Abstract
Long noncoding RNAs (lncRNAs) are transcripts that do not code for proteins, but nevertheless exert regulatory effects on various biochemical pathways, in part via interactions with proteins, DNA, and other RNAs. LncRNAs are thought to regulate transcription and other biological processes by acting, for example, as guides that target proteins to chromatin, scaffolds that facilitate protein–protein interactions and complex formation, and orchestrators of phase-separated compartments. The study of lncRNAs has reached an exciting time, as recent advances in experimental and computational methods allow for genome-wide interrogation of biochemical and biological mechanisms of these enigmatic transcripts. A better appreciation for the biochemical versatility of lncRNAs has allowed us to begin closing gaps in our knowledge of how they act in diverse cellular and organismal contexts, including development and disease.
INTRODUCTION
DNA, RNA, and proteins play critical roles in living cells, but RNA is uniquely able to both carry genetic information in the form of nucleotide sequence and also perform biochemical functions due to its ability to fold into complex tertiary structures. The long-held view that biochemical pathways are the domain of proteins has been reconsidered in recent years, as more roles have been attributed to once-thought non-functional RNA molecules that are not translated into proteins [1]. This paradigm shift was in part spurred by advances in large-scale analyses of gene expression, first provided by tiling oligonucleotide microarrays followed by even more powerful next generation sequencing technologies, which revealed active transcription in ever-growing swaths of noncoding regions of the genome [2–6]. Only a very small portion of this widespread noncoding transcription could be assigned to conserved RNAs with well-known functions, such as ribosomal RNAs, transfer RNAs, small nuclear and nucleolar RNAs, and microRNAs. Most of the remaining noncoding transcripts were grouped in a category comprising RNAs longer than 200 nucleotides, puzzlingly lacking an obvious function, that were originally termed simply “large” or “long” noncoding RNAs (lncRNAs) [7–9].
Thousands of lncRNAs have been annotated in animals, including various mammals [6, 9–12], zebrafish [13], and insects [14–16]. Although plant and yeast genomes also contain lncRNAs, this review focuses on studies in humans, mice, and fruit flies, with only passing mentions of lncRNAs from plants and fungi, which have been reviewed elsewhere [17–19].
Some lncRNAs had previously been identified by conventional methods. Arguably, the first molecule in this category was found in 1990, when the imprinted H19 transcript was noted to lack a long open reading frame and association with the translational machinery, suggesting that the RNA itself was functional, not an encoded protein product [20, 21]. Similarly, the lncRNA XIST, which is critical for X chromosome inactivation [22, 23], was discovered in 1991 and reported to lack coding potential [24, 25]. However, the functional classification of most lncRNAs has been hindered thus far by low expression levels, a lack of sequence conservation, and incomplete annotation in many organisms. In recent years, improved biochemical, genetic, and computational methods have given researchers more tools to study lncRNAs and their regulatory roles, to dissect their biochemical mechanisms of action, and to understand their contributions to cancer [26, 27] and other diseases [28, 29].
Many reviews have been written on various aspects of lncRNA biology [30–39]. In this one, we focus on relatively recent new insights into the function of nuclear lncRNAs in animals, with a particular focus on mechanistic studies and newly developed biochemical methods.
Biochemical mechanisms of lncRNA function
The pioneering papers reporting the existence of lncRNA on a broad scale, as well as the results of the first perturbation studies, suggested that this broad category of transcripts might share a common biological function in gene regulation [9, 40–42], as do known classes of small noncoding RNAs, such as microRNAs, and piwi-associated RNAs [43]. However, it has become clear that the only thing that all lncRNAs have in common is their lack of coding potential. Many different subtypes of lncRNAs exist, such as promoter-associated, enhancer-derived, antisense, and interleaving [30], each likely carrying out diverse functions in different molecular contexts. Even broader diversity exists among the interleaving lncRNA subtype (lincRNAs), which might possess a variety of biochemical and cellular functions and will likely require individual characterization. Nonetheless, several central themes are emerging for potential broad molecular roles that lncRNAs might be playing in the cell. In this review, we mainly elaborate on lncRNAs that have been proposed to regulate chromatin structure and transcription; however, roles for lncRNA in a diverse set of crucial processes such as alternative splicing [44–46], translation [47], and genome stability [48, 49] have been identified, and more surely remain to be discovered. Below we review new evidence in support of two previously proposed [50] biochemical mechanisms of lncRNA function, chromatin recruitment and protein complex scaffolding, as well as their emerging role as potential agents for nuclear organization and phase separation. We emphasize that these mechanisms of action need not be mutually exclusive, as the same lncRNA might have different biochemical functions (Figure 1).
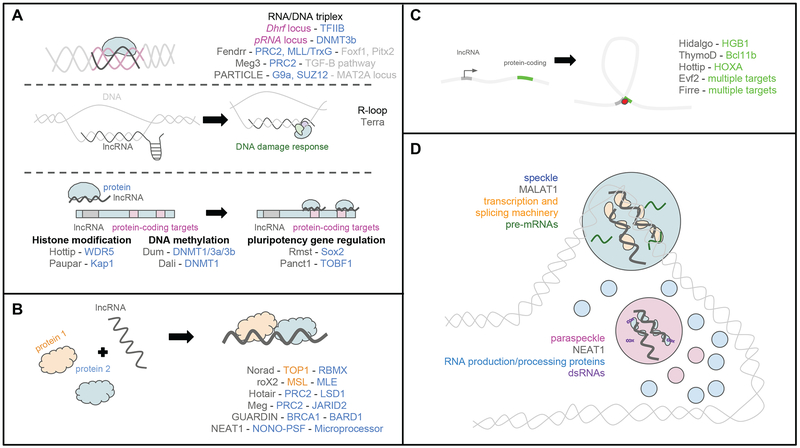
Figure 1. Biochemical functions of lncRNAs.
(A) Some lncRNAs form RNA/DNA triplexes (top), providing a potential mechanism for specific targeting of proteins to chromatin. LncRNAs discussed in the text are highlighted along with their associated proteins and target genes. LncRNAs could also recognize target regions on chromatin via Watson-Crick interactions with the DNA, forming R-loops (middle). TERRA forms an R-loop at its own locus at the telomeric region of chromosomes, activating the DNA damage response. Some lncRNAs are thought to guide chromatin-modifying complexes and transcription factors to target genes (bottom).
(B) A number of lncRNAs act as scaffolds to facilitate interactions between proteins, as in the NORAD-TOP1-RBMX complex that promotes genome stability and the rOX2-MSL-MLE complex involved in dosage compensation in Drosophila. Some lncRNAs have multiple biochemical functions, as with the scaffolding lncRNA GUARDIN, which also promotes genome stability through sequestering miR-23a.
(C) Some lncRNAs may affect gene expression by regulating genome. HIDALGO, ThymoD and HOTTIP regulate looping interactions at their own loci, while EVF2 and FIRRE are organizers of genome architecture on a larger scale. Some of these lncRNAs are aided by architectural proteins, such as CTCF and cohesin.
(D) MALAT1 and NEAT1 lncRNAs are important for assembly of two phase-separated bodies in the nucleus, speckles and paraspeckles, respectively. Speckles contain proteins involved in transcription and splicing; MALAT1 regulates the localization and distribution of these proteins. NEAT1 acts as a scaffold to bring together members of the RNA production and processing machinery in paraspeckles.
Chromatin guides
Among the first molecular function assigned to lncRNAs was acting as a guide to recruit protein complexes, in particular those that participate in gene regulation, to their target sites on chromatin [50, 51] (Figure 1A). Various models have been proposed for how lncRNAs localize to specific sites on chromatin to facilitate protein recruitment, either proximal to their transcribed locus or at distal loci. LncRNA-mediated targeting of proteins to chromatin is composed of two distinct processes: the interaction of a lncRNA with the DNA sequence on one side and with protein effectors on the other.
Finding targets on DNA
LncRNAs can target specific sequences on chromatin either via direct association with DNA or indirectly as part of a protein–RNA complex. As previously hypothesized [52, 53], sequence-driven recognition of genomic target sites by lncRNAs could occur either at intact double stranded DNA (triplex mode) or by displacing a DNA strand (R-loop mode). DNA–RNA triplexes (Figure 1A) are well-characterized structures formed by the sequence-specific interaction of an RNA strand with the major groove of a double-stranded DNA helix [54, 55]. These structures are stabilized by noncanonical Hoogsteen hydrogen bonding between an RNA base and the canonical Watson-Crick base pair of DNA. Triplexes can be either parallel or antiparallel, based on the orientation of the RNA strand relative to the target DNA strand [54]. One example of a lncRNA binding to chromatin via a triplex interaction occurs at the human DHFR locus, where an upstream ncRNA forms an RNA–DNA triplex at the promoter, preventing transcription by disrupting the pre-initiation complex [56]. Another example is found at the rDNA locus in mouse, from which ribosomal RNAs are transcribed. Here, lncRNAs from the sense (pRNA) and antisense (PAPAS) strand were shown to form triplexes locally at its promoter and to recruit to the locus the DNA methyltransferase DNMT3B and the chromatin remodeling complex NuRD, respectively [57, 58], with the result of repressing rDNA transcription. Additional lncRNAs reported to form triplexes at distal loci include the developmental regulator FENDRR, the imprinted lncRNA MEG3, and the irradiation-responsive PARTICLE [59–62], all of which were also reported to recruit the Polycomb repressive complex PRC2 (but see below for a discussion on the complexity of PRC2–RNA interactions). Despite these intriguing examples, clear rules have yet to emerge that would allow prediction of lncRNAs binding to genomic targets at distal loci based on the formation of DNA–RNA triplexes.
An alternative means by which lncRNAs could be recruited to chromatin is by forming DNA–RNA hybrid structures called R-loops (Figure 1A), whereby a complementary RNA directly pairs to DNA by displacing one of its strands [63]. This process normally occurs co-transcriptionally, when a nascent transcript binds to its template DNA. A well-characterized example is the Terra family of lncRNAs in S. cerevisiae, which regulate telomere length and cell senescence via R-loop formation [64]. Other studies in yeast have reported that lncRNA-mediated R-loops can occur post-transcriptionally at distal loci, but whether this is the case in higher organisms remains unknown [65, 66]. Emerging new technologies to map R-loops with high resolution and sensitivity in mammalian cells [67–70] might help answering this important question.
Some lncRNAs utilize spatial proximity to localize to specific chromatin sites. For example, local chromatin topology directs the spreading of XIST on the X chromosome and insertion of an Xist transgene at a different genomic locus changes its early localization pattern [71]. Whether this proximity model can be extended to other lncRNAs is not known.
Chromatin proteins that bind to lncRNAs
Typically, lncRNA are more dynamically expressed during development, and maintain more spatial specificity than protein-coding genes [10, 11]. Together with their ability to guide proteins to chromatin, this makes lncRNA prime candidates for factors that contribute target specificity to epigenetic regulators, which often lack DNA-binding domain, during development and in different cell types. A role for lncRNAs in recruiting the epigenetic silencer PRC2 to chromatin has been hypothesized for many years, though questions remain about the specificity of lncRNA binding and mechanisms of recruitment [39, 72, 73]. One of the first lncRNAs classified as functional was HOTAIR, which was shown to silence HOXD10 by facilitating the recruitment of the epigenetic silencing complex PRC2 through direct interactions [51]. Although following studies cast some doubt on the biochemical details of the initial model [74] and have not come to an agreement on the phenotypic consequences of the genetic deletion of Hotair in mice [75–78], the pioneering work on HOTAIR spurred a search for other lncRNA–protein interactions that might underpin the recruitment of transcriptional regulators to chromatin, which in many cases pointed back to PRC2 and its affinity for various lncRNAs.
