Supplemental Digital Content is available in the text
Abstract
High expression of the leukemia-associated gene meningioma-1 (MN1) is frequently found at diagnosis of acute myeloid leukemia (AML) and associates with adverse outcomes. The presence of measurable residual disease (MRD) in complete remission (CR) indicates high risk of relapse and worse outcome in AML patients. However, the prognostic impact of MN1 expression levels as MRD marker has not been evaluated. Digital droplet polymerase chain reaction (ddPCR) is a novel technique allowing sensitive and specific absolute gene expression quantification. We retrospectively analyzed 124 AML patients who received allogeneic hematopoietic stem cell transplantation (HSCT) in CR or CR with incomplete peripheral recovery. Absolute MN1 copy numbers in peripheral blood were assessed prior to HSCT (median 7; range 0–29 days) using ddPCR. High pre-HSCT MN1/Abelson murine leukemia viral oncogene homolog 1 gene (ABL1) copy numbers associated with a higher cumulative incidence of relapse after HSCT and—in relapsing patients—shorter time to relapse. In multivariable analysis, high pre-HSCT MN1/ABL1 copy numbers remained an independent prognosticator for relapse after HSCT. Patients with the highest pre-HSCT MN1/ABL1 copy numbers also had the highest risk of relapse. MN1 copy number assessment also added prognostic information to nucleophosmin 1 gene (NPM1) mutation- and brain and acute leukemia, cytoplasmic (BAALC) and Wilm's tumor gene 1 (WT1) expression-based MRD evaluation. Our study demonstrates the feasibility of the novel ddPCR technique for MN1/ABL1 copy number assessment as a marker for MRD. Evaluation of MN1/ABL1 copy numbers allows the identification of patients at high risk of relapse, independently of other diagnostic risk factors and MRD markers.
Introduction
For optimal and personalized treatment approaches in acute myeloid leukemia (AML), a reliable risk stratification at diagnosis and during disease course is required.1–3 Evaluation of measurable residual disease (MRD) during or after therapy may facilitate risk-adapted treatment decisions for individual AML patients.2–5 In today's clinical routine, AML MRD evaluation mostly relies on multiparameter flow cytometry (MFC) which is limited due to complex analyses performed in specialized laboratories6 and quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays. qRT-PCR is largely restricted to patients harboring stable and determinable fusion transcripts or specific, recurrent gene mutations, for example, mutated nucleophosmin 1 gene (NPM1).2,4,7–9 Next-generation sequencing studies showed AML to be composed of genetically different clones.1,3,10 Some subclones may acquire resistance mechanism during disease course and promote relapse molecularly distinct from the AML at diagnosis.1,10,11 Thus, the inclusion of gene expression analyses in an MRD marker panel may improve the sensitivity of MRD detection in AML patients. Recently, gene expressions of Wilm's tumor gene 1 (WT1)12,13 and brain and acute leukemia, cytoplasmic (BAALC)14–16 were shown to provide informative MRD data in AML remission.
The gene meningioma-1 (MN1) was found highly expressed in primitive (CD34-positive) hematopoietic cells and is downregulated during cell differentiation.17 Elevated levels were described in acute leukemias of myeloid and lymphoid lineage17 and shown to induce proliferation and inhibit myeloid differentiation.18,19 At diagnosis, high MN1 expression was linked to shorter overall survival (OS) and shorter disease-free survival in younger and older AML patients with normal cytogenetics.17,20,21 The feasibility of MN1 expression levels as MRD marker at a defined point in CR has not yet been evaluated. Only one study in 31 AML patients showed that MN1 levels during disease course parallel disease-specific alterations (ie, NPM1 mutations and fusion transcripts CBFB-MYH11 and RUNX1-RUNX1T).22 By contrast, low MN1 expression levels were found in the peripheral blood and bone marrow of healthy individuals. Thus, high bone marrow or blood MN1 expression might have potential use for MRD monitoring.22 Although allogeneic hematopoietic stem cell transplantation (HSCT) has been indicated as the consolidation therapy offering the highest chance of sustained CR in AML patients,3,23 detectable MRD prior to HSCT associates with worse outcomes.8,14,24 This may be especially true in reduced intensity or nonmyeloablative (NMA) conditioning regimens, which are increasingly used to allow HSCT in older or comorbid individuals.25–27 Here, we evaluated the prognostic impact of MN1/Abelson murine leukemia viral oncogene homolog 1 gene (ABL1) copy numbers prior to NMA-HSCT. We adopted the novel digital droplet PCR (ddPCR) technique that allows absolute copy number quantification without the need of standard curves.6,28 The use of peripheral blood enabled a rapid and easily repeatable approach for MRD measurement with high patient convenience. Our study is the first to evaluate the use of MN1 expression levels as a prognostic factor in CR in a larger patient cohort.
