Abstract
Supplemental Digital Content is available in the text
Several oncogenic fusion genes causing activation of the Abelson1 (ABL1) nonreceptor tyrosine kinase have been identified in leukemia. The most common fusion gene is BCR–ABL1 which is found in chronic myeloid leukemia (CML) and in 25% of adult B-cell acute lymphoblastic leukemia (B-ALL) patients.1,2 More recently, several variant ABL1 fusions have been identified in BCR–ABL1-like B-ALL.3 The BCR–ABL1 fusion is rarely found in T-cell acute lymphoblastic leukemia (T-ALL),4 but variant ABL1 fusions including ETV6–ABL1, NUP214–ABL1, and EML1–ABL1 have been described in about 6% of T-ALL cases.5–9
The BCR–ABL1 and ETV6–ABL1 fusions are constitutively activated tyrosine kinases depending on homo-oligomerization and interaction with the GRB2 adaptor for maximal kinase activity and full leukemogenic properties in vivo. Both fusion kinases induce a CML-like myeloproliferative disease (MPD) in bone marrow transplantation experiments.10,11 GRB2 binding is critical for the induction of a MPD in bone marrow transplant models: a BCR–ABL1 mutant lacking GRB2 binding was unable to induce short latency MPD in mice, but rather caused T-ALL or B-ALL with long latency.1 The NUP214–ABL1 fusion is a weaker oncoprotein, and induces T-ALL with a relatively long latency in a mouse bone marrow transplantation model.12 Unlike BCR–ABL1 and ETV6–ABL1, this fusion kinase does not rely on oligomerization for its oncogenic properties, but on its localization in the nuclear pore complex.12
In previous work, we have studied a T-ALL case with a cryptic translocation resulting in a fusion of EML1 to ABL1.8 We have shown that the EML1–ABL1 fusion protein is a constitutively active, imatinib sensitive tyrosine kinase that transforms Ba/F3 cells to growth factor independence.8 In this study, we investigated the in vivo oncogenic properties of EML1–ABL1 using a mouse bone marrow transplant model, and we analyzed the role of the coiled-coil domain and the interaction with GRB2 for EML1–ABL1 activity and transformation.
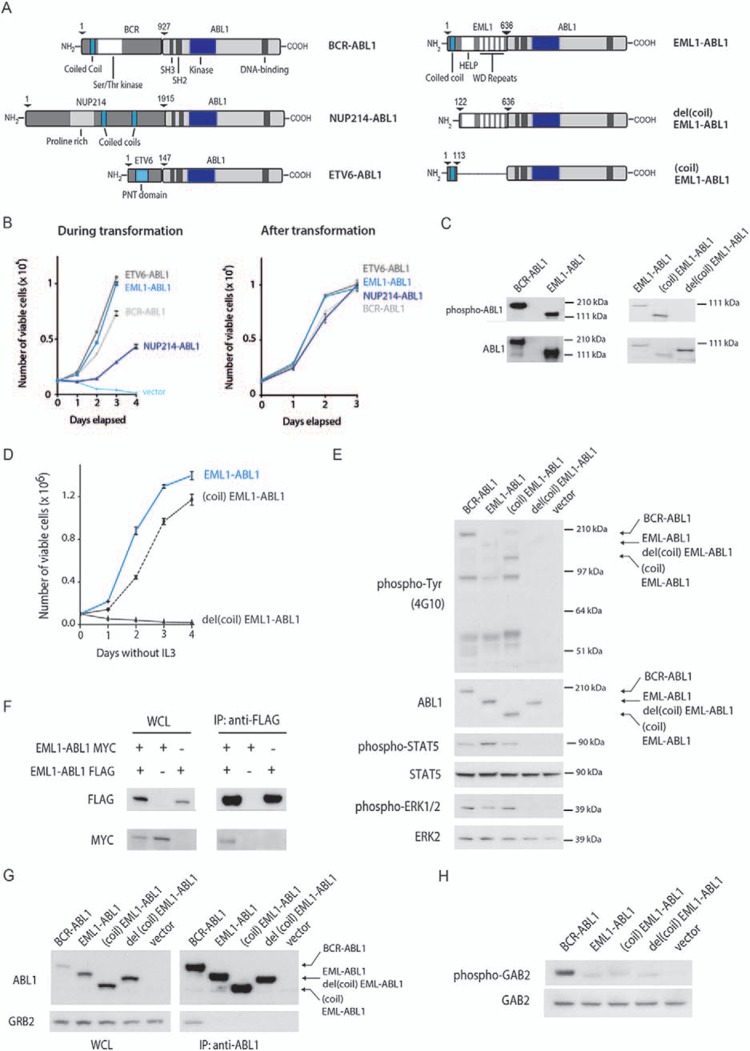
To characterize the EML1–ABL1 fusion protein in more detail, we compared the transformation rate of Ba/F3 cells expressing different ABL1 fusions (Fig. 1A). EML1–ABL1 and ETV6–ABL1 transformed Ba/F3 cells with the highest rate, followed by BCR–ABL1, while Ba/F3 cells expressing a NUP214–ABL1 fusion presented with a slower transformation kinetics. After transformation, Ba/F3 cells expressing the different ABL1 fusions displayed comparable growth rates (Fig. 1B). We consistently detected stronger autophosphorylation of BCR–ABL1 compared to EML1–ABL1 upon expression of the fusions in Ba/F3 and HEK293T cells (Fig. 1C and E).
