Abstract
Supplemental Digital Content is available in the text
Mature lymphoid malignancies are the most common hematological cancers, with approximately 93,500 B non-Hodgkin lymphomas (NHL), 17,500 Hodgkin lymphomas, 14,000 chronic lymphocytic leukemia (CLL), and 3300 T-NHL new cases being diagnosed every year in Europe.1,2 Their optimal management requires integrated morphological and immunophenotypic analysis of cell and tissue, together with chromosome and molecular analyses. The standardization of these techniques has improved their reproducibility and consequently their inclusion in evolving classifications of mature lymphoid malignancies.3
High throughput sequencing techniques provide useful diagnostic, prognostic, and theranostic information for the individualized clinical management of patients with mature lymphoid malignancies, but much controversy exists regarding panel design. In order to harmonize sequencing panels for clinical use, the French LYSA (LYmphoma Study Association) and GBMHM (Groupe de Biologistes Moléculaires des Hémopathies Malignes) cooperative groups established consensus minimal panels for B and T lineage lymphomas, which are presented here. An earlier, French-language, version of this consensus has been published previously.4
The arrival of high throughput next-generation sequencing (NGS) techniques represents a new opportunity but also a new challenge for the diagnosis of lymphoid malignancies.5 Compared to polymerase chain reaction and Sanger sequencing, it comes with additional specificities.
Spatial and temporal heterogeneity
While the histological appearance of most cancers is relatively homogeneous, regardless of the biopsy site, there is considerably more intraindividual molecular variability.6,7 Similarly, longitudinal studies have confirmed the suspected Darwinian evolution of cancer, with temporal evolution in clonal architecture.8 NGS has considerably improved our ability to detect subclonal alterations compared with Sanger sequencing (detection threshold around 10%), as long as sufficient sequencing depth is achieved (at least 500 reads/specific nucleotide). However, the analysis of the spatial heterogeneity requires the analysis of sample DNA from different sites, which is now achievable by analysis of circulating tumor (ct) DNA. The analysis of ct-DNA by NGS is a noninvasive procedure, which might also provide important prognostic information with longitudinal follow-up.9
Difficulties in interpreting variants
Pioneering studies in tumor oncogenesis were based on a relatively simple model whereby molecular alterations were necessary and sufficient for malignant transformation.10 Several observations suggest that this is an over-simplification, such as the existence of cancers with no apparent recurrent mutation11 or the disappointing results of targeted therapy based only on molecular abnormalities.12,13 This implies that a molecular abnormality must be interpreted in its global context, including the (WHO defined) histological subtype, immunophenotype, and somatic genetic structural and numerical abnormalities.3 Of note, in routine clinical practice, most of the NGS tumor analyses are not paired with analysis of constitutional genome, meaning that the somatic nature of the mutations is not formally demonstrated.
Technical difficulties
The validity of NGS results depends on the quality of the nucleic acids and their preanalytic treatment, particularly but not exclusively, tissue fixation. For example, sequencing errors can be due to cytosine deamination during formol fixation and paraffin embedding.14,15 It is consequently preferable, whenever possible, to work with fresh frozen tissue or extemporaneously extracted DNA. Each NGS step can influence the results: nucleic acid extraction techniques, library constitution and enrichment, sequencing technologies, internal quality controls, bioinformatic filtering, and biological interpretation. The existence of several NGS platforms allows the comparison of different approaches, but also makes harmonization and standardization indispensable, in order to provide homogeneous, reliable results throughout Europe. The Harmony program (https://www.harmony-alliance.eu) is developing Europe-wide databases of clinical and biological data, including NGS results. It is of paramount importance that the molecular data submitted to Harmony have been proven to be reproducible, via participation of the providers in national or European external quality assessment (EQA) programs, ideally in combination with continuing medical education (CME). Whether such EQA/CME should be national or European depends on the frequency of the analysis and practical logistics. As examples, the European Euro-MRD (http://www.euromrd.org) group, a member of the ESLHO (http://www.eslho.org) EHA-scientific working group (https://ehaweb.org/research/scientific-working-groups/) organizes EQA/CME for immunogenetic quantification of Minimal Residual Disease quantification in lymphoid malignancies, the UK NEQUAS system (https://ukneqas.org.uk) organizes stand-alone EQA for many molecular tests and the aforementioned French GBMHM (https://sites.google.com/site/gbmhmassociation/home) organizes national EQA/CME for the most common molecular tests in hematological malignancies, including lymphoid and myeloid NGS. The increasing interaction between EHA scientific working groups will facilitate harmonized, optimized approaches to EQA and CME.