In fact, several subunits of PRC2 as well as PRC1 (another well-studied Polycomb repressive complex [79]) have been reported to bind to RNA in mammals, including EZH2, JARID2, SUZ12, CBX7, and SCML2 [80–87]. Similarly, the plant lncRNAs COLDAIR, and COLDWRAP, which are involved in seasonal regulation of flowering in Arabidopsis thaliana, were observed to interact with PRC2 [88, 89]. In animals, members of the trithorax group of proteins, which counter Polycomb-mediated epigenetic silencing [90] can also bind to lncRNAs. HOTTIP is a lncRNA transcribed from the Hoxa locus that binds to WDR5, a subunit of the H3K4 methyltransferase MLL complex [91, 92]. Other examples of lncRNAs that guide deposition of epigenetic histone marks are AIR, which binds to the G9a H3K9 methyltransferase [93], and PAUPAR, which affects levels of the repressive H3K9me3 mark at target promoters by interacting with KAP1/TRIM28 [94, 95].
In addition to regulating the deposition of histone modifications, lncRNAs can affect another key epigenetic modification, DNA methylation, via the recruitment of DNA methyltransferases (DNMTs). In addition to the pRNA from the rDNA locus mentioned above [57], the lncRNAs ecCEBPA and DALI repress methylation at neighboring gene by interacting with and inhibiting DNMT1 [96, 97]. Furthermore, many lncRNAs involved in imprinting regulate DNA methylation at their associated protein-coding genes and might do so by directly or indirectly recruiting DNMTs [98].
In addition to guiding chromatin-modifying complexes to their targets, lncRNAs can participate in the recruitment of transcription factors, although these proteins also have a built-in ability to recognize DNA motifs embedded in the genomic sequence. The intronic lncRNA PANCT1 was originally identified in a screen for positive regulators of the pluripotency factor Pou5f1/Oct4 [99]. PANCT1 binds to the protein product that originates from its own locus, TOBF1, and increases its occupancy at target genes on chromatin. Interestingly, promoters for these genes are enriched for the Oct-Sox sequence motif, which is also embedded in the lncRNA itself. Rescue of TOBF1 binding can be achieved with overexpression of wild type PANCT1, but not a mutant with a scrambled Oct-Sox motif [100], demonstrating that the lncRNA can function at distal loci and suggesting that PANCT1 recognizes target sequence on DNA directly, perhaps by forming an R-loop (see above). The lncRNA RMST is another effector of cell fate choice that facilitates binding of SOX2 to the promoters of neurogenic transcription factors via direct association [101]. Finally, some lncRNAs prevent the recruitment of transcription factors to chromatin, as in the example of the lncRNA PANDA, which inhibits chromatin binding of the transcription factor NF-YA [102].
RNA binding is a property shared by many chromatin-modifying complexes and chromatin-associated proteins [40, 41, 103, 104], suggesting that the list of protein–RNA interactions with an epigenetic role is destined to grow. However, years of research on RNA-mediated regulation of chromatin function have taught us that isolated reports of lncRNA–protein interactions correlated with transcriptional effects might not be sufficient to draw strong conclusions on the biochemical and biological function of a lncRNA. Fortunately, emerging technologies will provide us with the ability to detect and manipulate lncRNA–protein interactions with ever increasing resolution and accuracy (see below) and will enable scientists in the field to perform rigorous rescue and tethering experiments using mutant proteins defective in RNA binding as controls. These experiments will allow us to confirm or refute proposed models for the function of lncRNAs within chromatin-associated complexes.
Scaffolds for complexes
A diverse collection of lncRNAs scaffold interactions between proteins and protein complexes (Figure 1B), affecting epigenetic regulation of gene expression, genome stability, nuclear organization, and microRNA biogenesis.
Drosophila achieves dosage compensation of the sex chromosome by doubling the transcriptional output from the single male X chromosome. This function is carried out by the dosage compensation complex (DCC), which is formed by the histone acetyltransferase Males-absent on first (MOF), four different Male sex lethal proteins (MSL1–4) and the RNA helicase Maleless (MLE) [105]. In addition to these protein components, spreading of DCC to the entire X chromosome requires the presence of at least one of two lncRNAs, roX1 and roX2 [106, 107]. Protein-RNA crosslinking revealed that two stem loops on roX2 act as a switch for the binding of MSL2: when MLE is bound to roX2, one of the stem-loops is unwound, allowing for the formation of the other stem-loop and promoting binding of MSL2 [108]. The presence of RNA, is required to stabilize the association of MLE with the other DCC proteins [109], suggesting the possibility that roX2 (and perhaps roX1) provides a scaffolding functions within this essential complex.
Another epigenetic regulator, PRC2, whose complex relationship with RNA was briefly discussed above, has also been shown to interact with scaffolding lncRNAs. HOTAIR was shown to mediate interactions between PRC2 and the histone demethylase LSD1 [110], whereas the imprinted lncRNA MEG3 was shown to facilitate interactions of the core PRC2 subunit EZH2 with the accessory subunit JARID2 [83].
The lncRNA NORAD was first shown to interact with PUMILIO proteins and proposed to sequester them to promote the stability and translation of PUMILIO mRNAs targets [111]. However, casting doubt on these initial conclusions, RAP-MS (described later) and biochemical studies revealed that NORAD is required for the assembly of a novel complex, termed NARC1, which includes RBMX, TOP1, ALYREF and the PRPF19–CDC5L complex, all of which had previously known roles in suppressing genomic instability [49]. The formation of this complex requires NORAD, which directly binds to RBMX. NORAD depletion causes replication-fork velocity reduction and cell-cycle defects, phenotypes similar to deletion of TOP1 or RBMX, two proteins in the NORAD-dependent complex, thus indicating a role in genome stability for the NORAD transcript. The Norad knockout phenotype of segregation defects was rescued by the reintroduction of the full-length transcript, confirming its ability to act at the RNA level.
Other scaffolding lncRNAs include NEAT1, which organizes paraspeckles, a nuclear subdomain involved in splicing regulation [112] (see below) and regulates the interaction between paraspeckle proteins and the Microprocessor complex, thus enhancing pri-microRNA processing [113], and the lncRNA GUARDIN, which promotes heterodimerization between BRCA1 and BARD1. This interaction stabilizes BRCA1 by preventing its polyubiquitination [114]. GUARDIN can also sequester miR-23a, stabilizing the shelterin complex member TRF2 [114], demonstrating the ability of a single lncRNA to act via multiple non-exclusive biochemical mechanisms.
Overall, experimental evidence is growing for a pervasive scaffolding function of lncRNAs, which was previously postulated [50, 115]. In addition to the candidate-based studies reviewed above, a recent proteome-wide analysis revealed that ~20% of cellular protein complexes are sensitive to RNase A treatment [116], suggesting that RNA is a component of many mature complexes or at a minimum participates in their assembly. What portion of these scaffolding RNAs are lncRNAs remains to be determined.
Architects of the genome
Another increasingly appreciated function of lncRNAs is the regulation of chromosome conformation [32] (Figure 1C). Xist-mediated repression is associated with a general reshaping of the X chromosome in space [117, 118] and with its relocalization to the nuclear lamina, possibly via a direct interaction of Xist with the lamin B receptor (LBR) [119–121]. Regulating enhancer-promoter contacts also provides a mechanism for controlling gene expression locally. HOTTIP was implicated as a factor in chromatin looping at the Hoxa locus [91], and two “activatory” lncRNAs, ncRNA-a3 and ncRNA-a7 facilitate the formation of chromatin loops between their own locus and target genes, dependent on their interaction with the Mediator complex [122].
A class of lncRNAs that may share this function is chromatin-enriched lncRNAs, or cheRNAs [123, 124]. Many cheRNAs overlap enhancers but have several properties that distinguishes them from canonical enhancer-associated RNAs [125], including polyadenylation, H3K4me3-marked promoters, and a specific strand bias. HIDALGO is a cheRNA derived from read-through transcription of the hemoglobin gene Hgb1, and is required for the interaction between Hgb1 and its enhancer during K562 differentiation. Silencing of Hidalgo using CRISPR interference (CRISPRi) attenuated the contacts between Hgb1 and its two interacting partners, which include the Hidalgo promoter, and reduced Hgb1 expression. Antisense oligonucleotide-mediated depletion of HIDALGO recapitulated this effect, demonstrating a role for the RNA.
The phenomenon of lncRNAs activating genes near the site of their transcription is widespread [126], perhaps suggesting that the facilitation of looping either by the act of transcription or by the lncRNA is responsible for local regulation of gene expression. Indeed, lncRNAs that act locally are enriched at the boundaries of topological associated domains [127, 128], suggesting a role for lncRNAs in regulating local genome architecture.
One way lncRNAs might shape chromatin conformation is by interacting with known architectural proteins, such as cohesin and CTCF [129, 130]. The lncRNA ThymoD activates expression of the T-cell specific gene Bcl11b by facilitating a promoter-enhancer loop [131]. ThymoD is transcribed from a downstream control region of Bcl11b and is required for interaction between Bcl11b and its enhancer. Attenuation of ThymoD transcription led to reduced CTCF and cohesin occupancy at the Bcl11b control region, abolishing the interaction and reducing Bcl11b expression. Another example is EVF2, transcribed from an ultraconserved super-enhancer, which maintains the gene expression program in mouse interneurons [132]. EVF2 localizes with cohesin subunits at targets genes, regulating their interactions of the super-enhancer from which it is transcribed.
The FIRRE lncRNA has been proposed to function as an organizer of interchromosomal interactions as well as a facilitator of correct nuclear localization of the inactive X chromosome. FIRRE localizes to the X chromosome from which it is transcribed, but also to distal autosomes, bringing these regions into nuclear proximity with each other [133]. FIRRE binds both cohesin and CTCF on the inactive X, contributing to its anchoring to the nucleolus [134]. Knockdown of FIRRE caused deregulation of both X-linked and autosomal genes, perhaps due to the loss of H3K27me3 on the inactive X chromosome. Interestingly, although CTCF occupancy and long scale interactions on the X chromosome are disrupted by Firre knockout, the overall structure of topologically associated domains (TADs) is unchanged [135].
The RNA binding activity of CTCF was initially characterized in the context of its interaction with the antisense lncRNA WRAP53 [136], and it was mapped to an RNA-binding region that was recently shown to be required for a large fraction of CTCF-dependent loops, suggesting the possibility that many more lncRNAs might control genome organization by interacting via CTCF [137, 138].