Figure 1.
EML1–ABL1 kinase activity and transformation in vitro is dependent on the coiled-coil domain, but not on GRB2/GAB2 interaction. (A) Schematic representation of BCR–ABL1, NUP214–ABL1, ETV6–ABL1, EML1–ABL1, del(coil)EML1–ABL1, and (coil)EML1–ABL1 protein structure. (B) Left: Proliferation of Ba/F3 cells expressing different ABL1 fusions in the absence of IL3. Day 0 on the curve represents the day on which IL3 was removed. Before transformation, cells were either selected using puromycin or sorted based on GFP expression. Right: Growth rate after transformation of Ba/F3 cells expressing the different ABL1 fusions. All measurements were performed in technical triplicates. Data represent mean ± SD of a representative experiment. Growth curves were repeated at least 3 times. (C) Western blot analysis of autophosphorylation (antiphospho-ABL1 antibody) and expression levels (anti-ABL1 antibody) of indicated BCR–ABL1 and EML1–ABL1 fusions in HEK293T cells. (D) Proliferation of Ba/F3 cells expressing indicated constructs in the absence of IL3. Day 0 on the curve represents the day on which IL3 was removed. All measurements were performed in technical triplicates. Data represent mean ± SD of a representative experiment. Growth curves were repeated at least 3 times. (E) Western blot analysis of whole cell lysates of Ba/F3 cells expressing indicated ABL1 fusions. General tyrosine phosphorylation and expression levels of ABL1 fusions were assessed with antiphosphotyrosine (4G10) and anti-ABL1 antibodies respectively. Phosphorylation levels of ABL1 downstream signaling targets STAT5 and ERK1/2 were analyzed with phospho-specific antibodies against these proteins. (F) Coimmunoprecipitation (co-IP) experiment showing oligomerization of EML1–ABL1 molecules. HEK293T cells were transfected with the indicated constructs. Expression of the corresponding proteins is shown on the Western blots on whole cell lysates (WCL) on the left, the blots on the right correspond to the samples after anti-FLAG IP. (G) Anti-ABL1 IP of BCR–ABL1 and indicated EML1–ABL1 variants expressed in HEK293T cells. Expression of the ABL1 fusion proteins is shown on the Western blots on whole cell lysates (WCL) on the left, the blots on the right correspond to the samples after anti-ABL1 IP. Co-IP of GRB2 with these ABL1 fusions was investigated by probing the Western blot with anti-GRB2 antiserum. (H) Western blot analysis of GAB2 phosphorylation and expression levels on whole cell lysates of Ba/F3 cells expressing indicated ABL1 fusions.
