Abstract
Acute myeloid leukemia (AML) is a genetically heterogeneous disease driven by a limited number of cooperating mutations. There is a long-standing debate as to whether AML driver mutations occur in hematopoietic stem or in more committed progenitor cells. Here, we review how different mouse models, despite their inherent limitations, have functionally demonstrated that cellular origin plays a critical role in the biology of the disease, influencing clinical outcome. AML driven by potent oncogenes such as mixed lineage leukemia fusions often seem to emerge from committed myeloid progenitors whereas AML without any major cytogenetic abnormalities seem to develop from a combination of preleukemic initiating events arising in the hematopoietic stem cell pool. More refined mouse models may serve as experimental platforms to identify and validate novel targeted therapeutic strategies.
Introduction
Acute myeloid leukemia (AML) is characterized by an accumulation of poorly differentiated myeloid cells and functional insufficiency of the hematopoietic system. Despite continuous advances in treatment, the majority of the patients still relapse and ultimately die of the disease.1 AML is a clinically and genetically heterogeneous disease driven by functional cooperation of a relatively small number of mutations.2 In addition to genetics and other factors, such as the patient's age and health status, the observed heterogeneity may also be the consequence of different cellular origins. It was the shift from a purely stochastic model toward a more hierarchical organization model of leukemia driven by a small population of cells, also referred as leukemia-initiating cells (LIC) or leukemic stem cells (LSC) that particularly raised interest in the role of cellular origin in the biology and clinical course of AML. Studies in genetically modified mice and xenografts of patient-derived cells (PDX) in immune deficient mice led to the hypothesis that AML is the product of cooperating genetic alterations in the hematopoietic stem cell (HSC) pool. The combination of improved multicolor flow cytometry with high-throughput “next-generation” sequencing (NGS) technologies revealed a complex interplay of genomic and epigenetic alterations that seem to be necessary to transform normal hematopoietic stem and progenitor cells (HSPC) into preleukemic states that may ultimately progress to AML. More recent studies in transgenic mouse strains and PDX models combined with cross-species transcriptomics suggested that AML in mice and humans in most cases originates from a continuum of early multipotent to more differentiated hematopoietic progenitor cells. However, there is increasing evidence that in about 10% to 20% of patients, AML may originate from more immature cells that are most likely part of cell pool that we call today long-term HSC (LT-HSC). Modeling of HSC-derived AML driven by a strong oncogene in mice has revealed a particularly invasive and drug-resistant phenotype associated with a genetic signature that also characterizes human AML with poor outcome. However, in AML lacking any predominant oncogenic driver mutations developing from clonal hematopoiesis and/or myelodysplasia (MDS) with one or several preleukemic mutations in cells from the HSC compartment, the definition of the cellular origin remains a challenge. Here, we summarize some of the key contributions that illustrate how mouse models have provided critical insights into the role of the cellular origin of AML (Table 1). Collectively many of these studies underline the importance of the cellular origin of AML not only for prognosis but also for personalized therapeutic strategies, particularly in AML subtypes that are driven by very potent oncogenes. However, several studies have also identified important limitations to consider when modeling the cellular origin of AML arising from multiple preleukemic mutations in which the ultimate driver is difficult to define.
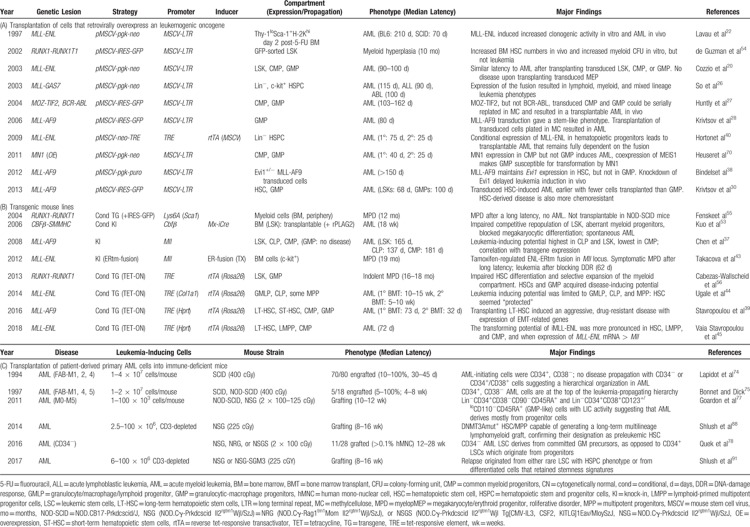
Table 1.
