Supplemental Digital Content is available in the text
Abstract
The clinical use of histone deacetylase inhibitors (HDACi) for the treatment of bone marrow failure and hematopoietic malignancies has increased dramatically over the last decades. Nonetheless, their effects on normal myelopoiesis remain poorly evaluated. Here, we treated cord blood derived CD34+ progenitor cells with two chemically distinct HDACi inhibitors MS-275 or SAHA and analyzed their effects on the transcriptome (RNA-seq), epigenome (H3K27ac ChIP-seq) and functional and morphological characteristics during neutrophil development. MS-275 (entinostat) selectively inhibits class I HDACs, with a preference for HDAC1, while SAHA (vorinostat) is a non-selective class I/II HDACi. Treatment with individual HDACi resulted in both overlapping and distinct effects on both transcriptome and epigenome, whereas functional effects were relatively similar. Both HDACi resulted in reduced expansion and increased apoptosis in neutrophil progenitor cells. Morphologically, HDACi disrupted normal neutrophil differentiation what was illustrated by decreased percentages of mature neutrophils. In addition, while SAHA treatment clearly showed a block at the promyelocytic stage, MS-275 treatment was characterized by dysplastic features and skewing towards the monocytic lineage. These effects could be mimicked using shRNA-mediated knockdown of HDAC1. Taken together, our data provide novel insights into the effects of HDAC inhibition on normal hematopoietic cells during neutrophil differentiation. These findings should be taken into account when considering the clinical use of MS-275 and SAHA, and can be potentially utilized to tailor more specific, hematopoietic-directed HDACi in the future.
Introduction
Hematopoietic lineage choice and progression are complex and dynamic processes, tightly regulated through epigenetic changes and the expression of specific transcription factors and regulatory cytokines.1–9 Over the last decade, many epigenetic modulators regulating hematopoiesis have been identified, including networks that affect DNA methylation, micro RNA expression, and post transcriptional modifications such as histone acetylation.10–13 Histone acetylation is dynamically regulated by histone acetyltransferases (HATs), which are required for the deposition of the acetyl group, and histone deacetylases (HDACs), which are required for the removal of the acetyl group. This epigenomic modification associates with active regulatory DNA and may influence gene expression by chromatin remodeling and the subsequent recruitment of transcriptional regulators.14 In addition, several studies have shown that HATs and HDACs also (de)acetylate non-histone proteins such as transcription factors, which play an important role in hematopoietic differentiation.15–22
Given the transcriptional changes associated with neoplastic growth, the use of “epigenetic drugs” has dramatically increased as (adjuvant) treatment of (hematological) malignancies. These drugs include HDAC inhibitors (HDACi) which induce a variety of cellular consequences, including apoptosis, cell cycle arrest, differentiation, and autophagy.23–28 Furthermore, HDACi tend to have more pronounced effects on malignant cells than on healthy cells, making them excellent candidates for clinical use.29,30 In addition, the utility of HDACi are also explored in healthy human hematopoietic stem/progenitor cells derived from cord blood to increase the self-renewal capacity for their use in clinical transplantations.31,32 Nevertheless, the effects of HDACi treatment on lineage choice and progression in normal hematopoietic cells remains surprisingly underexplored.
The potent but distinct HDACi MS-275 and SAHA are well studied in cancer cell models, but their effect on normal hematopoiesis is not yet known. MS-275 has mainly been studied for the use in cutaneous T-cell lymphoma, breast and colon cancer.33–36 SAHA is used for treatment of multiple myeloid malignancies and other tumor types, including melanoma. These HDACi belong to different structural groups, thereby having different affinity and selectivity for HDACs.37 The benzamide MS-275 (entinostat) selectively inhibits class I HDACs, with a preference for HDAC1 and to a lesser extent HDAC3, while the hydroxamic acid SAHA non-selectively inhibits class I/II HDACs.37 Although their differences in inhibitory selectivity, both compounds are known to increase acetylation on histones H3 and H4, induce p21 expression and activate apoptotic pathways in malignant cells.38–42
In this study, we investigated the effects of HDAC inhibition on normal hematopoiesis using two distinct HDACi, MS-275 and SAHA. We performed transcriptomic, epigenomic, functional and morphological analysis during neutrophil development in the presence and absence of the HDACi MS-275 and SAHA. HDACi treatment resulted in both overlapping and differential effects on the transcriptome and epigenome, with similar functional outcomes. Both HDACi resulted in reduced expansion and increased apoptosis in neutrophil progenitor cells. Morphologically, HDACi disrupted normal neutrophil differentiation which was illustrated by decreased percentages of mature neutrophils. In addition, while SAHA treatment clearly showed a block at the promyelocytic stage, MS-275 treatment was characterized by dysplastic features and skewing towards the monocytic lineage. These effects could be mimicked using an shRNA-knockdown of HDAC1, suggesting a role for HDAC1 in determining cell fate decisions throughout neutrophil differentiation. Taken together, our data provide novel insights into the effects of HDAC inhibition on cell state transitions during normal hematopoiesis which may have consequences for their clinical use.
Results
Treatment with MS-275 and SAHA differentially regulates gene expression during neutrophil development
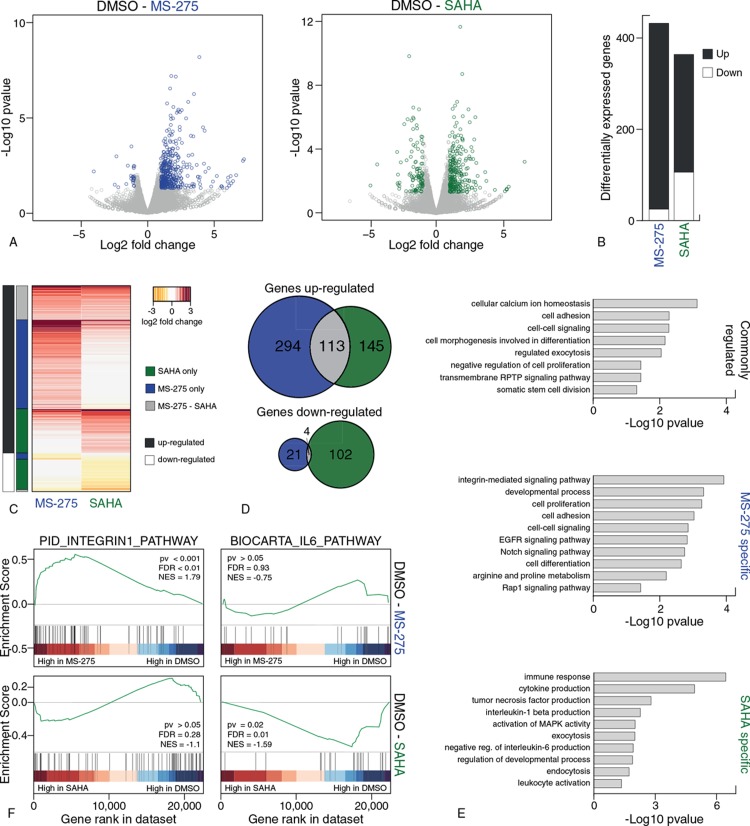
To evaluate the effects of HDAC inhibition during normal hematopoiesis, we differentiated CD34+ cells derived from umbilical cord blood towards mature neutrophils in the absence or presence of the class I specific HDACi MS-275 or the pan-HDACi SAHA. To analyze changes in the transcriptome caused by HDAC inhibition, CD34+ cells were differentiated for six days and treated overnight with MS-275, SAHA or DMSO, after which RNA-sequencing was performed (Supplemental Table 1, Supplemental Digital Content 1). Differential RNA expression analysis (DESeq2) of HDACi treatment compared to control DMSO resulted in 432 and 364 genes that were significantly differentially expressed (log2 fold change > 1, P < 0.05) after MS-275 and SAHA treatment respectively (Fig. 1A and B, Supplemental Table 2, Supplemental Digital Content 1). MS-275 treatment resulted predominantly in an up-regulation of genes, and this was also observed after SAHA treatment, albeit to a lesser extent (Fig. 1B). A minority of the genes showed expression changes upon both HDACi treatments, although almost 45% (113/258) of the genes that increased expression upon SAHA treatment were also found to be upregulated upon MS-275 treatment (Fig. 1C and D). Functional analysis of the gene sets using gene ontology revealed that commonly regulated genes were predominantly involved in processes such as ‘cell morphogenesis involved in differentiation’ and ‘negative regulation of cell proliferation’, the HDACi-specific genes were associated with distinct processes (Fig. 1E). Genes involved in integrin signaling were found to be overrepresented in MS-275 regulated genes which was further confirmed using Gene Set Enrichment Analysis, where the integrin pathway was enriched in MS-275 treated cells compared to DMSO and this was not observed upon SAHA treatment (Fig. 1F). Genes regulated by addition of SAHA were involved in cytokine production (Fig. 1E) including downregulation of the interleukin-6 production gene set, in contrast to no significant expression changes upon MS-275 treatment (Fig. 1F). Thus, besides overlapping transcriptional effects, HDACi treatment induced compound-specific changes illustrated by an anti-inflammatory gene expression profile upon SAHA treatment, and increased expression of genes involved in integrin signaling following MS-275 treatment.
