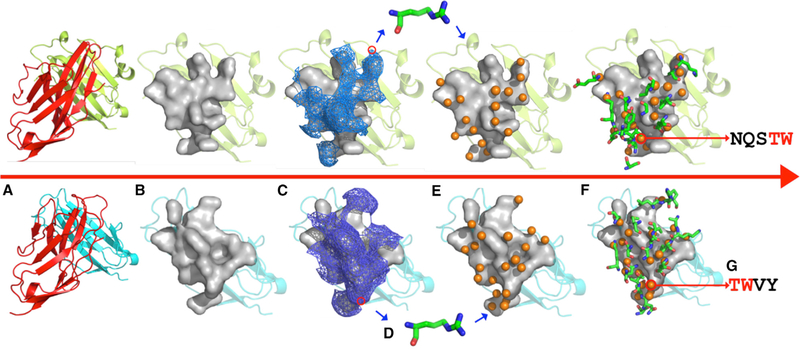
Figure 2. Schema of ProtLID Algorithm.
Major steps of the algorithm are shown: (A) input protein complexes(upper PD-1 [red]:PD-L1 [green], lower PD-1 [red]:PD-L2 [blue]), (B) identification of interface residues in target protein (in gray surface representation), (C) generation of mesh over interacting atoms of the interface residues of target proteins, (D) amino acid probes are placed in each mesh point and undergo extensive MD simulation to determine their preferencesover the surface, (E) prediction of rs-pharmacophores (orange spheres) from the analysis of MD simulation, consisting of the position and functional atom types (FAs), and (F) comparison of suggested FAs after com paring alternative rs-pharmacophores for different cognate ligands, (G) identifying differences of residue preferences between the two pharmacophores.