Results
MN1/ABL1 copy numbers in AML patients and healthy individuals
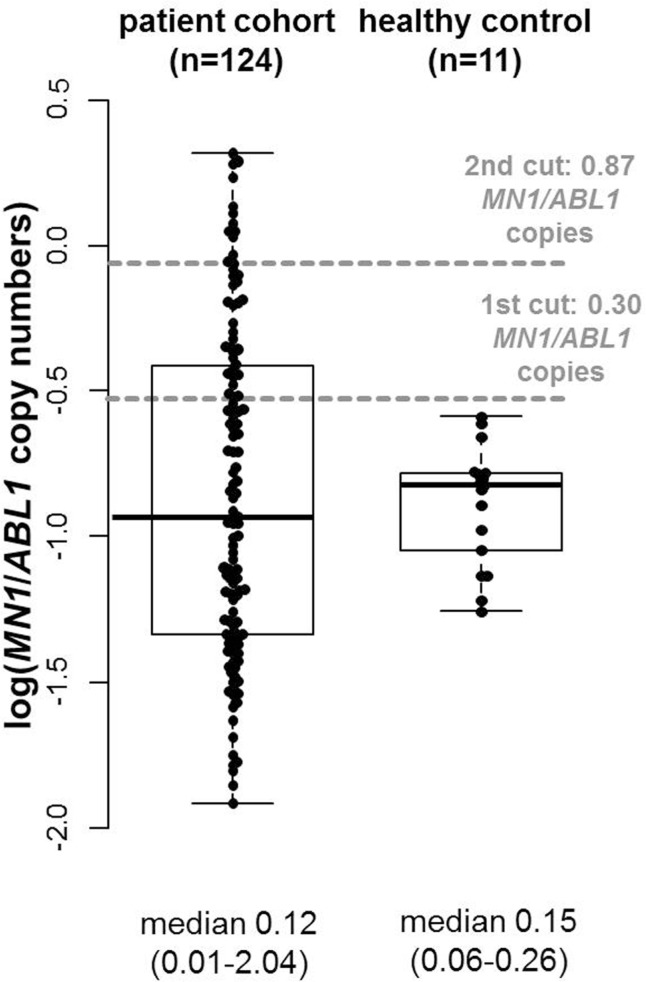
In the patient cohort in complete remission (CR) or CR with incomplete peripheral recovery (CRi; median 7, range 0–29 days) prior to allogeneic HSCT, median blood MN1/ABL1 copy numbers were 0.12 (range 0.01–2.04). In the healthy controls, we observed a median blood MN1/ABL1 copy number of 0.15 (range 0.06–0.26). Overall, AML patients in CR or CRi and the healthy control did not differ significantly in MN1/ABL1 copy numbers (P = 0.97, Fig. 1) and were evenly matched in sex (P = 1) while the healthy control was younger than the AML patients prior to HSCT (P = 0.01, Supplementary Table S2, Supplemental Digital Content). For further analyses, a 0.2992 pre-HSCT MN1/ABL1 copy numbers cutoff was used to define patients with high (n = 39, 31%) or low (n = 85, 69%) pre-HSCT MN1/ABL1 copy numbers in peripheral blood.
Figure 1.
Comparison of pre-HSCT MN1/ABL1 copy numbers in AML patients (n = 124) and healthy controls (n = 17).ABL1 = Abelson murine leukemia viral oncogene homolog 1 gene, AML = acute myeloid leukemia, HSCT = hematopoietic stem cell transplantation, MN1 = meningioma-1 gene.
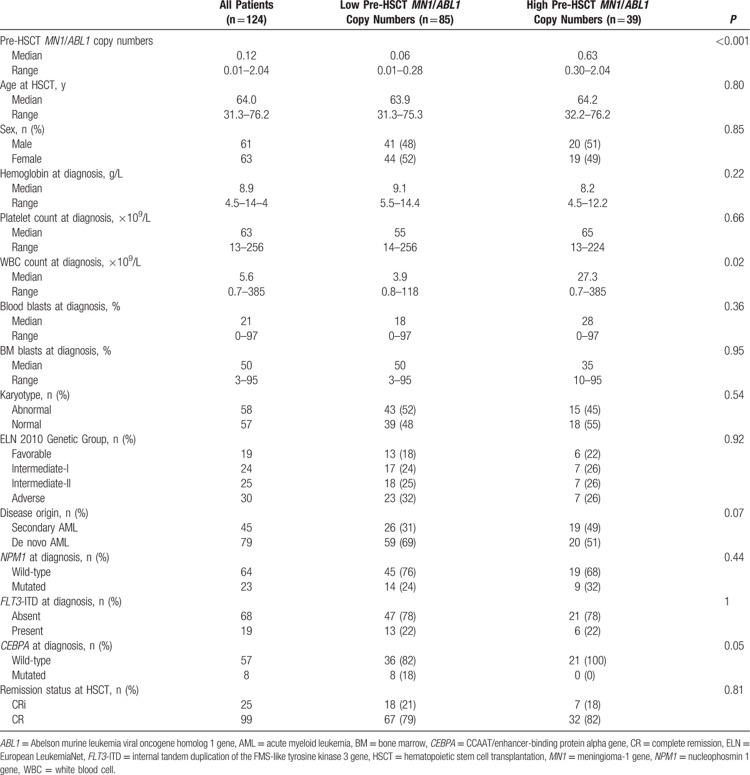
Associations of high pre-HSCT MN1/ABL1 copy numbers
Patients with high pre-HSCT MN1/ABL1 copy numbers had a trend for more secondary or treatment-related AML at diagnosis (P = 0.07). At diagnosis, patients with high pre-HSCT MN1/ABL1 copy numbers also had a trend for a higher CD34+/CD38− cell burden (P = 0.10), a higher white blood count (P = 0.02) and no patient with high pre-HSCT MN1/ABL1 copy numbers was CEBPA mutated (P = 0.05, Table 1). There were no associations of the pre-HSCT MN1/ABL1 copy numbers and other clinical, cytogenetic, molecular, or immunophenotypic characteristics at diagnosis (Table 1, Supplementary Table S1, Supplemental Digital Content). Pre-HSCT MN1/ABL1 copy numbers did also not associate with any tested pre-HSCT characteristics (Supplementary Table S1, Supplemental Digital Content).
Table 1.