Organizers of phase separation
Phase separation allows for functional compartmentalization in the cell, resulting in droplets where key factors are concentrated, thereby facilitating biochemical processes. Proteins with intrinsically disordered domains, or low complexity domains, interact to form “hubs,” which are ensembles of phase-separated molecules with hydrogel-like properties [139]. The assembly of stress granules, a phase-separated membrane-less organelle, is facilitated by the presence of RNA [140]. Many proteins with disordered regions interact with RNA [104, 141, 142], suggesting that the diversity of lncRNA sequences, expression patterns, and protein-binding properties might contribute to specifying compositionally and functionally distinct phase-separated compartments.
Paraspeckles and speckles are two such compartments that concentrate proteins in the nucleus in phase separated domains [143, 144] (Figure 1D). The lncRNA MALAT1 (also known as NEAT2) has long been known to associate with speckles in the nucleus [145], which contain members of transcription and splicing machinery [146]. MALAT1 regulates the localization and distribution of active splicing factors in speckles, thereby affecting the alternative splicing of pre-mRNAs and its acute depletion by knockdown is lethal in transformed HeLa cells [147], but a complete deletion is surprisingly compatible with mouse development [148], and therefore key questions about its function in vivo. In contrast, NEAT1 is known to be required for the formation of paraspeckles [143] which are almost completely abolished in NEAT1 knockdown cells [149]. Paraspeckles contain members of the DBHS protein family, which play a part in various aspects of RNA production and processing [150]. Both MALAT1 and NEAT1 were reported to localize to chromatin at active sites of transcription [151] and it is tempting to speculate that these lncRNAs might facilitate phase transition processes, which are increasingly appreciated to play a role in transcriptional regulation [152].
The functional domains of NEAT1 have recently been explored in great depth using systematic deletions of different regions of the transcript [112]. NEAT1 has two isoforms; the longer isoform is stabilized by a triple helix, and is required for paraspeckle formation. The middle domain of NEAT1 contains long repetitive sequences, and is necessary and sufficient (along with the terminal 5’ end and the stabilizing triple helix) for the formation of ordered paraspeckles. Treatment with the known disruptor of phase-separated structures 1,6-hexanediol dissolved intact paraspeckles and abolished the co-localization of NEAT1 with the essential paraspeckle protein NONO. In addition, cells expressing a deletion mutant of NEAT1 that lacks protein binding domains do not form normal paraspeckles. Tethering paraspeckle proteins to NEAT1 rescues normal paraspeckle formation; however, the NOPS dimer formation domain of NONO is required to successfully rescue the phenotype. These observations suggest that NEAT1 recruits NONO, which then initiates formation of the paraspeckles protein complexes, using NEAT1 as a scaffold.
Finally, it has been recently hypothesized that XIST might silence the X chromosomes by forming phase-separated “silence granules”, due to its interactions with disordered proteins, including FUS and hnRNPA2 [22, 153]. Although experimental evidence is required to support this suggestion, it provides an interesting new perspective on the biochemistry of XIST-mediated silencing.
“The company you keep” – techniques to study the lncRNA interactome
The common theme in the biochemical processes discussed above is that the molecular functions of lncRNAs emerge from their interactions with other proteins and DNA. Techniques to study protein–protein and DNA–protein interactions in both candidate-based and unbiased fashions have been available for decades, and have been coupled with high-throughput approaches in genomics and proteomics in recent years. Although similar approaches for lncRNA discovery have been lagged behind, the last few years have seen great advances in techniques to map protein–RNA and DNA–RNA interactions. The definitions for the many acronyms of methods mentioned in the following sections are collected in Table 1 along with the relevant citations.
Table 1.
Biochemical methods to identify lncRNA interactions.
| Method | Full name | Use | Citation |
|---|---|---|---|
| DNA–RNA | |||
| CHAR-seq | Chromatin-Associated RNA sequencing | genome-wide DNA–RNA contacts | Bell et al., Elife 2018 |
| CHART | Capture Hybridization Analysis of RNA Targets | candidate RNA localization on chromatin | Simon et al., PNAS 2011 |
| ChIRP | Chromatin Isolation by RNA Purification | candidate RNA localization on chromatin | Chu et al., Mol Cell 2011 |
| dChIRP | domain-specific Chromatin Isolation by RNA Purification | candidate RNA localization on chromatin | Quinn et al., Nat Biotechnol 2014 |
| GRID-seq | Global RNA Interactions with DNA by deep sequencing | genome-wide DNA–RNA contacts | Li et al., Nat Biotechnol 2017 |
| MARGI | Mapping RNA-genome Interactions | genome-wide DNA–RNA contacts | Sridhar et al., Curr Biol 2017 |
| RAP | RNA Antisense Purification | candidate RNA localization on chromatin | Engreitz et al., Science 2013 |
| RNA-DamID | RNA-DNA adenine methylase identification | candidate RNA localization on chromatin | Cheetham et al., Nat Struct Mol Biol 2018 |
| SPRITE | Split Pool Recognition of Interactions by Tag Extension | genome-wide DNA–RNA contacts | Quinodoz et al., Cell 2018 |
| protein–RNA | |||
| IPL | in vivo Proximity Labeling | proximity labeling for protein–RNA | Beck et al., J Proteome Res 2014 |
| CARIC | click Chemistry-assisted RNA Interactome Capture | proteome-scale identification of all RBPs | Huang et al., PNAS 2018 |
| CLIP | ultraviolet Crosslinking and Immunoprecipitation | discovery of RNA interactors of a candidate protein | Ule et al., Science 2003 |
| eCLIP | enhanced CLIP | discovery of RNA interactors of a candidate protein | Van Nostrand et al., Nat Methods 2016 |
| GoldCLIP | Gel-omitted ligation-dependent CLIP | discovery of RNA interactors of a candidate protein | Gu et al., Genom Prot Bioinf 2018 |
| HITS-CLIP | High-Throughput Sequencing of RNA isolated by CLIP | discovery of RNA interactors of a candidate protein | Licatalosi et al., Nature 2008 |
| iCLIP | individual nucleotide resolution CLIP | discovery of RNA interactors of a candidate protein | König et al., Nat Struct Mol Biol 2010 |
| iDRIP | identification of Direct RNA Interacting Proteins) | discovery of protein interactors of a candidate RNA | Minajigi et al., Science 2015 |
| RaPID | RNA-protein Interaction Detection | discovery of protein interactors of a candidate RNA | Ramanathan et al., Nat Methods 2018 |
| RBDmap | RNA-binding Domain Mapping on a proteome scale | proteome-scale identification of polyA-bound RBPs and domains | Castello et al., Mol Cell 2016 |
| RBR-ID | Proteomic Identification of RNA-binding Regions | proteome scale identification of RBPs and domains | He et al., Mol Cell 2016 |
| RICK | newly transcribed RNA Interactome capture with Click chemistry | proteome scale identification of nascent RNA-associated RBPs | Bao et al., Nat Methods 2018 |
| RIP-seq | RNA Immunoprecipitation and sequencing | discovery of RNA interactors of a candidate protein | Zhao et al., Mol Cell 2010 |
| uvCLAP | Ultraviolet Crosslinking and Affinity Purification | discovery of RNA interactors of a candidate protein | Maticzka et al., Nat Commun 2018 |
| XRNAX | protein-crosslinked RNA extraction | proteome scale identification of RBPs and domains | Trendel et al., Cell 2018 |
| RNA–RNA | |||
| CLASH | Crosslinking Ligation and Sequencing of Hybrids | candidate protein-associated RNA-RNA interactions | Helwak et al., Cell 2013 |
| COMRADES | Crosslinking of Matched RNAs and Deep Sequencing | identification of RNA duplex interactions of candidate RNAs | Ziv et al., Nat Methods 2018 |
| LIGR-seq | LIGation of interacting RNA and high-throughput sequencing | trasncriptome-wide identification of RNA duplexes | Sharma et al., Mol Cell 2016 |
| PARIS | Psoralen Analysis of RNA Interactions and Structures | transcriptome-wide identification of RNA duplexes | Lu et al., Cell 2016 |
| RAP-RNA | RAP for RNA-RNA interactions of a target RNA | intermolecular RNA interactions of candidate RNA | Engreitz et al., Cell 2013 |
Mapping lncRNAs on chromatin
The biochemical functions of lncRNAs go hand in hand with their subcellular localization. While some lncRNAs are exported to the cytoplasm, many remain in the nucleus, where they can interact with DNA and protein complexes. LncRNAs are generally enriched in the nucleus compared to protein-coding RNAs [6, 154], and recent efforts fusing typically cytoplasmic transcripts to different regions of known nuclear lncRNAs have identified sequence motifs responsible for lncRNA nuclear retention [155, 156]. These motifs demonstrated higher conservation than other lncRNA regions, suggestive of functionality [156]. One such motif found was an Alu-derived sequence, called SIRLOIN for SINE-derived nuclear RNA localization [155]. The nuclear retention of SIRLOIN-containing RNAs was mediated by the RNA-binding protein HNRNPK.
The longstanding hypothesis that lncRNAs participate in gene regulation [30, 157, 158] is supported by the observation that many of them are enriched in the chromatin fraction [123, 124]. Mapping lncRNAs along the genome is thus critical for understanding this suspected function. Various methods have been developed toward this end, including CHART, ChIRP, and RAP [71, 159, 160]. These methods identify genome-wide lncRNA binding sites by chemical crosslinking followed by the capture of target RNAs using antisense biotinylated probes (Figure 2A). DNA that co-purifies with the lncRNA of choice is sequenced and regions of enrichment are identified, similar to existing strategies to localize protein on chromatin, such as chromatin immunoprecipitation. While conceptually similar, these methods differ in probe design, crosslinking strategies, and DNA elution conditions. RAP uses long 120 nt probes that tile the entire RNA sequence, whereas CHART and ChIRP opt for shorter 20–25 nt probes. ChIRP employs glutaraldehyde crosslinking to fix DNA–RNA contacts, which can potentially capture interactions over long distance, in contrast to CHART’s usage of formaldehyde, which crosslinks across shorter distances. RAP uses both formaldehyde and disuccinimidyl glutarate to capture both shorter- and longer-range contacts [161]. dChIRP is a further refinement of the ChIRP method, which probes individual lncRNA domains, providing a higher resolution map of interactions [162].
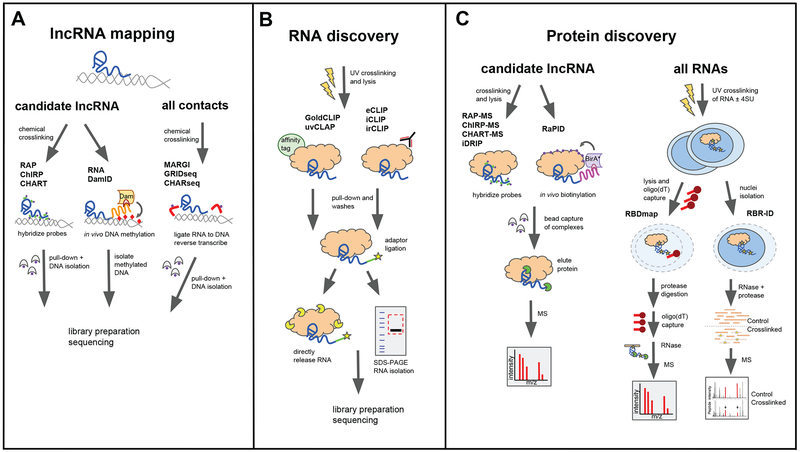
Figure 2. Methods for mapping the lncRNA interactome.
