Abstract
Proteasomes are multienzyme complexes that maintain protein homeostasis (proteostasis) and important cellular functions through the degradation of misfolded, redundant, and damaged proteins. It is well established that aging is associated with the accumulation of damaged and misfolded proteins. This phenomenon is paralleled by declined proteasome activity. When the accumulation of redundant proteins exceed degradation, undesirable signaling and/or aggregation occurs and are the hallmarks of neurodegenerative diseases and many cancers. Thus, increasing proteasome activity has been recognized as a new approach to delay the onset or ameliorate the symptoms of neurodegenerative and other proteotoxic disorders. Enhancement of proteasome activity has many therapeutic potentials but is still a relatively unexplored field. In this perspective, we review current approaches, genetic manipulation, posttranslational modification, and small molecule proteasome agonists used to increase proteasome activity, challenges facing the field, and applications beyond aging and neurodegenerative diseases.
Graphical Abstract
1. INTRODUCTION
Precise and accurate regulation of important biological activities for self-maintenance requires the cellular protein pool to be in a continuous flux.1 This protein quality control is maintained by the protein homeostasis (proteostasis) network, which consists of the protein synthesis machinery (the ribosomes), protein folding complexes (the chaperones), and two proteolytic systems: the proteasomal and the lysosomal (autophagy) systems.2
Molecular chaperones assist in cotranslational folding of nascent polypeptides attached to the ribosome, thereby preventing them from nonproductive interactions and aggregation.3 Furthermore, chaperones recognize unfolded or aberrant proteins and assist them in regaining stability. Proteins with damages beyond repair are eliminated through the proteolytic pathways to avoid proteotoxicity due to aggregation or undesirable interactions.4,5
The plasticity and crosstalk among the protein synthesis, protein folding, and protein degradation systems sustain proteostasis under different cellular and environmental conditions such as xenobiotic and oxidative stress, cellular growth, and differentiation. The activities of the different proteostasis systems are functionally connected, and compensatory strategies are in place to elude proteostasis failure if the activity of one or more of the network components deteriorates.6–8 However, as we age, malfunctioning of the protein homeostasis network is inevitable and interferes with crucial signaling pathways and is often associated with multiple human diseases.9,10 Modulation of intracellular protein concentration via regulation of the proteolytic machineries has long been validated as promising milieu for the development of treatments for different human diseases such as neurodegeneration, cancer, and autoimmunity.9,11–15
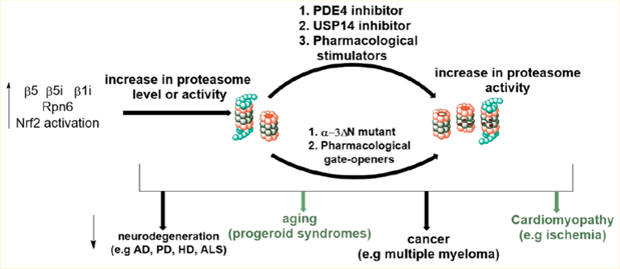
The proteasome is the cell’s first defense mechanism against accumulating proteotoxic stresses induced by oxidative damage. The 20S proteasome complex is capable of directly targeting oxidatively damaged proteins to detoxify the cell.16–18 Reactive oxygen species, from exogenous sources, the mitochondrial respiratory chain, and other cellular metabolism, accumulate as we age, causing substantial damage to proteins and other macromolecules.19,20 Upon oxidative damage, proteins unfold and expose hydrophobic regions which makes them prone to aggregation. Thus, to combat increasing levels of oxidatively damaged proteins, an increase in the 20S proteasome complex is generated by disassembly of the 26S proteasome.21,22 However, when accumulation of damaged proteins exceed degradation, deregulation of proteostasis and proteotoxic stress occurs, which are the hallmarks of several neurological disorders, including Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS, also called Lou Gehrig’s disease).9,10,23–27 Furthermore, decline in 26S proteasome activity and accumulation of ubiquitinylated proteins have been observed in post-mortem brains of AD patients.28 This suggests that both 20S and 26S are essential for the degradation of these aggregation-prone proteins.
Genetic manipulation of the proteasome proteolytic systems in animal models of different neurodegenerative disorders suggest that stimulating the activities of the proteolytic systems could be an effective strategy to treat these disorders.29,30 In aged individuals, who are the major victim of this disease class, the proteasome exists mainly as the latent 20S,19,31–33 thus making 20S a better target for these diseases.34,35 Here, we discuss approaches that have been implemented in increasing both 20S and 26S proteasome-mediated proteolysis, challenges facing the field, and applications beyond aging and neurodegenerations.
1.1. Structure of the Human Proteasome.
In eukaryotic cells, the ubiquitin proteasome system (UPS) is the major selective proteolytic system that regulates the concentration of proteins involved in numerous cellular processes.12 At the center of the UPS is the proteasome. The human 26S proteasome consist of a barrel-shaped 20S core particle (CP) capped by one or two 19S regulatory particles (RPs) also called PA700.36–38 The 20S CP is a threonine protease that consists of four stacked rings. The two inner β-rings contain three catalytic subunits (β5, β2, and β1) that display chymotrypsin-like (CT-L), trypsin-like (Tryp-L), and caspase-like (Casp-L) activity, respectively.39–42 The outer α-rings serve as gated channels that regulate substrate entry and product exit from the inner catalytic chambers. These outer rings also act as docking surfaces for the 19S RP. The α and β-rings are each composed of heteroheptameric subunits; α1–α7 and β1–β7, respectively. The 26S proteasome is formed when the 28-subunit CP is docked on one or both ends by the ATPase 19S cap (PA700). The 19S RP is responsible for 20S gate opening, substrate recognition and binding, unfolding, and threading of ubiquitinated substrates into the 20S CP.38,43–46 A number of other non-ATPase regulatory particles such as the 11S complex (PA28) and PA200 (Blm10 in yeast) also reversibly associate with the 20S CP by docking onto the α-rings.47–50
The 19S RP consist of two subcomplexes, the base that interacts directly with the 20S and a peripheral lid.51,52 The base is comprised of hexameric AAA–ATPase subunits, Rpt1– Rpt6, and tetrameric non-ATPase subunits, Rpn1, Rpn2, Rpn10, and Rpn13. The ATPase activity in the base is essential for protein substrate unfolding, gate opening, and translocation of substrate into the 20S core. The lid is made of nine non-ATPase subunits; Rpn3, Rpn5–Rpn9, Rpn11, Rpn12, and Sem1. The lid, specifically the Rpn11 subunit, functions as a deubiquitinase.53 Rpn13, Rpn10, and other reversibly associated proteins, such as radiation sensitivity abnormal 23 (Rad23) and dual-specificity protein kinase 2 (Dsk2) serve as ubiquitin receptors that direct polyubiquitinated proteins to the proteasome.54,55 The Rpn subunits also create docking site(s) for other proteins including the proteasome associated deubiquitinating enzymes, ubiquitin specific peptidase 14 (USP14), and ubiquitin C-terminal hydrolase 37 (UCH37).56,57
In immune cells and during an immune response, or in response to treatments with cytokines, such as interferon-γ (IFN-γ) or tumor necrosis factor-α (TNF-α), the constitutive catalytic 20S proteasome subunits β1, β2, and β5 are swapped with the inducible subunits LMP2 (β1i), MECL-1 (β2i), and LMP7 (β5i), respectively, forming the immunoproteasome (i20S).58 The immunoproteasome has an altered substrate binding pocket which results in cleavage pattern optimized for generating peptides for presentation on the major histocompatibility complex (MHC) class I molecules.59–62 The i20S can also associate with the IFN-γ-inducible 11S (PA28) regulatory complex on one end and 19S on the other end to form a hybrid proteasome, or it can associate with the 11S on both ends.63 The so-called thymoproteasome, with β1i, β2i, and β5t catalytic subunits has also been identified in cortical epithelial cells of the thymus.64 The t20S is believed to play critical role in the positive selection of CD8+ T cells.
1.2. The 26S Proteasome and Ubiquitin-ATP-Dependent Proteolysis.
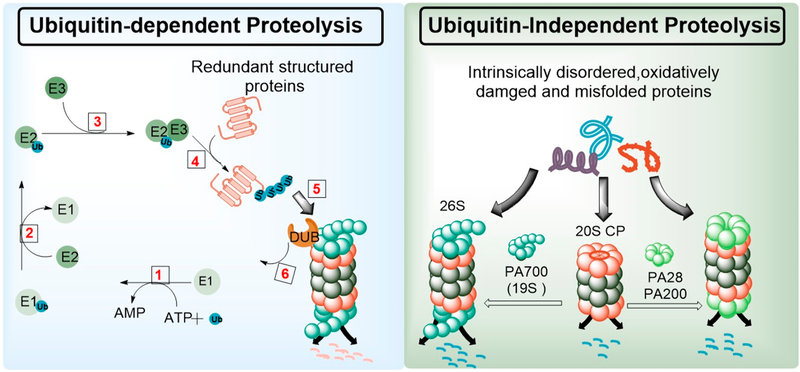
The 26S proteasome mainly targets structured proteins for degradation, although a certain fraction of misfolded and intrinsically disordered proteins (IDPs) are also degraded by the 26S in an ubiquitin-independent manner (Figure 1).65–68 Structured proteins due for degradation are tagged with chains of polyubiquitin which serve as a degron for their turnover.69 Polyubiquitinated proteins are recognized by the ubiquitin receptors, Rpn10 and Rpn13.54,55 The ubiquitin tag is then removed by deubiquitinases such as the Rpn11, USP14, and UCH37.57 The ATPase activities of the 19S base then unfolds and directs the protein into the catalytic chamber for degradation.65,70,71
Figure 1.
The proteasome degradation system. Structured proteins with polyubiquitin tags are degraded by the 26S proteasome in an ATP dependent manner (left). Partially unfolded, intrinsically disordered, and oxidatively damaged proteins are directly degraded by the 26S, free 20S, or 20S capped by non-ATPase caps such as PA28 and PA200, without the need for polyubiquitination (right).
