Abstract
Iron homeostasis ensures adequate iron for biological processes while preventing excessive iron accumulation, which can lead to tissue injury. In mammalian systems, iron availability is controlled by the interaction of the iron-regulatory hormone hepcidin with ferroportin, a molecule that functions both as the hepcidin receptor as well as the sole known cellular exporter of iron. By reducing iron export through ferroportin to blood plasma, hepcidin inhibits the mobilization of iron from stores and the absorption of dietary iron. Among the many processes requiring iron, erythropoiesis is the most iron-intensive, consuming most iron circulating in blood plasma. Under conditions of enhanced erythropoiesis, more iron is required to provide developing erythroblasts with adequate iron for heme and hemoglobin synthesis. Here the hormone erythroferrone, produced by erythroblasts, acts on hepatocytes to suppress hepcidin production, and thereby increase dietary iron absorption and mobilization from stores. This review focuses on the discovery of erythroferrone and recent advances in understanding the role of this hormone in the regulation of iron homeostasis during states of increased erythropoietic demand. Gaps in our understanding of the role of erythroferrone are highlighted for future study.
Iron homeostasis and regulation by hepcidin
Most iron in vertebrates is destined to become a component of hemoglobin, where it is essential for the transport of oxygen to all tissues. In healthy humans, approximately 60% of total-body iron is contained within circulating erythrocytes1 and each day the synthesis of new erythrocyte hemoglobin requires about 80%2 of the iron obtained from a combination of dietary sources and the recycling of senescent erythrocytes. When iron supply is inadequate, erythrocyte protein synthesis and development are altered in a regulated manner3 resulting in microcytic anemia and reduced oxygen-carrying capacity. To ensure an adequate supply of iron for erythropoiesis and other iron-requiring processes, tightly orchestrated regulatory mechanisms control dietary iron absorption and the efflux of iron from stores in splenic and hepatic macrophages and hepatocytes. Iron lacks a quantitatively significant route of excretion from the body, therefore the absorption of new iron from the diet must be balanced with daily losses and the mobilization of stored iron to preserve systemic iron homeostasis. Ferroportin, the only identified mammalian iron export protein, delivers to plasma dietary iron from enterocytes, as well as stored iron from hepatocytes and macrophages of the reticuloendothelial system.4,5 The common role of ferroportin in both iron absorption and iron recycling allows for the streamlined regulation of iron availability through the modulation of ferroportin abundance in enterocytes, macrophages, and hepatocytes. The master regulator of ferroportin, and therefore whole-body iron homeostasis, is hepcidin, a peptide hormone produced primarily by hepatocytes.6,7 Hepcidin binds to ferroportin and reduces cellular iron export by 2 mechanisms: the hepcidin-induced conformational change in ferroportin initiates its ubiquitination and degradation7 and the binding of hepcidin to ferroportin occludes iron transport.8 At the organismal level, decreased ferroportin activity causes decreased iron absorption and reduced mobilization from stores. Hepcidin expression is regulated by iron status, increasing with elevated serum iron concentrations and liver iron stores and decreasing in response to iron deficiency.9–11 Mutations in genes involved in the regulation of hepcidin expression, or in the gene encoding hepcidin itself,12–18 may result in inappropriately low hepcidin levels, causing excessive iron accumulation and tissue damage in iron-accumulating tissues. Depending on the timing, severity, and extent of iron loading, iron-induced tissue injuries include hepatic fibrosis and cirrhosis,19 endocrinopathies such as diabetes,20 and cardiomyopathy.21 On the other end of the spectrum, inappropriately elevated hepcidin concentrations, also resulting from genetic mutations,22 can cause iron-deficiency anemia that does not improve in response to iron supplementation, referred to as iron-refractory iron-deficiency anemia (IRIDA).
Erythroid regulators of hepcidin and the discovery of erythroferrone
Early studies of iron absorption in the 1950s,23 predating the identification of the specific proteins known today to be involved in iron homeostasis, indicated that iron absorption greatly increased in response to many, but not all, types of anemia.24 Anemia and the resulting renal hypoxia increase erythropoietin (EPO) production25 and thereby promote the production of new red blood cells. Increased erythropoiesis generates a large iron demand because erythrocytes contain about 1 mg of iron per mL of packed red cells, much more than other cells in the body. Therefore, the ability to rapidly mobilize and absorb additional iron in response to increased erythropoietic activity would expedite recovery in response to hematopoietic stress, such as blood loss or hypoxia. Later studies indicated that liver hepcidin expression decreases in response to anemia and that this suppression of hepcidin was dependent on the presence of functional erythroid precursors, as hepcidin suppression in response to anemia was not observed in mice treated with either γ irradiation or chemicals toxic to developing erythrocytes.26,27 Initially factors such as growth differentiation factor 15 (GDF15) and twisted gastrulation BMP signaling modulator 1 (TWSG1) were identified as possible candidates for this erythroid regulator of hepcidin.28,29 However, bone marrow expression of TWSG1 is unaffected by phlebotomy and mice lacking GDF15 do not differ in their degree of hepcidin suppression in response to bleeding compared with wild-type mice,30 arguing against a major role for these proteins in driving the erythroid-mediated suppression of hepcidin during conditions of erythropoietic stress.