NGS panel choice
In addition to technological considerations, the choice of target genes or panels is a crucial step in the development of appropriate diagnostic approaches. Although the availability of genome wide (whole-genome or whole-exome) approaches will become increasingly available, targeted analyses allow optimization of quality (read depth, variant characterization, reporting timelines, etc.), and cost, and are a pragmatic option for lymphoma diagnostics in the foreseeable future. An optimal panel must reply to diagnostic (both positive and differential), prognostic and theranostic requirements for the different lymphoma subtypes, while remaining sufficiently simple to allow for uniform application.
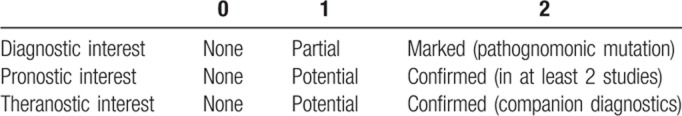
An exhaustive literature search was performed by a duo of clinical/molecular experts for each of the principal mature lymphoid subtypes. These included: diffuse large B cell lymphoma (DLBCL), follicular lymphoma (FL), mantle cell lymphoma, marginal zone lymphoma (MZL), hairy cell lymphoma (HCL), lymphoplasmocytic lymphoma, Waldenström macroglobulinemia (WM), and CLL for B cell malignancies. T cell subtypes included angioimmunoblastic T cell lymphoma (AITL) and other nodal lymphomas derived from T Follicular helper cells (TFH-PTCL), peripheral T cell lymphoma—not-otherwise specified, anaplastic large cell lymphoma, intestinal T lymphomas associated with an enteropathy or not (monomorphic epithelial intestinal T lymphoma), hepatosplenic T lymphoma, nasal type extranodal NK/T lymphoma, Sezary syndrome, adult T cell lymphoma/leukemia, large granular lymphocytes leukemia, and T prolymphocytic leukemia. For each of these entities, genes found to be mutated or affected by copy number variation (CNV) in more than 5% of cases were noted. The chromosomal location, number of exons, and known function were noted for each gene, as were the frequency of mutations (gain or loss of function, hotspot or diffuse) and CNV in each of the aforementioned entities. Finally, a clinical relevance score was attributed to each category (Table 1).
Table 1.
Clinical Relevance Scores
It is to be noted that this process was undertaken prior to publication of the joint Association for Molecular Pathology, American College of Medical Genetics and Genomics, American Society of Clinical Oncology, and College of American Pathologists’ recommendations16 and did not use strictly identical criteria. Our score 2 corresponds to their “Tier 1” recommendations, score 1 to “Tier 2” and our score 0 to “Tiers 3 and 4.”
This generated a compiled list of genes, which was circulated to GBMHM and LYSA experts for each disease subtype, who selected a minimal panel for their disease entity. The individual panels were then compared in order to select 2 consensus panels for mature B and T lymphoid disorders.
Among the numerous recurrent molecular abnormalities identified in mature lymphoid malignancies, 95 were considered to be of at least potential interest in one of the diagnostic/prognostic/theranostic categories and 29 to be of confirmed interest (score 2). As such, genes such as KMT2D/MLL2, although frequently mutated in mature B cell malignancies, were not selected due to the lack of clear evidence of their clinical significance, in order to maintain the number of selected genes within the limits of a panel to be used by most diagnostic sequencing platforms. ATM was also excluded because of its size and the difficulty in distinguishing somatic and germline mutations in the absence of analysis of nontumor DNA.