Ubiquitin is a small protein (~8 kDa) of 76 amino-acids, consisting of seven lysine (K) residues at positions 6, 11, 27, 29, 33, 48, and 63 through which it can be attached to protein substrates (Figure 1).72,73 Although polyubiquitination of substrate proteins at K-48 of ubiquitin serves as the primary degron for degradation via the 26S proteasome, ubiquitin ligation at other lysine residues, such as K-63, have been shown to direct proteins toward autophagy-mediated proteolysis.74 A well-defined series of enzymes, ubiquitin ligases (E1, E2, and E3) coordinate the attachment of mono- and polyubiquitin to proteins. Ubiquitin is first activated in an ATP-dependent reaction by an E1 ubiquitin-activating enzyme, to which it becomes attached by a thioester bond. Subsequently, the activated ubiquitin is transferred to the active site cysteine of the E2 ubiquitin-conjugating enzyme. Ubiquitin-protein ligase (E3), together with E2 catalyze the transfer of ubiquitin onto the protein that is destined for degradation (Figure 1, left panel).75–78
1.3. The 20S Proteasome and Ubiquitin-ATP-Independent Proteolysis.
Unlike the 26S proteasome which primarily degrades polyubiquitinated proteins,39 the 20S directly degrades misfolded, oxidatively damaged, and intrinsically disordered proteins and does not require the unfoldase activity of the 19S base. Furthermore, 20S-mediated proteolysis does not require polyubiquitination of its substrates (Figure 1 right panel).67,68,79 The 20S proteasome exist mainly in the closed-gate/latent conformation in which access to the catalytic chamber is occluded by converging N-terminal residues of the alpha subunits.39,80 The 20S-mediated proteolysis is thought to involve a direct interaction of the protein substrate with the alpha-3 subunit, which brings about a conformational change and subsequent degradation of the target protein.81 A number of non-ATPase regulatory particles such as the 11S complex (PA28) and Blm10 (PA200) also reversibly associate with the 20S CP by docking onto the α-rings. This interaction induces an open-gate conformation that promotes ubiquitin- and ATP-independent proteolysis.48–50 Therefore, small molecules that can induce open-gate conformations of the 20S proteasome can thus mimic ATP-and ubiquitin-independent proteolysis seen with non-ATPase caps.
2. MECHANISMS OF INCREASING PROTEASOME-MEDIATED PROTEOLYSIS
2.1. Proteasome Activation by Agonist Induced Conformational Alteration.
2.1.1. Peptide-Based Protea-some Agonists.
Endogenous protein activators such as the 19S, PA200, and PAN (from archaeal species) contain a conserved C-terminal hydrophobic-tyrosine-any amino acid (Hb-Y-X) motif that triggers 20S gate opening upon ATP binding.43 These Hb-Y-X motifs are inserted into intersubunit pockets created by neighboring α subunits. In these pockets, interactions with conserved residues are believed to result in a rotation in the α subunits and a displacement of a reverse-turn loop that maintains the open-gate conformer. Conceivably, C-terminal peptides derived from the Hb-X-Y motifs of Rpt2 and Rpt5 subunits of the 19S proteasome were found to enhance 20S-mediated peptide and protein degradation in vitro.44,82 More recently, proline- and arginine-rich (PR) peptides previously reported as allosteric proteasome inhibitors83,84 were modified with C-terminal Hb-X-Y residues to achieve activating properties.85 PA26 and PA28 lacking the Hb-Y-X motif activate the 20S proteasome through a mechanism distinct from that of ATPase activators and does not involve α subunit rotation.43 Thus, the diversity in structure and mechanism of proteasome activation by pharmacological agents and endogenous activators suggest the presence of different allosteric pockets that can be targeted for modulation of proteasome activities.
Synthetic peptide called proteasome-activating peptide 1 (PAP1) has also been reported to increase the CT-L proteasome activity via a gate opening mechanism in the α-ring of the 20S CP.86 This peptide protected fibroblasts from hydrogen peroxide-induced oxidative stress. Most importantly, PAP1 prevented the aggregation of superoxide dismutase 1 (SOD1) in a cellular model of amyotrophic lateral sclerosis (ALS). Similarly, Kisselev et al.87 reported evidence of hydrophobic peptide-induced gate opening in the α-ring of the 20S CP.
Although clinically used proteasome inhibitors are peptide-based,88 peptide drugs and peptide-based proteasome activators are limited by intrinsic peptide properties such as poor membrane permeability and metabolic instability.89 Furthermore, peptide activators baring the Hb-Y-X motifs may also compete with endogenous proteasome activators (19S, 11S), with possibilities of intricate cellular outcomes.
2.1.2. Small Molecule Proteasome Agonist.
Tanaka et al.90 in 1988 demonstrated that the eukaryotic 20S proteasome existed mainly in the latent state and could be activated in biochemical assays with low concentrations of sodium dodecyl sulfate (0.04–0.08% SDS) or with poly-lysine. SDS is believed to act via partial denaturation of the 20S and is characterized by inhibition at concentrations greater than 0.08%.80 SDS-like proteasome activation has also been observed with lipids,91 fatty acids,92 and the natural product oleuropein.93 Although the activity-response of these molecules preclude their use in physiologically relevance systems, they are invaluable in vitro tools.
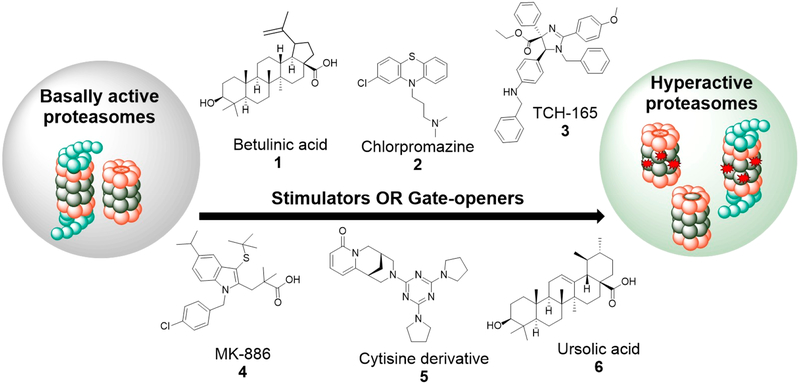
Progress toward the discovery of drug-like proteasome agonist has been slow, but a few have been identified. Molecules that interact with either the 20S or both the 20S and 26S proteasome, function through two main mechanisms, namely: (1) Gate-openers: Molecules that promote substrate entry into the catalytic pocket through allosteric interactions that induce gate-opening in the alpha ring of the 20S proteasome. (2) Stimulators: Molecules that promote 20S and/or 26S-mediated degradation through allosteric interactions that enhance substrate binding and/or degradation in one or more catalytic sites. Among the first reported 20S agonists is the triterpenoid, betulinic acid (Figure 2, compound 1),94 which was reported to specifically enhance the CT-L activity of the 20S proteasome. Unfortunately, chemical modifications to improve activity resulted in proteasome inhibitors with complicated structure activity relationships (SAR).
Figure 2.
Small molecule proteasome agonists: Small molecules that directly interact with the proteasome increase the activity of the enzyme either though gate-opening or through allosteric interaction that increase substrate binding and/or processing.
In a search for novel activators, Jones et al.95 screened the NIH clinical collection (NCC) and Prestwick library and identified chlorpromazine (Figure 2, compound 2) and other phenothiazines as 20S agonists that specifically enhanced 20S CT-L activity while promoting the degradation of intrinsically disordered proteins (IDPs), such as α-synuclein and tau in biochemical and cellular assays. Chemical modifications of chlorpromazine abolished its dopamine D2 activity while maintaining a significant proteasome stimulating ability. Coincidentally, methylene blue, a structural analogue of chlorpromazine has been demonstrated to reduce the levels of amyloid beta (Aβ) and rescues early cognitive deficit by increasing proteasome activity in a mouse model of AD.96 This further supports the robustness of phenothiazines as proteasome activators in AD therapy.
In a more mechanistic study, Njomen et al.97 identified the imidazoline TCH-165 (Figure 2, compound 3) and its analogues as 20S activators that induce gate-opening in the alpha ring of the 20S proteasome, as observed by atomic force microscopy (AFM). TCH-165 was found to enhance the degradation of IDPs in biochemical and cellular assays. In cells, TCH-165 also shifted the 20S–26S equilibrium toward the 20S, presumably by competing with the 19S cap for 20S binding. Interestingly, degradation of ubiquitinated proteins was largely maintained by single capped 20S.
In an effort to establish secondary assays for validation of “true” activators, Trader et al. screened a small library of the NCC and discovered MK-886 (Figure 2, compound 4)98 and AM-404 as new classes of 20S activators capable of enhancing the degradation of α-synuclein in cell culture. In a parallel effort, Coleman and Trader99 identified two 20S activators and a cytisine derivative (Figure 2, compound 5) as stimulator of both 20S and 26S. Among the 20S activators was the natural product, ursolic acid (Figure 2, compound 6), another triterpenoid and an analogue of betulinic acid. The mechanism of proteasome activation by ursolic acid99 appears to be distinct from that of betulinic acid,94 further supporting the complexity associated with these class of molecules and proteasome activators, in general.
Although some of these molecules have already been classified in literature35 as stimulators and gate-openers, it should be noted that imidazolines97 are the only class of 20S agonist with biophysical data (AFM)97 supporting their gate-opening activity.
2.2. Proteasome Activation by Modulation of Posttranslational Modification.
2.2.1. Small Molecule Kinase Modulators.
Proteasome activation has also been realized through upstream modulation of kinases, with ensuing post-translational modification of proteasome subunits. The human 26S proteasome undergoes reversible phosphorylation in response to the ever-changing pathophysiological state of the cells.100 More than 455 phosphorylation sites have been identified in the human 26S proteasome, and they play different roles ranging from modulation of 26S assembly, stability, and activity. For example, osmotic stress inhibits proteasome activity via p38-MAPK-dependent phosphorylation of Rpn2.101
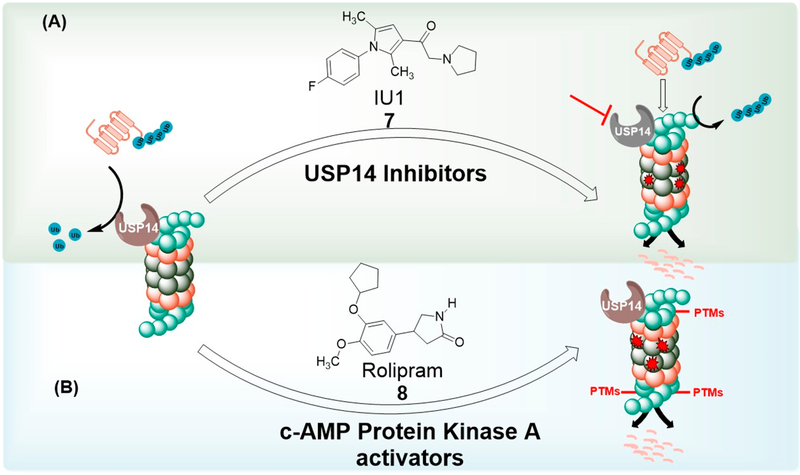
Phosphorylation of the proteasome Rpt6 subunit by cAMP-dependent protein kinase A (PKA) has been reported to upregulate 26S assembly and proteasome activity in vitro and in vivo.102,103 Rolipram (Figure 3, compound 8), a small molecule inhibitor of phosphodiesterase type 4 (PDE4),104 was found to raise the level of cAMP in the brain of mice, activated PKA and presumably increased proteasome activity through subunit (Rpt6) phosphorylation. Most importantly, rolipram promoted the clearance of abnormal tau and improved cognition in mouse model of Alzheimer’s disease.105
Figure 3.