(A) Outline of methods to study lncRNA localization on chromatin. RAP, ChIRP, and CHART (left) use chemical crosslinking and biotinylated probes to capture a candidate lncRNA crosslinked to chromatin and identify its genomic targets by sequencing the associated DNA. In RNA DamID (middle) cells express a lncRNA of interest fused to MS2 stem-loop sequences and a E. coli adenine methyltransferase (Dam) fused to the MS2 coat protein. This results in methylation at lncRNA-bound genomic sites in vivo. Methylated DNA is then isolated with a methylation-sensitive restriction enzyme and sequenced. MARGI, GRIDseq, and CHARseq (right) use a biotinylated oligonucleotide bridge to ligate RNA to DNA in close proximity, followed by affinity purification of ligated complexes, library preparation, and sequencing.
(B) Schematic of methods used to identify RNA interactors of a given RBP. GoldCLIP and uvCLAP use affinity handles to purify crosslinked protein–RNA complexes, followed by adaptor ligation and direct elution of bound RNAs for sequencing. eCLIP, iCLIP, and irCLIP use protein-specific antibodies, SDS-PAGE, membrane transfer, and membrane excision to isolate RNAs for sequencing.
(C) Outline of methods to identify lncRNA-protein interactions. Left: methods to detect protein interactors of a candidate lncRNA. RAP-MS, ChIRP-MS, and CHART-MS chemically crosslink RNA to protein, use biotinylated probes to capture target lncRNA-protein complexes, and digest with RNase to release bound proteins for mass spectrometry; RaPID targets the BirA* biotin ligase to a stem-loop modified RNA to biotinylate protein interactors in live cells. Right: methods to identify new RBPs that bind to RNA in cells. RBDmap (left) and RBR-ID (right) use UV to crosslink RNA to protein in live cells in the presence of absence of 4SU. RBDmap lyses cells and employs two rounds of oligo(dT) capture to enrich for polyadenylated transcripts (left). RBR-ID isolates cell nuclei to identify all nuclear RNA-binding proteins. Both methods include RNAse digestion and protease digestion steps to generate short crosslinked peptides that can be analyzed by MS (bottom).
These methods have yielded robust results in select contexts, for example in the characterization of early spreading of XIST on the X chromosome [71, 163] and the identification of chromatin binding sites for MALAT1 and NEAT1 [151], but they suffer from high levels of background signal and contamination from crosslinked interactions [161], and alternative strategies might be required in cases where the biological material is limiting or the lncRNA of interest is expressed at low levels.
More recently, RNA-DamID was developed to map lncRNA-occupied sites in vivo [164]. This method is based on the principle of targeted DamID, in which the bacterial adenine methylase Dam is expressed in cells as a fusion to a protein of interest, and methylates adenines in the vicinity of its genomic targets [165]. In RNA-DamID, a lncRNA of interest fused to MS2 stem loops and the Dam enzyme fused to MS2 coat protein (MCP) are co-expressed in live cells. The stem-loop–MCP interaction recruits Dam to the lncRNA, and, therefore, to its binding sites on chromatin, which are methylated and subsequently identified by selective digestion with a methylation-sensitive restriction enzyme followed by sequencing (Figure 2A). RNA-DamID was applied to map tissue-specific binding of roX RNAs in Drosophila embryos with increased accuracy and sensitivity compared to previous methods, by identifying fewer false-positive peaks of roX binding and reducing the amount of required input material 100-fold [164]. The drawback of this method is the demand for genetic manipulation of cells and tagging the lncRNA of interest with MS2 stem-loops, preventing its usage in certain cases, such as patient-derived samples.
The techniques above are designed to map the localization of only a single lncRNA. It has now become possible to map the distribution of all chromatin-associated RNAs in an unbiased manner with the development of MARGI, CHAR-seq, GRID-seq, and more recently SPRITE [166–169]. MARGI, CHAR-seq, and GRID-seq use chemical crosslinking followed by a biotinylated oligonucleotide bridge to ligate DNA and RNA in close proximity, forming an DNA–RNA chimera that is captured and sequenced after reverse transcription (Figure 2A). SPRITE does not use proximity ligation but instead employs a split-and-pool strategy to sequentially label interacting DNA and RNA with barcoded tags, which are identified after sequencing by matching reads that contain the same combination of barcodes [168]. These methods have thus far uncovered local and long-distance DNA–RNA contacts in both human and Drosophila, and could shed light on the potential roles of lncRNAs in orchestrating 3D genome architecture, but, as with any unbiased approach, suffer from the limitations of being skewed toward detecting the most abundant species.
Defining protein–RNA interactions
Identifying the proteins that interact with a given lncRNA is critical for understanding its function. Protein–RNA interactions can be interrogated in vivo and in vitro by various binding assays, including RNA immunoprecipitation (RIP) with or without crosslinking, electro-mobility shift assays (EMSA), filter-binding assays, and fluorescence anisotropy measurements. These however, can only test candidate interactions where both the protein and the RNA are known. Several unbiased methods have been developed to discover new protein–RNA interactions and we divide them into those that identify all RNAs bound to a protein of choice (“RNA discovery”, Figure 2B) or choose a lncRNA and identify all proteins that bind to it (“protein discovery”, Figure 2C).
RNA discovery
Historically, RNA discovery methods were developed first. Immunoprecipitation in RNA-preserving conditions was coupled first to custom-designed microarrays interrogating large sets of RNAs simultaneously [9, 41, 170–172] and then with unbiased deep sequencing (RIP-seq, [173]). However, native RNA immunoprecipitation-based methods, lacking stringent washing steps, are known to suffer from exceedingly high noise levels [174], especially when combined with PCR-mediated amplification and sensitive deep sequencing. More recent alternatives are all based on variants of CLIP and its high-throughput version, HITS-CLIP (also known as CLIP-seq), which were originally developed to study factors regulating alternative splicing in the brain [175, 176]. CLIP entails UV-mediated crosslinking of RNA to its interacting proteins in vivo followed by immunoprecipitation of a protein of interest, separation on an SDS-PAGE gel and transfer to nitrocellulose membrane. Once the presence of crosslinked protein–RNA complexes on the membrane is confirmed at a position compatible with the expected molecular weight of the protein of interest, they can be excised and the eluted RNA can be processed for deep sequencing (Figure 2B).
Because of the covalent nature of protein–RNA crosslinking, some of the biochemical steps in CLIP are performed in extremely stringent conditions (i.e. SDS denaturation during SDS-PAGE) but the initial forms of this method were still troubled by high levels of noise, as well as poor yields [177]. Some of these issues were addressed in subsequent improvements. iCLIP introduced a circular ligation step after reverse transcription (RT) during library preparation, which allowed recovery of the truncated cDNAs resulting from early termination of RT at the crosslink site, increasing yields and providing single-base resolution information regarding the position of the presumptive protein–RNA interaction [178]. eCLIP further refined the iCLIP protocol and included a strategy to sequence size-matched input material in parallel [179], which is in our opinion a necessary step to calculate enrichment of true RNA interactors.
Other variants of CLIP have been developed to address the issue of low yields at the biochemical level, by replacing the highly inefficient and time-consuming membrane transfer and elution steps with in-solution purification strategies. GoldCLIP and uvCLAP are both based on epitope tags that allow for purifications in extremely stringent conditions [180, 181]. The GoldCLIP method involves fusion of the protein of interest to the bacterial HaloTag, which covalently attaches to chloroalkane-conjugated beads during purifications, whereas uvCLAP introduced an affinity tag that allows for in vivo biotinylation of the protein of choice followed by streptavidin-mediated purification of the crosslinked complexes under denaturing conditions. uvCLAP was utilized to study the nuclear RNA helicase DHXF9, revealing that it binds to RNAs containing inverted Alu repeats, proving the method’s practical advantages [182]. However, both GoldCLIP and uvCLAP rely on molecular engineering of the protein of interest to introduce the affinity tag, and therefore might not be suitable to investigate protein-RNA interactions in all contexts.
Finally, proximity labeling approaches [183] are now becoming available for RNA discovery. These are experimental strategies conceptually different from those described above, which all rely on some type of affinity purification of a protein–RNA complex. Instead, proximity labeling converts the physical proximity of potential RNA interactors to a protein of choice in vivo into a chemical labeling that can then be used to recover and identify the RNAs. The first of these approaches was in vivo proximity labeling (IPL), which utilized photoactivatable biotin to tag RNAs in the proximity of a protein of choice [184]. More recently, proximity labeling techniques developed to study protein–protein interactions have also been adapted for RNA discovery. APEX, which uses an engineered ascorbate peroxidase 2 to biotinylate targets [185] has now been used to biotinylate proximal RNAs in vivo [186, 187]. Alternatively, BioID uses a modified form of the BirA biotin protein ligase to biotinylate target molecules [188] and has recently been applied to RNA with the name of RaPID [189]. It is important to note that, like all proximity labeling strategies, these methods reveal proximity in vivo and not physical interaction and therefore caution should be used in inferring biochemical relationships between the protein of choice and the newly identified RNAs without additional validation steps.
Protein discovery
The first techniques for protein discovery (i.e., to identify proteins bound to a lncRNA) have been adaptations of the hybridization capture methods CHART, ChIRP, and RAP described above, whereby a protein elution step was added after the purification of crosslinked protein-RNA complexes followed by mass spectrometry (MS) for protein identification. Several methods based on these principles have been developed to date, including CHART-MS, ChIRP-MS, RAP-MS, and iDRIP [120, 121, 151, 190] (Figure 2C). ChIRP-MS, RAP-MS, and iDRIP were used to identify interactors of the Xist lncRNA in mouse embryonic stem cells, including the transcriptional repressor SPEN, which was functionally validated. RAP-MS was also applied to study the role of the lncRNA Sammson in promoting melanoma cell survival by stabilizing mitochondrial protein p32 [26], and the orchestration of a protein–RNA complex essential for genome stability by the lncRNA Norad [49].
Proximity-based methods are also available for protein discovery. For example, RaPID is an adaptation of BioID that uses the bacterial biotin ligase mutant BirA* to biotinylate the protein interactors of a given RNA motif in live cells [189] (Figure 2C). The RNA of interest is fused to bacteriophage BoxB stem loops and coexpressed with BirA* fused to the BoxB-binding λN peptide. The BirA*–λN fusion binds strongly to the BoxB stem loops and biotinylates proteins associated with the target RNA motif in vivo, which are purified and analyzed by MS.
Identification of new RNA-binding proteins and domains
In addition to candidate-based studies of proteins known to bind lncRNAs, several methods have been developed to identify new RNA-binding proteins in an empirical manner and, more recently, map their RNA-binding domains. Pioneered by the Hentze group, these approaches initially relied on protein–RNA crosslinking followed by affinity purification of polyadenylated transcripts and identification of crosslinked proteins by MS [142, 191–193]. A refinement of this method, termed RBDmap, allowed for the simultaneous identification of RNA-binding proteins and the mapping of the interaction site at peptide resolution [141]. We developed RBR-ID, a similar method, but based on a slightly different principle, which allowed us to identify a larger scope of RNA-binding proteins, including those that bind non-polyadenylated transcripts, and to map their RNA-binding regions in vivo [104]. Several other methods have been developed along these same lines, including CARIC and RICK, which combine metabolic labeling of RNAs and biotin conjugation for purification of crosslinked RNA and protein [194, 195], and XRNAX, which captures crosslinked RNA-protein complexes in the insoluble TRIZOL interphase during purification for downstream sequencing or MS [196].