Modeling the Cellular Origin of AML in Mice
From clinical observations to transgenic mouse models
Pioneer studies by Phil Fialkow revealed that in chronic myeloid leukemia (CML) patients hematopoietic cells from multiple lineages carried the Philadelphia chromosome (the morphological correlate of the t(9;22)(q34;q11) translocation leading to expression of the BCR-ABL fusion) suggesting an origin high up in the hierarchy, most likely in stem cells. Expression of the same isotype of the polymorphic X-linked glucose-6-phosphate dehydrogenase in CML and AML cells led him to conclude that both malignancies may originate from multipotent cells within the HSC pool.3,4 Later, flow cytometer-assisted cell sorting combined with fluorescent in situ hybridization made possible the visualization of AML-associated cytogenetic aberrations in selected cells, which further supported a stem cell origin.5,6 Improved molecular tools facilitated the cloning of a large number of genetic alterations from AML blasts such as fusion oncogenes that turned out to be hallmarks of biologically distinct AML subtypes.7 The imminent question whether a given AML mutation might be a driver of the disease, initiated efforts to model AML, mostly in mice (Fig. 1). However, expression of AML-associated fusions as transgenes in the murine hematopoietic system by oocyte injections of randomly integrated expression cassettes turned out to be very challenging, as the regulatory elements of a given vector significantly influenced the resulting phenotype.8–11 Homologous recombination strategies ultimately led to the establishment of mice that developed AML upon expression of the respective mutations from their natural promoters.12
Figure 1.
Strategies to model AML in mice. There are 2 major approaches to model AML in mice. A particular AML-associated genetic mutation (eg, t(9;11) leading to expression of an MLL-AF9 fusion gene) may be expressed in the hematopoietic system of the mouse either by retroviral transduction of BM cells followed by transplantation, or by engineering the mutation in the germline (either randomly integrated or targeted to the proper locus allowing constitutive or inducible expression, respectively). To model AML with primary patient cells, leukemic cells (or enriched fractions thereof) are transplanted into sublethally irradiated immunodeficient mice of which NOD-scid-IL2rγnull (NSG) mice are currently the most frequently used strain. AML = acute myeloid leukemia, BM = bone marrow.
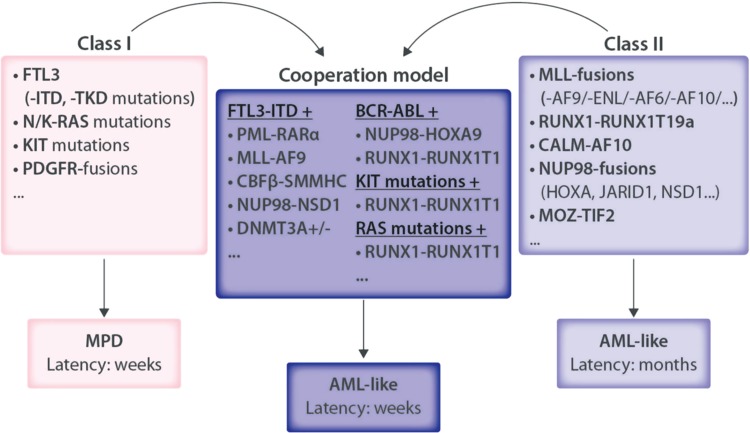
To facilitate the expression of a potential driver of leukemogenesis, David Baltimore's lab developed a protocol for efficient retroviral transduction of bone marrow (BM) cells followed by transplantation into irradiated syngeneic recipients. This approach allowed them to demonstrate that expression of the BCR-ABL fusion gene induces a CML-like disease in mice.13 A large number of subsequent studies revealed that retroviral expression of CML- and AML-associated mutations, which lead to constitutive activity of tyrosine kinases and related signaling mediators, induces a lethal myeloproliferative disorder (MPD) when retrovirally expressed in the mouse BM.14,15 By contrast, expression of AML-associated fusion genes involving transcription factors or epigenetic regulators was often not sufficient to phenocopy the disease in mice, with the exception of some fusions including the those affecting the mixed lineage leukemia (MLL) gene that potently induced AML, acute lymphoblastic leukemia (ALL), or MLL.16–23 Identification of coexisting mutations initiated another wave of studies demonstrating that coexpression of AML-associated fusion oncogenes involving transcription factors (such as the core binding factor [CBF] composed of RUNX1 and CBFbeta, the retinoic acid receptor alpha (RARA), or the MLL histone methyltransferase) together with mutated constitutively active tyrosine kinases such as FLT3, JAK2, or c-KIT, or mutants of signaling mediators such as the RAS-GTPases were sufficient to rapidly induce AML with full penetrance. These findings led Gary Gilliland and colleagues to formulate a genetic AML model based on the cooperation of 2 functional classes of mutations24 (Fig. 2).