Clinical Characteristics According to Pre-HSCT MN1/ABL1 Copy Numbers (High vs Low, 0.30 Cut), n = 124
Prognostic impact of pre-HSCT MN1/ABL1 copy numbers
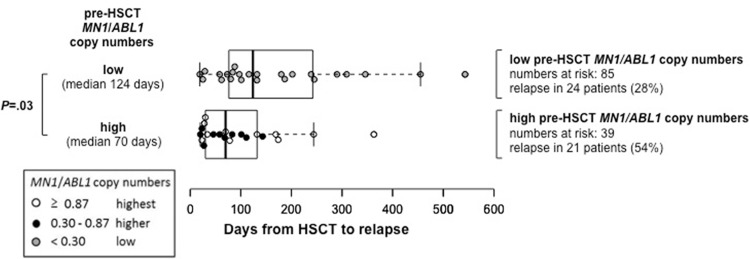
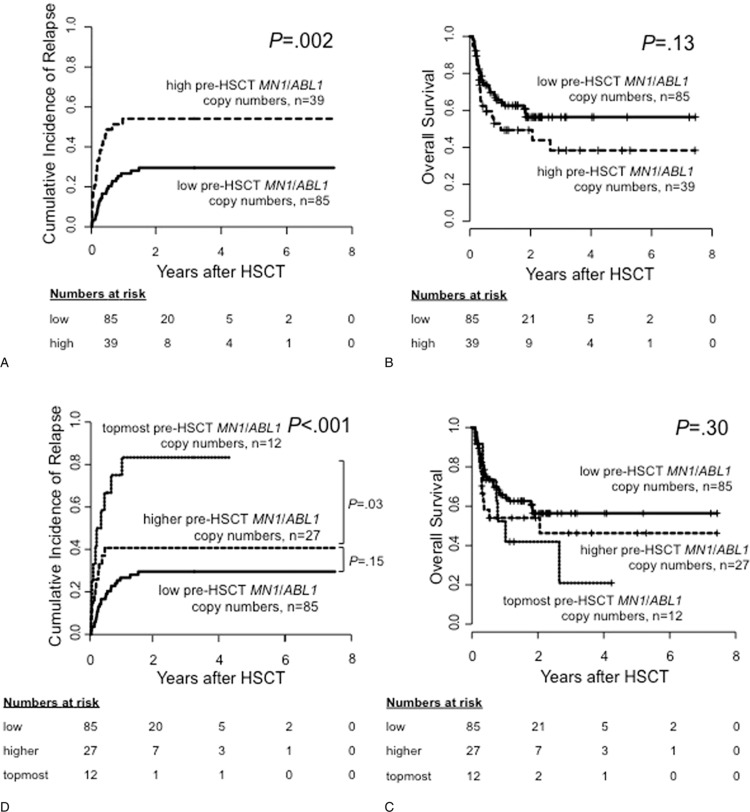
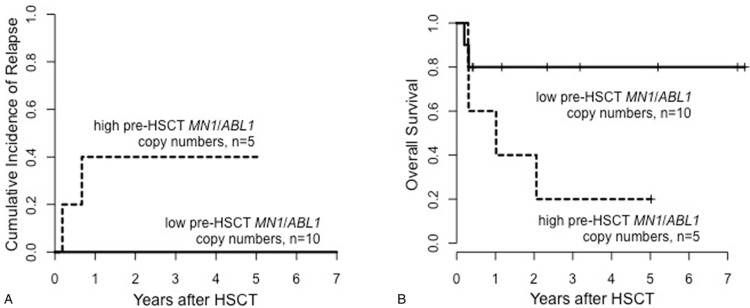
Considering only patients who relapsed after HSCT, patients with high pre-HSCT MN1/ABL1 copy numbers had a shorter time from HSCT to relapse compared with patients with low pre-HSCT MN1/ABL1 copy numbers (median 70, range 20–363 days vs median 124, range 19–543 days, P = 0.03, Fig. 2). Patients with high pre-HSCT MN1/ABL1 copy numbers had a significantly higher cumulative incidence of relapse (CIR, P = 0.002, Fig. 3A) which—despite a separation of the OS curves—did not translate into a significant shorter OS (P = 0.13, Fig. 3B). By contrast, there was no difference in nonrelapse mortality (NRM) between patients with high or low pre-HSCT MN1/ABL1 copy numbers (P = 0.28, Supplementary Fig. S1, Supplemental Digital Content). Similar effects on CIR and OS were also observed when we restricted our analyses to patients with normal karyotype (CIR, P = 0.005 and OS, P = 0.21; Supplementary Fig. S2, Supplemental Digital Content), de novo AML (CIR, P = 0.009 and OS, P = 0.006; Supplementary Fig. S3, Supplemental Digital Content), excluding patients receiving HSCT in CRi (CIR, P = 0.01 and OS, P = 0.10; Supplementary Fig. S4, Supplemental Digital Content) and in a landmark analysis of patients surviving longer than 100 days after HSCT (CIR, P = 0.02 and OS, P = 0.11; Supplementary Fig. S5, Supplemental Digital Content). In multivariable analysis, high pre-HSCT MN1/ABL1 copy numbers retained their prognostic impact on CIR after adjustment for European LeukemiaNet (ELN) 2010 genetic group (Table 2). None of the tested variables were significantly associated with OS in multivariable analysis in this set of patients.
Figure 2.
Time from HSCT to relapse according to high (median 70, range 20–363) or low (median 124, range 19–543) pre-HSCT MN1/ABL1 copy numbers, 0.30 cut, in patients suffering relapse after HSCT (n = 45).ABL1 = Abelson murine leukemia viral oncogene homolog 1 gene, HSCT = hematopoietic stem cell transplantation, MN1 = meningioma-1 gene.
Figure 3.
Outcome according to pre-HSCT MN1/ABL1 copy numbers for the whole cohort (n = 124). According to high versus low, 0.30 cut, (A) cumulative incidence of relapse and (B) overall survival; and according to the topmost versus higher versus low, 0.87 and 0.30 cut, (C) cumulative incidence of relapse and (D) overall survival. ABL1 = Abelson murine leukemia viral oncogene homolog 1 gene, HSCT = hematopoietic stem cell transplantation, MN1 = meningioma-1 gene.
Table 2.