Collectively, these methods have led to the realization that in addition to the several hundred “canonical” RNA-binding proteins, characterized by possessing one or more well-characterized RNA-binding domains (e.g. RRM, KH, dsRBD; [197]), hundreds—perhaps thousands—more form protein–RNA interactions in vivo, whose functional significance remains unexplored. We propose that in many cases these interactions might be taking place with lncRNAs and that characterizing them will greatly contribute to our understanding of lncRNA function. Furthermore, “targeted” versions of RBR-ID and RBDmap have been developed that can map protein–RNA interactions at high resolution within a complex of choice (e.g. PRC2 [198]). These studies will be instrumental to design “separation-of-function” mutants to address the biochemical and biological roles of RNA interactions within these complexes and validate the proposed biological functions of lncRNAs (see above).
RNA–RNA interactions
While small noncoding RNAs—including micro RNAs, Piwi-interacting RNAs, and small nucleolar RNAs—are known to function largely via RNA–RNA base pairing [43], whether lncRNAs also participate in functional interactions with other RNAs is much less clear.
RAP-RNA was one of the first methods developed to identify the RNA interactors of specific lncRNAs. In RAP-RNA, cellular RNA is chemically crosslinked with one of three crosslinking agents to enrich for direct, both direct and indirect, or only indirect RNA interactions [199]. This is followed by capture of a target lncRNA using antisense biotinylated probes. While RAP-RNA revealed that the abundant MALAT1 lncRNA interacts with pre-mRNAs at active chromatin loci, it recovered virtually no direct RNA interactors, suggesting that MALAT1 does not base pair with its mRNA targets.
CLASH is another recent method that has enabled the high throughput sequencing of direct RNA–RNA interactions [200]. CLASH requires the expression of a tagged “bait” protein in cells, followed by UV crosslinking of RNAs to protein. The tagged protein is pulled down with its crosslinked RNAs, which are ligated to form chimeric molecules that are subsequently sequenced. CLASH has been applied to identify global miRNA interactions in human cells, revealing widespread non-canonical binding of microRNAs to mRNAs and ncRNAs. CLASH may hold promise for identifying lncRNA–RNA interactions, but is limited by the requirement of a universal protein bait. Two additional methods—PARIS and LIGR-seq—allowed for the mapping of RNA duplexes in living cells through a combination of psoralen crosslinking and proximity ligation followed by sequencing [201, 202]. The resulting data can be used to infer RNA secondary structure as well as functional interactions. COMRADES, a similar method, uses tandem affinity purification to isolate RNA duplexes, and was developed alongside a new computational pipeline to predict and cluster RNA structures [203]. Because these methods identify global RNA interactions, it may be challenging to leverage them to study lncRNAs, many of which are lowly expressed.
Aiding biochemistry: genetic and computational tools to study lncRNAs
Although the many new methods discussed above have the potential to reveal a large number of new biochemical interactions of lncRNAs, often choosing which lncRNA to study is the most difficult question, and biochemistry alone might not provide the answer. The number of annotated lncRNAs in the human genome is in the order of nearly ten thousand [6], and in many cases the lncRNA itself has no discernible RNA-mediated function [204]. How, then, can the needle of a new functional lncRNA be found in the haystack of thousands of potentially non-functional annotations? New genetic and computational tools might provide the answer.
Genome-wide forward genetic screens
Forward genetic screens in mammalian cells have become feasible due to the power and versatility of CRISPR-based tools, which allow for both loss- and gain-of-function screening strategies in a pooled format [205] (Figure 3A) allowing the direct interrogation of thousands of lncRNAs in a relatively manageable experiment.
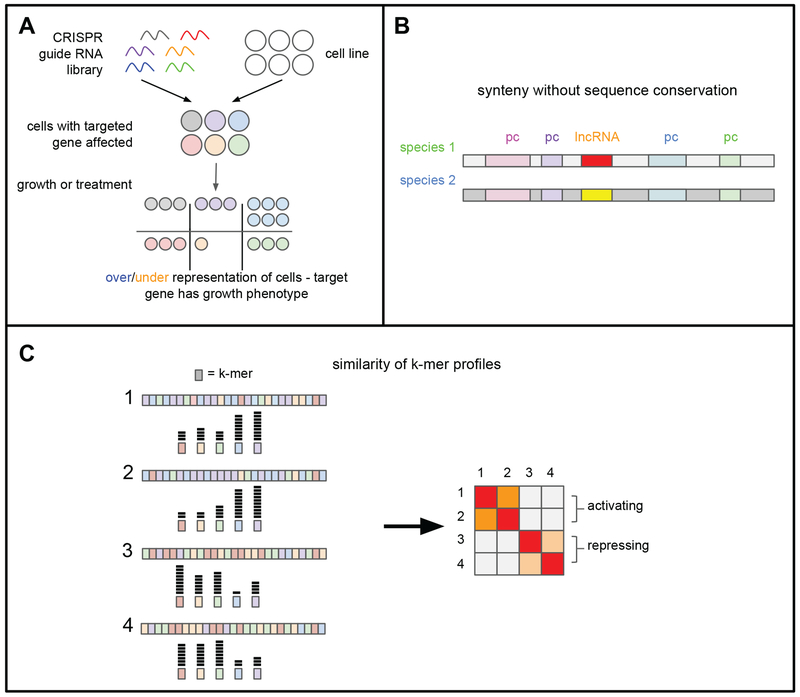
Figure 3. Functional characterization of lncRNAs.
(A) CRISPR screens have been used to identify functional lncRNAs. A guide RNA library is created against a set of lncRNA genes and transfected into cells, with the goal of targeting one gene in each cell. Different proteins can be targeted to the locus of interest, including Cas9 (knockout screens), a combination of NLS-dCas9-VP64 and MS2-p65-HSF1 (synergistic activation mediator, “SAM”; CRISPR activation screens), or dCas9-KRAB repressor (CRISPR interference screens). Following selection of successfully transfected cells, cells are allowed to proliferate, with the possible addition of a treatment such as a drug. After a period of growth, the composition of guide RNAs in the final pool of cells is compared to that in the starting population. Enriched or depleted guides indicate a phenotype for the targeted gene, depending on the direction of enrichment and the type of screen.
(B) Some lncRNAs exhibit synteny without conservation. In these cases, two lncRNAs that occupy the same genomic position in different species do not have significant sequence similarity, suggesting that these lncRNAs might act locally to regulate transcription of neighboring orthologous protein-coding genes.
(C) LncRNAs can be grouped functionally by their k-mer profile. In this example, activating lncRNA 1 and 2 have similar k-mer profiles, while repressing lncRNA 3 and 4 have similar k-mer profiles.
The first lncRNA loss-of-function screen utilized CRISPR interference [206] to identify lncRNAs required for survival in various cell lines [207]. Consistent with the marked cell type specificity of these noncoding transcripts, 89% of lncRNAs with a growth phenotype were specific to one cell type, compared to 55% of protein-coding mRNAs, and no single lncRNA was required for survival in all cell types. Critical pathways such as translation, DNA replication, and post-transcriptional regulation were perturbed by lncRNA repression. Functional lncRNAs identified by this screen have a set of distinct characteristics: expression level, distance from an enhancer, and number of exons were predictive of a growth phenotype following their repression. A gain-of-function screen identified lncRNAs that conferred resistance to BRAF inhibitors in melanoma cells [126]. One hit from this screen was the lncRNA EMICERI, which is responsible for the activation of the nearby protein-coding gene Mob3b, part of the vemurafenib resistance-associated Hippo signaling pathway. Most lncRNA hits that mediated BRAF resistance acted by affecting the expression of nearby genes, adding to the number of examples of lncRNAs that regulate targets locally.
Genetic screens have also been used to identify lncRNAs with positive or negative effects on growth in HuH7.5 liver cancer cells [208], lncRNAs that affect a specific pathway [209], and antisense lncRNAs that regulate cell viability following Ara-C chemotherapy in acute myeloid leukemia (AML) [210]. The AML screen found one coding/noncoding pair, GAS6/GAS6-AS2, that also had correlated expression in a large database of cancer cells treated with Ara-C. Overexpression of the lncRNA GAS6-AS2 caused the upregulation of its cognate protein-coding gene GAS6 locally but also induced expression of the GAS6 receptor gene, AXL, distally, suggesting that this lncRNA might act via multiple biochemical mechanisms.
Computational approaches to infer lncRNA function
Classic tools used to infer the functional importance of protein sequence, namely its evolutionary conservation, are less powerful in the context of lncRNAs, which are not under pressure to maintain a coding frame. However, rather than maintaining primary sequence through evolution, lncRNAs may conserve their function through positional homology, k-mer content, or secondary structure.
Many lncRNAs have synteny without sequence conservation, perhaps indicating a regulatory role that does not depend on sequence. For these genes, sequence conservation exists in protein-coding genes and genomic sequences flanking the lncRNA locus, but not in the lncRNA itself (Figure 3B) [211].
Also suggested is a conservation of short sequence motifs, or k-mers, as the frequency of certain k-mers may impact the ability of a protein to bind to the transcript [212]. The k-mer profiles of lncRNAs from different functional groups, such as those that repressive or activate transcription locally, tend to cluster together (Figure 3C). The localization and binding ability of proteins can be predicted from k-mer profiles. The dynamics of lncRNA biogenesis can also be used to infer functional classes of lncRNAs. Measuring transcription, splicing, degradation, localization, and translation dynamics of protein-coding and noncoding RNAs recovered known characteristics of lncRNAs, including slower splicing, synthesis, and processing rates, paired with higher degradation rates [213]. The function of lncRNAs were assigned by grouping lncRNAs with protein-coding genes of known function that had similar characteristics. This produced protein-coding/lncRNA groups with a wide range of metabolic properties and distinct likely functions.
Another common strategy for identifying lncRNAs and their function is correlating the expression of lncRNAs and protein-coding genes. Increasing amounts of sequencing data from various tissues and conditions allow for the detection of highly variable genes and the identification of frequently coexpressed genes. The function of a lncRNA can be inferred from the identity of protein-coding genes that have a similar transcriptional profile [10]. In non-model organisms, where sophisticated genetic tools are not always readily available, the approach of using large sequencing datasets to identify correlated lncRNAs and protein-coding genes has promise to prioritize the study of functional lncRNAs.
Finally, comparative genomics and evolutionary analyses of lncRNA sequence conservation have provided information on potential lncRNA function. Co-expression networks of lncRNAs and protein-coding genes that are expressed in multiple organisms helped identify lncRNAs involved in spermatogenesis, synaptic transmission, and muscle functions in the heart [10]. Although lncRNAs evolve more rapidly than protein-coding genes, lncRNA promoters contain more transcription factor binding sites than random intergenic regions do, and are particularly enriched for motifs recognized by homeobox transcription factors. The age of a lncRNA affects the likelihood of its active regulation, as the Polycomb protein SUZ12 and the pluripotency transcription factor OCT4 associate more with older, more conserved lncRNAs [10].