Enhancement of proteasome by modulation of posttranslational modifications (PTMs): (A) Inhibition of USP14 prevents ubiquitin chain trimming, thereby promoting 26S-mediated protein degradation. (B) Phosphorylation of 19S Rpn6 subunit through activation of c-AMP PKA enhances 26S-mediated proteolysis.
PKA-mediated phosphorylation of the non-ATPase 19S subunit Rpn6 has also been shown to enhance the degradation of ubiquitinated and aggregation-prone proteins such as tau in different cell lines.106 These studies validated the involvement of the 26S proteasome in the degradation of partially unfolded proteins (or intrinsically disordered proteins, IDPs), further supporting the significance of targeting either the 20S and/or 26S in the quest for increasing proteasome-mediated IDP clearance.
2.2.2. Small Molecule Deubiquitinase Inhibitors.
The 19S proteasome subunit Rpn11 has deubiquitinase activity that recycles polyubiquitin by removing them en bloc from protein substrates that are committed for degradation.53 Additional deubiquitinases USP14 and UCH37 reversibly interact with the proteasome and can trim ubiquitin chains independent of substrate commitment to degradation.56,57,107
Lee et al.108 demonstrated that ubiquitin chain trimming by USP14 inhibits substrate degradation by the proteasome. In the same study, small molecule inhibitor of USP14, IU1 (Figure 3, compound 7), was found to enhance the degradation of different proteasome substrates, including tau. IU1 also promoted the degradation of oxidized proteins, thereby conferring a cytoprotective effect toward oxidative stress in HEK cells.
Interestingly, inhibition of USP14 by siRNA knockdown or use of its small molecule inhibitor led to accumulation of polyubiquitinylated proteins with increased apoptosis of multiple myeloma cells.109 This translated to extended survival in xenograft models of multiple myeloma. In a parallel study, the small molecule inhibitor of USP14 and UCH37, b-AP15, also promoted the accumulation of polyubiquitin and delayed tumor progression in a mouse model of multiple myeloma110 as well as four different models of solid tumors.111 These intricate outcomes seen with USP14 inhibition might be a result of its many diverse cellular functions. Thus, targeting USP14 represents an exciting approach that may effectively translate in combating multiple types of cancers.
Furthermore, increasing proteasome-mediated proteolysis by inhibition of USP14 blocks autophagy flux.112 The mechanism of autophagy inhibition appears to involve enhanced proteasomal degradation of the autophagosome–lysosome fusion protein, UV radiation resistance-associated gene (UVRAG). This result is consistent with Choi et al.’s113 observation that proteasome activation via generation of open-gate mutant inhibited autophagy flux. Given the interplay between the two proteolytic systems, one is tempted to hypothesize that the antitumor effcacy of deubiquitinase inhibitors is directly related to autophagy inhibition and secondary to proteasome activation. However, genetic and pharmacological inhibition of the same USP14 was found to correct an in vivo model of impaired mitophagy114 and also increased autophagy flux through suppression of K-63 ubiquitination of Beclin 1.115 These complex observations further suggest the need to address the effect of increased proteasome activity at the level of molecules used, mechanism of increasing proteasome activity, and other targets of the small molecule that may be involved. For example, gate opening induced by deletion of α−3 N-terminal and by small molecule gate openers are anticipated to generate different conformationally active states of the proteasome, with ensuing disparity in protein degradation profile.
2.3. Proteasome Activation by Genetic Manipulation.
2.3.1. Upregulation of Proteasome Activity through Subunit Overexpression.
Pioneer work involving upregulation of proteasome activity through genetic manipulation were first illustrated by Gaczynska et al.116,117 In these studies, overexpression of β5i (LMP7) subunit in lymphoblasts and HeLa cells resulted in increased CT-L and Tryp-L activities, while overexpression of β1i (LMP2) only elevated the Tryp-L like activity. Therefore, increasing the activity of the immunoproteasome could also increase antigen processing thereby boosting immune response. In 2005, Chondogianni and co-workers118 published a more in-depth study on proteasome activation via subunit upregulation: Stable over-expression of β5 catalytic subunit in two established (WI38/T and HL60) and primary (IMR90) human fibroblast cell lines resulted in elevated levels of other β-subunits and increased level of assembled proteasome, with a concomitant increase in all three proteasome proteolytic activities. Consistent with the role of the proteasome, β5-overexpressing cell lines conferred protection against several oxidation-induced proteotoxic stresses, via enhanced degradation of oxidized proteins. A few parallel studies have also found β5-overexpression to protect against oxidative stress in human lens epithelial cells119 and to prevent replicative senescence of human bone marrow stromal cells.120 These findings further corroborate the role of the proteasome in combating diverse classes of proteotoxic disorders.
2.3.2. Proteasome Upregulation through Nrf2 Activation.
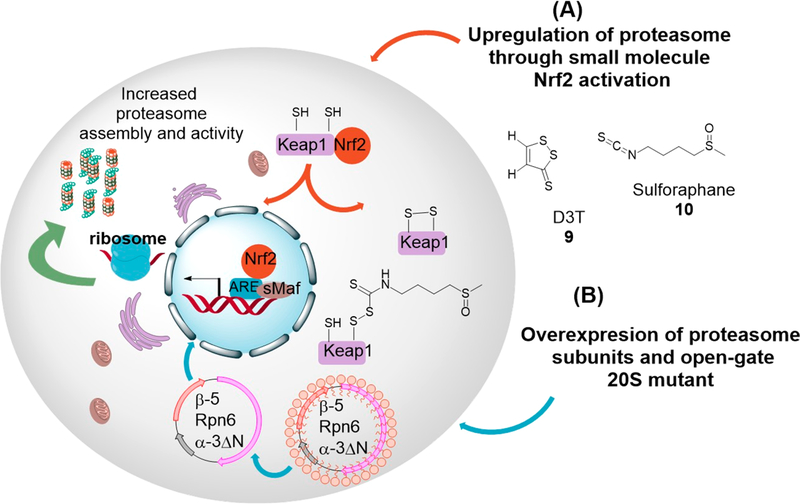
The transcription factor (nuclear factor (erythroid-derived 2)-like 2 (Nrf2), is a basic leucine zipper121 that controls the expression of antioxidant enzymes including glutathione S-transferase, NADPH quinone oxidoreductase 1122 as well as proteasome subunits.123 Accordingly, overexpression of the proteasome has also been achieved through small molecule activators of Nrf2. Kwak et al.123 demonstrated that the indirect antioxidant 3H-1,2-dithiole-3-thione (D3T), (Figure 4, compound 9) upregulated both 20S and 19S proteasome subunits as well as proteasome activity only in Nrf2 positive fibroblast. A more recent article showed that activation of Nrf2 with D3T significantly improved cognitive deficits in a mouse model of Alzheimer’s disease and dramatically reduced the level of insoluble amyloid beta (Aβ), as well as oxidative stress.124 The triterpenoid, 18α- glycyrrhetinic acid (18α-GA), has also been shown to delay replicative senescence in human fibroblast through Nrf2-mediated upregulation of proteasome subunit and activity.125 In Caenorhabditis elegans, 18α-GA also decreased Aβ deposition and delayed the progress of AD while promoting proteasome-dependent life span extension.126 Increased proteasome activity has also been attained by activating Nrf2 with the food additive tert-butylhydroquinone (t-BHQ) and food supplement sulforaphane. The increase in proteasome activity with t-BHQ and sulforaphane was accompanied by delayed differentiation, increased self-renewal and pluripotency in human embryonic stem cells (hESCs), and in induced pluripotent stem cell (iPS-IMR90).127 Similar activities have been observed by others in neuroblastoma cells, where sulforaphane (Figure 4, compound 10) -Nrf2-mediated upregulation of proteasome activity protected against oxidative stress.128
Figure 4.
Genetic upregulation of proteasome activity: (A) Activation of Nrf2 through small molecule oxidation and/or conjugation of keap1 cysteine residues promotes Nrf2-driven proteasome expression and activity. (B) Overexpression of proteasome β−5 and Rpn6 subunits increases 26S assembly and activity, while overexpression of N-terminal truncated α−3 subunit results in hyperactive open-gate 20S and 26S proteasomes.
2.3.3. Cap-Upregulation and Generation of Open-Gate Mutants.
The 20S proteasome exists mainly in its latent state conformation in which entrance into the catalytic chamber is occluded by converging N-terminal of the α-subunits.41,90,129 Proteins must access these gated channels to be degraded by the 20S core particle. Sensibly, one genetic approach to increase proteasome activity has been aimed at increasing accessibility through these gated channels. Certainly, over-expression of 19S Rpn6 subunit led to enrichment of assembled 26S and increased proteasome activity in embryonic stem cells.130 Meanwhile, enhancement of proteasome activity via overexpression of PA28α was found to protect against proteinopathy and ischemia/reperfusion injury in mice.131 These findings further highlight the importance of proteasome activation beyond neurodegenerative diseases and aging.
In a more recent study, Choi et al.113 took a unique approach toward genetic upregulation of proteasome activity. Choi et al. showed that deleting the N-terminal of α−3 subunit generates an α−3ΔN mutant that is incorporated into fully assembled 20S and 26S proteasomes. These α−3ΔN mutant proteasomes showed increased degradation of about two hundred 20S and 26S protein substrates, including tau and α-synuclein. Furthermore, the enhanced degradation of α-synuclein significantly delayed the formation of α-synuclein aggregates. Significant resistance against oxidative stress was also observed with cells carrying the open-gate mutant.
Most of these genetic approaches have only been explored at the in vitro levels (Figure 4), with little or no supporting animal data. However, one could envision the delivery of proteasome β5 subunit, 19S Rpn6, and/or α−3ΔN as gene therapy for the treatment of several diseases.
2.4. Proteasome Activation via Unnknown Mechanisms.
Pyrazolones have previously been reported to have neuroprotective function in a cellular model of ALS, with significant increase in the median survival time of an ALS transgenic mouse model.132–134 More recently, the activity of pyrazolone in the ALS model has been linked to proteasome activation.135 This conclusion was based on enhanced degradation of model proteasome substrates in cells, as well as affnity pull-down of Rpt2 and Rpt3 as interacting partners of pyrazolone. However, given that the activities of these compounds were not assayed with purified proteasome, it is not clear if they are direct proteasome agonists, modulators of endogenous proteasome activators (based on the pull-down assay with 19S subunits) or upstream modulators of proteasome posttranslational modifiers. Thus, in this review, and based on our classification, we categorize pyrazolones as enhancers of proteasome activity with unknown mechanism of action.