Screening of mouse bone marrow mRNA for genes differentially regulated in response to phlebotomy revealed the upregulation of the ERFE mRNA levels at time points preceding hepcidin suppression by phlebotomy.31 The gene Erfe was originally referred to as Fam132b and the product encoded by this gene was previously identified as a secreted protein and member of the C1q/TNF-related protein (CTRP) family, CTRP15, also termed myonectin.32 For the sake of clarity, in this review the gene will be referred to by its current HUGO nomenclature as ERFE (or Erfe for the mouse gene specifically) and its protein product as erythroferrone or ERFE. Follow-up experiments indicated that while Erfe is expressed to varying degrees in nonerythropoietic tissues, among the tissues analyzed, Erfe mRNA abundance increased after hemorrhage or erythropoietin stimulation only in the bone marrow and spleen,31 the primary sites of erythropoiesis and stress erythropoiesis in mice.33
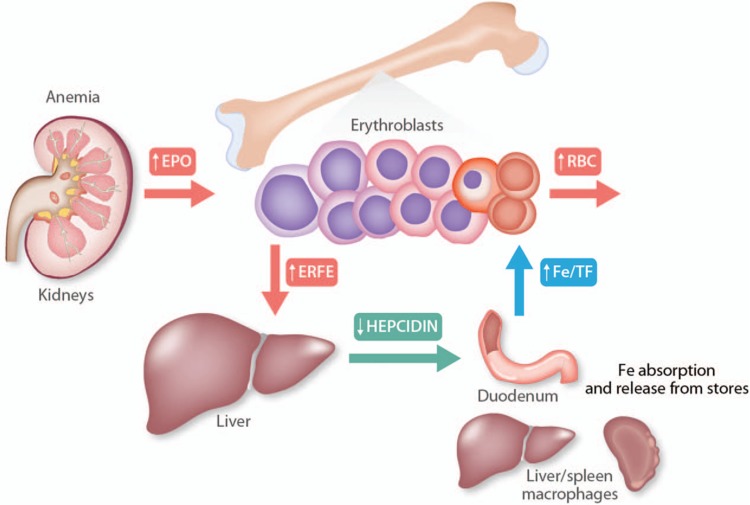
Studies in Erfe knockout mice showed that the loss of ERFE impairs the erythropoietic response during physiologic conditions such as rapid growth or after the pathological stress of blood loss, in which erythropoiesis is constrained by decreased iron availability, but that ERFE is not required for resting erythropoiesis in adult mice. Thus, ERFE is a mediator of the response to erythropoietic stress, suppressing hepcidin to promote the mobilization of stored iron and the absorption of dietary iron, so that the increased iron demands of developing erythrocytes can be met (Fig. 1). Specifically, relative to wild-type mice, mice lacking ERFE had reduced hemoglobin levels shortly after weaning, a time of rapid growth, and failed to suppress hepcidin in response to bleeding31 leading to impaired iron mobilization and delayed recovery from blood loss. In response to EPO, either endogenously produced after phlebotomy or administered by injection, erythroblasts at all stages of differentiation increased their expression of Erfe. This process was dependent on the activity of the STAT5 transcription factor and disruption of STAT5 signaling by inhibitor treatment prevented the induction of Erfe expression in response to EPO in vitro.31 The regulation of hepatocyte hepcidin expression by ERFE is likely direct, not requiring intermediate signaling by an additional cell type, as evidenced by in vitro experiments using primary hepatocytes and hepatocyte-derived cell lines in the absence of other hepatic nonparenchymal cell populations.31,34 The specific receptors that mediate the response of hepatocytes to ERFE are not yet known.
Figure 1.
Erythroferrone in systemic iron homeostasis. In response to anemia, increased EPO production by the kidney stimulates erythroblasts to increase the production of erythroferrone (ERFE), both because EPO increases the number of erythroblasts and because EPO increases the synthesis of ERFE by each erythroblast. Circulating ERFE acts directly on hepatocytes to suppress hepcidin production, leading to reduced plasma concentration of hepcidin. Low levels of circulating hepcidin allow the efflux of stored iron, primarily from macrophages and hepatocytes, as well as increased dietary iron absorption, so that more iron is loaded onto transferrin. Increased flows of plasma holotransferrin then deliver iron to erythroblasts for augmented heme and hemoglobin synthesis.
The role of erythroferrone in iron overload associated with ineffective erythropoiesis
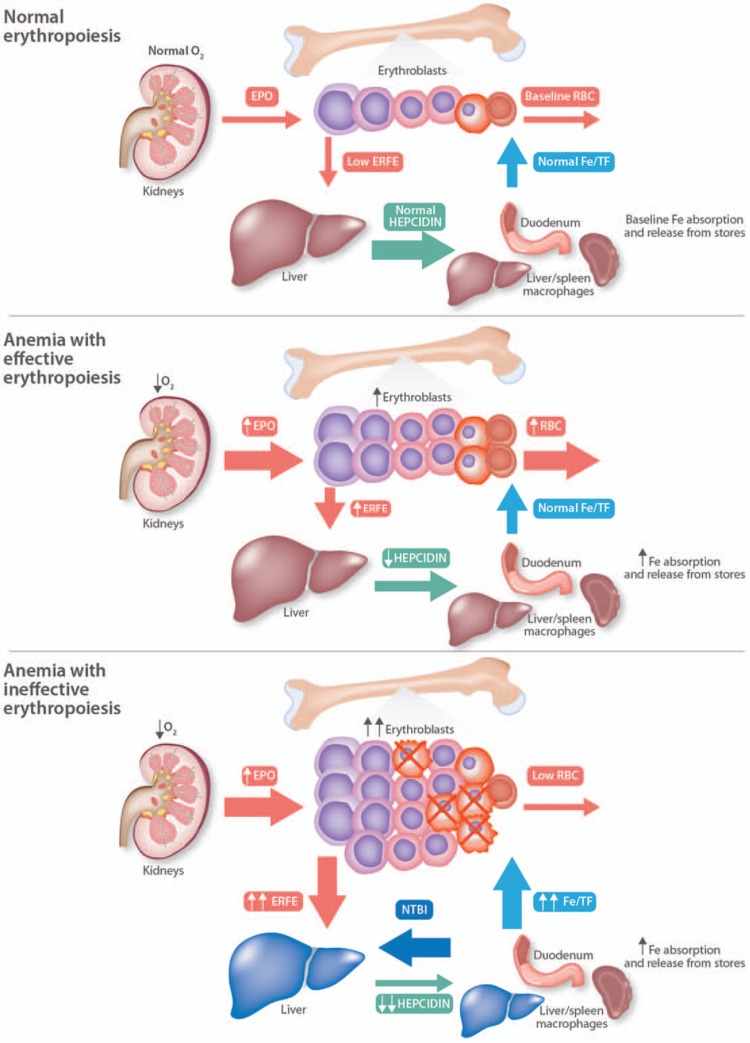
