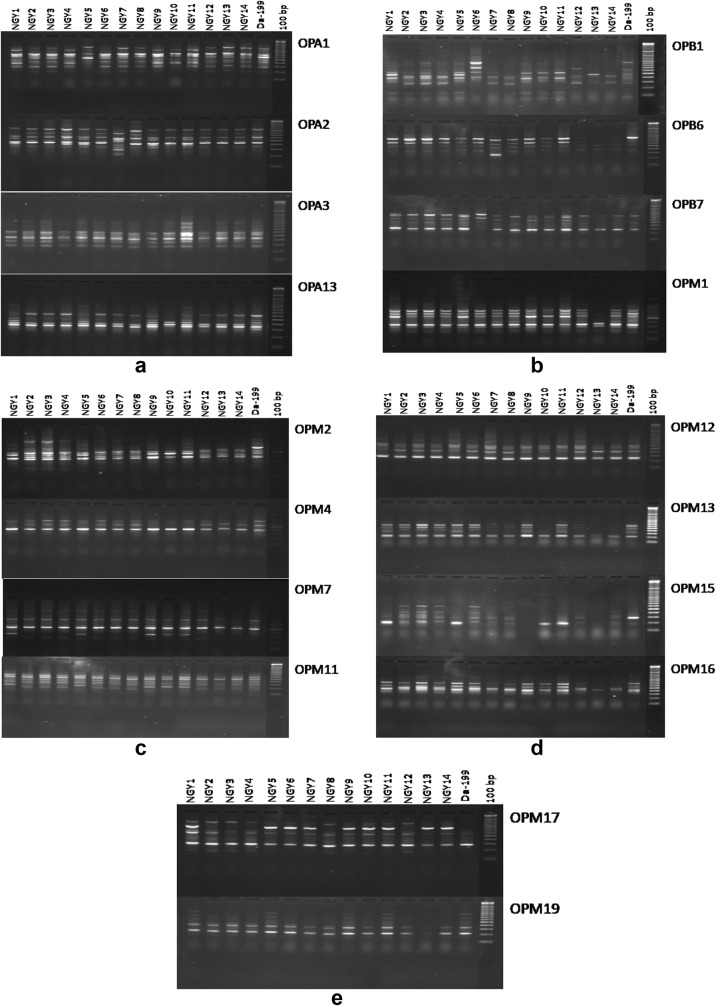
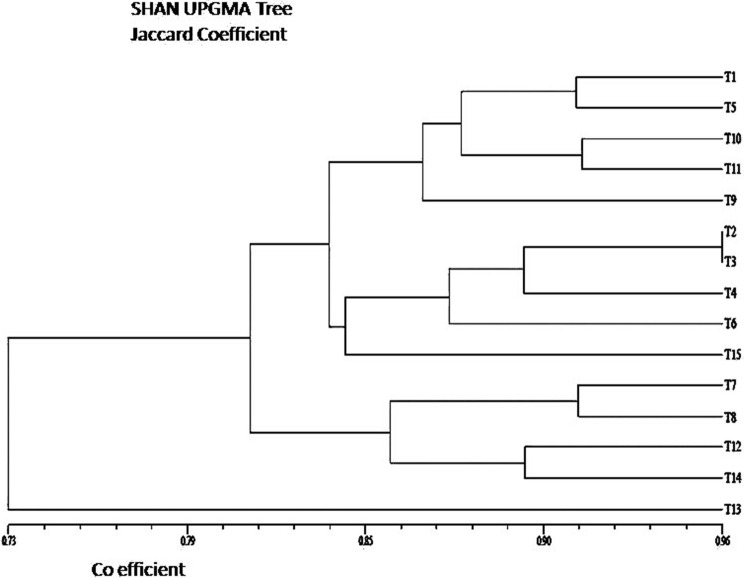
The fifteen different types of genotypes of yam has been displayed in Fig. 2. A total of 18 random RAPD primers were used for genomic profiling [(Fig. 3a–e) and Table 2] which produced reproducible banding pattern and the unitary data was exploited for phylogenetic analysis. The data was analysed by NTSYS-pc version 2.11 and the Jaccards coefficient was generated. The similarity matrix (Table 3) was subjected to UPGMA to generate the dendrogram from the RAPD fragments generated (Fig. 4). The highest similarity was found between two entries namely NGY-2 and NGY-3 (96%) and the least similarity was found between NGY-6 and NGY-13 (67%). The dendrogram displayed, reveals two major clusters along with the differentiated single entry NGY- 13, which was completely out grouped. The entries NGY- 1, NGY-2, NGY-3, NGY-4, NGY-5, NGY-6, NGY-9, NGY-10, NGY-11 and Da-199 formed the first sub cluster and NGY-7, NGY-8, NGY-12, NGY-14 formed the second sub cluster. A second hierarchical clustering had been done on the basis of aforementioned nine different biochemical parameters for fifteen different yam genotypes (Fig. 5). According to this analysis, two main clusters had been formed, of which the first cluster contained the three different genotypes like NGY-7, NGY-6 and NGY-9 whereas the second cluster had again been divided on two sub clusters. The first sub cluster included five varieties like NGY- 10, NGY- 8, NGY-12, NGY-1, NGY-13 and the second and last sub cluster contained remaining seven genotypes namely NGY-3, Da-199, NGY-4, NGY-14, NGY-2, NGY-5, NGY-11. The lowest Euclidian distance was found between NGY-13 and NGY-1 pair (1.62) followed by NGY-12 and NGY-1(2.62). For the