In another study by Leestemaker et al.,136 a chemical genetic screen was used to identify PD169316, a p38 MAPK inhibitor, as enhancer of 26S proteasome activity. Although the exact mechanism behind 26S activation by p38 MAPK inhibitor could not be deciphered, the authors also demonstrated a decrease in the level of α-synuclein as well as protection against toxic α-synuclein in primary mouse neurons. Most importantly, the authors also developed activity-based probes that bind to the proteasome catalytic sites in an activity-dependent manner, thereby allowing for the screening of proteasome modulators.136
3. CHALLENGES AND CONSIDERATIONS
3.1. Crosstalk with Autophagy and Intricate Outcomes.
The proteolytic arm of the protein homeostasis network is sustained by both the proteasome and the autophagy pathways. At the protein level, autophagy seems to be instigated to compensate for cellular stresses not addressed by the proteasomal clearance system.137 A well-balanced crosstalk between the two catabolic pathways ensures efficient maintenance of energy. In support of this, several studies have demonstrated that proteasome inhibition activates the autophagy pathway.8,138–140 Conversely, acute inhibition of autophagy also activates the proteasome, although chronic autophagy inhibition hinders proteasome-mediated proteolysis.7,141 Proteasome activation is therefore anticipated to reduce (the need for) autophagic clearance of proteins. Indeed, such phenomena were observed with the hyperactive open-gate 20S proteasome113 as well as when proteasome activity was enhanced by targeting USP14112 or with imidazolinebased 20S agonist (unpublished data from our lab). These observations could be specific to the type of agonist, mechanism of proteasome activation, and cell type, making the outcome of this crosstalk more difficult to predict. This is further illustrated by the case of USP14, where activation of 26S proteasome through genetic and pharmacological manipulation triggers conflicting outcomes in autophagic flux.112,114,115
3.2. Drug Repurposing Challenges.
Current paradigm in identifying small molecule proteasome activators has been to screen clinical collections of drugs.95,98,99,136 Hits from these libraries would typically be optimized to eliminate activity at their previous targets while maintaining activity at the new target (proteasome).95,142 This optimization and repurposing of hits from clinical library is challenging and has in part contributed to the slow progress in identifying small molecule proteasome agonist. Perhaps, identifying the “true” binding pockets of current 20S agonists through proteomic or crystallographic studies would allow for a streamline, computer-aided, structure-based drug design that could overcome this problem. While this may sound easy on paper, it could be quite a daunting task given the numerous binding sites and mechanisms of activation of this humongous 2.5 MDa complex. Nonetheless, this knowledge is anticipated to be invaluable in driving the field forward.
3.3. Lack of Standard Assays for Efficacy Studies.
Canonical proteasome activity assays use peptide substrates specific for each proteasome catalytic sites (CT-L, Suc-LLVYAMC; Tryp-L, Boc-LRR-AMC; Casp-L, Z-LLE-AMC). These peptides are conjugated with a C-terminal fluorophore that is activated upon proteasomal cleavage.40,143 The small size of these peptides allows them to navigate the narrow gate of latent 20S proteasome, resulting in high and varying baseline 20S proteasome activity. Furthermore, even this so-called standard assay is not standard as different laboratories use different substrate to enzyme ratios as well as different buffer systems. Moreover, detergent-like molecules that can relax this narrow 20S gate just enough to increase peptide substrate accessibility to the catalytic core appear as positive hit in these peptide assays. These non-bona fide agonists fail in assays that utilize more physiologically relevant protein substrates.98 These limitations result in ambiguous comparison of potency for proteasome agonists, from batch to batch and from lab to lab. Furthermore, these primary screens employ purified 20S and fluorogenic peptides specific to just one 20S catalytic site, thereby ignoring the allosteric interactions between the three catalytic sites that drive protein degradation.144 As such, stimulators of the other catalytic sites as well as molecules that act upstream of the proteasome are also missed in this screen.
The quest for proteasome agonists has been paralleled by the development of new assays to address these deficiencies and validate this new approach of targeting proteotoxic disorders. Among these assays are: (1) The IDPs based biochemical assays of Njomen et al.97 and Jones et al.,95 which address the need for physiologically relevant substrates. (2) The larger peptide mass spectrometry-based assay of Trader et al.,98 which eliminates non-bona fide activators. (3) The peptide-FRET reporter assay of Coleman and Trader,99 which addresses the problem of high background activity, and (4) the proteasome activity-based probe of Leestemaker et al.,136 which addresses the need for a high-throughput cellular system. Each of these methods described has one or more of the following limitations, namely, the use of peptides or small molecule probes instead of protein substrates of interest, tedious, and not high-throughput compatible, and/or measures the state of activeness of the proteasome as opposed to its ability to degrade specific proteotoxic proteins. Therefore, there is still the need for a cell-based and protein-based assay that is amenable to high-throughput screening.
3.4. Anticipated Toxicity of Proteasome Activators.
Enhancing proteasome activity reduces the proteotoxic burden cells experience upon aging. In humans,145,146 rodents,96,147,148 and cells,125,149–154 increased proteasome activity delays aging150,155–157 and results in longer lifespan158–160 by reducing proteotoxic pathologies. Supporting this idea, cells from human centenarians exhibit enhanced proteasome activity compared to cells from adults of different ages.145
Cellular studies using short-term exposure to proteasome agonists have thus far not illustrated inherent toxicity, however, no studies have yet defined any long-term effects.85 The lack of initial toxicity may be due to the protective roles of chaperones or other protein complexes, which greatly limit the access of the 20S proteasome to its targets, thus yielding proteolytic selectivity.34,68,161,162 In addition, the rate of proteasomal degradation of disordered regions is also dependent on the composition and length of specific disordered initiation sequences.163 Stimulation of proteasome activity is therefore anticipated to have a differential effect on the clearance of different proteins.
Proteasome inhibition is the mechanism of action of most chemotherapeutics targeting blood cancer such as multiple myeloma.164 Thus, a question that has become of concern is whether proteasome activators can cause cancer. Indeed, enhanced degradation of misfolded proteins through Nrf2-mediated upregulation of proteasome activity has been demonstrated to promote tumorigenesis.165 Other studies demonstrated that deletion of Keap1, a negative regulator of Nrf2, and Pten drive the progress of non-small-cell lung cancer.166,167 It should, however, be noted that Nrf2 activation drives the expression of many genes, including cyclin B and cyclin-dependent kinase 1(CDK1),168 which are involve in driving mitotic division.169 Thus, the effect of proteasome activation on tumorigenesis must be examined on a case by case basis.
4. CONCLUSION
Herein, we discussed different strategies that have been developed to increase proteasome activity while also reflecting on some challenges associated with the field. We further touched on the potential of proteasome activation being applicable to the reduction of proteotoxic stresses seen in aging, neurodegeneration, and other disorders. This means the appropriate mechanism of increasing proteasome activity will likely be contingent on the disease under consideration. Furthermore, just like any new field, the concept of proteasome activation has also prompted a lot of apprehensions such as tumor causing potential and effect on other proteolytic pathways. It is, however, important to keep in mind that the physiological outcome of proteasome activation will depend largely on the mechanism employed and how “specific” it is to the proteasome network. Most of all, this field is still at its infant stage with positive outcomes so far. Thus, the next few years should shine some light on some of these concerns as we move from basic proof of concept to effcacy validation in different animal models of diseases under consideration.
ACKNOWLEDGMENTS
Financial support for this work was provided by the National Institute of Allergy and Infectious Diseases (1R21AI117018-01A1) and the National Institute of General Medical Sciences (T32GM092715) of the National Institutes of Health. The authors also gratefully acknowledge financial support from Michigan State University.
ABBREVIATIONS USED
- AD
Alzheimer’s Disease
- AFM
atomic force microscopy
- ALS
amyotrophic lateral sclerosis
- Blm 10
bleomycin-sensitive
- Boc-LRR-AMC
tert-butoxyl-leucyl-arginyl-arginyl-7-amino-4-methylcoumarin
- Cdk1
cyclin dependent kinase 1
- CP
core particle
- D3T
3H-1,2-dithiole-3-thione
- Dsk2
dual-specificity protein kinase 2
- Hb-Y-X
hydrophobic-tyrosine-any amino acid
- HD
Huntington’s disease
- HL60
peripheral blood promyeloblast
- IDP
intrinsically disordered proteins
- IMR90
lung fibroblast
- LMP2
low molecular mass polypeptide 2
- LMP7
low molecular mass polypeptide 7
- MAPK
mitogen-activated protein kinase
- MECL-1
multicatalytic endopeptidase complex subunit 1
- MHC
major histocompatibility complex
- NCC
NIH (National Institutes of health) Clinical Collection
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- PA28
proteasome activator 28 subunit
Biography
Evert Njomen is currently working towards her Ph.D. degrees in Chemistry and Pharmacology at Michigan State University, under the supervision of Dr. Jetze Tepe. Her research focuses on the use of small molecule proteasome activators to understand the role of the ubiquitin-independent proteasome system in cellular function and the therapeutic potential of such perturbations. She received her B.Sc. degree in Biochemistry (2010) from the University of Buea (Cameroon) and a M.Sc. degree in Chemistry (2014) from Eastern Michigan University (EMU). At EMU, she studied peptide-based inhibitors of protein–protein interactions. Evert is interested in the use of chemical tools in exploring signaling pathways to better understand the fundamental mechanism underlying human diseases and the characterization of novel drug targets and candidates across several human diseases.