Figure 1.
Treatment with MS-275 and SAHA differentially regulates gene expression during neutrophil development. (A) Volcano plot of log2 fold changes in gene expression vs −log10 p value derived from DESeq2 analysis of DMSO vs MS-275 (left) and DMSO versus SAHA (right) of duplicate RNA-seq experiments. Colored dots (blue: MS-275, green: SAHA) indicate genes significantly differentially expressed compared to DMSO (log2 fold change > 1, p value < 0.05). (B) Number of significantly differentially expressed genes (log2 fold change > 1, p value < 0.05) upon MS-275 treatment and SAHA treatment (Supplemental Table 2, Supplemental Digital Content 1). (C) Heatmap of log2 fold changes (from DESeq2 analysis) of all differentially expressed genes after MS-275 and SAHA treatment. Columns on the left indicate shared genes (grey), MS-275 specific genes (blue) or SAHA-specific genes (green), and up-regulated genes (black) or down-regulated genes (white). (C) Overlap of differentially expressed genes upon HDACi treatment. (E) Significant overrepresented Gene Ontologies identified by DAVID Gene Ontology for each gene set: commonly regulated genes (top), MS-275 specific genes (middle) and SAHA specific genes (bottom). (F) Example of gene sets significantly differential expressed according to Gene Set Enrichment Analysis after MS-275 treatment (top) and SAHA treatment (bottom) treatment compared to DMSO control. EGFR = epidermal growth factor receptor, FDR = false discovery rate, MAPK = mitogen-activated protein kinase, NES = normalized enrichment score, pv = p-value, RPTP = receptor protein tyrosine phosphatase.
MS-275 modulation of H3K27acetylation correlates with changes in gene expression
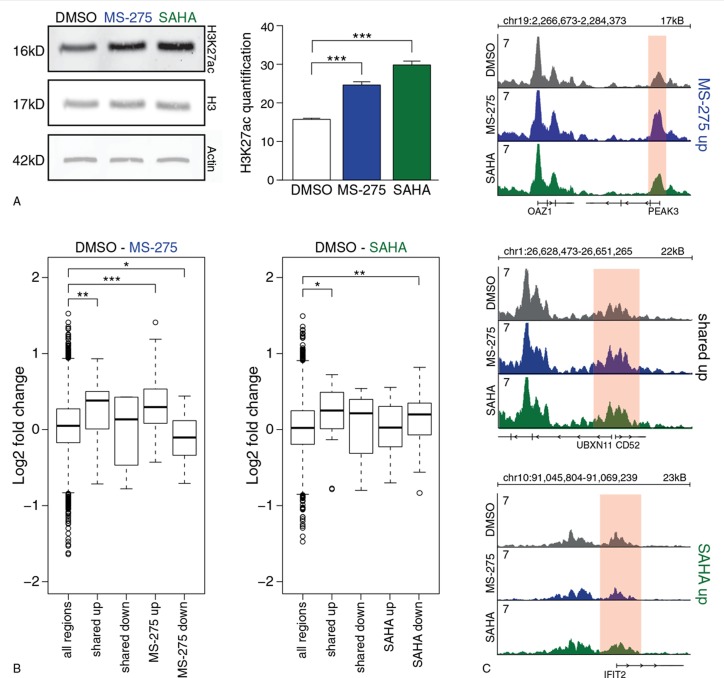
To link gene expression changes directly to changes in histone acetylation, we analyzed the enrichment of H3K27 acetylation (H3K27ac), which marks active non-coding regulatory DNA elements such as promoters and enhancers and is a substrate of HDACs. HDAC inhibition by MS-275 and SAHA resulted in a global increase in H3K27ac, most prominently following treatment with SAHA (Fig. 2A). To further analyze histone acetylation levels specifically on regions surrounding the differentially expressed genes, we analyzed H3K27ac on a genome wide scale using ChIP-sequencing. Analyzing promoters of differentially expressed genes, we observed significant differences in H3K27ac enrichment after both HDACi treatments compared to DMSO control (Fig. 2B). Promoters of up-regulated genes by MS-275 showed significant more H3K27ac enrichment compared to DMSO control (P < 0.001 for MS-275 specific genes, P = 0.008 for shared genes, Wilcoxon rank-sum test), while promoters of down-regulated genes showed lower H3K27ac enrichment (P = 0.039 for MS-275 specific genes only, Wilcoxon rank-sum test) (Fig. 2B and C). In contrast to MS-275, changes in H3K27ac enrichment following SAHA treatment did not correlate well with changes in gene expression. Promoters of commonly up-regulated genes were higher H3K27ac enriched after treatment (P = 0.013, Wilcoxon rank-sum test) and, surprisingly, promoters of SAHA down-regulated genes also showed higher H3K27ac enrichment (P = 0.002, Wilcoxon rank-sum test) (Fig. 2B and C). No significant differences were observed for promoters of SAHA up-regulated genes and commonly down-regulated genes (Fig. 2B and C). Thus, many of these genes are likely to be indirectly affected by HDACi.
Figure 2.
MS-275 modulation of H3K27acetylation correlates with changes in gene expression. (A) Quantification of three independent replicates of anti-H3K27acetylation Western Blot after overnight treatment at day 6 of CD34+ differentiation towards neutrophils. (B) Log2 fold changes of H3K27ac enrichment on promoters of commonly regulated genes, MS-275 specific (left) and SAHA specific differentially expressed genes (right) compared to DMSO control. Significance was calculated compared to all H3K27ac enriched regions. The boxes indicate the interquartile range with the center line representing the median value. The whiskers represent the maximum and minimum values and the outliers are plotted as dots. Significance is tested using a Wilcoxon rank-sum test, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (C) RPM normalized H3K27ac ChIP-seq tracks of promoters of genes that where differentially expressed in MS-275 specific (up), shared (middle) and SAHA specific (bottom). DMSO is visualized in black, MS-275 in blue and SAHA in green. Axis ranges from 0-7 RPM.