first sub cluster, the Euclidian distance between the first member (T7 = NGY-7 to T9 = NGY-9) was 12.45, followed by second sub cluster, for which the range of Euclidian distance was 9.60 (T10 = NGY-10 to T13 = NGY-13) and for third sub cluster, it was 13.66 (T3 = NGY-3 to T11 = NGY-11) (Table 4).
Fig. 2.
Tubers of different greater yam genotypes
Fig. 3.
RAPD profiles of different greater yam. Genotypes performed by eighteen different primers
Table 2.
List of eighteen different RAPD primers produced reproducible banding patterns and polymorphism generated in fifteen different genotypes of greater yam
| S. no. | Primer | Total no of amplicons | No of loci | No of polymorphic bands | No of monomorphic bands | Percent polymorphism (%) | RPI value |
|---|---|---|---|---|---|---|---|
| 1 | OPA-1 | 156 | 14 | 9 | 5 | 64.29 | 13.92 |
| 2 | OPA-2 | 132 | 15 | 10 | 5 | 66.67 | 14.91 |
| 3 | OPA-3 | 125 | 10 | 3 | 7 | 30.00 | 9.89 |
| 4 | OPA-13 | 132 | 11 | 4 | 7 | 36.37 | 10.90 |
| 5 | OPB-1 | 106 | 10 | 6 | 4 | 60.00 | 9.88 |
| 6 | OPB-6 | 82 | 8 | 5 | 3 | 62.50 | 7.84 |
| 7 | OPB-7 | 103 | 8 | 5 | 3 | 62.50 | 7.86 |
| 8 | OPM-1 | 121 | 10 | 6 | 4 | 60.00 | 9.88 |
| 9 | OPM-2 | 132 | 10 | 3 | 7 | 30.00 | 9.90 |
| 10 | OPM-4 | 103 | 9 | 3 | 6 | 33.34 | 8.86 |
| 11 | OPM-7 | 144 | 11 | 3 | 8 | 27.28 | 10.90 |
| 12 | OPM-11 | 92 | 10 | 5 | 4 | 50.00 | 8.86 |
| 13 | OPM-12 | 118 | 8 | 1 | 7 | 12.50 | 7.88 |
| 14 | OPM-13 | 94 | 7 | 5 | 2 | 71.42 | 6.86 |
| 15 | OPM-15 | 106 | 8 | 6 | 2 | 75.00 | 7.87 |
| 16 | OPM-16 | 121 | 9 | 3 | 6 | 33.34 | 8.88 |
| 17 | OPM-17 | 88 | 7 | 5 | 2 | 71.42 | 6.85 |
| 18 | OPM-19 | 133 | 10 | 3 | 7 | 30.00 | 9.90 |
| Total | 2088 | 175 | 85 | 89 | 876.63 | 171.83 | |
| Average | 116 | 9.722 | 4.722 | 4.944 | 48.70 | 9.55 |
Table 3.