To investigate whether the coiled-coil domain of EML1 is required and sufficient to initiate kinase activity and IL3 independent proliferation of Ba/F3 cells, we fused the coiled-coil domain of EML1 to ABL1 (Fig. 1A) and expressed this (coil)EML1–ABL1 construct in Ba/F3 and HEK293T cells. Expression of the (coil)EML1–ABL1 fusion protein resulted in IL3-independent proliferation of Ba/F3 cells, while a del(coil)EML1–ABL1 construct, lacking the coiled-coil domain of EML1 (Fig. 1A), was not able to transform Ba/F3 cells (Fig. 1D). In agreement with this, the (coil)EML1–ABL1 fusion protein, but not del(coil)EML1–ABL1, showed autophosphorylation in Ba/F3 and HEK293T cells and activated known downstream signaling proteins such as STAT5 and ERK1/2 in Ba/F3 cells (Fig. 1E). The necessity of the EML1 coiled-coil domain for EML1–ABL1 activity suggested that coiled-coil mediated oligomerization contributes to EML1–ABL1 activation. We confirmed oligomerization of EML1–ABL1 proteins by showing that MYC-tagged EML1–ABL1 could be coprecipitated with FLAG-tagged EML1–ABL1 (Fig. 1F).
For both BCR–ABL1 and ETV6–ABL1, binding of GRB2 to a phosphorylated tyrosine in respectively BCR or ETV6 was shown to be important for signaling through RAS and GAB2/PI3K/AKT. Moreover, interference with GRB2 association resulted in attenuation of their capacity to transform myeloid cells in vivo.11,13 In contrast, the NUP214–ABL1 fusion does not display detectable GRB2 interaction and GAB2 activation.12 We tested whether GRB2 binding may contribute to EML1–ABL1 activation and transformation by performing an anti-ABL1 coimmunoprecipitation. Although we could demonstrate interaction of GRB2 with BCR–ABL1, GRB2 interaction with EML1–ABL1 or (coil)EML1–ABL1 was undetectable (Fig. 1G). In addition, we analyzed phosphorylation of GAB2, an adaptor protein phosphorylated upon GRB2-mediated recruitment to BCR–ABL1.14 GAB2 phosphorylation was significantly stronger in BCR–ABL1 expressing Ba/F3 cells compared to EML1–ABL1 or (coil)EML1–ABL1 expressing cells (Fig. 1H). Altogether, these observations indicate that EML1–ABL1 does not activate GRB2/GAB2 signaling and that the oncogenic properties of EML1–ABL1 are not dependent on GRB2/GAB2 signaling.
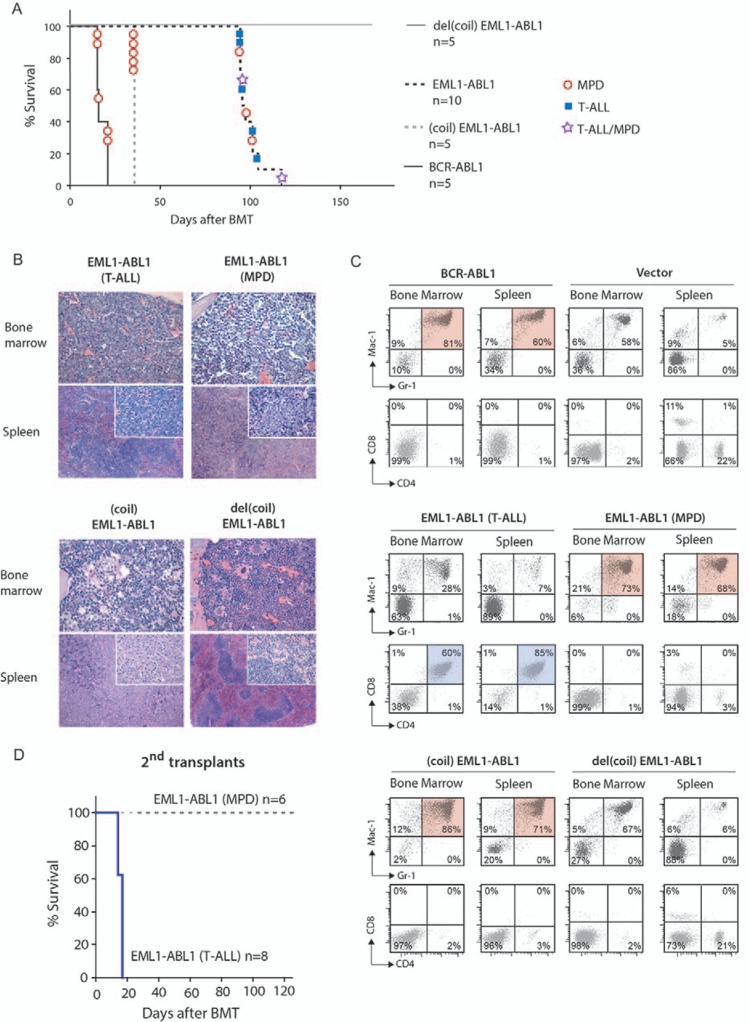
To compare the in vivo oncogenic properties of EML1–ABL1 and BCR–ABL1, mouse bone marrow cells were transduced with EML1–ABL1 or BCR–ABL1 and transplanted into irradiated syngeneic recipient mice. In agreement with previous reports, all 5 recipients of BCR–ABL1 transduced bone marrow cells developed a fatal CML-like MPD, 15 to 21 days after transplantation.15 In contrast, the 10 recipients of EML1–ABL1 transduced bone marrow cells developed a fatal disease after 96 to 119 days, a latency that was significantly longer compared to BCR–ABL1 (P < 0.001, Log-Rank test) (Fig. 2A, Supplemental Table 1, Supplemental Digital Content). Half (5/10) of the EML1–ABL1 transplanted animals developed T-ALL associated with enlargement of thymus and/or lymph nodes. Histologically, we found hypercellular