Figure 2.
Functional cooperation of AML-associated mutations as shown by the retroviral transduction transplantation model supported a model of AML as a product of 2 classes of mutations. Multiple studies have shown that transplantation of BM cells retrovirally expressing AML-associated mutations in tyrosine kinases and other signaling mediators that support proliferation and survival without altering differentiation (often referred as “class I mutations”) leads to lethal myeloproliferative diseases (MPD). By contrast, mutations in epigenetic regulators that mostly affect differentiation and/or self-renewal (“class II mutations”) may led to development of an AML-like disease, normally after a rather long latency. Coexpression of a class I with a class II mutation achieved by retroviral cotransduction or by transduction of BM cells harboring a particular transgene generally leads to rapid development of a full penetrant AML-like disease in mice. AML = acute myeloid leukemia, BM = bone marrow.
Modeling the cellular origin of MLL fusion AML in mice
Multiple studies have demonstrated that expression of several (but not all) MLL fusion genes (hallmarks of infant acute leukemia and of some de novo and therapy-related adult ALL and AML) is able to induce a leukemic phenotype in mice.25,26 To address whether activation of an MLL-fusion in stem or progenitor cells would affect the biology of the disease, researchers profited from improved cell enrichment strategies to retrovirally transduce defined cellular subpopulations of the hematopoietic hierarchy. Several groups found that expression of some of the most prevalent MLL fusions (eg, MLL-ENL, MLL-AF9) or other transcription factor fusions (eg, MOZ-TIF2) induced a similar AML phenotype in mice when expressed in lineage marker-depleted (Sca-1 and c-Kit-positive) BM stem and progenitors (LSK) cells, common myeloid progenitors (CMP), or granulocytic-macrophage progenitors (GMP).20,27,28 In sharp contrast, expression of BCR-ABL induced a CML-like disease only when expressed in LSK but not in more committed progenitor cells.27 Molecular characterization of CML progressing to blast crisis suggested the existence of leukemic GMP-like cells with alterations leading to aberrant self-renewal, indicating that cooperating mutations in progenitor cells may ultimately transform CML into AML.29 Although retroviral expression of the MLL-AF9 fusion in the LSK or GMP fraction resulted in a similar AML phenotype, some origin-related differences in disease latency, leukemia-initiating potential, and gene expression signatures were observed.30 Collectively, these observations suggested that the cell of origin might influence the transforming capacity of particularly potent leukemogenic oncogenes.
Despite proven utility in the determination of the transforming potential of a leukemia-associated mutation, retroviral transduction of BM cells is associated with several important limitations.31,32 First, efficient transduction depends on active cell proliferation: however, stimulation of BM cell growth with cytokines is inherently associated with cellular differentiation, resulting in a significant bias of the transduced cells. In addition, the number of LT-HSC that can be obtained from a single mouse is limited, and despite constant improvements in culture systems, it remains a challenge to expand these cells while maintaining their stem cell characteristics. Moreover, expression of the receptors that are necessary for viral transduction seems to depend on the differentiation status of the cells.33,34 Second, retroviral transgene expression levels are unpredictable, and may result in overexpression far beyond the levels observed in primary leukemic cells. Third, retroviral integration can lead to activation or suppression of potentially cooperating oncogenes or tumor suppressor genes that, respectively, support clonal expansion. Transduction with the frequently used murine stem cell virus (MSCV)-based expression vector seems to be sufficient to partly in vitro immortalize primary murine BM cells.35,36 Finally, transduction of the selected target cells might also be dependent on the titer of viral particles that, depending on the size and nature of the transgene, can be difficult to standardize.