In addition to promoter acetylation, H3K27ac on distal regulatory elements (DREs), including enhancers, can also be regulated by HDACi treatment. Therefore, the closest DRE of each differentially expressed gene was analyzed for changes in acetylation enrichment. Similar as observed on promoters, HDACi treatment with MS-275 resulted in increased levels of H3K27ac on DREs of up-regulated genes and in decreased levels on DREs of down-regulated genes although the magnitude of this effect was attenuated (Supplemental Fig. 1, Supplemental Digital Content 2). SAHA treatment induced a significant increase of H3K27ac enrichment only on DREs of commonly up-regulated genes (Supplemental Figure, Supplemental Digital Content 2). Thus, whereas MS-275 treatment resulted in specific changes in H3K27acetylation that correlated with the changes in gene expression, SAHA treatment overall did not show this connection suggesting a larger proportion of the differentially expressed genes are indirectly affected.
MS-275 and SAHA inhibit expansion and induce apoptosis during neutrophil differentiation
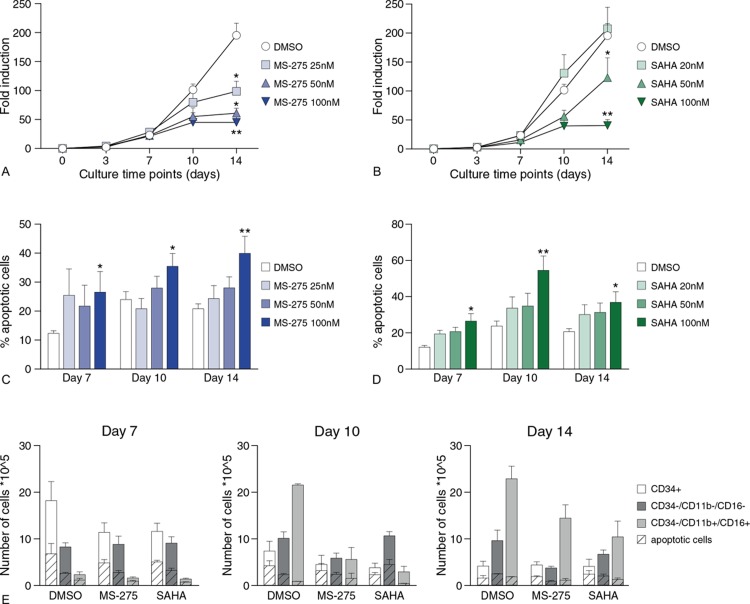
In addition to transcriptomic and epigenomic analysis, the functional effects of HDACi treatment regarding cell fate decisions were evaluated. Proliferation and apoptosis of neutrophil precursors were analyzed during neutrophil differentiation in the presence of HDACi. A concentration-dependent decrease was observed in the expansion of neutrophil progenitor cells upon treatment with MS-275 and SAHA (Fig. 3A and B). To evaluate whether decreased expansion was due to increased apoptosis, the fraction of cells that stained positive for Annexin V (early apoptosis) and propidium iodide (late apoptosis/necrosis) was analyzed. At all-time points, apoptosis was significantly increased in a concentration-dependent manner upon treatment with either MS-275 or SAHA (Fig. 3C and D). To further evaluate whether apoptosis occurred in a specific developmental stage during neutrophil differentiation, the number of Annexin V/propidium iodide-positive cells within the CD34+ and CD34- cell populations was measured. In addition, we separated the CD34-cell population in CD16/11b- and CD16/11b+ cells, reflecting the different stages through which the common myeloid progenitors evolve to mature neutrophils (Supplemental Fig. 2A, Supplemental Digital Content 2). Both HDACi induced apoptosis similarly throughout the three developmental stages, suggesting that apoptosis increased throughout whole neutrophil differentiation (Fig. 3E, Supplemental Fig. 2B, Supplemental Digital Content 2). Thus, MS-275 and SAHA decrease progenitor cell expansion during neutrophil differentiation, which is at least in part explained by an increase in apoptosis upon treatment with these drugs.
Figure 3.
MS-275 and SAHA inhibit expansion and induce apoptosis during neutrophil differentiation. (A, B) Fold induction of umbilical cord blood-derived CD34+ progenitor cells during neutrophil differentiation in the presence of 25 nM, 50 nM or 100 nM MS-275 (A), and 20 nM, 50 nM or 100 nM SAHA (B) and vehicle DMSO. Error bars represent SEM from three independent replicates, ∗P < 0.05, ∗∗P < 0.01. (C, D) Percentage of apoptotic cells (annexin-V / propidium iodide positive cells analyzed by FACS) of cells at days 7, 10 and 14 of neutrophil differentiation upon MS-275 treatment (C) and SAHA treatment (D). Error bars represent SEM from three independent replicates, ∗P < 0.05, ∗∗P < 0.01. (E) Absolute numbers (see Supplemental Fig. 2B, Supplemental Digital Content 2) for percentages) of viable and apoptotic cells (annexin-V / propidium iodide positive cells analyzed by FACS) after treatment with DMSO, 100 nM MS-275 or 100 nM SAHA at days 7, 10 and 14 of neutrophil differentiation in specific cell populations; i.e. CD34+ cells, including HSC and early progenitor cells; CD34-/CD16-/CD11b- cells, including immature neutrophils (promyelocytes, myelocytes, metamyelocytes; and CD34-/CD16+/CD11b+ cells, including mature neutrophils (banded and segmented). Error bars represent SEM from three independent replicates.