Because there was little overlap in the selected genes in mature B and T cell malignancies, we chose to separate the 2 panels, with laboratories being left free to use them in independent or combined libraries, depending on local priorities, constraints, and technical choices, such as capture versus amplicon approaches.
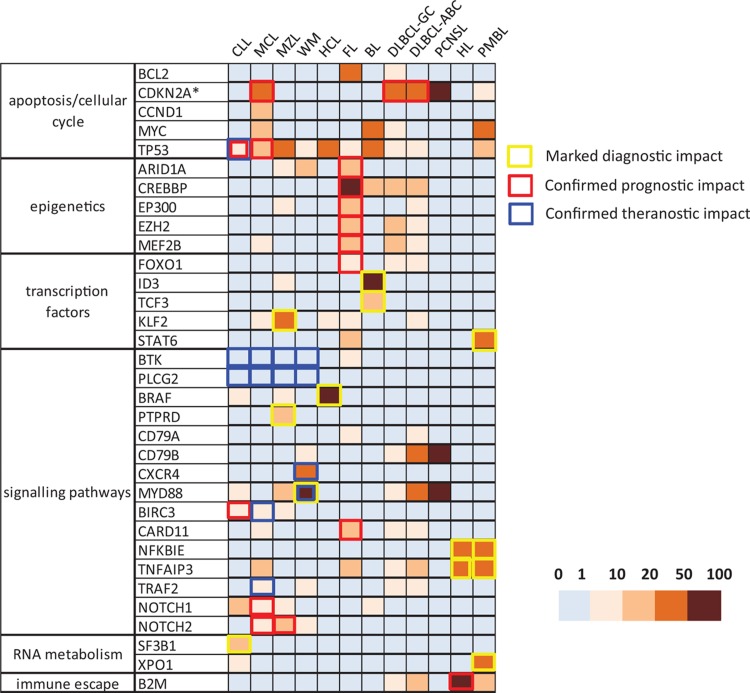
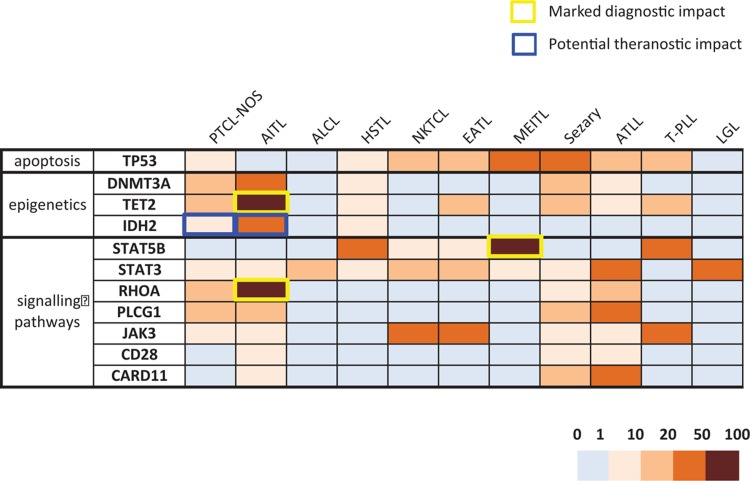
A final panel of 33 genes was identified for B cell lymphomas (Fig. 1 and Supplemental Table 1, Supplemental Digital Content 1) and a panel of 11 genes for T cell lymphomas (Fig. 2 and Supplemental Table 2, Supplemental Digital Content 2). This corresponds to approximately 504 exons for the B cell panel and 190 exons for the T cell panel if an exhaustive approach of analysis of all exons and nontranslated regions is chosen and 239 and 107 exons, respectively, if a restrictive approach covering only known mutation hotspots is adopted.