To overcome these limitations, researchers have generated a series of transgenic mouse models in which expression of a putative genetic AML driver is either controlled by the endogenous promoter or conditionally controlled from a heterologous promoter, for example, by a doxycycline (DOX)-inducible gene expression system. Rather surprisingly, transplantation of different subpopulations of the hematopoietic hierarchy from an MLL-AF9 knock-in mouse strain suggested that, in contrast to the retroviral model, the disease was induced by transplanting CMP, LSK, and common lymphoid progenitors (CLP) but not GMP. Interestingly, the leukemia-initiation potential of the different BM cell subpopulations seemed to correlate with the dose of the MLL-AF9 fusion, and the expression of the transcription factor EVI1, a putative direct downstream target.37 Retroviral expression of MLL-AF9 (“rMLL-AF9”) in murine LSK but not in more mature GMP was associated with high Evi1 expression, which turned out to be a general marker of poor prognosis associated with LT-HSC multilineage repopulating potential.38 The gene expression signatures of LSK-derived rMLL-AF9 leukemia were enriched for poor prognosis genes in human AML carrying MLL fusions suggesting that the cell of origin is able to determine clinically relevant subtypes of MLL-rearranged AML.30 To study the role of the cellular origin of MLL-AF9-mediated AML, we generated a DOX-inducible transgenic mouse (“iMLL-AF9”). DOX-induced iMLL-AF9 expression was dose-dependent and reached levels that were about 10-fold higher than endogenous Mll1 but 10 to 20 times lower than that achieved by rMLL-AF9 and led to a fully penetrant and reversible AML phenotype.39 Transplantation of naïve flow-sorted BM subpopulations into recipients on DOX revealed that the transfer of LT-HSC, short-term-HSC (ST-HSC), CMP, and GMP led to AML, whereas no disease was observed after transferring CLP, confirming earlier studies indicating that MLL-AF9 is able to transform HSC and more committed myeloid progenitors.15–19 Activation of the fusion in LT-HSC induced a more invasive disease than induction in ST-HSC or more committed downstream progenitor cells, further underlining that the cellular origin is a critical determinant for AML biology (Fig. 3A).
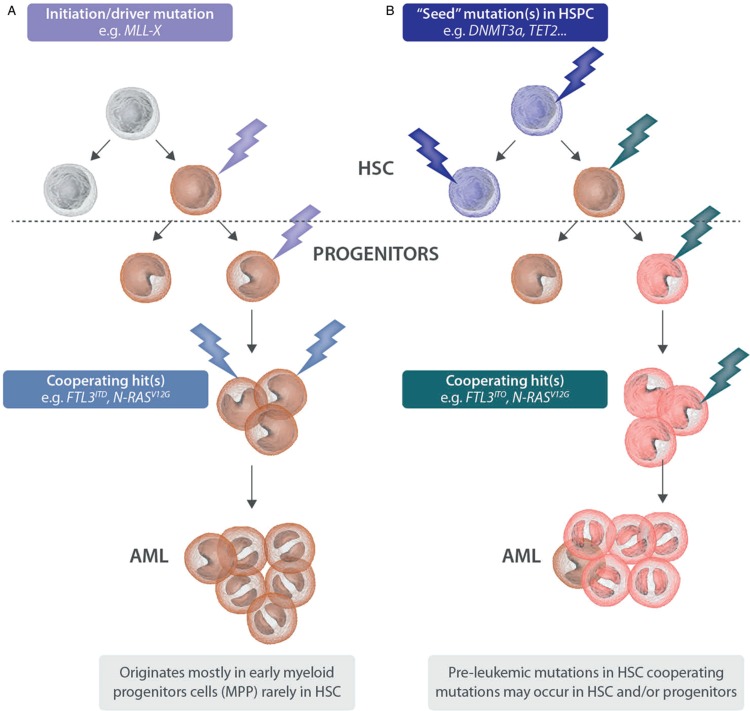
Figure 3.
Modeling the origin of AML. (A) Observations in transgenic mouse models suggest that AML driven by strong oncogenes such as MLL fusions mostly originates in multipotent progenitors: few cooperating mutations are needed to develop a symptomatic disease. In a minor fraction, the disease might occur from the HSC compartment leading to a particularly invasive and highly resistant phenotype. (B) A significant fraction of AML seems to be the product of cooperation of multiple mutations: early lesions in the HSC compartment provide a clonal advantage leading to a “preleukemic,” asymptomatic state; however, the gain of additional mutations (whether they occur in HSC or exclusively in more committed progenitor remains to be elucidated) is necessary to develop a symptomatic AML. AML = acute myeloid leukemia, BM = bone marrow, HSC = hematopoietic stem cell, MLL = mixed lineage leukemia.