MS-275 and SAHA differentially modulate neutrophil differentiation
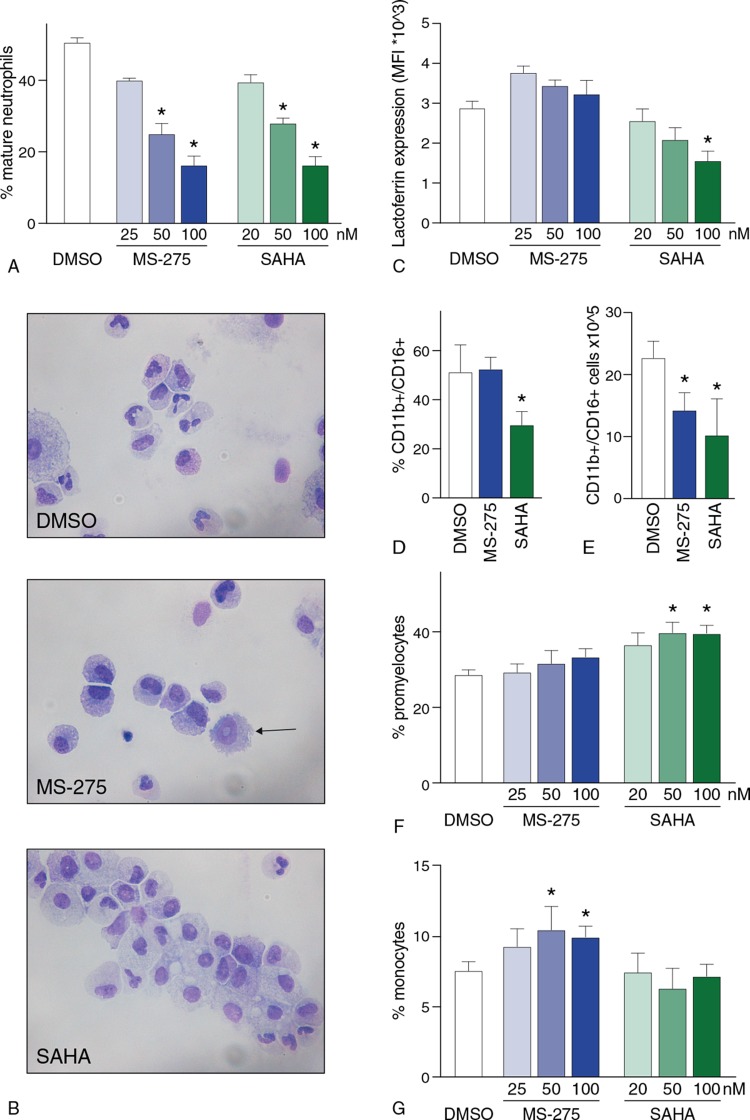
We subsequently analyzed whether MS-275 and SAHA treatment interfered with normal neutrophil differentiation by analysis of morphological features, neutrophil surface marker expression and intracellular lactoferrin staining. At day 14 of neutrophil development, cytospin analysis demonstrated a concentration-dependent decrease of mature neutrophils upon treatment with both MS-275 and SAHA (Fig. 4A and B, Supplemental Fig. 3A, Supplemental Digital Content 2). In addition, SAHA treatment showed a decrease in lactoferrin expression, a marker of secondary granule formation and reflecting functionally mature neutrophils (Fig. 4C). A decrease in the percentage of CD16/CD11b+ cells after terminal differentiation upon treatment with SAHA was observed, while this percentage remained similar upon MS-275 treatment compared to control DMSO (Fig. 4D). In addition, we observed a decrease in the absolute numbers of CD16/CD11b+ upon treatment with both MS-275 and SAHA (Fig. 4E), as expected by the observed decrease in expansion (Fig. 3). Furthermore, a relative accumulation of promyelocytes was observed upon SAHA-treatment, at the expense of further differentiation. This accumulation was also observed after treatment with MS-275, although not significant (Fig. 4B-F, Supplemental Fig. 3B, Supplemental Digital Content 2). On the other hand, MS-275 treatment resulted in an increased number of cells with dysplastic features, such as a ring-shaped nucleus and increased percentage of monocytes (Fig. 4B, 4G, Supplemental Fig. 3C, Supplemental Digital Content 2).
Figure 4.
MS-275 and SAHA differentially modulate neutrophil differentiation. (A) Percentage of morphologically mature neutrophils (either banded or segmented nuclei) at day 14 of HDACi treatment. Mature neutrophils were counted using cytospin analysis as described in the methods section. Absolute numbers can be found in Supplemental Fig. 3A, Supplemental Digital Content 2). Error bars represent SEM from three independent replicates, ∗P < 0.05. (B) Micrographs of cells at day 14 of neutrophil differentiation upon HDACi treatment (100 nM) or vehicle DMSO. A cell with dysplastic features is indicated with an arrow. Cells are stained according to May-Grünwald Giemsa staining. (C) Mean fluorescent intensity (MFI) after lactoferrin staining at day 14 of HDACi treatment analyzed by FACS. Error bars represent SEM from three independent replicates, ∗P < 0.05. (D-E) Percentages (D) and absolute numbers (E) of CD11b+/CD16+ cells at day 14 of neutrophil differentiation after treatment with DMSO, 100 nM MS-275 or 100 nM SAHA. Error bars represent SEM from three independent replicates, ∗P < 0.05. (F-G) Percentage of promyelocytes (F) and monocytes (G) at day 14 of neutrophil differentiation upon treatment with vehicle DMSO, MS-275 or SAHA. Absolute numbers can be found in Supplemental Fig. 3B-C, Supplemental Digital Content 2. Error bars represent SEM from three independent replicates, ∗P < 0.05.
Taken together, both treatment with MS-275 and SAHA disrupted normal neutrophil differentiation what was illustrated by decreased percentages of mature neutrophils. In addition, while SAHA treatment clearly showed a block at the promyelocytic stage, MS-275 treatment was characterized by dysplastic features and skewing towards the monocytic lineage.
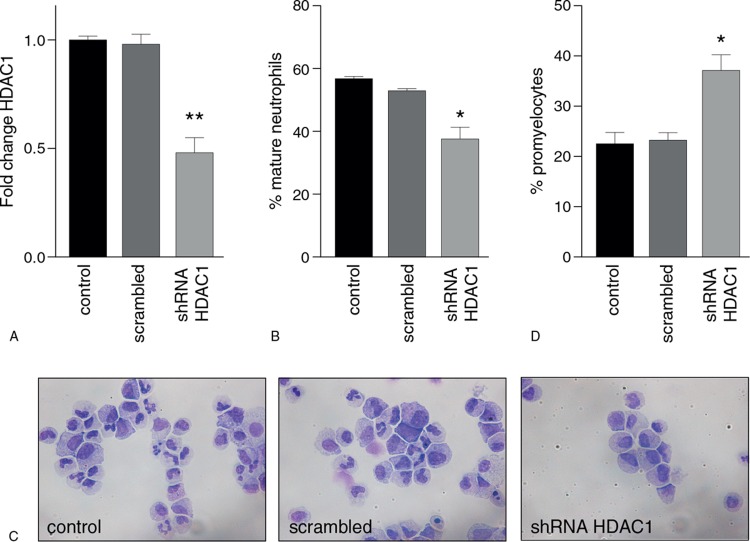
HDAC1 is required for neutrophil differentiation
Since both MS-275 and SAHA predominantly inhibit HDAC1 activity, the role of HDAC1 in terminal neutrophil differentiation was investigated by using an shRNA knockdown approach. CD34+ cells were transduced with an shRNA for HDAC1, resulting in a 50% decrease in mRNA expression (Fig. 5A), and differentiated into mature neutrophils. A significant decrease in the percentage of mature neutrophils at the end of differentiation was observed upon knockdown of HDAC1 (Fig. 5B and C). In addition, an increase in the percentage of promyelocytes was observed (Fig. 5C and D). These findings are similar to those observed upon treatment with the HDACi MS-275 and SAHA. Furthermore, the percentage of monocytes was also increased compared to the control (Supplemental Fig. 4, Supplemental Digital Content 2). However, upon transduction with the control shRNA, we also observed an increase in the percentage of monocytes, albeit without a decrease in mature neutrophils or an increase in promyelocytes. This therefore most likely reflects aspecific effects of retroviral manipulation of CD34+ progenitor cells. Summarized, we observed a similar outcome in morphological analysis upon HDAC1 knockdown compared with both HDACi, suggesting a role for HDAC1 during neutrophil differentiation which is affected by HDACi treatment.
Figure 5.
HDAC1 is required for neutrophil differentiation. (A) Fold change in HDAC1 expression, normalized for β2-microglobulin, after HDAC1 knockdown, scrambled and control. (B) Percentage of mature neutrophils after HDAC1 shRNA knockdown and scrambled control. Mature neutrophils were counted using cytospin analysis as described in the Materials and Methods. (C) Micrographs of cells at day 14 of neutrophil differentiation for control, scrambled and HDAC1 knockdown. Cells are stained according to May-Grünwald Giemsa staining. (D) Percentage of promyelocytes after HDAC1 shRNA knockdown and scrambled control. Error bars represent SEM from 3 independent replicates, ∗P < 0.05.