Because ERFE acts to suppress hepcidin in response to increased erythropoietic demand, it is a prime candidate to exert a similar role in conditions of dysfunctional erythropoiesis associated with iron overload (Fig. 2). Anemia in β-thalassemia intermedia and major, resulting from defective β-globin synthesis,35 is associated with decreased hepcidin expression,36 excessive iron absorption, and systemic iron overload.37 Deletion of Erfe in a mouse model of β-thalassemia (HbbHbbTh3/+)38 restores hepcidin expression to levels comparable with those measured in wild-type mice, and reduces tissue iron overload.39 The suppressive effect of ERFE on hepcidin expression in HbbHbbTh3/+ mice is most pronounced in young mice, before liver iron levels increase with age. In older HbbHbbTh3/+ mice hepcidin levels are comparable with wild-type mice but still inappropriately low relative to the amount of liver iron accumulated, which would normally increase hepcidin expression. The ability of ERFE to influence hepcidin in an animal model of chronically ineffective erythropoiesis also indicates that the action of ERFE influences hepcidin expression under long-term conditions as well as acute events that promote erythropoiesis, such as a single EPO injection or phlebotomy.31 Somewhat surprisingly, evidence suggests that in the context of ineffective erythropoiesis iron restriction can be beneficial, increasing hemoglobin as well as red cell quality and survival.40,41 However, while the absence of ERFE increases hepcidin levels, limiting not only iron accumulation but also available iron for erythropoiesis, in HbbTh3/+ mice this change does not improve anemia, perhaps because the resulting iron restriction is very mild.
Figure 2.
Erythroferrone in anemias with effective vs ineffective erythropoiesis. In anemias, decreased oxygen tension in the kidney drives increased erythropoietin production, which induces ERFE secretion by erythroblasts and thereby suppresses the hepatic production of hepcidin. In anemias with ineffective erythropoiesis, the erythroblast population is greatly expanded but many erythroblasts undergo apoptosis (indicated by X) before completing differentiation, so that relatively few erythrocytes are produced. Because there are many more erythroblasts when erythropoiesis is ineffective, ERFE production is further increased and the suppression of hepcidin is greater than in effective erythropoiesis. Very low hepcidin levels cause hyperabsorption of iron and the release of iron from macrophages. Transferrin saturation rises and the generation of nontransferrin-bound iron (NTBI) is increased. NTBI is taken up by the liver (depicted in blue) and other parenchymal organs, leading to organ damage.
In human β-thalassemia, serum ERFE concentrations are greatly increased, are transiently suppressed after erythrocyte transfusions, and inversely correlate with serum hepcidin concentrations,42 suggesting that ERFE contributes to the pathogenesis of iron overload by a similar mechanism as in the mouse model. Thus, future therapeutic interventions inhibiting the action of ERFE could be beneficial for preventing iron overload in β-thalassemia and other anemias with ineffective erythropoiesis. The contribution of ERFE to the pathogenesis of human β-thalassemia major could even be underestimated by the HbbTh3/+ mouse, as this mouse is a model of β-thalassemia intermedia. The HbbTh3/+ model generates much lower serum concentrations of ERFE than those measured in human β-thalassemia intermedia.39,42 Moreover, thalassemic mice continue to hyperabsorb iron even after iron accumulation in the liver raises hepcidin to wild-type concentrations, accumulating the additional iron in the spleen.39,43 This indicates that intestinal hyperabsorption of iron in murine β-thalassemia intermedia is resistant to inhibition by physiologic (but not therapeutic44) concentrations of hepcidin, perhaps because of upregulation of intestinal ferroportin by anemia and hypoxia.45 It is not known whether hepcidin resistance of similar severity develops in human β-thalassemia. These important differences between the mouse HbbTh3/+ thalassemia intermedia model and the human disease give hope that inhibition of ERFE in human β-thalassemia major could be even more effective than its ablation in the murine model of β-thalassemia intermedia. In transfusion-dependent patients, neutralization of ERFE is not expected to reverse iron overload from erythrocyte transfusions and would therefore have to be used in combination with iron chelators.