bone marrow and expansion of splenic white pulp due to infiltration of lymphoblastic cells (Fig. 2B). By flow cytometry, we detected a significant population of GFP+ CD4+/CD8+ cells in bone marrow and spleen, which was completely absent in vector control mice (Fig. 2C, Supplemental Table 2, Supplemental Digital Content). A CML-like MPD was observed in 30% (3/10) of recipients of EML1–ABL1 transduced bone marrow (Fig. 2A). This disorder was similar to the BCR–ABL1 associated disease with an increased myelopoiesis with precursors and excessive amounts of granulocytes in bone marrow and spleen (Fig. 2B). Immunophenotyping of bone marrow and spleen cells demonstrated significant populations of GFP+ Mac-1+/Gr-1+ myeloid cells in these organs (Fig. 2C). The remainder 20% (2/10) of mice developed a mixed T-ALL/MPD (Fig. 2A, Supplemental Table 2, Supplemental Digital Content). Tumor cells from mice that developed EML1–ABL1-induced leukemia did not show active Notch1 signaling on Western blot and did not harbor Notch1 gene mutations (data not shown).
Figure 2.
The EML1–ABL1 fusion induces T-ALL or MPD in a bone marrow transplantation model. (A) Kaplan–Meier survival plot of primary recipients of transduced bone marrow cells. For (coil)EML1–ABL1, all mice were sacrificed at day 36 after transplantation. For del(coil)EML1–ABL1, the mice were followed until day 170 after transplantation. On this moment, these mice did not display any sign of disease and were sacrificed. (B) Hematoxylin and eosin (H&E) stained bone marrow biopsies and spleen sections of mice transplanted with bone marrow cells expressing the indicated EML1–ABL1 constructs. The EML1–ABL1 (T-ALL) mice display hypercellular bone marrow infiltrated by lymphoblastic cells. Also the spleen architecture was partially or completely replaced by lymphoblastic cell infiltrate. Bone marrow and spleen of EML1–ABL1 (MPD) and (coil)EML1–ABL1 animals were infiltrated with myeloid precursors, granulocytes and plasma cells (only for (coil)EML1–ABL1). No histological abnormalities were detected in the del(coil)EML1–ABL1 animal. (C) Single cell suspensions from spleen and bone marrow were stained with antibodies against Mac-1 and Gr-1 or CD4 and CD8 and were subsequently analyzed by FACS. Dead cells were excluded from the analysis based on 7-AAD staining. The percentage of cells with each respective immunophenotype is indicated in each quadrant. The presence of a significant CD4+/CD8+ cell population in bone marrow and spleen cells of some of the EML1–ABL1 mice indicated these animals had developed T-cell leukemia. The BCR–ABL1, the (coil)EML1–ABL1 and some of the EML1–ABL1 transplanted mice displayed increased percentages of Mac-1+/Gr-1+ cells in bone marrow and spleen typically observed in myeloid diseases. (D) Kaplan–Meier survival plot of mice receiving 106 leukemic EML1–ABL1 T-ALL or MPD spleen cells isolated from diseased primary recipients. The mice transplanted with MPD cells were followed for 120 days. On day 120, these animals did not display any sign of disease and were sacrificed.