Discussion
In this study, we investigated the effects of HDAC inhibition on normal neutrophil differentiation by performing transcriptomic, epigenomic, functional and morphological analysis. CD34+ progenitor cells derived from umbilical cord blood were differentiated into mature neutrophils in the presence of class I specific HDACi MS-275 or pan-HDACi SAHA. HDAC inhibition generally resulted in decreased expansion rate, increased apoptosis, and decreased percentage of mature neutrophils. While SAHA treatment clearly showed a block at the promyelocytic stage, MS-275 treatment was characterized by dysplastic features and skewing towards the monocytic lineage. Since both HDACi inhibit HDAC1 activity, and knockdown of HDAC1 resulted in similar effects as HDACi treatments, our data suggest a role for HDAC1 in lineage progression and terminal neutrophil differentiation. This has indirectly been implicated in previous studies by using class I HDACi.43,44
Interestingly it appears that while HDACi-treatment results in distinct epigenetic and transcriptional changes with some overlap, functional phenotypic effects are generally similar. The differential effects on the transcriptome and epigenome can be explained in part by the fact that MS-275 preferentially targets HDAC1, whereas SAHA also inhibits class I/II HDACs. Naturally, nonspecific effects such as oxidative stress or DNA damage might play a role as well, but this was beyond the scope of our study.45 While our study is limited regarding sample size for the RNA-seq experiments, it does provide important insights into the transcriptional effects of the HDACi in terms of transcriptional regulation. Future experiments are now required to further confirm the effects on expression of individual genes. Genes down-regulated upon SAHA treatment were involved in cytokine production, specifically of tumor necrosis factor-alpha (TNF-α)-associated cytokines, and interleukins (IL) 1 and 6. This might indicate an anti-inflammatory response which has been observed in other studies.46 There is much debate about the role of interleukins and other cytokines in neutrophil development. In particular the direct effect of IL-6 on neutrophil function remains poorly understood, illustrated by conflicting evidence in the literature reporting that IL-6 can either delay, accelerate or have no effect on neutrophil apoptosis.47–50 On the other hand, HDAC inhibition using MS-275 resulted in increased expression of integrins, including CD11c and CD11d. Activation of integrins upon HDACi-treatment has also been previously described.51–53 While integrins, in multiple cells including neutrophils, are mostly known for their role in adhesion at the site of inflammation and transition to extravascular tissue, integrin signaling has also been proven to be of importance in other cellular processes, including degranulation, cytokine production, and activation of transcription through the nuclear transcription factor kappa B (NF-κB) pathway. In addition to its role in innate immunity, NF-κB signaling was shown to control a great variety of other well conserved cellular processes, including cell proliferation and apoptosis.54–57
Historically, two models concerning hematopoietic lineage commitment have been proposed. A deterministic model posits that lineage commitment and differentiation occurs through signaling by specific cytokines. This extrinsic motivation triggers uncommitted cells to differentiate into a given cell type. A second stochastic model describes that lineage commitment is driven by lineage specific transcription factors, that modulate expression of lineage specific genes including cytokines and cytokine receptors, making these cytokines select the fate of committed cells.2–7,58–64 Based on this, it is possible that upon treatment with SAHA, cytokine production is regulated through several HDACs, but independently of HDAC1 function, resulting in uniquely modulated gene expression profiles of these cytokines for SAHA compared to MS-275. Furthermore, this could well be the explanation for the differences observed in our functional studies regarding dysplastic features and differentiation blockage at the promyelocyte stage of neutrophil development.
In our study we evaluated the enrichment of H3K27ac, since this histone modification is a widely accepted mark for active gene regulatory elements.65 While changes in H3K27ac levels correlated with gene expression after MS-275 treatment, SAHA treatment also resulted in increased acetylation levels on promoters of down-regulated genes. This can potentially be explained by increased sensitivity of specific genomic regions to hyperacetylation, which results in the abolishment of transcription factor recruitment and proper chromatin remodeling, leading to decreased gene expression.66 ChIP-seq experiments with spike-in controls will further help evaluate global effects on H3K27ac, which is possibly affected more by SAHA compared to MS-275 treatment, as illustrated by the Western blot analysis (Fig. 2A). In addition, while our study was limited by the analysis of only one histone mark, acetylation of other histone residues or non-histone proteins are probably also of importance.65,67 This has been demonstrated in certain leukemia, where both hyperacetylation and hypoacetylation have been implicated in disease outcome.68–72 Apart from targeting acetylation on histones, HDACi are able to deacetylate non-histone proteins.70 For example, it has been shown that deacetylation of TAF9 by HDAC1 is required for expression of PU.1, a key regulator in hematopoietic lineage specification. In addition, acetylation of CCAAT/enhancer binding protein ε (C/EBPε) has been shown to be required for normal neutrophil development.71–73 These types of responses could also play a role in the differential changes in the transcriptome that we observe between treatment with MS-275 and SAHA.
Although HDACs are globally involved in the growth and differentiation of mammalian cells, relatively little is known about their specific roles in hematopoiesis, particularly in humans. It has been demonstrated that the expression of HDAC1 in hematopoiesis is dependent on the developmental stage of cells and plays a direct role in lineage choice.74 During myeloid development, HDAC1 expression is inhibited by CCAAT/enhancer binding proteins, while HDAC1 is activated by GATA-1 during erythro-megakaryocytic development. In relation to this, HDAC1-knockdown, or lower HDAC1 expression, resulted in impaired myeloid differentiation.74,75 This is in line with our study, demonstrating that inhibition of HDAC1 by two distinct HDACi resulted in impaired neutrophil differentiation.
Our data provide novel insights into the effects of HDAC inhibition on normal hematopoietic cells during neutrophil differentiation. Two distinct HDACi disrupt neutrophil development through alternative and overlapping effects on the transcriptome and epigenome, potentially mediated by HDAC1. These findings should be taken into account when considering the clinical use of MS-275 and SAHA, and can be potentially utilized to tailor more specific, hematopoietic-directed HDACi in the future.
Materials and Methods
Ethics approval and consent to participate
Umbilical cord blood (UCB) was collected after written informed consent was provided according to the Declaration of Helsinki. The use of UCB for this study was approved by the ethics committee of the University Medical Center Utrecht.
Isolation and culture of human CD34+ progenitor cells
CD34+ progenitor cells were isolated from UCB as described previously.44,73 Cells (1 × 105/mL) were cultured in Iscove's Modified Dulbecco's Medium (IMDM), and were differentiated towards neutrophils upon addition of SCF (50 ng/mL), FLT3 (50 ng/mL), IL-3 (100 nmol/mL), GM-CSF (100 nmol/mL) and G-CSF (30 ng/mL) on day 0. On day 3 and 7 only G-CSF was added. On days 3, 7, 10, and 14, cells were counted with trypan blue, and fresh medium was added to a density of 3 × 105/mL on day 3, and 5 × 105/mL on day 7 and onwards. The HDACi MS-275 (25, 50, and 100 nM) and SAHA (20, 50, and 100 nM) were freshly added with each change of medium.