Figure 1.
A heatmap representation of the prevalence of gene alterations in mature B lymphoid malignancies from the LYSA/GBMHM consensus panel. The ∗ symbol for CDKN2A underlines that this locus is altered by deletions (and not mutations). The borders of the squares are colored when the alteration has a clear clinical impact in a particular lymphoma subtype (diagnostic in yellow, prognostic in red, and theranostic in blue). ABC = activated B cell, BL = Burkitt lymphoma, CLL = chronic lymphocytic leukemia, DLBLCL = diffuse large B cell lymphoma, FL = follicular lymphoma, GC = germinal center, HCL = hairy cell lymphoma, HL = Hodgkin lymphoma, MCL = mantle cell lymphoma, MZL = marginal zone lymphoma, PCNSL = primary central nervous system lymphoma, PMBCL = primary mediastinal B cell lymphoma, WM = Waldenström macroglobulinemia.
Figure 2.
A heatmap representation of the prevalence of gene mutations in mature T lymphoid malignancies from the LYSA/GBMHM consensus panel. The borders of the squares are colored when the alteration has a clinical impact in a particular lymphoma subtype (diagnostic in yellow, theranostic in blue). AITL = angio-immunoblastic T lymphoma, ALCL = anaplastic large cell lymphoma, ATLL = adult T leukemia/lymphoma, EATL = enteropathy associated T lymphoma, HSTL = hepatosplenic T lymphoma, LGL = large granular lymphocytic leukemia, MEITL = monomorphic epitheliotropic intestinal T lymphoma, NKTCL = nasal type NK/T cell lymphoma, PTCL-NOS = peripheral T cell lymphoma, not otherwise specified, PTCL-TFH = nodal peripheral T cell lymphoma derived from TFH cells, Sezary = Sezary syndrome, T-PLL = T-prolymphocytic leukemia.
Genes of positive or differential diagnostic interest
As for most cancers, pathognomonic mutations are exceedingly rare in lymphoma, even if some molecular abnormalities are much more frequent in certain subtypes, thus representing useful diagnostic markers. This is well recognized for the BRAF V600E mutation in HCL, the SF3B1 K700E mutation in CLL, or the MYD88 L265P mutation in WM. In mature T lymphomas, RHOA (G17V), IDH2 (R172), and TET2 mutations are strongly associated with AITL and TFH-PTCL. In less clear-cut situations, mutational profiles should be interpreted in conjunction with other parameters, ideally within multidisciplinary tumor boards. Figure 1 is a graphic representation of the frequency of alterations in the 33 genes of the B cell panel in the different subgroups of mature B cell malignancies and Figure 2 shows equivalent data for the 11 genes of the T cell panel.
Genes with prognostic interest
The influence of mutation profiles on prognostic evaluation is evolving rapidly, partly due to evolving therapeutic practice. Examples in mature lymphoid malignancies include the m7 FLIPI score in FL17,18 or NOTCH1, SF3B1, and TP53 mutations in CLL, used in combination with cytogenetic abnormalities.19 Recent examples of NGS approaches with prognostic value include a complement to ABC/GCB subtyping of DLBCL,20–22 illustrating the need to update this type of panel regularly.
Genes of theranostic interest
This category includes genes whose mutational status already determines the choice of therapy, such as the use of ibrutinib in TP53-mutated CLL, and others which are likely to do so soon. The latter category includes targets such as EZH223 in FL, or CXCR4 in WM.24 It also includes genes for which mutations are selected in the presence of certain targeted treatments, such as BTK, PLCG2, or genes activating the alternative NF-kB pathway (BIRC3, TRAF2, TRAF3) in patients treated by ibrutinib.25,26 In T cell lymphomas, a biological rationale could justify treatment based on mutational profiles, such as the use of demethylating agents for lymphomas with epigenetic deregulation evidenced by mutations of TET2,27DNMT3A, and/or IDH2), or the use of specific IDH2 inhibitors, although this would obviously require prior rigorous evaluation.