Similarity matrix between each fifteen greater yam genotypes generated on the basis of RAPD band data analysis
| Genotypes | NGY-1 | NGY-2 | NGY-3 | NGY-4 | NGY-5 | NGY-6 | NGY-7 | NGY-8 | NGY-9 | NGY-10 | NGY-11 | NGY-12 | NGY-13 | NGY-14 | Da-199 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NGY-1 | 1.00 | ||||||||||||||
| NGY-2 | 0.86 | 1.00 | |||||||||||||
| NGY-3 | 0.86 | 0.96 | 1.00 | ||||||||||||
| NGY-4 | 0.89 | 0.91 | 0.89 | 1.00 | |||||||||||
| NGY-5 | 0.91 | 0.84 | 0.84 | 0.88 | 1.00 | ||||||||||
| NGY-6 | 0.80 | 0.89 | 0.89 | 0.84 | 0.82 | 1.00 | |||||||||
| NGY-7 | 0.81 | 0.88 | 0.84 | 0.86 | 0.80 | 0.85 | 1.00 | ||||||||
| NGY-8 | 0.77 | 0.87 | 0.83 | 0.86 | 0.78 | 0.82 | 0.91 | 1.00 | |||||||
| NGY-9 | 0.88 | 0.81 | 0.81 | 0.85 | 0.84 | 0.78 | 0.80 | 0.79 | 1.00 | ||||||
| NGY-10 | 0.87 | 0.84 | 0.83 | 0.85 | 0.87 | 0.81 | 0.84 | 0.83 | 0.85 | 1.00 | |||||
| NGY-11 | 0.88 | 0.83 | 0.84 | 0.85 | 0.88 | 0.83 | 0.81 | 0.80 | 0.89 | 0.91 | 1.00 | ||||
| NGY-12 | 0.81 | 0.84 | 0.81 | 0.82 | 0.78 | 0.78 | 0.89 | 0.87 | 0.77 | 0.82 | 0.79 | 1.00 | |||
| NGY-13 | 0.70 | 0.71 | 0.70 | 0.69 | 0.70 | 0.67 | 0.74 | 0.73 | 0.70 | 0.75 | 0.71 | 0.81 | 1.00 | ||
| NGY-14 | 0.77 | 0.81 | 0.79 | 0.77 | 0.77 | 0.76 | 0.84 | 0.83 | 0.75 | 0.81 | 0.78 | 0.90 | 0.89 | 1.00 | |
| Da-199 | 0.79 | 0.84 | 0.86 | 0.82 | 0.81 | 0.85 | 0.84 | 0.83 | 0.83 | 0.84 | 0.84 | 0.79 | 0.74 | 0.84 | 1.00 |
Fig. 4.
A Dendogram developed from UPGMA analysis showing genetic relationships between Dioscorea spp. by RAPD. T1 = NGY-1, T2 = NGY-2, T3 = NGY-3, T4 = NGY-4, T5 = NGY-5, T6 = NGY-6, T7 = NGY-7, T8 = NGY-8, T9 = NGY-9, T10 = NGY-10, T11 = NGY-11, T12 = NGY-12, T13 = NGY-13, T14 = NGY-14, T15 = Da-199
Fig. 5.
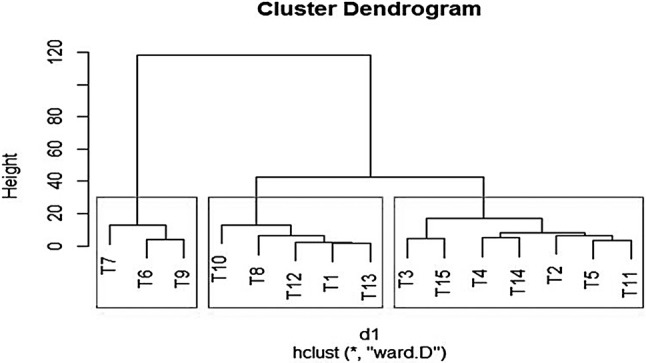
Hierarchical clustering based on different nutritional, antioxidant and antinutritional factors (diosgenin and tannin content) of fifteen genotypes of greater yam. T1 = NGY-1, T2 = NGY-2, T3 = NGY-3, T4 = NGY-4, T5 = NGY-5, T6 = NGY-6, T7 = NGY-7, T8 = NGY-8, T9 = NGY-9, T10 = NGY-10, T11 = NGY-11, T12 = NGY-12, T13 = NGY-13, T14 = NGY-14, T15 = Da-199
Table 4.