RNA-sequencing experiments
CD34+ cells were differentiated towards neutrophils for 6 days, followed by overnight treatment with MS-275 (100 nM), SAHA (100 nM) or vehicle DMSO. The next day, total RNA was extracted using the RNAeasy Kit (QIAGEN). RNA quality was tested on the Bioanalyzer (Agilent, Santa Clara, CA), and sample quality was optimal with RNA integrity number higher than 9.0. Sample preparation was performed using a Poly(A)Purist MAG Kit (Thermo Scientific) according to the manufacturer's instructions. Isolated mRNA was subsequently purified using an mRNA-ONLY Eukaryotic mRNA Isolation Kit (Epicentre Illumina, Madison, WI). Sequencing libraries were prepared using a SOLiD Total RNA-Seq Kit (Applied Biosystems Life Technologies) according to the standard protocol recommendations and sequenced on a SOLiD 5500 Wildfire sequencer to produce 50 bp reads as described previously.76 Multimapped and duplicate reads were removed using Samtools version 1.5. Reads in genes were counted using HTSeq.77 Noncoding genes (list from HUGO Gene Nomenclature Committee) were removed from the analysis. Read counts (prior to normalization) are deposited in Supplemental Table 1, Supplemental Digital Content 1. Library size normalization and differential gene expression analysis was performed using the DESeq2 package in R. Genes were significantly differentially expressed with a log2 fold change of > 1 and a p value < 0.05. Heatmaps were generated using the heatmap.2 function from gplots in R. Gene Ontology analysis tool DAVID (version 6.8, https://david.ncifcrf.gov)78,79 was used to identify overrepresented gene ontologies in our gene sets. To analyze complete gene sets that significantly changed upon HDACi treatment, we used the Gene Set Enrichment Analysis (https://software.broadinstitute.org/gsea/index.jsp) tool.
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) experiments
CD34+ cells were differentiated towards neutrophils for 6 days, followed by overnight treatment with MS-275 (100 nM), SAHA (100 nM) or vehicle DMSO. ChIP-seq was performed as described previously utilizing an anti-acetylated H3K27 antibody (ab4729, Abcam, Cambridge, MA).76 Chromatin was sheared, end-repaired, followed by ligation of sequencing adaptors and amplification of the library by ligation ± mediated PCR (LMPCR). After LMPCR, the library was purified, checked for the proper size range and for the absence of adaptor dimers on a 2% agarose gel, followed by sequencing on the SOLiD/AB sequencer (Applied Biosystems Life Technologies, Carlsbad, CA). Sequencing reads were mapped against the reference genome (hg19, NCBI3) using the BWA package (−c ± l 25 ± k 2 ± n 10).80 Non-uniquely placed reads were discarded. Statistical significant H3K27ac enriched regions compared to background were calculated for each sample using MACS2 version 2.1.1.2016030981 (p value < 10−5, extsize 300, local lambda 100,000) and extended to 2000 bp in length (+/− 1000 bp from peak center). Peaks from all samples were merged into one non-redundant list for further analysis, where overlapping peaks were merged. Reads in peaks were counted using Bedtools version 2.26.082 and fraction of reads in peaks per sample exceeded the 1% threshold used by the Encyclopedia of DNA Elements (ENCODE).83 Enriched regions that overlapped within 1000 bp from a transcriptional start site from the hg19 RefSeq list were annotated as promoters. For analysis of differences in H3K27ac enrichment, read counts were normalized for library size and peak size (reads per kilobase per million (RPKM)). Statistical analysis of differential H3K27acetylation enrichment was analyzed using a Wilcoxon rank-sum test in R. ChIP-seq panels were visualized in Integrated Genomics Viewer (IGV version 2.3.40, Broad Institute).84,85
Western blot analysis
Western blot analysis was performed using standard techniques. In brief, differentiating CD34+ progenitors were lysed in Laemmli buffer [0.12 mol/L Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, 0.05 μg/μL bromophenol blue, and 35 mmol/L β-mercaptoethanol], sonicated, and boiled for 5 minutes. Equal amounts of total lysate were analyzed by 12% sodium dodecylsulfate polyacrylamide gel electrophoresis. Proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA), incubated with blocking buffer (Tris buffered saline/Tween 20) containing 5% low-fat milk for 1 hour at room temperature before incubation with antibodies against acetyl-histone 3 (lysine 27) (Millipore, Billerica, MA) or histone 3 (Millipore, Billerica, MA) overnight at 4°C in a buffer containing Tris buffered saline/Tween 20 with 5% bovine serum albumin (BSA) (Sigma-Aldrich, Zwijndrecht, the Netherlands). Blots were subsequently incubated with peroxidase-conjugated secondary antibodies (Dako, Glostrup, Denmark) for 1 hour at room temperature. Chemiluminescence was used as a detection method according to the protocol of the manufacturer (Odyssey, Amersham Pharmacia, Amersham, UK).
Measurement of fold expansion and apoptosis
Cells were counted at days 3, 7, 10, and 14 to analyze fold expansion. Cells were stained with Trypan Blue (Thermo Fisher Scientific) and Trypan Blue negative cells were counted using a Fuchs-Rosenthal chamber. To measure apoptosis during HDACi treatment, cells were harvested at the indicated time points and washed with phosphate-buffered saline (PBS). Samples were subsequently incubated for 20 minutes with annexin V-fluorescein isothiocyanate (Bender MedSystems, Vienna, Austria) in binding buffer [10 mmol/L HEPES-NaOH (pH 7.4), 150 mmol/L NaCl, 2.5 mmol/L CaCl2] before being washed and resuspended in binding buffer containing 1 μg/mL propidium iodide (Bender MedSystems). Percentages of apoptotic cells were determined by FACS analysis (FACS Canto, Becton Dickinson, Alphen a/d Rijn, the Netherlands), as previously described.44,73
Flowcytometric analysis of myeloid progenitor cells
After 3, 7, 10 and 14 days of differentiation, cells were washed and resuspended in PBS/5% FCS (Hyclone) and subsequently incubated for 30 minutes on ice with a PE-conjugated CD34 antibody (Becton Dickinson), FITC-conjugated CD11b antibody (Becton Dickinson) and an APC-conjugated CD16 antibody (Becton Dickinson). After incubation, cells were again washed in PBS/5% FCS (Hyclone) and the percentages of CD34+, CD11b+ and CD16+ cells were determined by flow cytometry analysis (FACS Canto, Becton Dickinson).
Histochemical staining of hematopoietic cells
May-Grünwald Giemsa staining was used to analyze myeloid differentiation. After 14 days of neutrophil differentiation, cytospins were prepared from 5.0 × 104 differentiating granulocytes and were fixed in methanol for 3 minutes. After fixation, cytospins were stained in a 50% eosin methylene blue solution according to May-Grünwald (Sigma Aldrich, Seelze, Germany) for 15 min, rinsed in water for 10 seconds, and nuclei were counterstained with 10% Giemsa solution (Merck kGaA, Darmstadt, Germany) for 20 minutes. Neutrophil differentiation occurs through distinct stages from myeloblasts, promyelocytes I, promyelocytes II, myelocytes and metamyelocytes towards neutrophils with banded or segmented nuclei (Supplemental Figure 2A, Supplemental Digital Content 2). Mature neutrophils were characterized as cells containing either banded or segmented nuclei. Micrographs were acquired, after staining with May-Grünwald Giemsa solution, with an Axiostar plus microscope (Carl Zeiss, Sliedrecht, the Netherlands) fitted with an 100×/1.3 NA EC Plan Neofluor oil objective using Immersol 518F oil (Carl Zeiss), a Canon Powershot G5 camera (Canon Nederland, Hoofddorp, the Netherlands), and Canon Zoombrowser EX image acquisition software.