Multivariable Analysis for Patients Receiving HSCT (n = 124)
CD34 expression at diagnosis and pre-HSCT MN1/ABL1 copy numbers
Although MN1 was shown to be highly expressed in CD34-positive bone marrow cells,17 there are no studies reporting on MN1 as MRD marker in the context of CD34 expression status. In our study, data on CD34 status at diagnosis were available for 71 patients, 40 patients had CD34-positive and 31 patients had CD34-negative AML. Between patients with high or low pre-HSCT MN1/ABL1 copy numbers, we observed no significant differences of CD34 expression (P = 0.35) or CD34-positive disease at diagnosis (P = 0.80). Despite low patient numbers, a higher CIR was observed in patients with higher pre-HSCT MN1/ABL1 copy numbers when we restricted our analysis to patients diagnosed with CD34-positive AML (P = 0.001, Supplementary Fig. S6A, Supplemental Digital Content). By contrast, there were no significant differences in CIR according to pre-HSCT MN1/ABL1 copy numbers for the 31 patients with CD34-negative AML (P = 0.60, Supplementary Fig. S6B, Supplemental Digital Content). In 53 patients, information on the CD34 expression status at diagnosis was not available.
Prognostic impact of the topmost pre-HSCT MN1/ABL1 copy numbers
To evaluate whether within the group of patients with high pre-HSCT MN1/ABL1 copy numbers, the amount of pre-HSCT MN1/ABL1 copy numbers also impacts on outcome, a second optimal cutoff was applied. Subsequently, the patient cohort was divided into 3 groups according to pre-HSCT MN1/ABL1 copy numbers (0.30 and 0.87 cut): patients with low (n = 85), higher (n = 27), and the topmost (n = 12) pre-HSCT MN1/ABL1 copy numbers. Applying these cutoffs, a stepwise higher CIR was observed with increasing pre-HSCT MN1/ABL1 copy numbers (low vs higher, P = 0.15, higher vs topmost, P = 0.03, overall P < 0.001). However, despite the separation of these curves, again, no significant impact on OS was observed (Fig. 3C and D). Characteristics of patients with higher and the topmost pre-HSCT MN1/ABL1 copy numbers are shown in Supplementary Table S3 (Supplemental Digital Content).
Correlation of pre-HSCT MN1/ABL1 copy numbers with pre-HSCT BAALC/ABL1 copy number-, WT1 expression-, and NPM1 mutation-based MRD
Recently, our group showed the prognostic utility of pre-HSCT BAALC/ABL1 copy numbers and of pre-HSCT NPM1 for MRD assessment in AML patients undergoing allogeneic HSCT.8,14 Correlating MN1/ABL1 and BAALC/ABL1 copy numbers pre-HSCT, patients with high pre-HSCT MN1/ABL1 copy numbers also had higher pre-HSCT BAALC/ABL1 copy numbers (P < 0.001). Despite the correlation of pre-HSCT MN1/ABL1 and BAALC/ABL1 copy numbers (Pearson product moment correlation coefficient r = .79), 17 patients (14%) only showed high pre-HSCT expression of 1 of the 2 genes in peripheral blood. Next, we compared the Bayesian information criterion (BIC) for univariable models comprising the pre-HSCT MN1/ABL1 and BAALC/ABL1 copy number information alone or combined (both high vs one or both low) for their predictive value for CIR. Here, the model including the copy number information for both genes combined showed the lowest BIC (Supplementary Table S4, Supplemental Digital Content). This indicates that the evaluation of copy numbers of both genes has higher informative value for MRD assessment than pre-HSCT MN1/ABL1 and BAALC/ABL1 copy number information alone. For outcome analyses according to pre-HSCT BAALC/ABL1 copy numbers and the combination of pre-HSCT MN1/ABL1 and BAALC/ABL1 copy numbers, see Supplementary Information and Supplementary Figure S7 (Supplemental Digital Content).
For 111 of the 124 patients, pre-HSCT WT1/ABL1 expression levels were available. Pre-HSCT WT1/ABL1 expression and MN1/ABL1 copy numbers did not correlate well (Pearson product moment correlation coefficient r = .22, Supplementary Fig. S8, Supplemental Digital Content). However, using the published cutoff of previous work of our institution by Lange et al,13 patients with high pre-HSCT MN1/ABL1 copy numbers had significantly higher WT1/ABL1 expression (P < 0.001, Supplementary Table S1, Supplemental Digital Content). While patients with high pre-HSCT WT1/ABL1 expression had a significantly higher CIR (Supplementary Fig. S9A, Supplemental Digital Content), pre-HSCT MN1/ABL1 copy number assessment provided additional prognostic information to WT1/ABL1 expression (Supplementary Fig. S9B, Supplemental Digital Content). BIC comparison showed that the model with both pre-HSCT MN1 and WT1 expression provided higher informative value than the models with either MRD marker alone (Supplementary Table S5, Supplemental Digital Content).
For 20 of the 23 NPM1-mutated patients, information of pre-HSCT NPM1 MRD status was available (Supplementary Table S1, Supplemental Digital Content). Four of the 5 NPM1 MRD positive (MRDpos), patients also had high pre-HSCT MN1/ABL1 copy numbers. Two of them relapsed, while 2 patients died of NRM within 100 days after HSCT. One NPM1 MRDpos patient had low pre-HSCT MN1/ABL1 copy numbers and relapsed. In the 15 mutated NPM1 MRD negative (MRDneg) patients, 5 had high pre-HSCT MN1/ABL1 copy numbers. Of those, 2 patients relapsed, 1 patient died of NRM within 100 days after HSCT, and 2 patients are in continued remission. None of the 10 NPM1 MRD negative patients with low pre-HSCT MN1/ABL1 copy numbers relapsed. In the 15 mutated NPM1 MRDneg patients, we observed a clear separation of CIR (Fig. 4A) and OS (Fig. 4B) curves according to pre-HSCT MN1/ABL1 copy numbers, indicating higher CIR and shorter OS for patients with high pre-HSCT MN1/ABL1 copy numbers. In all 5 relapsing patients with information on MN1/ABL1 copy number and NPM1 MRD status pre-HSCT available, relapse could be predicted prior to HSCT by one (only NPM1 [n = 1], only MN1 [n = 2]), or both (n = 2) markers.
Figure 4.