The MLL-ENL fusion that results from t(11;19)(q23;p13) is mostly associated with ALL but has also been found in AML.28 Previous studies have shown that retroviral expression of MLL-ENL resulted in a very similar AML phenotype to that induced by the MLL-AF9 fusion.20,23,40–42 Further attempts modeling MLL-ENL-driven leukemia by knocking-in a tamoxifen (TX)-sensitive ENL-estrogen ligand binding domain (ER) fusion into the Mll locus resulted in an MPD: AML was only induced after chemically blocking the DNA damage response.43 Later, Bryder and colleagues established a DOX-regulated transgenic mouse model where MLL-ENL was regulated by a Tet operon expressed from the Col1a1 promoter.44 As activation of the fusion in different BM subpopulations followed by transplantation did not induce a disease they concluded that HSC and multipotent progenitors are inherently protected from malignant transformation by MLL-ENL. However, only cells in which MLL-ENL expression levels were equal to, or higher than, endogenous Mll1 were transformed by the fusion suggesting that the dose may be a critical denominator of the oncogenic fusion activity.44 We also established a DOX-inducible iMLL-ENL mouse line in which similar to iMLL-AF9 the Rosa26-controlled rtTA regulates the expression of the fusion integrated in the Hprt locus.45 Transplantation of different hematopoietic BM cell populations revealed that, in contrast to iMLL-AF9, the disease was only induced by HSC and early progenitors, but not GMP. DOX-induced iMLL-ENL mRNA expression levels exceeded expression of endogenous Mll1. We observed MLL-ENL fusion protein levels equal or exceeding MLL1 not only in iMLL-ENL+ leukemic cells, but also in t(11;19)+ human leukemic cell lines. Notably, we found that in primary leukemic cells from 5 out of 5 t(11;19)+ patients, the expression of MLL-ENL mRNA also surpassed MLL1. These observations are in line with recent work suggesting that the ratio of chimeric to wild-type MLL protein might be a critical determinant for maintenance of the transformed phenotype.46 This study also suggested that HSC or most likely bipotent lympho-myeloid progenitors might be the preferential targets of the MLL-ENL fusion. Notably transplantation of human cord blood HSPC retrovirally expressing MLL-ENL into NOD-SCID mice resulted in an acute leukemia-like phenotype with tumor cells expressing lymphoid markers without complete immunoglobulin gene rearrangements furthermore suggesting that HSC, CLP, or early B-cells are targets of MLL-ENL.47
Modeling the cellular origin of non-MLL fusion AML in mice
Modeling AML driven by CBF rearrangements such as the RUNX1-RUNX1T1 (initially referred to as AML1-ETO) or the CBFbeta-MYH11 fusion genes turned out to be rather difficult. Conventional knock-in models were hampered by embryonic lethality due to the dominant-negative impact of the fusion to wild-type RUNX1 and CBFbeta, both essential regulators of adult hematopoiesis.48–50 Conditional knock-in mice demonstrated that whereas expression of the CBFbeta-MYH11 fusion resulted in spontaneous AML after long latency, expression of RUNX1-RUNX1T1 seemed insufficient to induce an AML phenotype.51–53 The occurrence of tumors affecting different lineages including T-cells after chemical mutagenesis in RUNX1-RUNX1T1 knock-in mice suggested the cellular origin within the HSC compartment. In addition, retroviral RUNX1-RUNX1T1 overexpression resulted in a significantly increased number of LSK, further supporting the notion that this fusion might indeed affect the HSC compartment.54 Expression of a RUNX1-RUNX1T1 transgene under control of the Ly6A locus, which encodes the HSC marker Sca-1, resulted in a chronic MPD suggestive of some oncogenic activity in the HSC compartment.55 More recently, a DOX-inducible mouse model was generated in which the rtTA was integrated in the Rosa26 locus allowing RUNX1-RUNX1T1 expression in various blood lineages including LT-HSC. Transplantation of BM cells expressing iRUNX1-RUNX1T1 induced an MPD-like disease after a long latency. Transgenic GMP were able to further propagate the disease suggesting acquired self-renewal capacity of GMP.56 The presence of CBF fusions in perinatal blood spots on Guthrie cards from newborns, and the persistence of fusion-positive clones upon clinical remission suggest that these fusions might represent “preleukemic” mutations generally not sufficient to induce AML. Although cooperative mutations are necessary for development of the disease, AML blasts seem to be continuously dependent on these CBF fusions making them attractive targets for therapeutic intervention.57,58