The effect of erythroferrone in anemia of inflammation and in host defense
In anemia of inflammation, resulting from infection or chronic inflammatory processes, such as autoimmune diseases, IL-6 and other cytokines increase hepcidin production.46 Increased hepcidin concentrations reduce intestinal iron absorption and cause iron retention in hepatocytes and iron-recycling macrophages, decreasing plasma iron concentrations and contributing to anemia. The main targets of this innate immune response are “siderophilic” microbes, that is, microbes whose pathogenicity is enhanced by iron, especially nontransferrin-bound iron (NTBI). The hypoferremic response, mediated by hepcidin, decreases extracellular iron concentrations, minimizes the concentration of NTBI, and protects against infections with siderophilic pathogens.47,48 However, anemia that develops during infections causes renal hypoxia, increasing EPO concentrations and thereby stimulating ERFE production, suppressing hepcidin, and releasing iron from hepatocytes and iron-recycling macrophages, potentially also providing more iron for siderophilic pathogens. Indeed, in a mouse-model of malaria infection, the ablation of Erfe was associated with reduced parasitemia but more severe anemia in the late stage of the disease compared with wild-type mice.49 While an effect on mortality was not reported, the decrease in hepcidin and increase in serum iron in response to the action of ERFE may be detrimental in combating malaria or pathogens that benefit from the availability of iron. In a model of anemia of inflammation caused by heat-killed Brucella abortus, a pathogen that is inflammatory but no longer capable of proliferation, the iron mobilization and increased absorption mediated by ERFE had a net beneficial effect by promoting erythropoiesis and recovery from anemia.50 Thus, ERFE accelerates recovery from anemia by releasing more iron, at the potential cost of stimulating certain infections that have not yet been completely controlled by the immune system or effective treatment.
Erythroferrone as myonectin: a role in nutrient sensing/signaling
Myonectin was identified as a secreted protein abundantly expressed in skeletal muscle and encoded by the gene Fam132b (now Erfe).32 Although the protein forms secreted by muscle and by erythroblasts have not been directly compared, published data suggest that the proteins are very similar or identical. Investigation into the metabolic role of myonectin in lipid homeostasis and nutrient signaling has indicated that myonectin levels rise with nutrient availability and that myonectin promotes the uptake of nonesterified fatty acids (NEFA) by adipocyte and hepatocyte-derived cell lines.32 Myonectin is also reported to suppress hepatic autophagy, mediating crosstalk between the muscle and liver dictated by conditions of nutrient availability.51 Studies focusing on the metabolic role of myonectin differ from those directed toward the role of ERFE in iron metabolism in that metabolic studies report a baseline level of serum myonectin in mice orders of magnitude higher, even without stimulation by EPO, phlebotomy, or ineffective erythropoiesis, than iron-related studies in which serum ERFE levels are nearly undetectable at baseline39,52 (unpublished observations). The lack of phenotypic difference between mice lacking ERFE and their wild-type littermate controls in parameters related to iron status, except during periods of rapid growth, argues in favor of serum ERFE levels reaching physiologic significance only in response to erythropoietic stimuli.31 The discrepancy between assays detecting myonectin and ERFE may be due to methodological differences. Reports on the role of myonectin in lipid metabolism and nutrient sensing determined serum levels by the immunoblotting of serum samples compared with a standard curve of recombinant myonectin whereas iron-centric studies have quantified serum ERFE by using validated ELISAs that detected no analyte in Erfe−/− mice. The original myonectin assay was not tested in Erfe−/− mice and so it is not certain whether the detected species was in fact a product of the Erfe gene. In view of all these questions, we advocate a reexamination of the metabolic role of myonectin/ERFE using Erfe−/− mice as controls.
A unifying context for the role of ERFE as an erythroid hepcidin/iron regulator and as a potential metabolic regulator is suggested by studies that report improvements in glucose tolerance or insulin sensitivity after chronic treatment with EPO,53,54 which stimulates the production of ERFE. However, the direct contribution of ERFE to these changes in glucose homeostasis has not been determined and evidence for a direct effect of EPO on glucose homeostasis, mediated through EPO receptor signaling in white adipose tissue, has been reported.55 Studies using ERFE knockout mice will be required to distinguish the impact of ERFE on glucose homeostasis, under basal and EPO-stimulated conditions, from the direct action of EPO.
Mechanism of erythroferrone-mediated hepcidin suppression
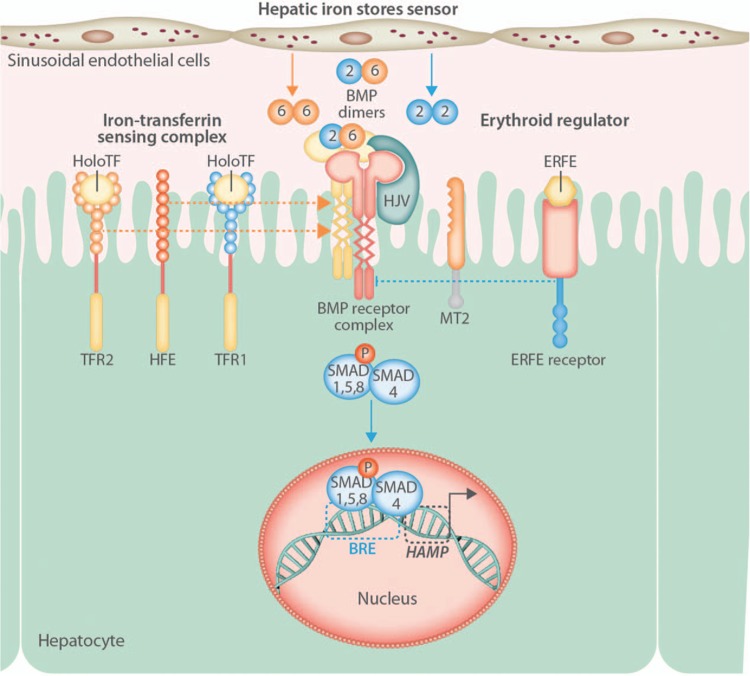