Perspectives
The rapid advances in our understanding of the impact of somatic mutation in lymphoid malignancies will make regular updating of consensus panels obligatory. The objectives of a given panel should also be borne in mind, since a limited “universal” panel, as described here, cannot replace more specific, extensive panels as, for example, recently described for DLBCL prognostic evaluation.21,22 The capacity of panels to evolve over time is also dependent on the technology used, with capture approaches evolving more easily than amplicon approaches.
The minimal panel described here is designed for platforms analyzing a wide variety of lymphoid disorders and can be complemented by additional targets of local interest, as long as such inclusion does not impact detection of the core target genes. Identification of a common core of minimal targets, and their diffusion and adoption by molecular diagnostic platforms providing results for cooperative clinical studies and for standardized patient care is important because it will allow:
-
(a)
external quality assessment of common targets;
-
(b)
the use of these targets for medico-economic evaluation of NGS techniques; and
-
(c)
the realization of retrospective national or international cooperative group studies using reproducible results, which have been proven to be robust in a multicenter setting through EQA programs.
The “universal” orientation panel described here could be considered excessive or inappropriate for routine hospital practice. One alternative would be development of a European network of reference laboratories for each subtype of lymphoid malignancy, who would perform more in-depth molecular analyses based on results of an even more restricted primary, local “universal lymphoid” NGS panel (or on initial orientation based purely on cell/tissue morphology and immunophenotype). Practical considerations such as reporting delays and the number of samples recruited by a given platform are important when deciding the most appropriate European set-up.
Reimbursement for NGS diagnostics in Europe varies significantly between European countries, with most undertaking to develop suitable models. In France, it is currently based on the volume sequenced, but more appropriate economic models that take into account variable transmission of results, depending on the clinical context, need to be developed. Medico-economic evaluation requires integration of consolidated costs (which are heavily dependent on the frequency of realization of a given panel), the impact of NGS on reduced use of classical tests and the therapeutic implications of more precise diagnostics. Assuming a unit cost of 600€/test and analysis of one-third of the 128,500 new cases of lymphoma and CLL, the global cost of diagnostic NGS in Europe (with no follow-up or relapse analysis) would be approximately 25 M€. It should, however, be emphasized that restricting use of innovative, costly, therapies on the basis of companion diagnostic identification of patients most likely to benefit can easily be cost-effective. These considerations clearly illustrate the need for appropriate guidelines for innovative tests and close clinical and laboratory interaction.
Patients requiring NGS analyses could be selected by a reference center after pluri-disciplinary tumor board evaluation of clinical, histological/morphological, immunophenotypic, cytogenetic, and molecular results and restricted to cases:
-
-
with diagnostic difficulties;
-
-
for prognostic evaluation when appropriate panels exist (most likely within the context of prospective cooperative multicenter trials);
-
-
with theranostic intent when the mutation status determines use of specific treatment, once again within the context of clinical trials and preferentially in relapsing/resistant patients.
It is important to emphasize that the attempt at harmonization of NGS panels described here must be accompanied by the use of standardized bioinformatics pipelines, preferably independent of those proposed by the sequencing platform manufacturers (often heterogeneous and poorly adapted to specific requirements), in order to optimize intercenter reporting reproducibility. Filtering of constitutional variants, the advantages, and inconveniences of different databases, cutoffs for variant allelic reporting and criteria for CNVs all need to be taken into consideration.28 Their harmonization will allow NGS results from different platforms to be exploited for individual patients.
Conclusions
The harmonization of diagnostic practice and quality control is a prerequisite for equal access to diagnostic precision across Europe. The consensus panel described here complements this approach by encouraging widespread use of a minimum set of shared targets for mature lymphoid malignancies. This will encourage interaction between different molecular genetic centers. While a minimal consensus panel is inevitably rapidly outdated, this approach provides a stepping stone to a common NGS language within mature lymphoid malignancies between platforms within and between countries.