Euclidian distance matrix between fifteen Dioscorea alata genotypes based on different nutritional (carbohydrate, starch, total soluble sugar, crude fat, crude fiber), antioxidant (anthocyanin, beta carotene) and antinutritional factors (diosgenin and tannin content) of fifteen genotypes of greater yam
| NGY-1 | NGY-2 | NGY-3 | NGY-4 | NGY-5 | NGY-6 | NGY-7 | NGY-8 | NGY-9 | NGY-10 | NGY-11 | NGY-12 | NGY-13 | NGY-14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NGY-2 | 11.74 | |||||||||||||
| NGY-3 | 19.01 | 7.90 | ||||||||||||
| NGY-4 | 9.89 | 7.86 | 12.75 | |||||||||||
| NGY-5 | 8.16 | 4.50 | 11.36 | 5.85 | ||||||||||
| NGY-6 | 27.77 | 38.96 | 45.90 | 34.54 | 35.53 | |||||||||
| NGY-7 | 19.59 | 30.81 | 37.82 | 26.63 | 27.41 | 8.86 | ||||||||
| NGY-8 | 5.62 | 10.57 | 17.00 | 6.50 | 7.83 | 29.12 | 21.22 | |||||||
| NGY-9 | 31.21 | 42.50 | 49.50 | 38.22 | 39.02 | 4.21 | 12.45 | 32.81 | ||||||
| NGY-10 | 9.71 | 19.71 | 26.24 | 14.71 | 16.18 | 20.13 | 12.88 | 9.61 | 23.74 | |||||
| NGY-11 | 6.09 | 7.07 | 13.66 | 5.70 | 3.52 | 32.86 | 24.78 | 5.26 | 36.42 | 13.47 | ||||
| NGY-12 | 2.62 | 11.46 | 18.56 | 8.90 | 8.10 | 27.66 | 19.65 | 4.05 | 31.20 | 8.79 | 5.80 | |||
| NGY-13 | 1.62 | 11.37 | 18.67 | 9.68 | 8.02 | 27.90 | 19.79 | 5.02 | 31.37 | 9.60 | 5.73 | 1.71 | ||
| NGY-14 | 12.83 | 5.64 | 8.44 | 5.08 | 5.86 | 38.78 | 30.91 | 10.14 | 42.40 | 18.80 | 7.32 | 11.97 | 12.46 | |
| NGY-15 | 16.41 | 6.00 | 4.48 | 9.91 | 8.63 | 43.23 | 35.26 | 14.36 | 46.83 | 23.38 | 10.81 | 15.86 | 16.02 | 5.14 |
Discussion
Carbohydrate content is an important parameter for tuber crops, and genotypes with low carbohydrate content may be useful for the consumption of the diabetic patients. The present study reveals the range of carbohydrate content is 87.85% to 51.87% in the fifteen yam genotypes (Table 1). In Nigerian yam, total carbohydrate was found 70.88–73.90% and depicted a lesser variation between different genotypes (Oko and Famurewa 2015), but in contrast, we obtained a large variation between different genotypes. A recent study also showed a very high amount of carbohydrate in another species of yam, Dioscoreapyrifolia (Sharlina et al. 2017) which illustrates the variability of carbohydrate content in tubers despite the relatedness of the genotypes.
It has been reported that in white yam (Dioscorearotundata), there is a great variation of starch content 33.9 to 75.7% (Oladeji et al. 2014) and 93.67% (Adegunwa et al. 2011) but for Dioscoreaalata, the starch content is 62.94% (Adegunwa et al. 2011), which was also similar (60.3-74.4%) with the results of Baah et al. (2009). Starch is known to account for about 80% on a dry weight basis of yam carbohydrate; which is also a dominant factor in determining the physicochemical, rheological and textural characteristics of yam food products (Moorthy 1994). Our results are also within the range (47% to 80.67%; Table 1) of the previously published works. From this study, NGY-3 (80.67%) and NGY-14(79.67%) may be developed as high starch lines.
Total soluble sugars or simply the sugar content is a significant character for yams as this character confers sweetness in the cooked tuber (Lebot et al. 2005). From the earlier studies, Dioscoreaalata, was shown to contain 2.13% (Adegunwa et al. 2011), 2.24 to 2.79% (Mitra 2012) while in Dioscoreaalata, a wide range of soluble sugar had also been detected earlier (Oladeji et al. 2014). In our experiments, we also obtained a wide range for this parameter (Table 1) and it can be suggested that NGY-1 should be isolated as a soluble sugar rich genotype that can be used for taste enhancement in food.