Most ERFE-related research thus far has focused on the physiologic effects of ERFE in the regulation of iron homeostasis. Less is currently known regarding the mechanism(s) by which the ERFE-mediated suppression of hepcidin occurs. Regulation of hepatocyte hepcidin expression is transcriptional and responds to iron status through the sensing of at least 2 inputs by hepatocytes: the iron-dependent secretion of bone morphogenetic proteins (BMPs) and the concentration of plasma holotransferrin (Fig. 3).56 Recent studies suggest that the iron-responsive source of BMPs (BMP2 and BMP6) are the neighboring hepatic sinusoidal endothelial cells.18,57 In hepatocytes, BMPs from sinusoidal endothelial cells, along with holotransferrin sensed by transferrin receptors 1 (TFR1) and 2 (TFR2), and the associated hemochromatosis protein HFE, activate pathways that converge on the phosphorylation of mothers against decapentaplegic (SMAD) homologs,17,58 specifically SMAD1/5/8 that form complexes with SMAD4. SMAD complexes acts as transcription factors and interact with BMP-responsive elements in the hepcidin promoter to stimulate hepcidin transcription.59,60 Initial reports indicated that mouse liver SMAD5 phosphorylation was unchanged in response to EPO or phlebotomy at a time point associated with maximal hepcidin suppression.31 However, more recent experiments using conditional knockout mice, in which the genes encoding both SMAD1 and SMAD5 had been deleted in hepatocytes, revealed that hepcidin suppression in response to EPO is mediated, in large part, through SMAD signaling and that treatment with ERFE suppresses SMAD1/5 phosphorylation in Hep3B cells, a hepatocyte-derived cell line.34 Additionally, hepcidin expression was not decreased in mouse hepatocytes lacking both SMAD1 and SMAD5 in response to treatment with ERFE. SMAD1 and SMAD5 likely have redundant functions in ERFE-mediated hepcidin suppression as liver hepcidin expression decreased in mice lacking either SMAD1 or SMAD5 in hepatocytes, to the same degree detected in wild-type mice, after treatment with EPO.34 The presence of SMAD8 is not sufficient to preserve ERFE signaling in the absence of SMAD1/5 but the effect of SMAD8 deletion on ERFE signaling has not been directly determined.
Figure 3.
Erythroferrone and the regulation of hepcidin synthesis in hepatocytes. Hepatic iron stores and the concentration of holotransferrin (HoloTF) regulate hepcidin (HAMP) transcription in hepatocytes. Dietary or parenteral iron loading results in increased circulating holotransferrin levels and increased secretion of bone morphogenetic protein (BMP) ligands by hepatic sinusoidal endothelial cells. BMP ligands interact with a BMP receptor complex comprised of type-1/2 BMP receptors, the BMP coreceptor hemojuvelin (HJV), and the serine protease matriptase-2 (MT2). HoloTF binds to transferrin receptors 1 (TFR1) and 2 (TFR2) dissociating HFE from TFR1 and facilitating stimulatory interactions of the BMP receptor complex with TFRs and HFE. The activation of BMP receptors causes phosphorylation of mothers against decapentaplegic (SMAD) homologs 1, 5, and 8. Phosphorylated SMAD1/5/8 form complexes with SMAD4 and translocate to the nucleus, where they act as transcription factors and interact with BMP-responsive elements (BRE) in the HAMP promoter, increasing hepcidin mRNA transcription. In response to erythropoietic stimulation, increased circulating levels of ERFE act, through an unknown receptor(s) and mechanism(s), to decrease SMAD1/5/8 phosphorylation in hepatocytes. Lower levels of phosphorylated SMAD1/5/8 result in less SMAD1/5/8 complex formation with SMAD4, diminishing its nuclear concentration, and decreasing HAMP mRNA transcription.
As evidence suggests that ERFE signaling occurs via the SMAD signaling pathway, various proteins that are known to regulate SMAD signaling can be considered logical candidates for the yet-unidentified ERFE receptor or other modifiers of ERFE signaling. The gene TMPRSS6 encodes the protein transmembrane protease serine 6, also known as matriptase 2 (MT2), a serine protease involved in the regulation of hepcidin expression.61 MT2 has attracted attention with regard to ERFE signaling because in Tmprss6−/− mice hepcidin was not suppressed in response to EPO.62 One postulated explanation for the lack of ERFE signaling in the absence of MT2 is that in this model SMAD signaling is hyperactivated to such an extent that the suppressive stimulus of ERFE is comparatively minor. This explanation is in line with data from mice fed high-iron diets, a treatment that increases SMAD phosphorylation,63 in which hepcidin suppression in response to bleeding is blunted.31 However, when Tmprss6−/− mice were combined with BMP6−/− mice, to decrease baseline SMAD phosphorylation and hepcidin expression to levels approximating those of wild-type mice, EPO treatment still failed to reduce liver hepcidin expression.62 Alone this evidence suggests an indispensable role for MT2 in ERFE signaling but is contradicted by data from isolated primary Tmprss6−/− hepatocytes that are responsive to ERFE in vitro,64 suggesting that the loss of ERFE signaling along with the absence of MT2 in vivo is attributable to other unidentified factors.
Phlebotomy also elicits a decrease in hepatic hepcidin expression in mice lacking hemojuvelin and TFR2,31 proteins involved in BMP/iron sensing and the regulation of SMAD activation,65,66 suggesting that these proteins are not required for ERFE signaling. Additionally, inhibition of the type-1 BMP receptor kinase activity by dorsomorphin marginally restores the ability of EPO to suppress hepatic hepcidin expression in Tmprss6−/− mice, arguing against the requirement for type-1 BMP receptors in ERFE signaling.67 The roles of other proteins involved in the regulation of SMAD activation/suppression have not yet been investigated in the context of ERFE-mediated hepcidin suppression.