Jetze J. Tepe is an Associate Professor at the Department of Chemistry and Pharmacology & Toxicology at Michigan State University. He received his Ph.D. in Chemistry from the University of Virginia (T. L. Macdonald) and completed his postdoctoral studies at Colorado State University (R. M. Williams). Research in his lab is focused on the synthesis of marine sponge metabolites and their unnatural drug-like analogues as antagonists or agonists of proteasome function. By developing new synthetic methodologies, drug-like derivatives of natural products are prepared and interrogated for their clinical significance in biochemical assays, in cell culture, and in animal models, with a therapeutic focus on multiple myeloma and neurodegenerative diseases.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Kaushik S; Cuervo AM Proteostasis and Aging. Nat. Med 2015, 21, 1406–1415. [DOI] [PubMed] [Google Scholar]
- (2).Hetz C; Glimcher LH Protein Homeostasis Networks in Physiology and Disease. Curr. Opin. Cell Biol 2011, 23, 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Frydman J Folding of Newly Translated Proteins in Vivo: The Role of Molecular Chaperones. Annu. Rev. Biochem 2001, 70, 603–647 [DOI] [PubMed] [Google Scholar]
- (4).Douglas PM; Summers DW; Cyr DM Molecular Chaperones Antagonize Proteotoxicity by Differentially Modulating Protein Aggregation Pathways. Prion 2009, 3, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Niforou K; Cheimonidou C; Trougakos IP Molecular Chaperones and Proteostasis Regulation during Redox Imbalance. Redox Biol. 2014, 2, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ji CH; Kwon YT Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol. Cells 2017, 40, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Korolchuk VI; Mansilla A; Menzies FM; Rubinsztein DC Autophagy Inhibition Compromises Degradation of Ubiquitin-Proteasome Pathway Substrates. Mol. Cell 2009, 33, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zheng Q; Su H; Tian Z; Wang X Proteasome Malfunction Activates Macroautophagy in the Heart. Am. J. Cardiovasc. Dis 2011, 1, 214–226. [PMC free article] [PubMed] [Google Scholar]
- (9).Halliwell B; Isacson O; Jenner P; McNaught KSP; Olanow CW Failure of the Ubiquitin-Proteasome System in Parkinson’s Disease. Nat. Rev. Neurosci 2001, 2, 589–594. [DOI] [PubMed] [Google Scholar]
- (10).Saez I; Vilchez D The Mechanistic Links Between Proteasome Activity, Aging and Age-Related Diseases. Curr. Genomics 2014, 15, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Calamini B; Morimoto RI Protein Homeostasis as a Therapeutic Target for Diseases of Protein Conformation. Curr. Top. Med. Chem 2013, 12, 2623–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Voges D; Zwickl P; Baumeister W The 26S Proteasome: A Molecular Machine Designed for Controlled Proteolysis. Annu. Rev. Biochem 1999, 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- (13).Voorhees PM; Dees EC; O’Neil B; Orlowski RZ The Proteasome as a Target for Cancer Therapy. Clin. Cancer Res 2003, 9, 6316–6325. [PubMed] [Google Scholar]
- (14).Ostrowska H The Ubiquitin-Proteasome System: A Novel Target for Anticancer and Anti-Inflammatory Drug Research. Cell. Mol. Biol. Lett 2008, 13 (3), 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Opattova A; Cente M; Novak M; Filipcik P The Ubiquitin Proteasome System as a Potential Therapeutic Target for Treatment of Neurodegenerative Diseases. Gen. Physiol. Biophys 2015, 34, 337–352. [DOI] [PubMed] [Google Scholar]
- (16).Jung T; Grune T The Proteasome and Its Role in the Degradation of Oxidized Proteins. IUBMB Life 2008, 60, 743–752. [DOI] [PubMed] [Google Scholar]
- (17).Grune T; Reinheckel T; Davies KJ Degradation of Oxidized Proteins in Mammalian Cells. FASEB J. 1997, 11, 526–534. [PubMed] [Google Scholar]
- (18).Sitte N; Merker K; Grune T Proteasome-Dependent Degradation of Oxidized Proteins in MRC-5 Fibroblasts. FEBS Lett. 1998, 440, 399–402. [DOI] [PubMed] [Google Scholar]
- (19).Grune T Oxidative Stress, Aging and the Proteasomal System. Biogerontology 2000, 1, 31–40. [DOI] [PubMed] [Google Scholar]
- (20).Breusing N; Arndt J; Voss P; Bresgen N; Wiswedel I; Gardemann A; Siems W; Grune T Inverse Correlation of Protein Oxidation and Proteasome Activity in Liver and Lung. Mech. Ageing Dev 2009, 130, 748–753. [DOI] [PubMed] [Google Scholar]
- (21).Reinheckel T; Sitte N; Ullrich O; Kuckelkorn U; Davies KJ; Grune T Comparative Resistance of the 20S and 26S Proteasome to Oxidative Stress. Biochem. J 1998, 335, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang X; Yen J; Kaiser P; Huang L Regulation of the 26S Proteasome Complex During Oxidative Stress. Sci. Signaling 2010, 3, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lam YA; Pickart CM; Alban A; Landon M; Jamieson C; Ramage R; Mayer RJ; Layfield R Inhibition of the Ubiquitin-Proteasome System in Alzheimer’s Disease. Proc. Natl. Acad. Sci. U. S.A 2000, 97, 9902–9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ross CA; Poirier MA Protein Aggregation and Neurodegenerative Disease. Nat. Med 2004, 10, S10–S17. [DOI] [PubMed] [Google Scholar]
- (25).Soto C Unfolding the Role of Protein Misfolding in Neurodegenerative Diseases. Nat. Rev. Neurosci 2003, 4, 49–60. [DOI] [PubMed] [Google Scholar]
- (26).Keller JN; Hanni KB; Markesbery WR Impaired Proteasome Function in Alzheimer’s Disease. J. Neurochem 2000, 75, 436–439. [DOI] [PubMed] [Google Scholar]
- (27).Tofaris GK; Layfield R; Spillantini MG Alpha-Synuclein Metabolism and Aggregation Is Linked to Ubiquitin-Independent Degradation by the Proteasome. FEBS Lett. 2001, 509, 22–26. [DOI] [PubMed] [Google Scholar]
- (28).Perry G; Friedman R; Shaw G; Chau V Ubiquitin Is Detected in Neurofibrillary Tangles and Senile Plaque Neurites of Alzheimer Disease Brains. Proc. Natl. Acad. Sci. U. S. A 1987, 84, 3033–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bedford L; Hay D; Devoy A; Paine S; Powe DG; Seth R; Gray T; Topham I; Fone K; Rezvani N; Mee M; Soane T; Layfield R; Sheppard PW; Ebendal T; Usoskin D; Lowe J; Mayer RJ Depletion of 26S Proteasomes in Mouse Brain Neurons Causes Neurodegeneration and Lewy-like Inclusions Resembling Human Pale Bodies. J. Neurosci 2008, 28, 8189–8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liu Y; Hettinger CL; Zhang D; Rezvani K; Wang X; Wang H The Proteasome Function Reporter GFPu Accumulates in Young Brains of the APPswe/PS1dE9 Alzheimer’s Disease Mouse Model. Cell. Mol. Neurobiol 2014, 34, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zabel C; Nguyen HP; Hin SC; Hartl D; Mao L; Klose J Proteasome and Oxidative Phoshorylation Changes May Explain Why Aging Is a Risk Factor for Neurodegenerative Disorders. J. Proteomics 2010, 73, 2230–2238 [DOI] [PubMed] [Google Scholar]
- (32).Vigouroux S; Briand M; Briand Y Linkage between the Proteasome Pathway and Neurodegenerative Diseases and Aging. Mol. Neurobiol 2004, 30, 201–221. [DOI] [PubMed] [Google Scholar]
- (33).Tomaru U; Takahashi S; Ishizu A; Miyatake Y; Gohda A; Suzuki S; Ono A; Ohara J; Baba T; Murata S; Tanaka K; Kasahara M Decreased Proteasomal Activity Causes Age-Related Phenotypes and Promotes the Development of Metabolic Abnormalities. Am. J. Pathol 2012, 180, 963–972. [DOI] [PubMed] [Google Scholar]
- (34).Opoku-Nsiah KA; Gestwicki JE Aim for the Core: Suitability of the Ubiquitin-Independent 20S Proteasome as a Drug Target in Neurodegeneration. Transl. Res 2018, 198, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Coleman RA; Trader DJ All About the Core: A Therapeutic Strategy to Prevent Protein Accumulation with Proteasome Core Particle Stimulators. ACS Pharmacol. Transl. Sci 2018, 1, 140–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Xie Y Structure, Assembly and Homeostatic Regulation of the 26S Proteasome. J. Mol. Cell Biol 2010, 2, 308–317. [DOI] [PubMed] [Google Scholar]
- (37).Huang X; Luan B; Wu J; Shi Y An Atomic Structure of the Human 26S Proteasome. Nat. Struct. Mol. Biol 2016, 23, 778–785. [DOI] [PubMed] [Google Scholar]
- (38).da Fonseca PCA; Morris EP Structure of the Human 26S Proteasome: Subunit Radial Displacements Open the Gate into the Proteolytic Core. J. Biol. Chem 2008, 283, 23305–23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Tanaka K; Yoshimura T; Kumatori A; Ichihara A; Ikai A; Nishigai M; Kameyama K; Takagi T Proteasomes (Multi-Protease Complexes) as 20 S Ring-Shaped Particles in a Variety of Eukaryotic Cells. J. Biol. Chem 1988, 263, 16209–16217. [PubMed] [Google Scholar]
- (40).Groll M; Heinemeyer W; Jäger S; Ullrich T; Bochtler M; Wolf DH; Huber R The Catalytic Sites of 20S Proteasomes and Their Role in Subunit Maturation: A Mutational and Crystallographic Study. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 10976–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Unno M; Mizushima T; Morimoto Y; Tomisugi Y; Tanaka K; Yasuoka N; Tsukihara T The Structure of the Mammalian 20S Proteasome at 2.75 Å Resolution. Structure 2002, 10, 609–618. [DOI] [PubMed] [Google Scholar]
- (42).Groll M; Bochtler M; Brandstetter H; Clausen T; Huber R Molecular Machines for Protein Degradation. ChemBioChem 2005, 6, 222–256. [DOI] [PubMed] [Google Scholar]
- (43).Rabl J; Smith DM; Yu Y; Chang S-C; Goldberg AL; Cheng Y Mechanism of Gate Opening in the 20S Proteasome by the Proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Smith DM; Chang S-C; Park S; Finley D; Cheng Y; Goldberg AL Docking of the Proteasomal ATPases’ Carboxyl Termini in the 20S Proteasome’s Alpha Ring Opens the Gate for Substrate Entry. Mol. Cell 2007, 27, 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chu-Ping M; Vu JH; Proske RJ; Slaughter CA; DeMartino GN Identification, Purification, and Characterization of a High Molecular Weight, ATP-Dependent Activator (PA700) of the 20 S Proteasome. J. Biol. Chem 1994, 269, 3539–3547. [PubMed] [Google Scholar]