Outcome according to pre-HSCT MN1/ABL1 copy numbers, high versus low, 0.30 cut, in mutated NPM1 MRDneg patients (n = 15). (A) Cumulative incidence of relapse and (B) overall survival. ABL1 = Abelson murine leukemia viral oncogene homolog 1 gene, HSCT = hematopoietic stem cell transplantation, MN1 = meningioma-1 gene, MRD = measurable residual disease, NPM1 = nucleophosmin 1 gene.
Discussion
Evaluation of MRD during or after AML therapy is of growing importance as it allows dynamic and personalized risk stratification.2 Thus, MRD assessment in AML patients is increasingly integrated in clinical trials and daily routine.3,4,29 However, AML relapse might be mediated by clones that gained additional mutations or subclones genetically distinct to the AML clone that caused the initial leukemia.1,3,10 Consequently, evaluation of more than 1 MRD marker might help to improve risk-adapted treatment.
The transcription factor MN1 was first described in the rare t(12;22)(p13;q11) AML as fusion partner of TEL resulting in a fusion protein with oncogenic potential.30MN1 is overexpressed in AML with inv(16)(p13q22),18,22,31 high EVI1 expression,31 and some cases with normal karyotype.17,22 In the latter, a strong association of high MN1 expression at diagnosis with adverse outcomes in younger and older AML patients was described.17,20,21 By contrast, low MN1 expression levels were found in the bone marrow and peripheral blood of healthy individuals.22 Assessing MN1 expression as a potential MRD marker, Carturan et al22 demonstrated MN1 expression to be parallel to simultaneously evaluated fusion gene transcript levels. However, the feasibility of MN1 copy number measurement for MRD assessment in morphologic CR in a larger AML patient cohort has not been evaluated. In this study, we retrospectively analyzed peripheral blood of AML patients in hematologic CR prior to allogeneic HSCT for consolidation therapy. Using ddPCR, absolute MN1/ABL1 copy numbers were assessed in all patients and a healthy control cohort. The determined optimal cutoff to differentiate between patients with high or low pre-HSCT MN1/ABL1 copy numbers was also higher than the 2-fold standard deviation over the median of the healthy control cohort, allowing a reliable distinction to a physiological MN1 expression background. While we observed no differences in pre-HSCT clinical characteristics such as numbers of chemotherapy cycles or remission status (Table 1, Supplementary Table S1, Supplemental Digital Content), which matches the literature on other MRD markers,8,14 patients with high pre-HSCT MN1/ABL1 copy numbers had a significantly higher risk of relapse compared with patients with low pre-HSCT MN1/ABL1 copy numbers (P = 0.002, Fig. 3A). Despite a separation of the OS curves, OS was not significantly different according to pre-HSCT MN1/ABL1 copy number status, likely due to the restricted patient number in our study.
In multivariable analysis, the impact of high pre-HSCT MN1/ABL1 copy numbers on relapse was shown to be independent from other known risk factors in AML (Table 2). We also observed that the time from HSCT to relapse was significantly shorter in relapsing patients with high pre-HSCT MN1/ABL1 copy numbers than in patients with low pre-HSCT MN1/ABL1 copy numbers (P = 0.03, Fig. 2). In the group of patients with high pre-HSCT MN1/ABL1 copy numbers (n = 39), we evaluated if increasing MN1/ABL1 copy numbers also associated with a higher relapse risk. Patients with the topmost pre-HSCT MN1/ABL1 copy numbers (>0.87) also had the highest risk of suffering relapse after HSCT (P < 0.001, Fig. 3C): 1 year after HSCT, in patients with the topmost pre-HSCT MN1/ABL1 copy numbers CIR was 71% compared with 41% in patients with higher (0.30–0.87) and only 27% in patients with low (<0.30) pre-HSCT MN1/ABL1 copy numbers. Again, despite no significant differences in OS, a separation of the OS curves was observed according to pre-HSCT MN1/ABL1 copy numbers using both cutoffs (Fig. 3D). We conclude that not only a “higher than normal” MN1 copy number correlates with a higher relapse risk but that the absolute amount of MN1/ABL1 copy numbers may also provide additional prognostic information. A correlation of higher MRD levels with higher relapse risk has also recently been described for MFC MRD assessment.32
Carturan et al22 suggested MN1 as a possible MRD marker with particular benefit in 45% of AML cases lacking other suitable genetic MRD markers. It remains to be investigated for which subset of AML patients MN1 copy number analysis for MRD detection will be most informative. In our study, subgroup analyses of patients with normal karyotype (Supplementary Fig. S2, Supplemental Digital Content) or de novo disease (Supplementary Fig. S3, Supplemental Digital Content) showed resembling impact on outcome as in the whole cohort. In previous reports, high MN1 expression at diagnosis was linked to immature AML subtypes with higher CD34 expression.17,33 Thus, we investigated the possibility to predict relapse within the patient cohorts with CD34-positive AML (n = 40) and CD34-negative AML (n = 31) at diagnosis. Here, we observed a strong impact of high pre-HSCT MN1/ABL1 copy numbers on CIR in patients with CD34-positive AML (P = 0.001, Supplementary Fig. 6A) while no impact was observed in patients with CD34-negative AML (P = 0.60, Supplementary Fig. 6B). This suggests that evaluation of pre-HSCT MN1/ABL1 copy numbers might be of higher value in patients with an immature CD34-positive AML phenotype. However, these subanalyses were restricted by limited patient numbers. Furthermore, NPM1-mutated AML was enriched in the CD34-negative cohort (53% of patients), which might explain low relapse rates and the missing prognostic impact of pre-HSCT MN1/ABL1 copy numbers in these patients. Larger trials should evaluate for which subgroups of patients MN1 assessment in CR is of the highest prognostic significance.