Future directions
Identification of an erythroferrone receptor
Many fundamental questions remain unanswered regarding the mechanism(s) of ERFE signaling. To date no putative receptor for ERFE has been identified and little is known about the dependence of ERFE activity on its oligomerization and posttranslational modification. Other members of the CTRP family are demonstrated to interact with ERFE, and ERFE produced by overexpressing cells is reported to present as oligomeric complexes of varying size, similar to other CTRP proteins.32 Unpublished observations from our lab suggest that purified monomeric ERFE is less potent at suppressing hepcidin expression compared with unpurified ERFE found in overexpressing-cell supernatants, suggesting that the formation of oligomeric complexes may enhance ERFE signaling. Another area that has not been extensively investigated is the effect of ERFE on other cell populations besides hepatocytes. To date, ERFE signaling has focused primarily on hepatocytes and adipocytes,31,32,51 it remains to be seen whether ERFE signaling affects other cell types or if the effect of ERFE is restricted to select populations of cells. The discovery that ERFE suppresses SMAD signaling provides a metric to assess ERFE signaling in cell populations that do not express substantial amounts of hepcidin, potentially providing a new approach to identify elements of the ERFE signaling pathway and explore the effects of ERFE signaling in other cell populations. The action of ERFE in suppressing hepcidin production by nonhepatocyte cell populations is also of interest in light of recent evidence that hepcidin locally produced by cardiomyocytes regulates the iron status of these cells in an autocrine or paracrine fashion.68 If ERFE can modulate hepcidin expression in cell populations such as cardiomyocytes, where the effects of hepcidin are autocrine or paracrine rather than endocrine, then it may affect iron metabolism in cell types that are under the control of local, rather than systemic, hepcidin production. Other cell populations such as pancreatic β-cells,69 adipocytes,70 gastric parietal cells,71 and various cells types in the kidney72 are also reported to express hepcidin and warrant future examination of the effect of ERFE on local hepcidin production and tissue iron homeostasis.
Therapeutic control of erythroferrone signaling
Based on the lack of hepcidin suppression by erythropoietin in Tmprss6−/− mice,62 supraphysiological amounts of ERFE may be required to lower inappropriately high hepcidin in anemia of inflammation or iron-refractory iron deficiency anemia. Whether this is a feasible and safe approach to treating these disorders remains to be demonstrated. At the other end of the spectrum, anemias with ineffective erythropoiesis and low hepcidin leading to iron overload, dramatic improvements in lifespan and iron-overload related complications have been brought about by the use of chelation therapy, for example, in patients with β-thalassemia major. However, chronic iron accumulation still occurs and heart failure, attributed to cardiac iron loading, is a major cause of death in patients with β-thalassemia major who are noncompliant or do not tolerate current chelators.73 Evidence that ERFE is elevated in β-thalassemia, in both mice and humans,39,42 and that ERFE contributes to hepcidin suppression and the accompanying iron overload observed in this condition, makes ERFE a candidate target for therapeutic inhibition, especially because it is a “picomolar” hormone that can be neutralized by appropriate long-acting antibodies. Several anti-ERFE antibodies have been generated making these logical candidates for testing to confirm an inhibitory effect on ERFE signaling and determine the phenotypic effect in models of ineffective erythropoiesis associated with inappropriately low hepcidin and iron accumulation. High-throughput screening of small-molecule libraries to identify chemical inhibitors of ERFE signaling may also be a useful approach if preliminary testing of ERFE inhibition suggests a beneficial therapeutic effect. Such treatment may reduce excessive iron accumulation and the accompanying complications in patients with β-thalassemia, either in combination with chelators in β-thalassemia major, or alone in β-thalassemia intermedia where iron overload is largely or wholly dependent on the hyperabsorption of dietary iron.
Acknowledgments
The authors acknowledge the many discussions with our colleagues Leon Kautz and Elizabeta Nemeth that contributed importantly to the concepts discussed in this manuscript.
Footnotes
Citation: Coffey R, Ganz T. Erythroferrone: An Erythroid Regulator of Hepcidin and Iron Metabolism. HemaSphere, 2018;2:2. http://dx.doi.org/10.1097/HS9.0000000000000035
Funding: NIH/NIDDK R01 DK 065029 (Ganz) and NIH/NHLBI U54HL119893 (Palazzolo) to T.G., NIH/NHLBI T32 HL072752 (Dubinett) to R.C.
Disclosure: T.G. and UCLA have filed patent applications on the composition and use of erythroferrone. UCLA has issued licenses on the use of erythroferrone. T.G. is a scientific cofounder and consultant to Silarus Pharma and Intrinsic LifeSciences on projects involving erythroferrone.
R.C. and T.G. have written and edited the manuscript.