Concluding remarks/future outlook
We have witnessed an acceleration of new technologies and strategies to discover and characterize functional lncRNAs, several of which are reviewed above. Looking ahead, continued improvements to biochemical and genomic technologies will allow for the functional interrogation of lncRNAs in specific tissues and cell-types, as well as the identification of novel lncRNAs with important biological roles.
Given their very specific and restricted expression patterns, we believe that great insight into lncRNAs function will be offered by single-cell sequencing technologies [31]. Indeed it has already been shown that at least in the case of some lncRNAs their presumptive lower expression level in bulk samples was a consequence of the smaller population of cells in heterogenous mixtures that expressed them. A collection of 81 single-cell transcriptomes revealed that lncRNA are activated during reprogramming and suppress lineage-specific genes on a single-cell level [214]. Another study of 226 cells from radial sections of the mouse neocortex identified cell type-specific lncRNAs [214]. Single-cell sequencing has revealed different lncRNA profiles among fibroblast cell types [215] and between neurons and glia in Drosophila [216]. Although these studies are informative, higher-throughput studies may provide more information about the specificity and regulatory potential of lncRNA in the context of a whole tissue or organism.
Evolutionary studies of lncRNAs have been aided by new annotations of lncRNAs in a variety of model and non-model organisms, facilitated by new and improved genome assemblies. For example, new lncRNA annotations in Xenopus tropicalis and the analysis of their expression patterns suggested functional roles in tissue identity during development [217]. Annotations of lncRNAs have become available in multiple social insect species [15, 16], where alternative developmental trajectories give rise to individuals with identical genomes but vastly different morphological, physiological, and behavioral phenotypes. We analyzed lncRNA expression in Harpegnathos saltator, an ant that maintains reproductive plasticity in workers during a unique caste transition [218]. Major transcriptional changes occur in the brain during this event, including differential regulation of lncRNAs [16, 219]. As CRISPR-mediated editing of the germline has been achieved in Harpegnathos ants [220], these newly discovered caste-specific transcripts are prime targets for the functional investigation of lncRNAs potentially involved in brain function and behavior.
Another emerging frontier in lncRNA research is the analysis of their post-transcriptional chemical modification [221]. Pioneering studies on XIST showed that its modification with m6A is required for the silencing of several X-linked genes [222]. DCI, a protein required for XIST-mediated transcriptional silencing, binds preferentially at m6A residues. As folding of lncRNAs is thought to be key to their function, it will be of great interest to determine if chemical modification that affect their three-dimensional structure contribute to their biological and biochemical versatility.
Given the many known and suspected roles of lncRNAs in various cellular processes, it should not come as a surprise that dysregulation of lncRNAs can lead to disease or aberrant phenotypes. LncRNA are increasingly being considered as therapeutic targets [223, 224]. As mentioned above, genetic screens have detected many lncRNAs involved in cancer progression [126, 209, 210]. Additionally, organisms that transmit or cause disease can affect how the immune system recognizes threats through regulation of lncRNAs in their own transcriptome or in host cells. The genomes of mosquitoes and of the malaria parasites harbor lncRNAs [225–228], and the parasite Toxoplasma gondii affects the lncRNA profile of its host [229]. The annotation of lncRNAs in more organisms will lead to better understanding of their function and will provide new avenues to study lncRNA-related diseases and their treatment.
ACKNOWLEDGMENTS
The authors thank M. Owens and R. Warneford-Thomson for critical reading of the manuscript. R.B. acknowledges support from the NIH (DP2MH107055, R01GM127408), the Searle Scholars Program (15-SSP-102), the March of Dimes Foundation (1-FY-15-344), a Linda Pechenik Montague Investigator Award, and the Charles E. Kaufman Foundation (KA2016-85223). E.J.S. was supported in part by an NIH training grant (T32HG000046). A.P. was supported in part by an NIH training grant T32 HD083185.
REFERENCES
- 1.Cech TR and Steitz JA (2014) The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell. 157, 77–94 [DOI] [PubMed] [Google Scholar]
- 2.Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP and Gingeras TR (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science. 296, 916–919 [DOI] [PubMed] [Google Scholar]
- 4.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y, Consortium, F., I, R. G. E. R. G. P. and Team, I. I. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 420, 563–573 [DOI] [PubMed] [Google Scholar]
- 5.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M and Snyder M (2004) Global identification of human transcribed sequences with genome tiling arrays. Science. 306, 2242–2246 [DOI] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J and Guigo R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattick JS (2005) The functional genomics of noncoding RNA. Science. 309, 1527–1528 [DOI] [PubMed] [Google Scholar]
- 8.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H and Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 316, 1484–1488 [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL and Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 458, 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F and Kaessmann H (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 505, 635–640 [DOI] [PubMed] [Google Scholar]
- 11.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A and Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pervouchine DD, Djebali S, Breschi A, Davis CA, Barja PP, Dobin A, Tanzer A, Lagarde J, Zaleski C, See LH, Fastuca M, Drenkow J, Wang H, Bussotti G, Pei B, Balasubramanian S, Monlong J, Harmanci A, Gerstein M, Beer MA, Notredame C, Guigo R and Gingeras TR (2015) Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nature communications. 6, 5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A and Schier AF (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL and Ponting CP (2012) Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome biology and evolution. 4, 427–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayakodi M, Jung JW, Park D, Ahn YJ, Lee SC, Shin SY, Shin C, Yang TJ and Kwon HW (2015) Genome-wide characterization of long intergenic non-coding RNAs (lincRNAs) provides new insight into viral diseases in honey bees Apis cerana and Apis mellifera. BMC Genomics. 16, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields EJ, Sheng L, Weiner AK, Garcia BA and Bonasio R (2018) High-Quality Genome Assemblies Reveal Long Non-coding RNAs Expressed in Ant Brains. Cell reports. 23, 3078–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ariel F, Romero-Barrios N, Jegu T, Benhamed M and Crespi M (2015) Battles and hijacks: noncoding transcription in plants. Trends Plant Sci 20, 362–371 [DOI] [PubMed] [Google Scholar]
- 18.Chekanova JA (2015) Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27, 207–216 [DOI] [PubMed] [Google Scholar]
- 19.Yamashita A, Shichino Y and Yamamoto M (2016) The long non-coding RNA world in yeasts. Biochimica et biophysica acta. 1859, 147–154 [DOI] [PubMed] [Google Scholar]
- 20.Bartolomei MS, Zemel S and Tilghman SM (1991) Parental imprinting of the mouse H19 gene. Nature. 351, 153–155 [DOI] [PubMed] [Google Scholar]
- 21.Brannan CI, Dees EC, Ingram RS and Tilghman SM (1990) The product of the H19 gene may function as an RNA. Mol Cell Biol 10, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galupa R and Heard E (2018) X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation. Annu Rev Genet 52, 535–566 [DOI] [PubMed] [Google Scholar]
- 23.Sahakyan A, Yang Y and Plath K (2018) The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol 28, 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 349, 38–44 [DOI] [PubMed] [Google Scholar]
- 25.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S and Rastan S (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 71, 515–526 [DOI] [PubMed] [Google Scholar]
- 26.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, Rogiers A, Hermans E, Baatsen P, Aerts S, Amant F, Van Aelst S, van den Oord J, de Strooper B, Davidson I, Lafontaine DL, Gevaert K, Vandesompele J, Mestdagh P and Marine JC (2016) Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 531, 518–522 [DOI] [PubMed] [Google Scholar]
- 27.Hua JT, Ahmed M, Guo H, Zhang Y, Chen S, Soares F, Lu J, Zhou S, Wang M, Li H, Larson NB, McDonnell SK, Patel PS, Liang Y, Yao CQ, van der Kwast T, Lupien M, Feng FY, Zoubeidi A, Tsao MS, Thibodeau SN, Boutros PC and He HH (2018) Risk SNP-Mediated Promoter-Enhancer Switching Drives Prostate Cancer through lncRNA PCAT19. Cell. 174, 564–575 e518 [DOI] [PubMed] [Google Scholar]
- 28.Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR and Ghosh S (2016) A long noncoding RNA associated with susceptibility to celiac disease. Science. 352, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo Sardo V, Chubukov P, Ferguson W, Kumar A, Teng EL, Duran M, Zhang L, Cost G, Engler AJ, Urnov F, Topol EJ, Torkamani A and Baldwin KK (2018) Unveiling the Role of the Most Impactful Cardiovascular Risk Locus through Haplotype Editing. Cell. 175, 1796–1810 e1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonasio R and Shiekhattar R (2014) Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48, 433–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deveson IW, Hardwick SA, Mercer TR and Mattick JS (2017) The Dimensions, Dynamics, and Relevance of the Mammalian Noncoding Transcriptome. Trends Genet 33, 464–478 [DOI] [PubMed] [Google Scholar]
- 32.Engreitz JM, Ollikainen N and Guttman M (2016) Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 17, 756–770 [DOI] [PubMed] [Google Scholar]
- 33.Kopp F and Mendell JT (2018) Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 172, 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutenberg-Schoenberg M, Sexton AN and Simon MD (2016) The Properties of Long Noncoding RNAs That Regulate Chromatin. Annu Rev Genomics Hum Genet 17, 69–94 [DOI] [PubMed] [Google Scholar]
- 35.Ulitsky I (2016) Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet 17, 601–614 [DOI] [PubMed] [Google Scholar]
- 36.Quinn JJ and Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17, 47–62 [DOI] [PubMed] [Google Scholar]
- 37.Goff LA and Rinn JL (2015) Linking RNA biology to lncRNAs. Genome Res 25, 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Froberg JE and Lee JT (2014) Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci 39, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long Y, Wang X, Youmans DT and Cech TR (2017) How do lncRNAs regulate transcription? Sci Adv 3, eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE and Lander ES (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 477, 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES and Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 106, 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science. 338, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 43.Holoch D and Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero-Barrios N, Legascue MF, Benhamed M, Ariel F and Crespi M (2018) Splicing regulation by long noncoding RNAs. Nucleic Acids Res 46, 2169–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercer T, Dinger M and Mattick J (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet [DOI] [PubMed] [Google Scholar]
- 46.Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ and Mattick JS (2014) The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular psychiatry. 19, 486–494 [DOI] [PubMed] [Google Scholar]
- 47.Yoon JH, Abdelmohsen K and Gorospe M (2013) Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 425, 3723–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, Payumo AY, Peres-da-Silva A, Broz DK, Baum R, Guo S, Chen JK, Attardi LD and Chang HY (2016) An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet 48, 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP, Subramanian V, Chen J, Schenone M, Guttman M, Carr SA and Lander ES (2018) The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 561, 132–136 [DOI] [PubMed] [Google Scholar]
- 50.Wang KC and Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell. 43, 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 129, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonasio R, Tu S and Reinberg D (2010) Molecular signals of epigenetic states. Science. 330, 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung T and Chang HY (2010) Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 7, 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Syed J and Sugiyama H (2016) RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem Biol 23, 1325–1333 [DOI] [PubMed] [Google Scholar]
- 55.Roberts RW and Crothers DM (1992) Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 258, 1463–1466 [DOI] [PubMed] [Google Scholar]
- 56.Martianov I, Ramadass A, Serra Barros A, Chow N and Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 445, 666–670 [DOI] [PubMed] [Google Scholar]
- 57.Schmitz KM, Mayer C, Postepska A and Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24, 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Z, Senturk N, Song C and Grummt I (2018) lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev 32, 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grote P and Herrmann BG (2013) The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol 10, 1579–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M and Herrmann BG (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E and Kanduri C (2015) MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nature communications. 6, 7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson MJ and Anastasov N (2015) PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell reports. 11, 474–485 [DOI] [PubMed] [Google Scholar]