We transplanted bone marrow cells derived from EML1–ABL1 associated T-ALL or MPD to 8 and 6 secondary recipient mice respectively. After 14 to 17 days, all animals transplanted with cells derived from T-cell leukemias developed again T-ALL. The disease was characterized by leukocytosis, splenomegaly and in 3/8 animals we observed paralysis of hind limbs. In contrast, animals transplanted with cells from EML1–ABL1 associated MPD did not develop any disease in a period of 120 days, in agreement with previous mouse models showing that MPD is not transplantable (Fig. 2D).
We also conducted the bone marrow transplant experiment with the EML1–ABL1 deletion variants (coil)EML1–ABL1 and del(coil)EML1–ABL1 (Fig. 1A). As expected, the 5 animals receiving del(coil)EML1–ABL1 transduced bone marrow cells did not develop any disease, since this deletion mutant has no kinase activity. Remarkably, all 5 recipients of (coil)EML1–ABL1 developed fatal CML-like MPD with leukocytosis, splenomegaly, and hemorrhages, and this after a latency of only 36 days. This latency was significantly longer than for the BCR–ABL1 associated disease but significantly shorter than for the EML1–ABL1 associated disease (P = 0.002 and 0.0002, respectively, Log-Rank test) (Fig. 2A). Histologically, bone marrow and spleen were infiltrated by maturing granulocytes and plasma cells (Fig. 2B). By immunophenotyping, we could demonstrate massive presence of GFP+ Mac1+/Gr1+ myeloid cells in bone marrow or spleen (Fig. 2C, Supplemental Table 2, Supplemental Digital Content). Taken together, our data show that the coiled-coil domain of EML1 is required and sufficient for the in vivo transforming properties of EML1–ABL1.
In summary, our results demonstrate that oligomerization of ABL1 is sufficient to activate its tyrosine kinase activity and to drive transformation of hematopoietic progenitor cells in vivo, but it also indicates that other factors further influence the strength of kinase activity and the capabilities to drive lymphoid versus myeloid transformation. For BCR–ABL1 and ETV6–ABL1, it was nicely demonstrated that GRB2 binding is important for myeloid cell transformation. In contrast, for EML1–ABL1, we show that GRB2 is not important, and that myeloid versus lymphoid transformation is influenced by the sites by which EML1 and ABL1 are fused together. When looking at the structure of EML1, we could not pinpoint any specific EML1-domain that could explain weaker kinase activity of EML1–ABL1 compared to (coil)EML1–ABL1. Since (coil)EML1–ABL1 induces only myeloid cell transformation, the drive toward lymphoid transformation in EML1–ABL1 might be due to the presence of specific domains in EML1. Furthermore, it is possible that full-length EML1–ABL1 interacts with proteins having an inhibitory effect on its kinase activity and that these interactions are not, or to a lesser extent, occurring for (coil)EML1–ABL1. Another possible explanation is that the conformation of the EML1–ABL1 protein is less favorable to support dimerization than that of (coil)EML1–ABL1.