Lactoferrin staining
To analyze lactoferrin levels after 14 days of neutrophil differentiation, cells were fixed in 100 μL 0.5% formaldehyde for 15 minutes at 37°C, after which the cells were permeabilized in 900 μL of ice-cold methanol for 30 minutes on ice. Cells were subsequently washed with PBS, resuspended with phycoerythrin (PE)-conjugated lactoferrin antibody (Immunotech, Marseille, France) and incubated for 20 minutes. Cells were then washed again and FACS analysis was performed (FACS Canto, Becton Dickinson), as described before.73
shRNA viral transduction of CD34+ progenitor cells
A lentiviral construct was used containing shRNA control (Sigma-Aldrich) or shRNA targeting HDAC1 (Sigma-Aldrich) and a GFP fluorescent gene in the pLKO.1 vector. HEK293FT cells were grown in 10-cm dishes at 37°C and 5% CO2, in DMEM supplemented with 10% FBS, 2 mM glutamine and antibiotics (50 U/ml penicillin and 50 mg/ml streptomycin). After 1 day, lentivirus was produced by co-transfection of 5 μg of pLKO.1 vector containing shRNA, 1.8 μg of lentiviral packaging vector pLP/VSVG, and 3.25 μg of lentiviral packaging vector psPAX2 overnight using 50 μl of PEI (Polysciences Inc., Warrington, PA). The next day, medium was replaced, and the cells were cultured for 24 hours. The supernatant containing virus was collected and filtered through a 0.2-μm filter. Transduction was performed by adding 1 ml of viral supernatant, 1 ml of culturing medium, and 8 μg/ml Polybrene to CD34+ progenitor cells. After 1 day, the cells were washed with PBS, and new culturing medium was added. Selection was achieved by fluorescence-activated cell sorting on GFP. Expression changes of HDAC1 upon knockdown were analyzed using RT-qPCR, and normalized for β2-microglobulin.
Statistical analysis
Statistical analysis what involved multiple concentrations of HDACi and control DMSO was performed using a one-way analysis of variance test, followed by a Dunnet multiple comparison test (comparison with the control) (Prism GraphPad Software). P values of 0.05 or less were considered significant (∗P < 0.05; ∗∗P < 0.01, ∗∗∗P < 0.001).
Supplementary Material
Supplementary Material
Footnotes
Citation: Govers AMAP, Wiggers CRM, van Boxtel R, Mokry M, Nieuwenhuis EES, Creyghton MP, Bartels M, Coffer PJ. Transcriptomic and Epigenomic Profiling of Histone Deacetylase Inhibitor Treatment Reveals Distinct Gene Regulation Profiles Leading to Impaired Neutrophil Development. HemaSphere, 2019;00:00–00. http://dx.doi.org/10.1097/HS9.0000000000000270.
AMAPG and CRMW contributed equally to this work.
MB and PJC contributed equally as senior authors.
AMAPG and RB performed experiments. CRMW and MM analyzed data. AMAPG, CRMW, MB and PJC wrote the manuscript. MPC and EESN provided intellectual input for the manuscript. All authors read and approved the manuscript.
This research was supported by the Dutch Cancer Society (DSC, Grant Number: 4855), and the Foundation Friends of the UMC Utrecht/Wilhelmina Children's Hospital.
The authors declare no conflicts of interest.
References
- 1.Kondo M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol Rev. 2010;238:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rieger MA, Hoppe PS, Smejkal BM, et al. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. [DOI] [PubMed] [Google Scholar]
- 3.Pina C, Fugazza C, Tipping AJ, et al. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14:287–294. [DOI] [PubMed] [Google Scholar]
- 4.Endele M, Etzrodt M, Schroeder T. Instruction of hematopoietic lineage choice by cytokine signaling. Exp Cell Res. 2014;329:207–213. [DOI] [PubMed] [Google Scholar]
- 5.Cvejic A. Mechanisms of fate decision and lineage commitment during haematopoiesis. Immunol Cell Biol. 2016;94:230–235. [DOI] [PubMed] [Google Scholar]
- 6.Hoppe PS, Schwarzfischer M, Loeffler D, et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535:299–302. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Kostadima M, Martens JHA, et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014;345: 1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho MSH, Medcalf RL, Livesey SA, et al. The dynamics of adult haematopoiesis in the bone and bone marrow environment. Br J Haematol. 2015;170:472–486. [DOI] [PubMed] [Google Scholar]
- 10.Kurkewich JL, Hansen J, Klopfenstein N, et al. The miR-23a∼27a∼24-2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 2017;13:e1006887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss CN, Ito K. A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. Int Rev Cell Mol Biol. 2017;334:99–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menon V, Ghaffari S. Transcription factors FOXO in the regulation of homeostatic hematopoiesis. Curr Opin Hematol. 2018;25:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Rooij LPMH, Chan DCH, Keyvani Chahi A, et al. Post-transcriptional regulation in hematopoiesis: RNA binding proteins take control. Biochem Cell Biol. 2019;97:10–20. [DOI] [PubMed] [Google Scholar]
- 14.Seto E, Yoshida M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheer C, Kratz C, Witt O, et al. Hematologic response to vorinostat treatment in relapsed myeloid leukemia of down syndrome. Pediatr Blood Cancer. 2016;63:1677–1679. [DOI] [PubMed] [Google Scholar]
- 16.Valiuliene G, Stirblyte I, Jasnauskaite M, et al. Anti-leukemic effects of HDACi Belinostat and HMTi 3-Deazaneplanocin A on human acute promyelocytic leukemia cells. Eur J Pharmacol. 2017;799:143–153. [DOI] [PubMed] [Google Scholar]
- 17.Burianova I, Kuzelova K, Mitrovsky O, et al. Histone deacetylase inhibitors in plasma cell leukemia treatment: effect of bone marrow microenvironment. Neoplasma. 2017;64:228–237. [DOI] [PubMed] [Google Scholar]
- 18.Lyberg K, Ali HA, Grootens J, et al. Histone deacetylase inhibitor SAHA mediates mast cell death and epigenetic silencing of constitutively active D816V KIT in systemic mastocytosis. Oncotarget. 2017;8:9647–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noack K, Mahendrarajah N, Hennig D, et al. Analysis of the interplay between all-trans retinoic acid and histone deacetylase inhibitors in leukemic cells. Arch Toxicol. 2017;91:2191–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morabito F, Voso MT, Hohaus S, et al. Panobinostat for the treatment of acute myelogenous leukemia. Expert Opin Investig Drugs. 2016;25:1117–1131. [DOI] [PubMed] [Google Scholar]
- 21.Rücker FG, Lang KM, Fütterer M, et al. Molecular dissection of valproic acid effects in acute myeloid leukemia identifies predictive networks. Epigenetics. 2016;11:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowe JM. AML in 2017: Advances in clinical practice. Best Pract Res Clin Haematol. 2017;30:283–286. [DOI] [PubMed] [Google Scholar]
- 23.Robert T, Vanoli F, Chiolo I, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong F, Miller KM. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res. 2013;750:23–30. [DOI] [PubMed] [Google Scholar]
- 25.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog. 2015;20:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. [DOI] [PubMed] [Google Scholar]
- 28.Sui X, Zhu J, Zhou J, et al. Epigenetic modifications as regulatory elements of autophagy in cancer. Cancer Lett. 2015;360:106–113. [DOI] [PubMed] [Google Scholar]
- 29.Chang J, Varghese DS, Gillam MC, et al. Differential response of cancer cells to HDAC inhibitors trichostatin A and depsipeptide. Br J Cancer. 2012;106:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolden JE, Shi W, Jankowski K, et al. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis. 2013;4:e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam YM, Chan YF, Chan LC, et al. Histone deacetylase inhibitors induce leukemia gene expression in cord blood hematopoietic stem cells expanded ex vivo. Int J Hematol. 2017;105:37–43. [DOI] [PubMed] [Google Scholar]