As each MRD assay (PCR vs MFC) and marker (fusion gene vs gene mutation vs gene expression) has distinct advantages and disadvantages, combining more than 1 marker for MRD assessment will presumably improve risk stratification.7,34,35 Thus, we also evaluated MN1 MRD results in the context of 3 other MRD markers available for our patient set. Previously, our institution and others were able to show that BAALC and WT1 may function as markers for residual disease in patients after chemotherapy as well as prior to HSCT.12–16 As expected, in the here presented cohort, high pre-HSCT BAALC/ABL1 copy numbers also associated with a higher CIR (P = 0.007) and a trend for shorter OS (P = 0.08, Supplementary Information). When we combined the information of pre-HSCT BAALC/ABL1 and MN1/ABL1 copy numbers (both high vs one or both low, Supplementary Fig. S7C and D, Supplemental Digital Content), our data suggested that evaluation of both genes might be more informative with respect to the risk of relapse after HSCT. Similarly, a high pre-HSCT WT1/ABL1 expression associated with higher CIR (Supplementary Fig. S9A, Supplemental Digital Content). Combining MRD information of WT1 and MN1 also provided additional prognostic information: patients with high expression of either of both markers had higher CIR than patients with low expression of both markers but lower CIR than patients with high expression of both markers (overall P < 0.001, Supplementary Fig. S9B, Supplemental Digital Content). One of the most established MRD markers in AML is NPM1 mutations, which are present in approximately 35% of AML patients at diagnosis.4,8,9,36 In our cohort, information on pre-HSCT NPM1 MRD status was available for 20 NPM1-mutated patients. In the 15 mutated NPM1 MRDneg patients pre-HSCT, we observed a clear separation of the CIR and OS curves (Fig. 4) according to pre-HSCT MN1/ABL1 copy numbers. Two of the five relapsing patients were mutated NPM1 MRDneg prior to HSCT but had high pre-HSCT MN1/ABL1 copy numbers. These patients may have relapsed with an NPM1-negative clone. Unfortunately no patient material for further analyses was available for these patients. MN1 is known to highly correlate with CD34-positive17,29 and NPM1 wild-type AML.17,20,21 By contrast, NPM1 mutations associate with CD34-negative leukemia.36 Thus, MN1 MRD assessment might complement NPM1 mutation-based MRD assessment.
In AML, MRD assessment prior to consolidating allogeneic HSCT is increasingly performed.14,29,32,37 However, in patients with persisting MRD, the question remains as to which treatment approach—for example, additional chemotherapy prior to HSCT, intensification of conditioning regimens or prophylactic donor lymphocyte infusions—would be feasible to improve outcomes and will have to be subject of future prospective clinical trials. These are also needed to evaluate whether patients with high pre-HSCT MN1/ABL1 copy numbers benefit from an allogeneic NMA-HSCT or will have to be lead to alternative treatment options.
Limitations of our study are the retrospective nature and limited patient numbers in the evaluated subgroups. Thus, prospective trials should validate the prognostic use of MN1 expression as novel and promising MRD marker.
In conclusion, our study is the first to show that assessment of MN1/ABL1 copy numbers is feasible for MRD evaluation in AML patients. Patients with high pre-HSCT MN1/ABL1 copy numbers had a significantly higher CIR and shorter time to relapse, independent of other known genetic and molecular factors at diagnosis or HSCT-related parameters. Patients with the topmost MN1/ABL1 copy numbers had the highest relapse incidence after HSCT, probably due to a higher residual disease burden in these patients. Our data also indicate that MN1 copy number assessment may have the potential to improve BAALC-, WT1-, and NPM1-based MRD assessment.
Materials and methods
Patients and treatment
We retrospectively analyzed 124 adult AML patients who received allogeneic HSCT at the University of Leipzig between September 2002 and December 2015. Median age at HSCT was 64.0 (range 31.3–76.2) years. For all patients, peripheral blood samples at a median of 7 (range 0–29) days prior to HSCT were available. Prior to HSCT, patients received age-dependent chemotherapy protocols (under or over 60 years), further details are given in the Supplementary Information. All patients were consolidated with HSCT in first (53%) or second CR (27%) or CRi (20%). All patients received NMA conditioning consisting of fludarabine 30 mg/m2 for 3 days and 2 Gy total body irradiation prior to HSCT25,38 followed by infusion of granulocyte colony stimulating factor-mobilized peripheral blood stem cells. Reasons for applying NMA conditioning as opposed to myeloablative conditioning were age over 50 years for patients receiving unrelated HSCT (n = 104), age over 55 years for patients receiving related HSCT (n = 18), previous autologous HSCT (n = 1), or active infection at HSCT (n = 1). Further patients’ characteristics are provided in Table 1 and Supplementary Table S1, Supplemental Digital Content. Written informed consent for participation in these studies was obtained in accordance with the Declaration of Helsinki. Median follow-up for patients alive was 1.8 years.
Healthy control cohort
Additionally, peripheral blood of a control cohort of 17 healthy volunteers was evaluated for absolute MN1/ABL1 copy numbers. The healthy individuals had a median age of 53.6 (range 32.5–82.0) years; their characteristics are shown in Supplementary Table S2, Supplemental Digital Content. Written informed consent was obtained for all healthy individuals.
ddPCR assessment of MN1/ABL1 copy numbers
Mononuclear cells were isolated from peripheral blood. RNA was extracted from 1 × 107 cells and processed to complementary DNA as previously described.35 Absolute MN1 copy numbers were assessed using a probe-based ddPCR assay (BioRad, Hercules, CA; Assay ID: dHsaCPE5040386) according to manufacturer's specifications. Absolute ABL1 copy numbers were assessed as previously described.14 ddPCR was performed on a QX100 platform (BioRad), and QuantaSoft software (BioRad) was used for raw data processing. With the droplet generator, each sample was divided into approximately 10,000 to 20,000 partitions (droplets). After PCR amplification the samples were placed into the droplet reader, where each droplet was read as positive or negative for the gene expression by issuing specific fluorescence signals (FAM and HEX). Redistribution according to the Poisson algorithm determined the absolute target copy number in the original sample.