References
- 1.Chapter 2 Gross and elemental content of reference man. Ann ICRP/ICRP Publ 1975; OS_23:273–334. [Google Scholar]
- 2.Finch CA, Deubelbeiss K, Cook JD, et al. Ferrokinetics in man. Medicine 1970; 49:17–54. [DOI] [PubMed] [Google Scholar]
- 3.Han AP, Yu C, Lu L, et al. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 2001; 20:6909–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knutson MD, Oukka M, Koss LM, et al. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci U S A 2005; 102:1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donovan A, Lima CA, Pinkus JL, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab 2005; 1:191–200. [DOI] [PubMed] [Google Scholar]
- 6.Zumerle S, Mathieu JR, Delga S, et al. Targeted disruption of hepcidin in the liver recapitulates the hemochromatotic phenotype. Blood 2014; 123:3646–3650. [DOI] [PubMed] [Google Scholar]
- 7.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004; 306:2090–2093. [DOI] [PubMed] [Google Scholar]
- 8.Aschemeyer S, Qiao B, Stefanova D, et al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood 2018; 131:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos E, Kautz L, Rodriguez R, et al. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology 2011; 53:1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer DM, Wilkins SJ, Becker EM, et al. Hepcidin expression inversely correlates with the expression of duodenal iron transporters and iron absorption in rats. Gastroenterology 2002; 123:835–844. [DOI] [PubMed] [Google Scholar]
- 11.Pigeon C, Ilyin G, Courselaud B, et al. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 2001; 276:7811–7819. [DOI] [PubMed] [Google Scholar]
- 12.Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 1996; 13:399–408. [DOI] [PubMed] [Google Scholar]
- 13.Lee PL, Beutler E, Rao SV, et al. Genetic abnormalities and juvenile hemochromatosis: mutations of the HJV gene encoding hemojuvelin. Blood 2004; 103:4669–4671. [DOI] [PubMed] [Google Scholar]
- 14.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest 2005; 115:2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 2003; 33:21–22. [DOI] [PubMed] [Google Scholar]
- 16.Camaschella C, Roetto A, Cali A, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 2000; 25:14–15. [DOI] [PubMed] [Google Scholar]
- 17.Meynard D, Kautz L, Darnaud V, et al. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet 2009; 41:478–481. [DOI] [PubMed] [Google Scholar]
- 18.Koch PS, Olsavszky V, Ulbrich F, et al. Angiocrine Bmp2 signaling in murine liver controls normal iron homeostasis. Blood 2017; 129:415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loreal O, Deugnier Y, Moirand R, et al. Liver fibrosis in genetic hemochromatosis. Respective roles of iron and non-iron-related factors in 127 homozygous patients. J Hepatol 1992; 16:122–127. [DOI] [PubMed] [Google Scholar]
- 20.McClain DA, Abraham D, Rogers J, et al. High prevalence of abnormal glucose homeostasis secondary to decreased insulin secretion in individuals with hereditary haemochromatosis. Diabetologia 2006; 49:1661–1669. [DOI] [PubMed] [Google Scholar]
- 21.Niederau C, Fischer R, Sonnenberg A, et al. Survival and causes of death in cirrhotic and in noncirrhotic patients with primary hemochromatosis. N Engl J Med 1985; 313:1256–1262. [DOI] [PubMed] [Google Scholar]
- 22.Finberg KE, Heeney MM, Campagna DR, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet 2008; 40:569–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bothwell TH, Pirzio-Biroli G, Finch CA. Iron absorption. I. Factors influencing absorption. J Lab Clin Med 1958; 51:24–36. [PubMed] [Google Scholar]
- 24.Finch C. Regulators of iron balance in humans. Blood 1994; 84:1697–1702. [PubMed] [Google Scholar]
- 25.Koury MJ, Bondurant MC. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science 1990; 248:378–381. [DOI] [PubMed] [Google Scholar]
- 26.Pak M, Lopez MA, Gabayan V, et al. Suppression of hepcidin during anemia requires erythropoietic activity. Blood 2006; 108:3730–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vokurka M, Krijt J, Sulc K, et al. Hepcidin mRNA levels in mouse liver respond to inhibition of erythropoiesis. Physiol Res 2006; 55:667–674. [DOI] [PubMed] [Google Scholar]
- 28.Tanno T, Bhanu NV, Oneal PA, et al. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med 2007; 13:1096–1101. [DOI] [PubMed] [Google Scholar]
- 29.Tanno T, Porayette P, Sripichai O, et al. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood 2009; 114:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casanovas G, Vujic Spasic M, Casu C, et al. The murine growth differentiation factor 15 is not essential for systemic iron homeostasis in phlebotomized mice. Haematologica 2013; 98:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kautz L, Jung G, Valore EV, et al. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet 2014; 46:678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seldin MM, Peterson JM, Byerly MS, et al. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem 2012; 287:11968–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenox LE, Shi L, Hegde S, et al. Extramedullary erythropoiesis in the adult liver requires BMP-4/Smad5-dependent signaling. Exp Hematol 2009; 37:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang CY, Core AB, Canali S, et al. Smad1/5 is required for erythropoietin-mediated suppression of hepcidin in mice. Blood 2017; 130:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarjour RA, Murad H, Moasses F, et al. Molecular update of beta-thalassemia mutations in the Syrian population: identification of rare beta-thalassemia mutations. Hemoglobin 2014; 38:272–276. [DOI] [PubMed] [Google Scholar]
- 36.Kattamis A, Papassotiriou I, Palaiologou D, et al. The effects of erythropoetic activity and iron burden on hepcidin expression in patients with thalassemia major. Haematologica 2006; 91:809–812. [PubMed] [Google Scholar]
- 37.Pippard MJ, Callender ST, Warner GT, et al. Iron absorption and loading in beta-thalassaemia intermedia. Lancet 1979; 2:819–821. [DOI] [PubMed] [Google Scholar]