- (46).Finley D; Chen X; Walters KJ Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci 2016, 41, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ustrell V; Hoffman L; Pratt G; Rechsteiner M PA200, a Nuclear Proteasome Activator Involved in DNA Repair. EMBO J. 2002, 21, 3516–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Stadtmueller BM; Hill CP Proteasome Activators. Mol. Cell 2011, 41, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sadre-Bazzaz K; Whitby FG; Robinson H; Formosa T; Hill CP Structure of a Blm10 Complex Reveals Common Mechanisms for Proteasome Binding and Gate Opening. Mol. Cell 2010, 37, 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Whitby FG; Masters EI; Kramer L; Knowlton JR; Yao Y; Wang CC; Hill CP Structural Basis for the Activation of 20S Proteasomes by 11S Regulators. Nature 2000, 408, 115–120. [DOI] [PubMed] [Google Scholar]
- (51).Sharon M; Taverner T; Ambroggio XI; Deshaies RJ; Robinson CV Structural Organization of the 19S Proteasome Lid: Insights from MS of Intact Complexes. PLoS Biol. 2006, 4, e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ehlinger A; Walters KJ Structural Insights into Proteasome Activation by the 19S Regulatory Particle. Biochemistry 2013, 52, 3618–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Verma R; Aravind L; Oania R; McDonald WH; Yates JR; Koonin EV; Deshaies RJ Role of Rpn11 Metalloprotease in Deubiquitination and Degradation by the 26S Proteasome. Science 2002, 298, 611–615. [DOI] [PubMed] [Google Scholar]
- (54).Husnjak K; Elsasser S; Zhang N; Chen X; Randles L; Shi Y; Hofmann K; Walters K; Finley D; Dikic I Proteasome Subunit Rpn13 Is a Novel Ubiquitin Receptor. Nature 2008, 453, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Elsasser S; Chandler-Militello D; Müller B; Hanna J; Finley D Rad23 and Rpn10 Serve as Alternative Ubiquitin Receptors for the Proteasome. J. Biol. Chem 2004, 279, 26817–26822. [DOI] [PubMed] [Google Scholar]
- (56).Yao T; Cohen RE A Cryptic Protease Couples Deubiquitination and Degradation by the Proteasome. Nature 2002, 419, 403–407. [DOI] [PubMed] [Google Scholar]
- (57).Lee MJ; Lee B-H; Hanna J; King RW; Finley D Trimming of Ubiquitin Chains by Proteasome-Associated Deubiquitinating Enzymes. Mol. Cell. Proteomics 2011, 10, R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Aki M; Shimbara N; Takashina M; Akiyama K; Kagawa S; Tamura T; Tanahashi N; Yoshimura T; Tanaka K; Ichihara A Interferon-Gamma Induces Different Subunit Organizations and Functional Diversity of Proteasomes. J. Biochem 1994, 115, 257–269. [DOI] [PubMed] [Google Scholar]
- (59).Basler M; Kirk CJ; Groettrup M The Immunoproteasome in Antigen Processing and Other Immunological Functions. Curr. Opin. Immunol 2013, 25, 74–80. [DOI] [PubMed] [Google Scholar]
- (60).Huber EM; Basler M; Schwab R; Heinemeyer W; Kirk CJ; Groettrup M; Groll M Immuno- and Constitutive Proteasome Crystal Structures Reveal Differences in Substrate and Inhibitor Specificity. Cell 2012, 148, 727–738. [DOI] [PubMed] [Google Scholar]
- (61).Sijts AJAM; Standera S; Toes REM; Ruppert T; Beekman NJCM; van Veelen PA; Ossendorp FA; Melief CJM; Kloetzel PM MHC Class I Antigen Processing of an Adenovirus CTL Epitope Is Linked to the Levels of Immunoproteasomes in Infected Cells. J. Immunol 2000, 164, 4500–4506. [DOI] [PubMed] [Google Scholar]
- (62).Toes REM; Nussbaum AK; Degermann S; Schirle M; Emmerich NPN; Kraft M; Laplace C; Zwinderman A; Dick TP; Müller J; Schonfisch B; Schmid C; Fehling HJ; Stevanovic S; Rammensee HG; Schild H Discrete Cleavage Motifs of Constitutive and Immunoproteasomes Revealed by Quantitative Analysis of Cleavage Products. J. Exp. Med 2001, 194, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).McCarthy MK; Weinberg JB The Immunoproteasome and Viral Infection: A Complex Regulator of Inflammation. Front. Microbiol 2015, 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Murata S; Sasaki K; Kishimoto T; Niwa S; Hayashi H; Takahama Y; Tanaka K Regulation of CD8+ T Cell Development by Thymus-Specific Proteasomes. Science 2007, 316, 1349–1353. [DOI] [PubMed] [Google Scholar]
- (65).Waxman L; Fagan JM; Goldberg AL Demonstration of Two Distinct High Molecular Weight Proteases in Rabbit Reticulocytes, One of Which Degrades Ubiquitin Conjugates. J. Biol. Chem 1987, 262, 2451–2457. [PubMed] [Google Scholar]
- (66).Ben-Nissan G; Sharon M Regulating the 20S Proteasome Ubiquitin-Independent Degradation Pathway. Biomolecules 2014, 4, 862–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Baugh JM; Viktorova EG; Pilipenko EV Proteasomes Can Degrade a Significant Proportion of Cellular Proteins Independent of Ubiquitination. J. Mol. Biol 2009, 386, 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Erales J; Coffino P Ubiquitin-Independent Proteasomal Degradation. Biochim. Biophys. Acta, Mol. Cell Res 2014, 1843, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Johnson ES; Bartel B; Seufert W; Varshavsky A Ubiquitin as a Degradation Signal. EMBO J. 1992, 11, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Etlinger JD; Goldberg AL A Soluble ATP-Dependent Proteolytic System Responsible for the Degradation of Abnormal Proteins in Reticulocytes. Proc. Natl. Acad. Sci. U. S. A 1977, 74, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ciechanover A; Heller H; Elias S; Haas AL; Hershko A ATP-Dependent Conjugation of Reticulocyte Proteins with the Polypeptide Required for Protein Degradation. Proc. Natl. Acad. Sci. U. S. A 1980, 77, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Akutsu M; Dikic I; Bremm A Ubiquitin Chain Diversity at a Glance. J. Cell Sci. 2016, 129 (5), 875–880. [DOI] [PubMed] [Google Scholar]
- (73).Vijay-Kumar S; Bugg CE; Cook WJ Structure of Ubiquitin Refined at 1.8Å resolution. J. Mol. Biol 1987, 194, 531–544. [DOI] [PubMed] [Google Scholar]
- (74).Kraft C; Peter M; Hofmann K Selective Autophagy: Ubiquitin-Mediated Recognition and Beyond. Nat. Cell Biol 2010, 12, 836–841. [DOI] [PubMed] [Google Scholar]
- (75).Hershko A; Ciechanover A The Ubiquitin System. Annu. Rev. Biochem 1998, 67, 425–479. [DOI] [PubMed] [Google Scholar]
- (76).Komander D; Rape M The Ubiquitin Code. Annu. Rev. Biochem 2012, 81, 203–229. [DOI] [PubMed] [Google Scholar]
- (77).Pickart CM Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem 2001, 70, 503–533. [DOI] [PubMed] [Google Scholar]
- (78).Weissman AM Ubiquitin and Proteasomes: Themes and Variations on Ubiquitylation. Nat. Rev. Mol. Cell Biol 2001, 2, 169–178. [DOI] [PubMed] [Google Scholar]
- (79).Pickering AM; Davies KJA Degradation of Damaged Proteins - The Main Function of the 20S Proteasome. Prog. Mol. Biol. Transl. Sci 2012, 109, 227–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Tanaka K; Yoshimura T; Ichihara A Role of Substrate in Reversible Activation of Proteasomes (Multi-Protease Complexes) by Sodium Dodecyl Sulfate. J. Biochem 1989, 106, 495–500. [DOI] [PubMed] [Google Scholar]
- (81).Biran A; Myers N; Adler J; Broennimann K; Reuven N; Shaul YA 20S Proteasome Receptor for Degradation of Intrinsically Disordered Proteins. bioRxiv 2017, 210898. [Google Scholar]
- (82).Gillette TG; Kumar B; Thompson D; Slaughter CA; DeMartino GN Differential Roles of the COOH Termini of AAA Subunits of PA700 (19 S Regulator) in Asymmetric Assembly and Activation of the 26 S Proteasome. J. Biol. Chem 2008, 283, 31813–31822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gaczynska M; Osmulski PA; Gao Y; Post MJ; Simons M Proline- and Arginine-Rich Peptides Constitute a Novel Class of Allosteric Inhibitors of Proteasome Activity. Biochemistry 2003, 42, 8663–8670. [DOI] [PubMed] [Google Scholar]
- (84).Anbanandam A; Albarado DC; Tirziu DC; Simons M; Veeraraghavan S Molecular Basis for Proline- and Arginine-Rich Peptide Inhibition of Proteasome. J. Mol. Biol 2008, 384, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Giżyńska M; Witkowska J; Karpowicz P; Rostankowski R; Chocron ES; Pickering AM; Osmulski P; Gaczynska M; Jankowska E Proline- and Arginine-Rich Peptides as Flexible Allosteric Modulators of Human Proteasome Activity. J. Med. Chem 2019, 62, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Dal Vechio FH; Cerqueira F; Augusto O; Lopes R; Demasi M Peptides That Activate the 20S Proteasome by Gate Opening Increased Oxidized Protein Removal and Reduced Protein Aggregation. Free Radical Biol. Med 2014, 67, 304–313. [DOI] [PubMed] [Google Scholar]
- (87).Kisselev AF; Kaganovich D; Goldberg AL Binding of Hydrophobic Peptides to Several Non-Catalytic Sites Promotes Peptide Hydrolysis by All Active Sites of 20 S Proteasomes. Evidence for Peptide-Induced Channel Opening in the Alpha-Rings. J. Biol. Chem 2002, 277, 22260–22270. [DOI] [PubMed] [Google Scholar]
- (88).Micale N; Scarbaci K; Troiano V; Ettari R; Grasso S; Zappalà M Peptide-Based Proteasome Inhibitors in Anticancer Drug Design. Med. Res. Rev 2014, 34, 1001–1069. [DOI] [PubMed] [Google Scholar]
- (89).Otvos L; Wade JD Current Challenges in Peptide-Based Drug Discovery. Front. Chem 2014, 2, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Tanaka K; Yoshimura T; Kumatori A; Ichihara A; Ikai A; Nishigai M; Kameyama K; Takagi T Proteasomes (Multi-Protease Complexes) as 20 S Ring-Shaped Particles in a Variety of Eukaryotic Cells. J. Biol. Chem 1988, 263, 16209–16217. [PubMed] [Google Scholar]