- 32.Chaurasia P, Gajzer DC, Schaniel C, et al. Epigenetic reprogramming induces the expansion of cord blood stem cells. J Clin Invest. 2014;124:2378–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Copeland A, Buglio D, Younes A. Histone deacetylase inhibitors in lymphoma. Curr Opin Oncol. 2010;22:431–436. [DOI] [PubMed] [Google Scholar]
- 34.Lemoine M, Younes A. Histone deacetylase inhibitors in the treatment of lymphoma. Discov Med. 2010;10:462–470. [PubMed] [Google Scholar]
- 35.Connolly RM, Rudek MA, Piekarz R. Entinostat: a promising treatment option for patients with advanced breast cancer. Futur Oncol. 2017;13:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapani D, Esposito A, Criscitiello C, et al. Entinostat for the treatment of breast cancer. Expert Opin Investig Drugs. 2017;26:965–971. [DOI] [PubMed] [Google Scholar]
- 37.Ververis K, Hiong A, Karagiannis TC, et al. Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents. Biologics. 2013;7:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jóna A, Khaskhely N, Buglio D, et al. The histone deacetylase inhibitor entinostat (SNDX-275) induces apoptosis in Hodgkin lymphoma cells and synergizes with Bcl-2 family inhibitors. Exp Hematol. 2011;39:1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21 CIP1/WAF1 1. Cancer Res. 2003;63:3637–3645. [PubMed] [Google Scholar]
- 40.Bracker TU, Sommer A, Fichtner I, et al. Efficacy of MS-275, a selective inhibitor of class I histone deacetylases, in human colon cancer models. Int J Oncol. 2009;4:909–920. [DOI] [PubMed] [Google Scholar]
- 41.Richon VM, Sandhoff TW, Rifkind RA, et al. Histone deacetylase inhibitor selectively induces p21 WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci. 2000;97:10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Zhao Y, Gou WF, et al. The anti-tumor effects and molecular mechanisms of suberoylanilide hydroxamic acid (SAHA) on the aggressive phenotypes of ovarian carcinoma cells. PLoS One. 2013;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawson ND, Berliner N. Neutrophil maturation and the role of retinoic acid. Exp Hematol. 1999;27:1355–1367. [DOI] [PubMed] [Google Scholar]
- 44.Bartels M, Geest CR, Bierings M, et al. Histone deacetylase inhibition modulates cell fate decisions during myeloid differentiation. Haematologica. 2010;95:1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Leite de Oliveira R, Huijberts S, et al. An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell. 2018;173:1413–1425. [DOI] [PubMed] [Google Scholar]
- 46.Leoni F, Zaliani A, Bertolini G, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci. 2002;99:2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrante A. Activation of neutrophils by interleukins-1 and -2 and tumor necrosis factors. Immunol Ser. 1992;57:417–436. [PubMed] [Google Scholar]
- 48.Biffl WL, Moore EE, Moore FA, et al. Interleukin-6 suppression of neutrophil apoptosis is neutrophil concentration dependent. J Leukoc Biol. 1995;58:582–584. [DOI] [PubMed] [Google Scholar]
- 49.Afford SC, Pongracz J, Stockley RA, et al. The induction by human interleukin-6 of apoptosis in the promonocytic cell line U937 and human neutrophils. J Biol Chem. 1992;267:21612–21616. [PubMed] [Google Scholar]
- 50.McNamee JP, Bellier PV, Kutzner BC, et al. Effect of pro-inflammatory cytokines on spontaneous apoptosis in leukocyte sub-sets within a whole blood culture. Cytokine. 2005;31:161–167. [DOI] [PubMed] [Google Scholar]
- 51.Wedel S, Hudak L, Seibel JM, et al. Molecular targeting of prostate cancer cells by a triple drug combination down-regulates integrin driven adhesion processes, delays cell cycle progression and interferes with the cdk-cyclin axis. BMC Cancer. 2011;11:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsaur I, Hudak L, Makarevic J, et al. Intensified antineoplastic effect by combining an HDAC-inhibitor, an mTOR-inhibitor and low dosed interferon alpha in prostate cancer cells. J Cell Mol Med. 2015;19:1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin K-T, Yeh S-H, Chen D-S, et al. Epigenetic activation of alpha4, beta2 and beta6 integrins involved in cell migration in trichostatin A-treated Hep3B cells. J Biomed Sci. 2005;12:803–813. [DOI] [PubMed] [Google Scholar]
- 54.Luo D, McGettrick HM, Stone PC, et al. The roles of integrins in function of human neutrophils after their migration through endothelium into interstitial matrix. PLoS One. 2015;10:e0118593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kettritz R, Choi M, Rolle S, et al. Integrins and cytokines activate nuclear transcription factor-kappaB in human neutrophils. J Biol Chem. 2004;279:2657–2665. [DOI] [PubMed] [Google Scholar]
- 56.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berton G, Lowell CA. Integrin signalling in neutrophils and macrophages. Cell Signal. 1999;11:621–635. [DOI] [PubMed] [Google Scholar]
- 58.Metcalf D. Hematopoietic cytokines. Blood. 2008;111:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Metcalf D. Lineage commitment of hemopoietic progenitor cells in developing blast cell colonies: influence of colony-stimulating factors. Proc Natl Acad Sci U S A. 1991;88:11310–11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nature reviews. Genetics. 2000;1:57–64. [DOI] [PubMed] [Google Scholar]
- 61.Metcalf D. On Hematopoietic Stem Cell Fate. Immunity. 2007;26:669–673. [DOI] [PubMed] [Google Scholar]
- 62.Metcalf D, Laabi Y. Lineage commitment and maturation induction in normal and leukemic preprogenitor cells. Ann N Y Acad Sci. 2001;938:278–291. [DOI] [PubMed] [Google Scholar]
- 63.Dahl R, Walsh JC, Lancki D, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. [DOI] [PubMed] [Google Scholar]
- 64.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Creyghton MP, Cheng AW, Welstead CG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci. 2010;107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bode KA, Schroder K, Hume DA, et al. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122:596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segre CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol. 2011;2011: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valiuliené G, Treigytè G, Savickiené J, et al. Histone modifications patterns in tissues and tumours from acute promyelocytic leukemia xenograft model in response to combined epigenetic therapy. Biomed Pharmacother. 2016;79:62–70. [DOI] [PubMed] [Google Scholar]
- 69.Maroschik B, Gürtler A, Krämer A, et al. Radiation-induced alterations of histone post-translational modification levels in lymphoblastoid cell lines. Radiat Oncol. 2014;9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ocker M. Deacetylase inhibitors - focus on non-histone targets and effects. World J Biol Chem. 2010;1:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jian W, Yan B, Huang S, et al. Histone deacetylase 1 activates PU.1 gene transcription through regulating TAF9 deacetylation and transcription factor IID assembly. FASEB J Off Publ Fed Am Soc Exp Biol. 2017;31:4104–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vishwakarma BA, Nguyen N, Makishima H, et al. Runx1 repression by histone deacetylation is critical for Setbp1-induced mouse myeloid leukemia development. Leukemia. 2016;30:200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bartels M, Govers AM, Fleskens V, et al. Acetylation of C/EBPe is a prerequisite for terminal neutrophil differentiation. Blood. 2015;125:1782–1792. [DOI] [PubMed] [Google Scholar]
- 74.Wada T, Kikuchi J, Nishimura N, et al. Expression levels of histone deacetylases determine the cell fate of hematopoietic progenitors. J Biol Chem. 2009;284:30673–30683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilting RH, Yanover E, Heideman MR, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Boxtel R, Gomez-Puerto C, Mokry M, et al. FOXP1 acts through a negative feedback loop to suppress FOXO-induced apoptosis. Cell Death Differ. 2013;20:1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 79.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Landt S, Marinov G. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.