- 63.Santos-Pereira JM and Aguilera A (2015) R loops: new modulators of genome dynamics and function. Nat Rev Genet 16, 583–597 [DOI] [PubMed] [Google Scholar]
- 64.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K and Luke B (2013) Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20, 1199–1205 [DOI] [PubMed] [Google Scholar]
- 65.Wahba L, Gore SK and Koshland D (2013) The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2, e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE and Tran EJ (2016) Regulated Formation of lncRNA-DNA Hybrids Enables Faster Transcriptional Induction and Environmental Adaptation. Mol Cell. 61, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan Q, Shields EJ, Bonasio R and Sarma K (2018) Mapping native R-loops genome-wide using a targeted nuclease approach. bioRxiv, 457226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dumelie JG and Jaffrey SR (2017) Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. eLife. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X and Chedin F (2016) Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell. 63, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H, Zhou Y, Zhang DE and Fu XD (2017) R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell. 68, 745–757 e745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K and Guttman M (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 341, 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brockdorff N (2013) Noncoding RNA and Polycomb recruitment. RNA. 19, 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ringrose L (2017) Noncoding RNAs in Polycomb and Trithorax Regulation: A Quantitative Perspective. Annu Rev Genet 51, 385–411 [DOI] [PubMed] [Google Scholar]
- 74.Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B and Margueron R (2017) PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 36, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amandio AR, Necsulea A, Joye E, Mascrez B and Duboule D (2016) Hotair Is Dispensible for Mouse Development. PLoS Genet 12, e1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA and Chang HY (2013) Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell reports. 5, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L, Helms JA and Chang HY (2016) Comment on “Hotair Is Dispensable for Mouse Development”. PLoS Genet 12, e1006406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Selleri L, Bartolomei MS, Bickmore WA, He L, Stubbs L, Reik W and Barsh GS (2016) A Hox-Embedded Long Noncoding RNA: Is It All Hot Air? PLoS Genet 12, e1006485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simon JA and Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10, 697–708 [DOI] [PubMed] [Google Scholar]
- 80.Rosenberg M, Blum R, Kesner B, Maier VK, Szanto A and Lee JT (2017) Denaturing CLIP, dCLIP, Pipeline Identifies Discrete RNA Footprints on Chromatin-Associated Proteins and Reveals that CBX7 Targets 3’ UTRs to Regulate mRNA Expression. Cell Syst 5, 368–385 e315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, Ule J and Jenner RG (2016) The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 26, 896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y and Reinberg D (2014) Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. eLife. 3, e02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A and Reinberg D (2014) Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 53, 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaneko S, Son J, Shen SS, Reinberg D and Bonasio R (2013) PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20, 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davidovich C, Zheng L, Goodrich KJ and Cech TR (2013) Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20, 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cifuentes-Rojas C, Hernandez AJ, Sarma K and Lee JT (2014) Regulatory Interactions between RNA and Polycomb Repressive Complex 2. Mol Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao J, Sun BK, Erwin JA, Song JJ and Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 322, 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heo JB and Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 331, 76–79 [DOI] [PubMed] [Google Scholar]
- 89.Kim DH and Sung S (2017) Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev Cell 40, 302–312 e304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schuettengruber B, Bourbon HM, Di Croce L and Cavalli G (2017) Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 171, 34–57 [DOI] [PubMed] [Google Scholar]
- 91.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA and Chang HY (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 472, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M and Chang HY (2014) Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 3, e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R and Fraser P (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 322, 1717–1720 [DOI] [PubMed] [Google Scholar]
- 94.Pavlaki I, Alammari F, Sun B, Clark N, Sirey T, Lee S, Woodcock DJ, Ponting CP, Szele FG and Vance KW (2018) The long non-coding RNA Paupar promotes KAP1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J. 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL and Ponting CP (2014) The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 33, 296–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL and Tenen DG (2013) DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 503, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW and Ponting CP (2014) The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife. 3, e04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee JT and Bartolomei MS (2013) X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 152, 1308–1323 [DOI] [PubMed] [Google Scholar]
- 99.Chakraborty D, Kappei D, Theis M, Nitzsche A, Ding L, Paszkowski-Rogacz M, Surendranath V, Berger N, Schulz H, Saar K, Hubner N and Buchholz F (2012) Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat Methods. 9, 360–362 [DOI] [PubMed] [Google Scholar]
- 100.Chakraborty D, Paszkowski-Rogacz M, Berger N, Ding L, Mircetic J, Fu J, Iesmantavicius V, Choudhary C, Anastassiadis K, Stewart AF and Buchholz F (2017) lncRNA Panct1 Maintains Mouse Embryonic Stem Cell Identity by Regulating TOBF1 Recruitment to Oct-Sox Sequences in Early G1. Cell reports. 21, 3012–3021 [DOI] [PubMed] [Google Scholar]
- 101.Ng SY, Bogu GK, Soh BS and Stanton LW (2013) The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 51, 349–359 [DOI] [PubMed] [Google Scholar]
- 102.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ and Chang HY (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.D GH, Kelley DR, Tenen D, Bernstein B and Rinn JL (2016) Widespread RNA binding by chromatin-associated proteins. Genome Biol 17, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He C, Sidoli S, Warneford-Thomson R, Tatomer DC, Wilusz JE, Garcia BA and Bonasio R (2016) High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol Cell. 64, 416–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Samata M and Akhtar A (2018) Dosage Compensation of the X Chromosome: A Complex Epigenetic Assignment Involving Chromatin Regulators and Long Noncoding RNAs. Annu Rev Biochem 87, 323–350 [DOI] [PubMed] [Google Scholar]
- 106.Franke A and Baker BS (1999) The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 4, 117–122 [DOI] [PubMed] [Google Scholar]
- 107.Figueiredo ML, Kim M, Philip P, Allgardsson A, Stenberg P and Larsson J (2014) Non-coding roX RNAs prevent the binding of the MSL-complex to heterochromatic regions. PLoS Genet 10, e1004865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ilik IA, Maticzka D, Georgiev P, Gutierrez NM, Backofen R and Akhtar A (2017) A mutually exclusive stem-loop arrangement in roX2 RNA is essential for X-chromosome regulation in Drosophila. Genes Dev 31, 1973–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, Chang HY and Akhtar A (2013) Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 51, 156–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E and Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science. 329, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y and Mendell JT (2016) Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 164, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G and Hirose T (2018) Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol Cell. 70, 1038–1053 e1037 [DOI] [PubMed] [Google Scholar]
- 113.Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, Li H, Gou LT, Zhang Y, Wang Y, Yeo GW, Zhou Y and Fu XD (2017) NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol 24, 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD and Wu M (2018) GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol 20, 492–502 [DOI] [PubMed] [Google Scholar]
- 115.Spitale RC, Tsai MC and Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 6, 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mallam AL, Sae-Lee W, Schaub JM, Tu F, Battenhouse A, Jang YJ, Kim J, Finkelstein IJ, Marcotte EM and Drew K (2018) Systematic discovery of endogenous human ribonucleoprotein complexes. bioRxiv, 480061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E and Dekker J (2016) Structural organization of the inactive X chromosome in the mouse. Nature. 535, 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E and de Laat W (2011) The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P and Guttman M (2016) Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 354, 468–472 [DOI] [PubMed] [Google Scholar]
- 120.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K and Guttman M (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 521, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W and Lee JT (2015) Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA and Shiekhattar R (2013) Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 494, 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Werner MS and Ruthenburg AJ (2015) Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell reports. 12, 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Werner MS, Sullivan MA, Shah RN, Nadadur RD, Grzybowski AT, Galat V, Moskowitz IP and Ruthenburg AJ (2017) Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat Struct Mol Biol 24, 596–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim TK and Shiekhattar R (2015) Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 162, 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng YY, Yoon CH, Boehm JS, Lander ES and Zhang F (2017) Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 548, 343–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Amaral PP, Leonardi T, Han N, Vire E, Gascoigne DK, Arias-Carrasco R, Buscher M, Pandolfini L, Zhang A, Pluchino S, Maracaja-Coutinho V, Nakaya HI, Hemberg M, Shiekhattar R, Enright AJ and Kouzarides T (2018) Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol 19, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tan JY, Smith AAT, Ferreira da Silva M, Matthey-Doret C, Rueedi R, Sonmez R, Ding D, Kutalik Z, Bergmann S and Marques AC (2017) cis-Acting Complex-Trait-Associated lincRNA Expression Correlates with Modulation of Chromosomal Architecture. Cell reports. 18, 2280–2288 [DOI] [PubMed] [Google Scholar]
- 129.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J and Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature. 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Phillips JE and Corces VG (2009) CTCF: master weaver of the genome. Cell. 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, Wiest DL and Murre C (2017) Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell. 171, 103–119 e118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cajigas I, Chakraborty A, Swyter KR, Luo H, Bastidas M, Nigro M, Morris ER, Chen S, VanGompel MJW, Leib D, Kohtz SJ, Martina M, Koh S, Ay F and Kohtz JD (2018) The Evf2 Ultraconserved Enhancer lncRNA Functionally and Spatially Organizes Megabase Distant Genes in the Developing Forebrain. Mol Cell. 71, 956–972 e959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A and Rinn JL (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21, 198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, Noble WS, Shendure J and Disteche CM (2015) The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol 16, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barutcu AR, Maass PG, Lewandowski JP, Weiner CL and Rinn JL (2018) A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nature communications. 9, 1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Saldana-Meyer R, Gonzalez-Buendia E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F and Reinberg D (2014) CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev 28, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hansen AS, Hsieh T-HS, Cattoglio C, Pustova I, Darzacq X and Tjian R (2018) An RNA-binding region regulates CTCF clustering and chromatin looping. bioRxiv, 495432 [Google Scholar]
- 138.Saldana-Meyer R, Rodriguez-Hernaez J, Nishana M, Jacome-Lopez K, Nora EP, Bruneau BG, Furlan-Magaril M, Skok J and Reinberg D (2019) RNA interactions with CTCF are essential for its proper function. bioRxiv, 530014 [Google Scholar]
- 139.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science. 357 [DOI] [PubMed] [Google Scholar]
- 140.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T and Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J and Hentze MW (2016) Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol Cell. 63, 696–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J and Hentze MW (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 143.Fox AH, Nakagawa S, Hirose T and Bond CS (2018) Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem Sci 43, 124–135 [DOI] [PubMed] [Google Scholar]
- 144.Uversky VN (2017) Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 44, 18–30 [DOI] [PubMed] [Google Scholar]