Our data thus suggest that in addition to the binding of GRB2, also kinase activity and/or downstream signaling of ABL1 fusions are important determinants of disease phenotype. The mouse model for EML1–ABL1-induced T-ALL is of interest, as it comes with less technical challenges as compared to our previously described NUP214–ABL1 T-ALL model, which are related to the large size of NUP214–ABL1 and associated low transduction efficiencies.12 This EML1–ABL1 model can now be explored to study cooperating events in T-ALL pathogenesis and for testing novel therapies.
Supplementary Material
Footnotes
Citation: Vanden Bempt M, Mentens N, Vandenberghe P, Cools J, De Keersmaecker K. EML1–ABL1 Is Activated by Coiled-Coil-Mediated Oligomerization and Induces T-Cell Acute Lymphoblastic Leukemia or Myeloproliferative Disease in a Mouse Bone Marrow Transplant Model. HemaSphere, 2018;2:2. http://dx.doi.org/10.1097/HS9.0000000000000032
Funding/support: This work is supported by grants from the “Fonds voor Wetenschappelijk Onderzoek-Vlaanderen” and Kom op Tegen Kanker (J.C.). M.V.B. holds a PhD fellowship strategic basic research from the Research Foundation—Flanders (FWO).
Author contributions: M.V.B., J.C., and K.D.K. designed and performed research, analyzed data, and wrote the paper; N.M. performed research; P.V. provided reagents and expertise and wrote the paper.
The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Ren R. Mechanisms of BCR–ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 2005; 5:172–183. [DOI] [PubMed] [Google Scholar]
- 2.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol 2015; 12:344–357. [DOI] [PubMed] [Google Scholar]
- 3.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014; 371:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagemeijer A, Graux C. ABL1 rearrangements in T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer 2010; 49:299–308. [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos P, Ridge SA, Boucher CA, et al. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res 1995; 55:34–38. [PubMed] [Google Scholar]
- 6.Van Limbergen H, Beverloo HB, van Drunen E, et al. Molecular cytogenetic and clinical findings in ETV6/ABL1-positive leukemia. Genes Chromosomes Cancer 2001; 30:274–282. [PubMed] [Google Scholar]
- 7.Graux C, Cools J, Melotte C, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet 2004; 36:1084–1089. [DOI] [PubMed] [Google Scholar]
- 8.De Keersmaecker K, Graux C, Odero MD, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32). Blood 2005; 105:4849–4852. [DOI] [PubMed] [Google Scholar]
- 9.De Keersmaecker K, Lahortiga I, Graux C, et al. Transition from EML1-ABL1 to NUP214-ABL1 positivity in a patient with acute T-lymphoblastic leukemia. Leukemia 2006; 20:2202–2204. [DOI] [PubMed] [Google Scholar]
- 10.Million RP, Van Etten RA. The Grb2 binding site is required for the induction of chronic myeloid leukemia-like disease in mice by the Bcr/Abl tyrosine kinase. Blood 2000; 96:664–670. [PubMed] [Google Scholar]
- 11.Million RP, Harakawa N, Roumiantsev S, et al. A direct binding site for Grb2 contributes to transformation and leukemogenesis by the Tel-Abl (ETV6-Abl) tyrosine kinase. Mol Cell Biol 2004; 24:4685–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Keersmaecker K, Rocnik JL, Bernad R, et al. Kinase activation and transformation by NUP214-ABL1 is dependent on the context of the nuclear pore. Mol Cell 2008; 31:134–142. [DOI] [PubMed] [Google Scholar]
- 13.Pendergast AM, Quilliam LA, Cripe LD, et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell 1993; 75:175–185. [PubMed] [Google Scholar]
- 14.Sattler M, Mohi MG, Pride YB, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 2002; 1:479–492. [DOI] [PubMed] [Google Scholar]
- 15.Million RP, Aster J, Gilliland DG, et al. The Tel-Abl (ETV6-Abl) tyrosine kinase, product of complex (9;12) translocations in human leukemia, induces distinct myeloproliferative disease in mice. Blood 2002; 99:4568–4577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.