- (91).Ruiz d Mena I; Mahillo E; Arribas J; Castaño JG Kinetic Mechanism of Activation by Cardiolipin (Diphosphatidylglycerol) of the Rat Liver Multicatalytic Proteinase. Biochem. J 1993, 296, 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Watanabe N; Yamada S Activation of 20S Proteasomes from Spinach Leaves by Fatty Acids. Plant Cell Physiol. 1996, 37, 147–151. [DOI] [PubMed] [Google Scholar]
- (93).Katsiki M; Chondrogianni N; Chinou I; Rivett AJ; Gonos ES The Olive Constituent Oleuropein Exhibits Proteasome Stimulatory Properties In Vitro and Confers Life Span Extension of Human Embryonic Fibroblasts. Rejuvenation Res. 2007, 10, 157–172. [DOI] [PubMed] [Google Scholar]
- (94).Huang L; Ho P; Chen C-H Activation and Inhibition of Proteasomes by Betulinic Acid and Its Derivatives. FEBS Lett. 2007, 581, 4955–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Jones CL; Njomen E; Sjögren B; Dexheimer TS; Tepe JJ Small Molecule Enhancement of 20S Proteasome Activity Targets Intrinsically Disordered Proteins. ACS Chem. Biol 2017, 12, 2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Medina DX; Caccamo A; Oddo S Methylene Blue Reduces Aβ Levels and Rescues Early Cognitive Deficit by Increasing Proteasome Activity. Brain Pathol. 2011, 21, 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Njomen E; Osmulski PA; Jones CL; Gaczynska M; Tepe JJ Small Molecule Modulation of Proteasome Assembly. Biochemistry 2018, 57, 4214–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Trader DJ; Simanski S; Dickson P; Kodadek T Establishment of a Suite of Assays That Support the Discovery of Proteasome Stimulators. Biochim. Biophys. Acta, Gen. Subj 2017, 1861, 892–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Coleman RA; Trader DJ Development and Application of a Sensitive Peptide Reporter to Discover 20S Proteasome Stimulators. ACS Comb. Sci 2018, 20, 269–276. [DOI] [PubMed] [Google Scholar]
- (100).Guo X; Huang X; Chen MJ Reversible Phosphorylation of the 26S Proteasome. Protein Cell 2017, 8, 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Lee S-H; Park Y; Yoon SK; Yoon J-B Osmotic Stress Inhibits Proteasome by P38 MAPK-Dependent Phosphorylation. J. Biol. Chem 2010, 285, 41280–41289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Zhang F; Hu Y; Huang P; Toleman CA; Paterson AJ; Kudlow JE Proteasome Function Is Regulated by Cyclic AMP-Dependent Protein Kinase through Phosphorylation of Rpt6. J. Biol. Chem 2007, 282, 22460–22471. [DOI] [PubMed] [Google Scholar]
- (103).Asai M; Tsukamoto O; Minamino T; Asanuma H; Fujita M; Asano Y; Takahama H; Sasaki H; Higo S; Asakura M; Takashima S; Hori M; Kitakaze M PKA Rapidly Enhances Proteasome Assembly and Activity in in Vivo Canine Hearts. J. Mol. Cell. Cardiol 2009, 46, 452–462. [DOI] [PubMed] [Google Scholar]
- (104).Park S-J; Ahmad F; Philp A; Baar K; Williams T; Luo H; Ke H; Rehmann H; Taussig R; Brown AL; Kim MK; Beaven MA; Burgin AB; Manganiello V; Chung JH Resveratrol Ameliorates Aging-Related Metabolic Phenotypes by Inhibiting CAMP Phosphodiesterases. Cell 2012, 148, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Myeku N; Clelland CL; Emrani S; Kukushkin NV; Yu WH; Goldberg AL; Duff KE Tau-Driven 26S Proteasome Impairment and Cognitive Dysfunction Can Be Prevented Early in Disease by Activating CAMP-PKA Signaling. Nat. Med 2016, 22, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Lokireddy S; Kukushkin NV; Goldberg AL CAMP-Induced Phosphorylation of 26S Proteasomes on Rpn6/PSMD11 Enhances Their Activity and the Degradation of Misfolded Proteins. Proc. Natl. Acad. Sci. U. S. A 2015, 112, E7176–E7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Koulich E; Li X; DeMartino GN Relative Structural and Functional Roles of Multiple Deubiquitylating Proteins Associated with Mammalian 26S Proteasome. Mol. Biol. Cell 2008, 19, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Lee B-H; Lee MJ; Park S; Oh D-C; Elsasser S; Chen P-C; Gartner C; Dimova N; Hanna J; Gygi SP; Wilson SM; King RW; Finley D Enhancement of Proteasome Activity by a Small-Molecule Inhibitor of USP14. Nature 2010, 467, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Wang X; Mazurkiewicz M; Hillert E-K; Olofsson MH; Pierrou S; Hillertz P; Gullbo J; Selvaraju K; Paulus A; Akhtar S; Bossler F; Khan AC; Linder S; D’Arcy P The Proteasome Deubiquitinase Inhibitor VLX1570 Shows Selectivity for Ubiquitin-Specific Protease-14 and Induces Apoptosis of Multiple Myeloma Cells. Sci. Rep 2016, 6, 26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Tian Z; D’Arcy P; Wang X; Ray A; Tai Y-T; Hu Y; Carrasco RD; Richardson P; Linder S; Chauhan D; Anderson KC A Novel Small Molecule Inhibitor of Deubiquitylating Enzyme USP14 and UCHL5 Induces Apoptosis in Multiple Myeloma and Overcomes Bortezomib Resistance. Blood 2014, 123, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).D’Arcy P; Brnjic S; Olofsson MH; Fryknäs M; Lindsten K; De Cesare M; Perego P; Sadeghi B; Hassan M; Larsson R; Linder S Inhibition of Proteasome Deubiquitinating Activity as a New Cancer Therapy. Nat. Med 2011, 17, 1636–1640. [DOI] [PubMed] [Google Scholar]
- (112).Kim E; Park S; Lee JH; Mun JY; Choi WH; Yun Y; Lee J; Kim JH; Kang M-J; Lee MJ Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018, 24, 732–743. [DOI] [PubMed] [Google Scholar]
- (113).Choi WH; de Poot SAH; Lee JH; Kim JH; Han DH; Kim YK; Finley D; Lee MJ Open-Gate Mutants of the Mammalian Proteasome Show Enhanced Ubiquitin-Conjugate Degradation. Nat. Commun 2016, 7, 10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Chakraborty J; von Stockum S; Marchesan E; Caicci F; Ferrari V; Rakovic A; Klein C; Antonini A; Bubacco L; Ziviani E USP14 Inhibition Corrects an in Vivo Model of Impaired Mitophagy. EMBO Mol. Med 2018, 10, No. e9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Xu D; Shan B; Sun H; Xiao J; Zhu K; Xie X; Li X; Liang W; Lu X; Qian L; Yuan J USP14 Regulates Autophagy by Suppressing K63 Ubiquitination of Beclin 1. Genes Dev. 2016, 30, 1718–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Gaczynska M; Rock KL; Spies T; Goldberg AL Peptidase Activities of Proteasomes Are Differentially Regulated by the Major Histocompatibility Complex-Encoded Genes for LMP2 and LMP7. Proc. Natl. Acad. Sci. U. S. A 1994, 91, 9213–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Gaczynska M; Goldberg AL; Tanaka K; Hendil KB; Rock KL Proteasome Subunits X and Y Alter Peptidase Activities in Opposite Ways to the Interferon-γ-Induced Subunits LMP2 and LMP7. J. Biol. Chem 1996, 271, 17275–17280. [DOI] [PubMed] [Google Scholar]
- (118).Chondrogianni N; Tzavelas C; Pemberton AJ; Nezis IP; Rivett AJ; Gonos ES Overexpression of Proteasome B5 Assembled Subunit Increases the Amount of Proteasome and Confers Ameliorated Response to Oxidative Stress and Higher Survival Rates.J. Biol. Chem 2005, 280, 11840–11850. [DOI] [PubMed] [Google Scholar]
- (119).Liu Y; Liu X; Zhang T; Luna C; Liton PB; Gonzalez P Cytoprotective Effects of Proteasome B5 Subunit Overexpression in Lens Epithelial Cells. Mol. Vis 2007, 13, 31–38. [PMC free article] [PubMed] [Google Scholar]
- (120).Lu L; Song H-F; Wei J-L; Liu X-Q; Song W-H; Yan B-Y; Yang G-J; Li A; Yang W-L Ameliorating Replicative Senescence of Human Bone Marrow Stromal Cells by PSMB5 Overexpression. Biochem. Biophys. Res. Commun 2014, 443, 1182–1188. [DOI] [PubMed] [Google Scholar]
- (121).Moi P; Chan K; Asunis I; Cao A; Kan YW Isolation of NF-E2-Related Factor 2 (Nrf2), a NF-E2-like Basic Leucine Zipper Transcriptional Activator That Binds to the Tandem NF-E2/AP1 Repeat of the Beta-Globin Locus Control Region. Proc. Natl. Acad. Sci. U. S. A 1994, 91, 9926–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Nguyen T; Yang CS; Pickett CB The Pathways and Molecular Mechanisms Regulating Nrf2 Activation in Response to Chemical Stress. Free Radical Biol. Med 2004, 37, 433–441. [DOI] [PubMed] [Google Scholar]
- (123).Kwak M-K; Wakabayashi N; Greenlaw JL; Yamamoto M; Kensler TW Antioxidants Enhance Mammalian Proteasome Expression through the Keap1-Nrf2 Signaling Pathway. Mol. Cell. Biol 2003, 23, 8786–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Cui Y; Ma S; Zhang C; Li D; Yang B; Lv P; Xing Q; Huang T; Yang GL; Cao W; Guan F Pharmacological Activation of the Nrf2 Pathway by 3H-1, 2-Dithiole-3-Thione Is Neuroprotective in a Mouse Model of Alzheimer Disease. Behav. Brain Res 2018, 336, 219–226. [DOI] [PubMed] [Google Scholar]
- (125).Kapeta S; Chondrogianni N; Gonos ES Nuclear Erythroid Factor 2-Mediated Proteasome Activation Delays Senescence in Human Fibroblasts. J. Biol. Chem 2010, 285, 8171–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Papaevgeniou N; Sakellari M; Jha S; Tavernarakis N; Holmberg CI; Gonos ES; Chondrogianni N 18α-Glycyrrhetinic Acid Proteasome Activator Decelerates Aging and Alzheimer’s Disease Progression in Caenorhabditis Elegans and Neuronal Cultures. Antioxid. Redox Signaling 2016, 25, 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Jang J; Wang Y; Kim H-S; Lalli MA; Kosik KS Nrf2, a Regulator of the Proteasome, Controls Self-Renewal and Pluripotency in Human Embryonic Stem Cells. Stem Cells 2014, 32, 2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Kwak M-K; Cho J-M; Huang B; Shin S; Kensler TW Role of Increased Expression of the Proteasome in the Protective Effects of Sulforaphane against Hydrogen Peroxide-Mediated Cytotoxicity in Murine Neuroblastoma Cells. Free Radical Biol. Med 2007, 43, 809–817. [DOI] [PubMed] [Google Scholar]