Supplementary Material
Supplementary Material
Footnotes
Citation: Sujobert P, Le Bris Y, de Leval L, Gros A, Merlio JP, Pastoret C, Huet S, Sarkozy C, Davi F, Callanan M, Thieblemont C, Sibon D, Asnafi V, Preudhomme C, Gaulard P, Jardin F, Salles G, Macintyre E. The Need for a Consensus Next-generation Sequencing Panel for Mature Lymphoid Malignancies. HemaSphere, 2018;00:00. http://dx.doi.org/10.1097/HS9.0000000000000169.
Funding/support: None.
Disclosure: The authors have indicated they have no potential conflicts of interest to disclose.
All authors contributed to establishment of the recommendations. PS and EM wrote the paper. All the authors approved the paper.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. [DOI] [PubMed] [Google Scholar]
- 2.Watson L, Wyld P, Catovsky D. Disease burden of chronic lymphocytic leukaemia within the European Union. Eur J Haematol. 2008;81:253–258. [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sujobert P, Bris YL, Leval L, et al. Definition of a minimal genes set for mature lymphoid blood diseases. Hématologie. 2018;1–2:27–59. [Google Scholar]
- 5.Rosenquist R, Rosenwald A, Du M-Q, et al. Clinical impact of recurrently mutated genes on lymphoma diagnostics: state-of-the-art and beyond. Haematologica. 2016;101:1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araf S, Wang J, Korfi K, et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia. 2018;32:1258–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtz DM, Scherer F, Jin MC, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol. 2018;36:2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 11.Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373:726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Tourneau C, Delord J-P, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. [DOI] [PubMed] [Google Scholar]
- 14.Guyard A, Boyez A, Pujals A, et al. DNA degrades during storage in formalin-fixed and paraffin-embedded tissue blocks. Virchows Arch Int J Pathol. 2017;471:491–500. [DOI] [PubMed] [Google Scholar]
- 15.Williams C, Pontén F, Moberg C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagn. 2017;19:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurinovic V, Kridel R, Staiger AM, et al. Clinicogenetic risk models predict early progression of follicular lymphoma after first-line immunochemotherapy. Blood. 2016;128:1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastore A, Jurinovic V, Kridel R, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015;16:1111–1122. [DOI] [PubMed] [Google Scholar]
- 19.Zent CS, Burack WR. Mutations in chronic lymphocytic leukemia and how they affect therapy choice: focus on NOTCH1, SF3B1, and TP53. Hematol Am Soc Hematol Educ Program. 2014;2014:119–124. [DOI] [PubMed] [Google Scholar]
- 20.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378:1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morschhauser F, Salles G, McKay P, et al. Interim report from a phase 2 multicenter study of tazeùetostat, an EZH2 inhibitor, in patients with relapsed or refractory B-cell non Hodgkin lymphomas. Hematol Oncol. 2017;35:24–25. [Google Scholar]
- 24.Treon SP, Tripsas CK, Meid K, et al. Ibrutinib in previously treated Waldenström's macroglobulinemia. N Engl J Med. 2015;372:1430–1440. [DOI] [PubMed] [Google Scholar]
- 25.Rahal R, Frick M, Romero R, et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med. 2014;20:87–92. [DOI] [PubMed] [Google Scholar]
- 26.Woyach JA, Furman RR, Liu T-M, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemonnier F, Dupuis J, Sujobert P, et al. Treatment with 5-azacytidine induces a sustained response in patients with angioimmunoblastic T-cell lymphoma. Blood. 2018;132:2305–2309. [DOI] [PubMed] [Google Scholar]
- 28.Guillermin Y, Lopez J, Chabane K, et al. What does this mutation mean? The tools and pitfalls of variant interpretation in lymphoid malignancies. Int J Mol Sci. 2018;19:E1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.