The PML-RARA fusion gene is the product of t(15;17)(q22;q21) and the molecular hallmark of acute promyelocytic leukemia (APL).59 Earlier attempts to model this disease in mice showed that expression of a PML-RARA transgene controlled by myeloid promoters such as human cathepsin G (CTSG) or S100A8/MRP8 was able to initiate a myeloproliferative disease with progression to an APL-like phenotype after long latency with incomplete penetrance.9,60,61 Retroviral expression of potentially collaborating oncogenes such as FLT3-ITD, PIM2, or mutDMNT3A in BM cells from CTSG-PML-RARA transgenic mice reduced the latency and increased the penetrance of the APL phenotype.62–64 However, the putative origin in myeloid progenitors was challenged by the expression of T-cell lineage associated transcripts, T-cell receptor (TCR) rearrangements as well as by frequent expression of lymphoid surface markers on t(15;17)+ APL cells.65 Interestingly, in about 20% of APL patients the blasts express CD34 and CD2 on their surface; however, whether these markers are indeed indicative of an HSC origin remains unclear.66,67 More recently Ley and colleagues showed that expression of a PML-RARA transgene (controlled by the Ctsg or Mrp8 promoter) might not be limited to myeloid progenitors but might also occur in HSC and early progenitor cells.68 Relatively few studies have addressed the role of the cellular origin of aberrantly expressed or mutated genes in AML. Using a classical retroviral expression transplantation approach, Heuser and colleagues found that CMP but not GMP are susceptible to transformation by the meningioma-1 (MN1) gene often found aberrantly highly expressed in AML.69 Interestingly, overexpression of the HOX-interacting homeodomain transcription factor MEIS1 rendered GMP susceptible to MN1-induced transformation by regulation of common target genes, which suggests that the cellular targets may change due to accumulation of functionally cooperating mutations.70
Modeling the cellular origin by transplantation of human AML cells into immune-deficient mice
Pioneer work revealed that immune-deficient mice can be repopulated not only with normal human myeloid cells but also with primary leukemic blasts of the lymphoid and myeloid lineage.71–73 Subsequent studies provided proof of concept that the leukemia-initiating capacity of primary human AML cells in immune deficient NOD-SCID mice could be attributed to the CD34+CD38− subpopulation containing HSC, suggesting a hierarchical organization.74,75 Following the hypothesis that LIC are part of the HSC compartment, scientists studied RUNX1-RUNX1T1+ AML. They found that RUNX1-RUNXT1 mRNA was expressed in mature monocytes and B cells as well as HSC, lymphoid progenitors, and myeloid progenitors during disease remission in patients strongly suggesting that RUNX1− RUNX1T1+ HSC are capable of self-renewal and differentiate into mature blood cells in vivo.76
Another landmark study addressed the origin of AML by xenotransplanting a large number of primary human samples into NSG (NOD-scid-IL2rγ−/−) mice.77 Eighty percent of the CD34+ AML cases contained 2 predominant cell populations with an immune phenotype of CD38−CD90−CD45RA+ or CD38+CD110+CD45RA+ both corresponding to normal early and more mature hematopoietic progenitor cells rather than HSC. Importantly, both cell populations had leukemia initiating potential upon serial transplantation. There seemed to be a hierarchical organization as CD38−CD45RA+ cells with a higher number of LIC gave rise to leukemic GMP (with less LIC) but not vice versa. This work suggested that the majority of CD34+ AML originate in hematopoietic progenitor cells rather than in the HSC compartment. In AML composed of >98% CD34− cells, there seem to be multiple CD34− and CD34+ LIC-containing populations. Even though the expression profiles of CD34− LIC seem very similar to those of normal CD34− GMP, these LIC express multiple normal stem cell transcriptional regulators, suggesting that the nature of the genetic/epigenetic driver events determine a disordered transcriptional program that results in LIC arrested in differentiation at either the progenitor or precursor stages of hematopoiesis.78