Crude fat is relevant as lipids contribute to the texture, flavour and aroma of foods, thereby prolonging satiety and facilitate the absorption of lipid-soluble vitamins. It also contributes to the palatability of the crops (FAO 2010). A range of crude fat content has been recorded in various types of yams, e.g. 6.14% for D. bulbifera (Shajeela et al. 2011), 1.59% in aerial yam in Ghana (Sanful and Engmann 2016), 18.26% to 28.54%) in Trifoliate Yam (Dioscoreadumetorum) (Akinoso et al. 2016), but in our study, we obtained the highest crude fat content in NGY-11 (2.32%) which is lesser in comparison to the other species of yams, conferring no extra addition of smoothness due to fat content in flour.
Fibre is important for preventing obesity by increasing food bowel movement and also helps in preventing diabetes, colon cancer and other ailments of the gastro-intestinal tract. Fibre consumption also lowers plasma cholesterol level in body and reduces the incidence of cardiovascular disease (Topping and Clifton 2001). In our study, the range of crude fibre was much higher than the earlier reports in False Yam (Icacinatrichantha), Flour (1.06 to 1.51%) (Umoh and Iwe 2014), 0.6–1.5% in greater yam (Bhandari et al. 2003) and in elephant foot yam (Lenka and Nedunchezhiyan 2011). In the present study, NGY-11, NGY-1 and NGY-5 genotypes (Table 1) with higher crude fibre content can be selected and cultivated specially for high crude fibre content.
The importance of anthocyanin is its usage as food colour in ice cream, lemon juice and hard candy in food industry. Also, in Dioscorea plantation, pests and diseases were not common and use of fertilizers was not a prerequisite, thereby yam has potential to act as an effective anthocyanin source. In our present study, a range of 1.01 to 3.25 mg/g anthocyanin content was observed, in contrast to earlier reports of 1.43 mg/g (Jose and Muhammed 2015). According to our result, NGY-4 (3.25 mg/g) can be suggested for this region to be grown as an anthocyanin rich genotype.
β-Carotenoid is a precursor for the synthesis of vitamin A. The β-carotenoids are converted in Vitamin A through a biochemical process in human body, hence the intake of β-carotenoid is essential from plant sources. Yellow-fleshed yams can provide appreciable amounts of carotenoids to benefit human nutrition and health as they also act as antioxidants (Ferede, et al. 2010). The earlier report of β-Carotenoid in Yam cultivars with a range of 1.7 mg/100 gm to 2.6 mg/100 gm (Mitra and Tarafdar 2008) was higher than our genotypes. This may be because of the richness in another antioxidant, anthocyanin, hindered the accumulation of carotenoids. Our study encourages the cultivation of NGY-6 and NGY-7 as β-Carotenoid rich genotpes but both containing lesser amounts of anthocyanin.
Diosgenin is an antinutrient and steroid sapogenin, consumption of which may cause harm to pregnant women, hence it is desirable that low diosgenin genotypes like NGY-7 to be recommended for the cultivation. On the other hand, as yam sapogenin is used for manufacturing of contraceptive pills, for the purpose of sapogenin production, the genotypes like NGY-1 and NGY-9 can be suggested (Table 1).
Tannin is considered as an antinutrient factor as it affects the nutritional value of food products by forming complex with protein (both substrate and enzyme) thereby inhibiting digestion and absorption (Osuntogun et al. 1989). In our experiment, we obtained much lower range of tannin content, 0.923 mg/100 g to 2.447 mg/100 g (Table 1 and Fig. 1a–d) compared to the others in Dioscoreaalata (0.21%) (Ezeocha and Ojimelukwe 2012). This is a good indication for lower tannin containing tubers and so all the genotypes are suitable for cultivation from this point of view.