- 38.Yang B, Kirby S, Lewis J, et al. A mouse model for beta 0-thalassemia. Proc Natl Acad Sci U S A 1995; 92:11608–11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kautz L, Jung G, Du X, et al. Erythroferrone contributes to hepcidin suppression and iron overload in a mouse model of beta-thalassemia. Blood 2015; 126:2031–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nai A, Pagani A, Mandelli G, et al. Deletion of TMPRSS6 attenuates the phenotype in a mouse model of beta-thalassemia. Blood 2012; 119:5021–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo S, Casu C, Gardenghi S, et al. Reducing TMPRSS6 ameliorates hemochromatosis and beta-thalassemia in mice. J Clin Invest 2013; 123:1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganz T, Jung G, Naeim A, et al. Immunoassay for human serum erythroferrone. Blood 2017; 130:1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardenghi S, Marongiu MF, Ramos P, et al. Ineffective erythropoiesis in beta-thalassemia is characterized by increased iron absorption mediated by down-regulation of hepcidin and up-regulation of ferroportin. Blood 2007; 109:5027–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casu C, Oikonomidou PR, Chen H, et al. Minihepcidin peptides as disease modifiers in mice affected by beta-thalassemia and polycythemia vera. Blood 2016; 128:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson ER, Taylor M, Xue X, et al. Intestinal HIF2alpha promotes tissue-iron accumulation in disorders of iron overload with anemia. Proc Natl Acad Sci U S A 2013; 110:E4922–E4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemeth E, Rivera S, Gabayan V, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest 2004; 113:1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arezes J, Jung G, Gabayan V, et al. Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe 2015; 17:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefanova D, Raychev A, Arezes J, et al. Endogenous hepcidin and its agonist mediate resistance to selected infections by clearing non-transferrin-bound iron. Blood 2017; 130:245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latour C, Wlodarczyk MF, Jung G, et al. Erythroferrone contributes to hepcidin repression in a mouse model of malarial anemia. Haematologica 2017; 102:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kautz L, Jung G, Nemeth E, et al. Erythroferrone contributes to recovery from anemia of inflammation. Blood 2014; 124:2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seldin MM, Lei X, Tan SY, et al. Skeletal muscle-derived myonectin activates the mammalian target of rapamycin (mTOR) pathway to suppress autophagy in liver. J Biol Chem 2013; 288:36073–36082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Choesang T, Bao W, et al. Decreasing TfR1 expression reverses anemia and hepcidin suppression in beta-thalassemic mice. Blood 2017; 129:1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caillaud C, Mechta M, Ainge H, et al. Chronic erythropoietin treatment improves diet-induced glucose intolerance in rats. J Endocrinol 2015; 225:77–88. [DOI] [PubMed] [Google Scholar]
- 54.Katz O, Stuible M, Golishevski N, et al. Erythropoietin treatment leads to reduced blood glucose levels and body mass: insights from murine models. J Endocrinol 2010; 205:87–95. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, Teng R, Di L, et al. PPARalpha and Sirt1 mediate erythropoietin action in increasing metabolic activity and browning of white adipocytes to protect against obesity and metabolic disorders. Diabetes 2013; 62:4122–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin L, Valore EV, Nemeth E, et al. Iron transferrin regulates hepcidin synthesis in primary hepatocyte culture through hemojuvelin and BMP2/4. Blood 2007; 110:2182–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canali S, Zumbrennen-Bullough KB, Core AB, et al. Endothelial cells produce bone morphogenetic protein 6 required for iron homeostasis in mice. Blood 2017; 129:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramey G, Deschemin JC, Vaulont S. Cross-talk between the mitogen activated protein kinase and bone morphogenetic protein/hemojuvelin pathways is required for the induction of hepcidin by holotransferrin in primary mouse hepatocytes. Haematologica 2009; 94:765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casanovas G, Mleczko-Sanecka K, Altamura S, et al. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med (Berl) 2009; 87:471–480. [DOI] [PubMed] [Google Scholar]
- 60.Verga Falzacappa MV, Casanovas G, Hentze MW, et al. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med (Berl) 2008; 86:531–540. [DOI] [PubMed] [Google Scholar]
- 61.Silvestri L, Pagani A, Nai A, et al. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab 2008; 8:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nai A, Rubio A, Campanella A, et al. Limiting hepatic Bmp-Smad signaling by matriptase-2 is required for erythropoietin-mediated hepcidin suppression in mice. Blood 2016; 127:2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corradini E, Meynard D, Wu Q, et al. Serum and liver iron differently regulate the bone morphogenetic protein 6 (BMP6)-SMAD signaling pathway in mice. Hepatology 2011; 54:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aschemeyer S, Gabayan V, Ganz T, et al. Erythroferrone and matriptase-2 independently regulate hepcidin expression. Am J Hematol 2017; 92:E61–E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Babitt JL, Huang FW, Wrighting DM, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 2006; 38:531–539. [DOI] [PubMed] [Google Scholar]
- 66.Corradini E, Rozier M, Meynard D, et al. Iron regulation of hepcidin despite attenuated Smad1,5,8 signaling in mice without transferrin receptor 2 or Hfe. Gastroenterology 2011; 141:1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu PB, Hong CC, Sachidanandan C, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 2008; 4:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lakhal-Littleton S, Wolna M, Chung YJ, et al. An essential cell-autonomous role for hepcidin in cardiac iron homeostasis. Elife 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kulaksiz H, Fein E, Redecker P, et al. Pancreatic beta-cells express hepcidin, an iron-uptake regulatory peptide. J Endocrinol 2008; 197:241–249. [DOI] [PubMed] [Google Scholar]
- 70.Bekri S, Gual P, Anty R, et al. Increased adipose tissue expression of hepcidin in severe obesity is independent from diabetes and NASH. Gastroenterology 2006; 131:788–796. [DOI] [PubMed] [Google Scholar]
- 71.Schwarz P, Kubler JA, Strnad P, et al. Hepcidin is localised in gastric parietal cells, regulates acid secretion and is induced by Helicobacter pylori infection. Gut 2012; 61:193–201. [DOI] [PubMed] [Google Scholar]
- 72.Kulaksiz H, Theilig F, Bachmann S, et al. The iron-regulatory peptide hormone hepcidin: expression and cellular localization in the mammalian kidney. J Endocrinol 2005; 184:361–370. [DOI] [PubMed] [Google Scholar]
- 73.Borgna-Pignatti C, Cappellini MD, De Stefano P, et al. Survival and complications in thalassemia. Ann N Y Acad Sci 2005; 1054:40–47. [DOI] [PubMed] [Google Scholar]