MN1/ABL1 cutoff definition
Using the R package “OptimalCutpoints”39 the optimal cutoff of 0.2992 absolute pre-HSCT MN1/ABL1 copies (high vs low) was determined to differentiate according to their relapse probability. To evaluate whether MN1/ABL1 quantification in patients with very high pre-HSCT MN1/ABL1 copy number allowed the identification of a very high-risk group, a second optimal cutoff of 0.8693 absolute MN1/ABL1 copy number was assessed in these patients and discriminated a cohort with higher (n = 27, 69% of patients with high pre-HSCT MN1/ABL1 copy numbers) or the topmost pre-HSCT MN1/ABL1 copy numbers (n = 12, 31% of patients with high pre-HSCT MN1/ABL1 copy numbers).
Flow cytometry, cytogenetics, and molecular markers
In patients with pretreatment bone marrow material available, cytogenetic analyses were performed centrally in our institution using standard banding techniques. In cases were no metaphases could be obtained, fluorescence in situ hybridization was used to screen for recurrent abnormalities (ie, del5/5q, del7/7q, trisomy 8, abn11q23, t(8;21), inv(16), and t(15;17) [n = 5]). At diagnosis, the presence of internal tandem duplication in the FLT3 gene (FLT3-ITD), mutations in the FLT3 tyrosine kinase domain (FLT3-TKD) and in the NPM1 and CEBPA genes were determined as previously described.40 Patients were grouped according to the ELN 2010 classification in 4 risk groups.41 For 71 patients with material available, the bone marrow CD34 and CD38 expression on mononuclear cells at diagnosis was determined as previously described.42 Patients were considered CD34-positive when more than 20% of blasts at diagnosis reacted with the CD34 antibody.43
Analysis of other MRD markers
For all patients, pre-HSCT BAALC/ABL1 copy numbers were evaluated by ddPCR as previously described.14 In 111 patients, WT1/ABL1 expression levels prior to HSCT were evaluated using quantitative PCR as previously described.13 For 20 patients with NPM1-mutated AML, pre-HSCT NPM1 MRD status was evaluated by ddPCR as previously described.8 The applicability of all 3 markers for MRD evaluation has been previously published by our institution.8,13,14
Definition of clinical endpoints and statistical analyses
All statistical analyses were performed using the R statistical software platform (version 3.4.3). CIR was calculated from HSCT to morphologic relapse and OS was calculated from HSCT to death from any cause. Associations of the pre-HSCT MN1/ABL1 copy numbers with baseline clinical, demographic, and molecular features were compared using the Kruskal-Wallis test and Fisher exact test for continuous and categorical variables, respectively. For OS, survival estimates were calculated using the Kaplan-Meier method and groups were compared with the log-rank test. CIR was calculated considering the competing risk NRM using the Fine and Gray model.
Acknowledgments
The authors thank Christel Müller, Daniela Bretschneider, Evelin Hennig, Dr Sabine Leiblein, Martina Pleß, Ulrike Bergmann, Janet Bogardt, Annette Jilo, and Dagmar Cron for their help in determining cytogenetic, morphologic, and immunological analyses; Christine Günther, Scarlett Schwabe, Ines Kovacs, and Kathrin Wildenberger for their help in sample processing and Dr Max Hubmann and Prof Ralf Burckhardt for their help in WT1 expression analysis.
Supplementary Material
Footnotes
Citation: Jentzsch M, Bill M, Grimm J, Schulz J, Beinicke S, Häntschel J, Goldmann K, Pönisch W, Franke G-N, Vucinic V, Cross M, Behre G, Lange T, Niederwieser D, Schwind S. Prognostic Impact of Blood MN1 Copy Numbers Before Allogeneic Stem Cell Transplantation in Patients With Acute Myeloid Leukemia. HemaSphere, 2018;00:00. http://dx.doi.org/10.1097/HS9.0000000000000167.
Funding/support: This study was supported by the Deutsche José Carreras Stiftung e.V. (04R/2016 [SS], PS15/15 [JG]), Verein Zusammen gegen den Krebs e.V. [SS], and Ein Herz für Kinder e.V. [SS].
Disclosure: The authors have indicated they have no potential conflicts of interest to disclose.
MJ and SS contributed to the design and analysis of this study and the writing of the manuscript, and all authors agreed on the final version. MJ, JS, JG, MB, JH, SB, and KG carried out the laboratory-based research; MJ and SS performed statistical analyses; and MJ, G-NF, WP, MC, VV, GB, TL, DN, and SS were involved directly or indirectly in the care of patients and/or sample procurement.