In order to reveal the relatedness between the different genotypes, an hierarchical classification had been carried in this study. RAPD markers have been successfully employed in the study of varietal genetic relationship among tuber crops (Gawande et al. 2015). Scientists have employed RAPD markers to study the genetic relationships and the intraspecific variability in African Yam bean (Popoola et al. 2017) and between different species. The dendrogram drawn on the basis of RAPD analysis produced more than 90% similarities for different pairs of entries including NGY-1 and NGY-5, NGY-2 and NGY-4, NGY-7 and NGY-8, NGY-10 and NGY-11, NGY-12 and NGY-14 (Fig. 4). According to our experiments, total loci detected were 175, out of which 89 were monomorphic with an average of 4.94 loci per primer and 85 were polymorphic with average of 4.72 loci per primer. The average polymorphism shown by the primers were 48.702%, with the highest of 75% with the primer OPM-15 and lowest of 30% by the primers OPA-3, OPM-2 and OPM-19. The RAPD primer index value (RPI) ranged from 6.85 to 14.90 with an average of 9.54 and the primer OPA-2 produced the highest number of polymorphic loci (10) out of 15 loci with total production of 132 amplicons (Table 2), hence this RAPD primer can be used for marker assisted selection. NGY-2 and NGY-3 were placed in the same subcluster at the same distance and showed the highest similarity 96% and thus they may be the clone of the same genotype to be cultivated in different places with different local name but actually consisted the same genotype which can be clarified further more with SSR or ISSR profiling. The results also demonstrated that the genotypes collected from different districts of south Gujarat region like Valsad, Navsari, Dang, Surat had higher similarity. Only the entry NGY-13 is distinctly different and it can potentially be used as a pre-breeding material for the introgression of the new genes. In Fig. 5, it is clear that the range of lowest (T10 = NGY-10 to T13 = NGY-13) Euclidian distance compared to the other two distances was 9.60 whereas in the similarity matrix of RAPD derived data, the similarity between T10 = NGY-10 and T13 = NGY-13 was only75%.The hierarchical cluster on the basis of various biochemical traits revealed considerable differences between the cluster in this study refer the same line of study by Mulualem et al. (2018). The clustering pattern revealed that genotypes of the south Gujarat origin were distributed into different groups, which gave an indication of lack of parallelism between clustering pattern and geographic distribution of genotypes. This may be explained as a result of differences in adaption to various environmental conditions.
Conclusion
The above mentioned study revealed highest carbohydrate content in NGY-3, fat and crude fiber content in NGY-11, anthocyanin in NGY-4, β-carotene in NGY-7. Lowest amount of antinutrients like tannin and diosgenin was found in NGY-6, and NGY-7 respectively. As the molecular studies revealed NGY-2 and NGY-3 having 96% similarities, they may be the duplicate of the same genotypes that can be studied further for better germplasm conservation. Hierarchical clustering of fifteen genotypes based on biochemical parameters though did not totally match with the molecular clustering but the two genotypes namely NGY-2 and NGY-3 became the candidate of the same cluster. In a nutshell, NGY-3, NGY-4, NGY-6, NGY-7 and NGY-11 can be exploited as different varieties for the aforesaid particular biochemical parameters as well as NGY-2 and NGY-3 may be further investigated for the duplicity of the same genotype. For south Gujarat zone, though NGY-4 has highest anthocyanin content but NGY-7 can be recommended for cultivation due to its better performance on account of yield, richness in anthocyanin and better adoption in the environment of south Gujarat.
Compliance with ethical standards
Conflict of interest
The authors declare that they do not have any conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kajal S. Patel, Email: kajalpatel99789@gmail.com
Nilima Karmakar, Email: nilimanau13@gmail.com.
Ketan D. Desai, Email: ketan_nau62@yahoo.in
Ajay V. Narwade, Email: narwadeajay_2000@yahoo.com
Gayacharan Chakravarty, Email: gayabio83@gmail.com.
Manoj Kanti Debnath, Email: mkanti1984@gmail.com.