- (129).Förster A; Whitby FG; Hill CP The Pore of Activated 20S Proteasomes Has an Ordered 7-fold Symmetric Conformation. EMBO J. 2003, 22, 4356–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Vilchez D; Boyer L; Morantte I; Lutz M; Merkwirth C; Joyce D; Spencer B; Page L; Masliah E; Berggren WT; Gage FH; Dillin A Increased Proteasome Activity in Human Embryonic Stem Cells Is Regulated by PSMD11. Nature 2012, 489, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Li J; Horak KM; Su H; Sanbe A; Robbins J; Wang X Enhancement of Proteasomal Function Protects against Cardiac Proteinopathy and Ischemia/Reperfusion Injury in Mice. J. Clin. Invest 2011, 121, 3689–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Benmohamed R; Arvanites AC; Kim J; Ferrante RJ; Silverman RB; Morimoto RI; Kirsch DR Identification of Compounds Protective against G93A-SOD1 Toxicity for the Treatment of Amyotrophic Lateral Sclerosis. Amyotrophic Lateral Scler. 2011, 12, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Chen T; Benmohamed R; Kim J; Smith K; Amante D; Morimoto RI; Kirsch DR; Ferrante RJ; Silverman RB ADME-Guided Design and Synthesis of Aryloxanyl Pyrazolone Derivatives To Block Mutant Superoxide Dismutase 1 (SOD1) Cytotoxicity and Protein Aggregation: Potential Application for the Treatment of Amyotrophic Lateral Sclerosis. J. Med. Chem 2012, 55, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Chen T; Benmohamed R; Arvanites AC; Ranaivo HR; Morimoto RI; Ferrante RJ; Watterson DM; Kirsch DR; Silverman RB Arylsulfanyl Pyrazolones Block Mutant SOD1-G93A Aggregation. Potential Application for the Treatment of Amyotrophic Lateral Sclerosis. Bioorg. Med. Chem 2011, 19, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Trippier PC; Zhao KT; Fox SG; Schiefer IT; Benmohamed R; Moran J; Kirsch DR; Morimoto RI; Silverman RB Proteasome Activation Is a Mechanism for Pyrazolone Small Molecules Displaying Therapeutic Potential in Amyotrophic Lateral Sclerosis. ACS Chem. Neurosci 2014, 5, 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Leestemaker Y; de Jong A; Witting KF; Penning R; Schuurman K; Rodenko B; Zaal EA; van de Kooij B; Laufer S; Heck AJR; Borst J; Scheper W; Berkers CR; Ovaa H Proteasome Activation by Small Molecules. Cell Chem. Biol 2017, 24, 725–736. [DOI] [PubMed] [Google Scholar]
- (137).Cohen-Kaplan V; Livneh I; Avni N; Cohen-Rosenzweig C; Ciechanover A The Ubiquitin-Proteasome System and Autophagy: Coordinated and Independent Activities. Int. J. Biochem. Cell Biol 2016, 79, 403–418. [DOI] [PubMed] [Google Scholar]
- (138).Zhu K; Dunner K; McConkey DJ Proteasome Inhibitors Activate Autophagy as a Cytoprotective Response in Human Prostate Cancer Cells. Oncogene 2010, 29, 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Tang B; Cai J; Sun L; Li Y; Qu J; Snider BJ; Wu S Proteasome Inhibitors Activate Autophagy Involving Inhibition of PI3K-Akt-MTOR Pathway as an Anti-Oxidation Defense in Human RPE Cells. PLoS One 2014, 9, No. e103364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Sha Z; Schnell HM; Ruoff K; Goldberg A Rapid Induction of P62 and GABARAPL1 upon Proteasome Inhibition Promotes Survival before Autophagy Activation. J. Cell Biol 2018, 217, 1757–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Wang C; Wang X The Interplay between Autophagy and the Ubiquitin-Proteasome System in Cardiac Proteotoxicity. Biochim. Biophys. Acta, Mol. Basis Dis 2015, 1852, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Coleman RA; Muli CS; Zhao Y; Bhardwaj A; Newhouse TR; Trader DJ Analysis of Chain Length, Substitution Patterns, and Unsaturation of AM-404 Derivatives as 20S Proteasome Stimulators. Bioorg. Med. Chem. Lett 2019, 29, 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Kisselev AF; Goldberg AL Monitoring Activity and Inhibition of 26S Proteasomes with Fluorogenic Peptide Substrates In Ubiquitin and Protein Degradation, Part A; Methods in Enzymology; Academic Press, 2005; Vol. 398, pp 364–378. [DOI] [PubMed] [Google Scholar]
- (144).Kisselev AF; Akopian TN; Castillo V; Goldberg AL Proteasome Active Sites Allosterically Regulate Each Other, Suggesting a Cyclical Bite-Chew Mechanism for Protein Breakdown. Mol. Cell 1999, 4, 395–402. [DOI] [PubMed] [Google Scholar]
- (145).Chondrogianni N; Petropoulos I; Franceschi C; Friguet B; Gonos ES Fibroblast Cultures from Healthy Centenarians Have an Active Proteasome. Exp. Gerontol 2000, 35, 721–728. [DOI] [PubMed] [Google Scholar]
- (146).Cabreiro F; Perichon M; Jatje J; Malavolta M; Mocchegiani E; Friguet B; Petropoulos I Zinc supplementation in the elderly subjects: effect on oxidized protein degradation and repair systems in peripheral blood lymphocytes. Exp. Gerontol 2008, 43, 483–487. [DOI] [PubMed] [Google Scholar]
- (147).Rodriguez KA; Edrey YH; Osmulski P; Gaczynska M; Buffenstein R Altered Composition of Liver Proteasome Assemblies Contributes to Enhanced Proteasome Activity in the Exceptionally Long-Lived Naked Mole-Rat. PLoS One 2012, 7, No. e35890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Rodriguez KA; Osmulski PA; Pierce A; Weintraub ST; Gaczynska M; Buffenstein R A Cytosolic Protein Factor from the Naked Mole-Rat Activates Proteasomes of Other Species and Protects These from Inhibition. Biochim. Biophys. Acta, Mol. Basis Dis 2014, 1842, 2060–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Kapetanou M; Chondrogianni N; Petrakis S; Koliakos G; Gonos ES Proteasome Activation Enhances Stemness and Lifespan of Human Mesenchymal Stem Cells. Free Radical Biol. Med 2017, 103, 226–235. [DOI] [PubMed] [Google Scholar]
- (150).Chondrogianni N; Sakellari M; Lefaki M; Papaevgeniou N; Gonos ES Proteasome Activation Delays Aging in Vitro and in Vivo. Free Radical Biol. Med 2014, 71, 303–320. [DOI] [PubMed] [Google Scholar]
- (151).Chondrogianni N; Georgila K; Kourtis N; Tavernarakis N; Gonos Efstathios S Enhanced Proteasome Degradation Extends Caenorhabditis Elegans Lifespan and Alleviates Aggregation-Related Pathologies. Free Radical Biol. Med 2014, 75, S18. [DOI] [PubMed] [Google Scholar]
- (152).Chondrogianni N; Gonos ES Proteasome Function Determines Cellular Homeostasis and the Rate of Aging. Adv. Exp. Med. Biol 2010, 694, 38–46. [DOI] [PubMed] [Google Scholar]
- (153).Chondrogianni N; Gonos ES Overexpression of HUMP1/POMP Proteasome Accessory Protein Enhances Proteasome-Mediated Antioxidant Defence. Exp. Gerontol 2007, 42, 899–903. [DOI] [PubMed] [Google Scholar]
- (154).Voutetakis K; Delitsikou V; Magouritsas MG; Gonos ES Anti-Ageing Properties of Khelma LongevityTM: Treatment of Human Fibroblasts Increases Proteasome Levels and Decreases the Levels of Oxidized Proteins. New Biotechnol. 2017, 38, 36–39. [DOI] [PubMed] [Google Scholar]
- (155).Gonos E Proteasome Activation Delays Aging and Protects against Proteotoxicity in Neurodegenerative Disease. Adv. Exp. Med. Biol 2015, 821, 7. [DOI] [PubMed] [Google Scholar]
- (156).Chondrogianni N; Voutetakis K; Kapetanou M; Delitsikou V; Papaevgeniou N; Sakellari M; Lefaki M; Filippopoulou K; Gonos ES Proteasome Activation: An Innovative Promising Approach for Delaying Aging and Retarding Age-Related Diseases. Ageing Res. Rev 2015, 23, 37–55. [DOI] [PubMed] [Google Scholar]
- (157).Gonos E Proteasome Activation as a Novel Anti-Aging Strategy. Free Radical Biol. Med 2014, 75, S7. [DOI] [PubMed] [Google Scholar]
- (158).Vilchez D; Morantte I; Liu Z; Douglas PM; Merkwirth C; Rodrigues APC; Manning G; Dillin A RPN-6 Determines C. Elegans Longevity under Proteotoxic Stress Conditions. Nature 2012, 489, 263–268. [DOI] [PubMed] [Google Scholar]
- (159).Kruegel U; Robison B; Dange T; Kahlert G; Delaney JR; Kotireddy S; Tsuchiya M; Tsuchiyama S; Murakami CJ; Schleit J; Sutphin G; Carr D; Tar K; Dittmar G; Kaeberlein M; Kennedy BK; Schmidt M Elevated Proteasome Capacity Extends Replicative Lifespan in Saccharomyces Cerevisiae. PLoS Genet. 2011, 7, No. e1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Mayor T; Sharon M; Glickman MH Tuning the Proteasome to Brighten the End of the Journey. Am. J. Physiol. -Cell Physiol 2016, 311, C793–C804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Tsvetkov P; Myers N; Moscovitz O; Sharon M; Prilusky J; Shaul Y Thermo-Resistant Intrinsically Disordered Proteins Are Efficient 20S Proteasome Substrates. Mol. BioSyst 2012, 8, 368–373. [DOI] [PubMed] [Google Scholar]
- (162).Tsvetkov P; Reuven N; Shaul Y The Nanny Model for IDPs. Nat. Chem. Biol 2009, 5, 778–781. [DOI] [PubMed] [Google Scholar]
- (163).Fishbain S; Inobe T; Israeli E; Chavali S; Yu H; Kago G; Babu MM; Matouschek A Sequence Composition of Disordered Regions Fine-Tunes Protein Half-Life. Nat. Struct. Mol. Biol 2015, 22, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Rastogi N; Mishra DP Therapeutic Targeting of Cancer Cell Cycle Using Proteasome Inhibitors. Cell Div. 2012, 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Chen L; Brewer MD; Guo L; Wang R; Jiang P; Yang X Enhanced Degradation of Misfolded Proteins Promotes Tumori-genesis. Cell Rep. 2017, 18, 3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Best SA; De Souza DP; Kersbergen A; Policheni AN; Dayalan S; Tull D; Rathi V; Gray DH; Ritchie ME; McConville MJ; Sutherland KD Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metab. 2018, 27, 935–943. [DOI] [PubMed] [Google Scholar]
- (167).Solis LM; Behrens C; Dong W; Suraokar M; Ozburn NC; Moran CA; Corvalan AH; Biswal S; Swisher SG; Bekele BN; Minna JD; Stewart DJ; Wistuba II Nrf2 and Keap1 Abnormalities in Non-Small Cell Lung Carcinoma and Association with Clinicopathologic Features. Clin. Cancer Res 2010, 16, 3743–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Lin Y; Sui L-C; Wu R-H; Ma R-J; Fu H-Y; Xu J-J; Qiu X-H; Chen L Nrf2 Inhibition Affects Cell Cycle Progression during Early Mouse Embryo Development. J. Reprod. Dev 2018, 64, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Gavet O; Pines J Activation of Cyclin B1–Cdk1 Synchronizes Events in the Nucleus and the Cytoplasm at Mitosis. J. Cell Biol 2010, 189, 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]