References
- 1.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. [DOI] [PubMed] [Google Scholar]
- 4.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–433. [DOI] [PubMed] [Google Scholar]
- 5.Hourigan CS, Gale RP, Gormley NJ, et al. Measurable residual disease testing in acute myeloid leukaemia. Leukemia. 2017;31:1482–1490. [DOI] [PubMed] [Google Scholar]
- 6.Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for “prime time”? Blood. 2014;124:3345–3355. [DOI] [PubMed] [Google Scholar]
- 7.Doubek M, Palasek I, Pospisil Z, et al. Detection and treatment of molecular relapse in acute myeloid leukemia with RUNX1 (AML1), CBFB, or MLL gene translocations: frequent quantitative monitoring of molecular markers in different compartments and correlation with WT1 gene expression. Exp Hematol. 2009;37:659–672. [DOI] [PubMed] [Google Scholar]
- 8.Bill M, Grimm J, Jentzsch M, et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann Hematol. 2018;97:1757–1765. [DOI] [PubMed] [Google Scholar]
- 9.Ommen HB, Schnittger S, Jovanovic JV, et al. Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood. 2010;115:198–205. [DOI] [PubMed] [Google Scholar]
- 10.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostergaard M, Olesen LH, Hasle H, et al. WT1 gene expression: an excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients—results from a single-centre study. Br J Haematol. 2004;125:590–600. [DOI] [PubMed] [Google Scholar]
- 13.Lange T, Hubmann M, Burkhardt R, et al. Monitoring of WT1 expression in PB and CD34(+) donor chimerism of BM predicts early relapse in AML and MDS patients after hematopoietic cell transplantation with reduced-intensity conditioning. Leukemia. 2011;25:498–505. [DOI] [PubMed] [Google Scholar]
- 14.Jentzsch M, Bill M, Grimm J, et al. High BAALC copy numbers in peripheral blood prior to allogeneic transplantation predict early relapse in acute myeloid leukemia patients. Oncotarget. 2017;8:87944–87954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber S, Haferlach T, Alpermann T, et al. Feasibility of BAALC gene expression for detection of minimal residual disease and risk stratification in normal karyotype acute myeloid leukaemia. Br J Haematol. 2016;175:904–916. [DOI] [PubMed] [Google Scholar]
- 16.Najima Y, Ohashi K, Kawamura M, et al. Molecular monitoring of BAALC expression in patients with CD34-positive acute leukemia. Int J Hematol. 2010;91:636–645. [DOI] [PubMed] [Google Scholar]
- 17.Heuser M, Beutel G, Krauter J, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–3905. [DOI] [PubMed] [Google Scholar]
- 18.Carella C, Bonten J, Sirma S, et al. MN1 overexpression is an important step in the development of inv(16) AML. Leukemia. 2007;21:1679–1690. [DOI] [PubMed] [Google Scholar]
- 19.Heuser M, Argiropoulos B, Kuchenbauer F, et al. MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in patients with AML. Blood. 2007;110:1639–1647. [DOI] [PubMed] [Google Scholar]
- 20.Schwind S, Marcucci G, Kohlschmidt J, et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood. 2011;118:4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langer C, Marcucci G, Holland KB, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2009;27:3198–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carturan S, Petiti J, Rosso V, et al. Variable but consistent pattern of meningioma 1 gene (MN1) expression in different genetic subsets of acute myelogenous leukaemia and its potential use as a marker for minimal residual disease detection. Oncotarget. 2016;7:74082–74096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta V, Tallman MS, Weisdorf DJ. Allogeneic hematopoietic cell transplantation for adults with acute myeloid leukemia: myths, controversies, and unknowns. Blood. 2011;117:2307–2318. [DOI] [PubMed] [Google Scholar]
- 24.McSweeney PA, Niederwieser D, Shizuru JA, et al. Hematopoietic cell transplantation in older patients with hematologic malignancies: replacing high-dose cytotoxic therapy with graft-versus-tumor effects. Blood. 2001;97:3390–3400. [DOI] [PubMed] [Google Scholar]
- 25.Araki D, Wood BL, Othus M, et al. Allogeneic hematopoietic cell transplantation for acute myeloid leukemia: time to move toward a minimal residual disease-based definition of complete remission? J Clin Oncol. 2016;34:329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muffly L, Pasquini MC, Martens M, et al. Increasing use of allogeneic hematopoietic cell transplantation in patients age 70 years and older in the United States. Blood. 2017;130:1156–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- 28.Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venditti A, Piciocchi A, Candoni A, et al. Risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia: results of the AML1310 trial of the GIMEMA group [abstract]. Haematologica. 2017;102 (s2):6–7. abstract S111. [Google Scholar]
- 30.Kawagoe H, Grosveld GC. MN1-TEL myeloid oncoprotein expressed in multipotent progenitors perturbs both myeloid and lymphoid growth and causes T-lymphoid tumors in mice. Blood. 2005;106:4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. [DOI] [PubMed] [Google Scholar]
- 32.Guolo F, Minetto P, Clavio M, et al. Combining flow cytometry and WT1 assessment improves the prognostic value of pre-transplant minimal residual disease in acute myeloid leukemia. Haematologica. 2017;102:e348–e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuser M, Wingen LU, Steinemann D, et al. Gene-expression profiles and their association with drug resistance in adult acute myeloid leukemia. Haematologica. 2005;90:1484–1492. [PubMed] [Google Scholar]
- 34.Zeijlemaker W, Meijer R, Kelder A, et al. Leukemic stem cell frequency combined with MRD is an important biomarker to predict relapse in acute myeloid leukemia. Results from a prospective H102 study [abstract]. Haematologica. 2017;102 (s2):7–8. abstract S113.27909216 [Google Scholar]
- 35.Lange T, Niederwieser DW, Deininger MW. Residual disease in chronic myeloid leukemia after induction of molecular remission. N Engl J Med. 2003;349:1483–1484. [DOI] [PubMed] [Google Scholar]
- 36.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. [DOI] [PubMed] [Google Scholar]
- 37.Getta BM, Devlin SM, Levine RL, et al. Multicolor flow cytometry and multigene next-generation sequencing are complementary and highly predictive for relapse in acute myeloid leukemia after allogeneic transplantation. Biol Blood Marrow Transplant. 2017;23:1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegenbart U, Niederwieser D, Sandmaier BM, et al. Treatment for acute myelogenous leukemia by low-dose, total-body, irradiation-based conditioning and hematopoietic cell transplantation from related and unrelated donors. J Clin Oncol. 2006;24:444–453. [DOI] [PubMed] [Google Scholar]
- 39.López-Ratón M, Rodríguez-Álvarez M, Cadarso-Suárez C, et al. OptimalCutpoints: an R package for selecting optimal cutpoints in diagnostic tests. J Stat Softw. 2014;61:1–36. [Google Scholar]
- 40.Bill M, Jentzsch M, Grimm J, et al. Prognostic impact of the European LeukemiaNet standardized reporting system in older AML patients receiving stem cell transplantation after non-myeloablative conditioning. Bone Marrow Transplant. 2017;52:932–935. [DOI] [PubMed] [Google Scholar]
- 41.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. [DOI] [PubMed] [Google Scholar]
- 42.Jentzsch M, Bill M, Nicolet D, et al. Prognostic impact of the CD34+/CD38− cell burden in patients with acute myeloid leukemia receiving allogeneic stem cell transplantation. Am J Hematol. 2017;92:388–396. [DOI] [PubMed] [Google Scholar]
- 43.Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia. 1995;9:1783–1786. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.