References
- Adegunwa MO, Alamu EO, Omitogun LA. Effect of processing on the nutritional contents of yam and cocoyam tubers. J Appl Biosci. 2011;46:3086–3092. [Google Scholar]
- Akinoso R, Olatoye KK, Ogunyele OO. Potentials of trifoliate yam (Dioscorea dumetorum) in noodles production. J Food Process Tecnol. 2016;7(8):1–6. [Google Scholar]
- AOAC . Official methods of analysis. 14. Washington: Association of Official Analytic Chemist; 1984. [Google Scholar]
- Baah FD, Maziya-Dixon B, Asiedu R, Oduro I, Ellis WO. Nutritional and biochemical composition of D. alata (Dioscorea spp.) tubers. J Food Agric Environ. 2009;7(2):373–378. [Google Scholar]
- Baccou JC, Lambert F, Sauvaire Y. Spectrophotometric method for the determination of total steroidal sapogenin. Analyst. 1977;102:458–465. doi: 10.1039/an9770200458. [DOI] [PubMed] [Google Scholar]
- Behera KK, Maharana T, Sahoo S, Prusti A. Biochemical quantification of protein, fat, starch, crude fibre, ash and dry matter content indifferent collection of greater yam (Dioscorea alata L.) found in Orissa. Nat Sci. 2009;7(7):24–32. [Google Scholar]
- Bhandari MR, Kasai T, Kawabata J. Nutritional evaluation of wild yam (Dioscorea spp.) tubers of Nepal. Food Chem. 2003;82(4):619–623. doi: 10.1016/S0308-8146(03)00019-0. [DOI] [Google Scholar]
- Contreras-Pacheco ML, Santacruz-Ruvalcaba F, Garcia-Fajardo JA, Jesus Sanchez JG, Ruız MAL, Estarron-Espinosa M, Castro-Castro A. Diosgenin quantification, characterisation and chemical composition in a tuber collection of Dioscorea spp. in the state of Jalisco, Mexico. Int J Food Sci Tecnol. 2013;48:2111–2118. [Google Scholar]
- de Amorim RC. Feature relevance in ward’s hierarchical clustering using the Lp norm. J Classif. 2015;32(1):46–62. doi: 10.1007/s00357-015-9167-1. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12(1):13–15. [Google Scholar]
- Durgawale TP, Durgawale PP, Khanwelkar CC. Quantitative estimation of tannins by HPLC. Der Pharm Lett. 2016;8(3):123–126. [Google Scholar]
- Eshel D. Non-chemical approaches for postharvest quality management of underground vegetables. Stewart Postharvest Rev. 2011;1(3):1–7. doi: 10.2212/spr.2011.1.3. [DOI] [Google Scholar]
- Ezeocha VC, Ojimelukwe PC. The impact of cooking on the proximate composition and anti-nutritional factors of water yam (Dioscorea alata) J Stored Prod Postharvest Res. 2012;3(13):172–176. [Google Scholar]
- FAO (2010) Fats and fatty acids in human nutrition-Report of an expert consultation. FAO Food and Nutrition, Paper 91, Food and Agriculture Organization of the United Nations, Rome [PubMed]
- Ferede R, Maziya-Dixon B, Alamu OE, Asiedu R. Identification and quantification of major carotenoids of deep yellow-fleshed yam (tropical Dioscorea dumetorum) J Food Agric Environ. 2010;8(3&4):160–166. [Google Scholar]
- Gawande PA, Nimbhorkar NV, Deshmukh VP, Thakare PV. Assessment of genetic relationships among Dioscorea spp. of Melghat Tiger Reserve Maharashtra of Central India, by using RAPD and its sequences. J Biosci Biotech. 2015;4(3):303–312. [Google Scholar]
- Hedge JE, Hofreiter BT (1962) Methods in carbohydrate chemistry, 17. In: Whistler RL, BeMiller JN (eds) Academic Press, New York, 420
- Hosseinian FS, Li W, Beta T. Measurement of anthocyanins and other phytochemicals in purple wheat. Food Chem. 2008;109(4):916–924. doi: 10.1016/j.foodchem.2007.12.083. [DOI] [PubMed] [Google Scholar]
- Jose A, Muhammed R. Extraction and evaluation of anthocyanin from Dioscorea alata (L.) for its application as a natural food colour. Int J Sci Technol. 2015;3(9):41–47. [Google Scholar]
- Karnjanawipagul P, Nittayanuntawech W, Rojsanga P, Suntornsuk L. Analysis of β-carotene in carrot by spectrophotometry. Mahidol Univ J Pharm Sci. 2010;37(1–2):8–16. [Google Scholar]