- 145.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB and Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Galganski L, Urbanek MO and Krzyzosiak WJ (2017) Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45, 10350–10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG and Prasanth KV (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 39, 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C and Spector DL (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell reports. 2, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Clemson C, Hutchinson J, Sara S, Ensminger A, Fox A, Chess A and Lawrence J (2009) An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fox AH and Lamond AI (2010) Paraspeckles. Cold Spring Harbor perspectives in biology. 2, a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY and Kingston RE (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 55, 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hnisz D, Shrinivas K, Young RA, Chakraborty AK and Sharp PA (2017) A Phase Separation Model for Transcriptional Control. Cell. 169, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cerase A, Armaos A, Cid-Samper F, Avner P and Tartaglia GG (2018) Xist lncRNA forms silencing granules that induce heterochromatin formation and repressive complexes recruitment by phase separation. bioRxiv, 351015 [Google Scholar]
- 154.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL and Raj A (2015) Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lubelsky Y and Ulitsky I (2018) Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 555, 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shukla CJ, McCorkindale AL, Gerhardinger C, Korthauer KD, Cabili MN, Shechner DM, Irizarry RA, Maass PG and Rinn JL (2018) High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mattick JS (2003) Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 25, 930–939 [DOI] [PubMed] [Google Scholar]
- 158.Rinn JL and Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI and Kingston RE (2011) The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 108, 20497–20502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chu C, Qu K, Zhong FL, Artandi SE and Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 44, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Simon MD (2016) Insight into lncRNA biology using hybridization capture analyses. Biochimica et biophysica acta. 1859, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A and Chang HY (2014) Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol 32, 933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE and Lee JT (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 504, 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheetham SW and Brand AH (2018) RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites. Nat Struct Mol Biol 25, 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Steensel B and Henikoff S (2000) Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18, 424–428 [DOI] [PubMed] [Google Scholar]
- 166.Bell JC, Jukam D, Teran NA, Risca VI, Smith OK, Johnson WL, Skotheim JM, Greenleaf WJ and Straight AF (2018) Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li X, Zhou B, Chen L, Gou LT, Li H and Fu XD (2017) GRID-seq reveals the global RNA-chromatin interactome. Nat Biotechnol 35, 940–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, Cai L, McDonel P, Garber M and Guttman M (2018) Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 174, 744–757 e724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sridhar B, Rivas-Astroza M, Nguyen TC, Chen W, Yan Z, Cao X, Hebert L and Zhong S (2017) Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr Biol 27, 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gilbert C, Kristjuhan A, Winkler GS and Svejstrup JQ (2004) Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 14, 457–464 [DOI] [PubMed] [Google Scholar]
- 171.Keene JD, Komisarow JM and Friedersdorf MB (2006) RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nature protocols. 1, 302–307 [DOI] [PubMed] [Google Scholar]
- 172.Tenenbaum SA, Carson CC, Lager PJ and Keene JD (2000) Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 97, 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M and Lee JT (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 40, 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mili S and Steitz JA (2004) Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 10, 1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC and Darnell RB (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 456, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A and Darnell RB (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science. 302, 1212–1215 [DOI] [PubMed] [Google Scholar]
- 177.Friedersdorf MB and Keene JD (2014) Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol 15, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM and Ule J (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17, 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, Stanton R, Rigo F, Guttman M and Yeo GW (2016) Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods. 13, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gu J, Wang M, Yang Y, Qiu D, Zhang Y, Ma J, Zhou Y, Hannon GJ and Yu Y (2018) GoldCLIP: Gel-omitted Ligation-dependent CLIP. Genomics Proteomics Bioinformatics. 16, 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Maticzka D, Ilik IA, Aktas T, Backofen R and Akhtar A (2018) uvCLAP is a fast and non-radioactive method to identify in vivo targets of RNA-binding proteins. Nature communications. 9, 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R and Akhtar A (2017) DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 544, 115–119 [DOI] [PubMed] [Google Scholar]
- 183.Kim DI and Roux KJ (2016) Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 26, 804–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Beck DB, Narendra V, Drury WJ 3rd, Casey R, Jansen PW, Yuan ZF, Garcia BA, Vermeulen M and Bonasio R (2014) In vivo proximity labeling for the detection of protein-protein and protein-RNA interactions. J Proteome Res 13, 6135–6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA and Ting AY (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 339, 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fazal FM, Han S, Kaewsapsak P, Parker KR, Xu J, Boettiger AN, Chang HY and Ting AY (2018) Atlas of Subcellular RNA Localization Revealed by APEX-seq. bioRxiv, 454470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Padron A, Iwasaki S and Ingolia N (2018) Proximity RNA labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. bioRxiv, 454066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Roux KJ, Kim DI, Raida M and Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of cell biology. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ramanathan M, Majzoub K, Rao DS, Neela PH, Zarnegar BJ, Mondal S, Roth JG, Gai H, Kovalski JR, Siprashvili Z, Palmer TD, Carette JE and Khavari PA (2018) RNA-protein interaction detection in living cells. Nat Methods. 15, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E and Chang HY (2015) Systematic discovery of Xist RNA binding proteins. Cell. 161, 404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C and Landthaler M (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 192.Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W, Krijgsveld J and Hentze MW (2015) The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nature communications. 6, 10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Conrad T, Albrecht AS, de Melo Costa VR, Sauer S, Meierhofer D and Orom UA (2016) Serial interactome capture of the human cell nucleus. Nature communications. 7, 11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bao X, Guo X, Yin M, Tariq M, Lai Y, Kanwal S, Zhou J, Li N, Lv Y, Pulido-Quetglas C, Wang X, Ji L, Khan MJ, Zhu X, Luo Z, Shao C, Lim DH, Liu X, Li N, Wang W, He M, Liu YL, Ward C, Wang T, Zhang G, Wang D, Yang J, Chen Y, Zhang C, Jauch R, Yang YG, Wang Y, Qin B, Anko ML, Hutchins AP, Sun H, Wang H, Fu XD, Zhang B and Esteban MA (2018) Capturing the interactome of newly transcribed RNA. Nat Methods. 15, 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Huang R, Han M, Meng L and Chen X (2018) Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc Natl Acad Sci U S A. 115, E3879–E3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Trendel J, Schwarzl T, Horos R, Prakash A, Bateman A, Hentze MW and Krijgsveld J (2018) The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell [DOI] [PubMed] [Google Scholar]
- 197.Lunde BM, Moore C and Varani G (2007) RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 8, 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhang Q, McKenzie NJ, Warneford-Thomson R, Gail EH, Flanigan SF, Owen BM, Lauman R, Levina V, Garcia BA, Schittenhelm RB, Bonasio R and Davidovich C ((in press)) RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nature Structural and Molecular Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M and Lander ES (2014) RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 159, 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Helwak A, Kudla G, Dudnakova T and Tollervey D (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 153, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR and Chang HY (2016) RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 165, 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sharma E, Sterne-Weiler T, O’Hanlon D and Blencowe BJ (2016) Global Mapping of Human RNA-RNA Interactions. Mol Cell. 62, 618–626 [DOI] [PubMed] [Google Scholar]
- 203.Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu ZY, Kwok CK, Qin CF, MacRae IJ, Goodfellow I, Marioni JC, Kudla G and Miska EA (2018) COMRADES determines in vivo RNA structures and interactions. Nat Methods. 15, 785–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 539, 452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shalem O, Sanjana NE and Zhang F (2015) High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP and Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152, 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS and Lim DA (2017) CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, Yuan P, Brown M, Liu XS and Wei W (2016) Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol 34, 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Koirala P, Huang J, Ho TT, Wu F, Ding X and Mo YY (2017) LncRNA AK023948 is a positive regulator of AKT. Nature communications. 8, 14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL, Provero P, Church GM, Clohessy JG and Pandolfi PP (2018) An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell. 173, 649–664 e620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP and Ulitsky I (2015) Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell reports. 11, 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kirk JM, Kim SO, Inoue K, Smola MJ, Lee DM, Schertzer MD, Wooten JS, Baker AR, Sprague D, Collins DW, Horning CR, Wang S, Chen Q, Weeks KM, Mucha PJ and Calabrese JM (2018) Functional classification of long non-coding RNAs by k-mer content. Nat Genet 50, 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mukherjee N, Calviello L, Hirsekorn A, de Pretis S, Pelizzola M and Ohler U (2017) Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol 24, 86–96 [DOI] [PubMed] [Google Scholar]
- 214.Kim DH, Marinov GK, Pepke S, Singer ZS, He P, Williams B, Schroth GP, Elowitz MB and Wold BJ (2015) Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell stem cell. 16, 88–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, Liu Z, Stripp B, Tang J, Liang J, Noble PW and Jiang D (2018) Single-Cell Deconvolution of Fibroblast Heterogeneity in Mouse Pulmonary Fibrosis. Cell reports. 22, 3625–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, Aibar S, Makhzami S, Christiaens V, Bravo Gonzalez-Blas C, Poovathingal S, Hulselmans G, Spanier KI, Moerman T, Vanspauwen B, Geurs S, Voet T, Lammertyn J, Thienpont B, Liu S, Konstantinides N, Fiers M, Verstreken P and Aerts S (2018) A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell. 174, 982–998 e920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Forouzmand E, Owens NDL, Blitz IL, Paraiso KD, Khokha MK, Gilchrist MJ, Xie X and Cho KWY (2017) Developmentally regulated long non-coding RNAs in Xenopus tropicalis. Dev Biol 426, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, Zhang P, Huang Z, Berger SL, Reinberg D, Wang J and Liebig J (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science. 329, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gospocic J, Shields EJ, Glastad KM, Lin Y, Penick CA, Yan H, Mikheyev AS, Linksvayer TA, Garcia BA, Berger SL, Liebig J, Reinberg D and Bonasio R (2017) The Neuropeptide Corazonin Controls Social Behavior and Caste Identity in Ants. Cell. 170, 748–759 e712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yan H, Opachaloemphan C, Mancini G, Yang H, Gallitto M, Mlejnek J, Leibholz A, Haight K, Ghaninia M, Huo L, Perry M, Slone J, Zhou X, Traficante M, Penick CA, Dolezal K, Gokhale K, Stevens K, Fetter-Pruneda I, Bonasio R, Zwiebel LJ, Berger SL, Liebig J, Reinberg D and Desplan C (2017) An Engineered orco Mutation Produces Aberrant Social Behavior and Defective Neural Development in Ants. Cell. 170, 736–747 e739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Roundtree IA, Evans ME, Pan T and He C (2017) Dynamic RNA Modifications in Gene Expression Regulation. Cell. 169, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M and Jaffrey SR (2016) m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 537, 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 224.Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL and Rigo F (2015) Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 518, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, Volkman S, Duraisingh M, Wirth D, Sabeti PC and Rinn JL (2011) A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Calderwood MS, Gannoun-Zaki L, Wellems TE and Deitsch KW (2003) Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem 278, 34125–34132 [DOI] [PubMed] [Google Scholar]
- 227.Epp C, Li F, Howitt CA, Chookajorn T and Deitsch KW (2009) Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 15, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jenkins AM, Waterhouse RM and Muskavitch MA (2015) Long non-coding RNA discovery across the genus anopheles reveals conserved secondary structures within and beyond the Gambiae complex. BMC Genomics. 16, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Menard KL, Haskins BE, Colombo AP and Denkers EY (2018) Toxoplasma gondii Manipulates Expression of Host Long Noncoding RNA during Intracellular Infection. Sci Rep 8, 15017. [DOI] [PMC free article] [PubMed] [Google Scholar]