- Lebot V, Malapa R, Molisale T, Marchand JL. Physicochemical characterization of yam (Dioscorea alata L) tubers from Vanuatu. Genet Resour Crop Evol. 2005;53(6):1199–1208. doi: 10.1007/s10722-005-2013-2. [DOI] [Google Scholar]
- Lenka A, Nedunchezhiyan M. Biochemical composition of two edible aroids. E-Planet. 2011;10(2):39–42. [Google Scholar]
- Martin FW (1974) Tropical yam and their potential. D. bulbifera. Agric. Learn Book No. 446. USDA
- Maynard AJ. Methods in food analysis. New York: Academic Press; 1970. p. 176. [Google Scholar]
- Mitra S. Nutritional status of orange-fleshed sweet potatoes in alleviating Vitamin A malnutrition through a food based approach. J Nutr Food Sci. 2012;2:160. [Google Scholar]
- Mitra S, Tarafdar J (2008) Present status and future prospects of elephant foot yam cultivation in west bengal. In: Palaniswami MS et al. (ed) National seminar on amorphophallus: innovative technologies, July 19–20, 2008, Patna, Bihar-abstract book, status papers and extended summery, pp 25–29
- Moorthy SN (1994) Tuber crop starches. Technical Bulletin Series 18. Central Tuber Crops Research Institute, Thiruvananthapuram, Kerala, India
- Mulualem T, Mekbib F, Hussein S, Gebre NE. Analysis of biochemical composition of yams (Dioscorea spp.) landraces from Southwest Ethiopia. Agrotechnology. 2018;7(1):1–8. doi: 10.4172/2168-9881.1000177. [DOI] [Google Scholar]
- Oko AO, Famurewa AC. Estimation of nutritional and starch characteristics of Dioscorea alata (water yam) varieties commonly cultivated in the South-Eastern Nigeria. Br J Appl Sci Technol. 2015;6(2):145–152. doi: 10.9734/BJAST/2015/14095. [DOI] [Google Scholar]
- Oladeji AE, Bussie MD, Christian OC, Roberts A. Physicochemical and bioactive properties of selected white yam (Dioscorea rotundata) varieties adapted to riverine areas of Nigeria. Afr J Food Sci. 2014;8(7):402–409. doi: 10.5897/AJFS2014.1154. [DOI] [Google Scholar]
- Osuntogun A, Adewusi SR, Adewusi A, Ogundiwin JO, Nwasike CC. Effect of cultivar, steeping, and malting on tannin, total polyphenol, and cyanide content of Nigerian sorghum. Cereal Chem. 1989;66:87–89. [Google Scholar]
- Popoola JO, Adebayo BM, Adegbite AE, Omonhinmin AC, Adewale BD. Fruit morphometric and RAPD evaluation of intraspecific variability in some accessions of African yam bean (Sphenostylis stenocarpa Hochst. ex. A. Rich. Harms) Annu Res Rev Biol. 2017;14(4):1–10. doi: 10.9734/ARRB/2017/34264. [DOI] [Google Scholar]
- Sanful RE, Engmann FN. Physico-chemical and pasting characteristics of flour and starch from aerial yam. Am J Food Sci Nutr. 2016;3(1):1–7. [Google Scholar]
- Shajeela PS, Mohan VR, Louis Jesudas L, Tresina-Soris P. Nutritional and antinutritional evaluation of wild yam (Dioscorea spp) Trop Subtrop Agroecosyst. 2011;14:723–730. [Google Scholar]
- Sharlina MSE, Yaacob WA, Lazim AM, Fazry S, Lim SJ, Abdullah S, Noordin A, Kumaran M. Physicochemical properties of starch from Dioscorea pyrifolia tubers. Food Chem. 2017;220:225–232. doi: 10.1016/j.foodchem.2016.09.196. [DOI] [PubMed] [Google Scholar]
- Timpa JD, Bruke JJ, Quisenberry JE, Wendt CW. Carbohydrate utilization under stress. Novosibresic-USSR. 1985;5:65–69. [Google Scholar]
- Topping DL, Clifton DL. SCFA and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Umetsu Y, Hirata KS, Kudo I. Spectrophotometric determination of saponins in Yucca extract used as food additive. J AOAC Int. 2000;836:1451–1454. [PubMed] [Google Scholar]
- Umoh EO, Iwe MO. Effects of processing on the nutrient composition of false yam (Icacina trichantha) flour. Off J Niger Inst Food Sci Technol. 2014;32(2):1–7. [Google Scholar]
- Williams JGK, Kubelik AR, Livak KJ, Antoni Rafalski J, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research. 1990;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]