NGS studies of primary human AML cells led to the identification of several recurrent mutations mostly affecting metabolic regulators such as IDH1/2, DNMT3A, or TET2. Sequencing of 200 AML genomes revealed that, in addition to fusion oncogenes, most AML patients carry about 10 to 15 coding single nucleotide variants at diagnosis.79 Earlier studies suggested that human HSPC naturally acquire about 5 to 10 potential mutations per year.80 DNA sequencing of peripheral blood cells from a large number of individuals not selected for any cancer or other hematologic diseases revealed molecular evidence for clonal hematopoiesis with somatic mutations in up to 10% of people over 65 years of age, of which mutations of DNMT3A, ASXL1, and TET2 were the most prevalent. Importantly, clonal hematopoiesis associated with distinct mutations turned out to be a strong risk factor with predictive power for the development of hematologic cancers.81–84 Very recent work suggested that the risk to develop AML can be predicted in healthy individuals by screening the preleukemic mutations that are significantly different from the age-related mutations.85 In fact, somatic mutations in IDH1, IDH2, TP53, DNMT3A, TET2, and spliceosome genes significantly increased the odds of developing AML.86 In the same context, other AML-related mutations affecting EZH2, CBP/EP300, CBL, JAK2, but also fusion oncogenes such as RUNX1-RUNX1T1 or CBFbeta-MYH11 have been found as “preleukemic” mutations in healthy individuals.87 Collectively these studies suggest that in a significant fraction of patients leukemogenesis can be considered as a continuum from normal to clonal hematopoiesis and to a preleukemic state from cells that carry particular epi/genetic variants initially conferring self-renewal but maintaining a normal differentiation potential. Upon the acquisition of additional alterations interfering with cellular differentiation progression to AML may occur (Fig. 3B). Such preleukemic states seem to represent a hematopoietic reservoir of mutant alleles that upon occurrence of the “matching” cooperating mutation will lead to the emergence of a dominant clone occurring not only in AML but also in a wide variety of hematologic malignancies such as MDS, MPD, CML, and lymphoid neoplasms.
Comparative sequencing of AML blasts at diagnosis and relapse using normal T-cells as surrogates for mutations originating from HSC allowed a better definition of preleukemic mutations. Comparing highly purified HSC, progenitor and mature cell populations, the Dick laboratory found that recurrent DNMT3A mutations exist at high allele frequency, but without coincident NPM1 mutations present in AML blasts.88 Therefore, the presence of DNMT3A mutations in both T-cells and AML cells, coupled with the lack of an NPM1 mutation in T-cells, suggested that DNMT3A mutations may represent drivers in ancestral HSC that give rise to both lineages. In addition, comparison of allele frequencies at diagnosis, remission, and relapse revealed an increased prevalence of the mutated allele at remission and relapse, suggesting that these preleukemic mutations do indeed represent a genetic reservoir for disease recurrence. Similarly, another study found preleukemic mutations in HSPC that were associated with long-term reconstitution in severely immune-compromised mice. They identified mutations in IDH2, DNMT3A, ASXL1, and IKFZ1 in HSPC, leukemic blasts, and also in T- and/or B-cells.89,90 Akin to Shlush et al, heterozygous DNMT3A and IDH1/2 mutations were among the most prevalent preleukemic alterations and not eradicated by chemotherapy. Most relapses contained the preleukemic mutations, some acquired additional mutations and some seemed to originate from different clones. More recently, combined genetic and functional analysis of purified subpopulations and PDX from paired diagnostic/relapse samples allowed the Dick laboratory to show that therapy-resistant cells were indeed already present at diagnosis. Relapse originated either from rare LSC with a stem/progenitor phenotype or from larger subclones of immunophenotypically more committed cells that retained a strong stemness-related gene expression signature.91
These functional studies were mostly based on PDX of highly selected AML patients into immune-deficient mouse strains such as NOD-SCID or NSG. They showed that in contrast to AML expressing a strong oncogenic driver such as an MLL fusion, the cellular origin might be difficult to define, and seems more a moving rather than a static target. In a given individual, several preleukemic mutations may occur in different, or even the same HSC (Fig. 3B). Depending on its nature the mutation may be eliminated, tolerated, or even selected toward clonal hematopoiesis. Collaborator mutations most likely predominantly occur in the highly dividing compartment of early multipotent (eg, lymphoid-primed multipotent progenitor cells) or more lineage restricted (eg, GMP) compartments leading to rapid expansion of one of few clones resulting in a symptomatic disease. In the minority of the cases, the initiation and/or the cooperating mutations might occur in the HSC compartment, which, similar to MLL-driven AML, may result in a more aggressive disease. It will be important to determine the combination of preleukemic mutations with the highest risk for progressing into de novo AML or inducing MDS/MPN that ultimately progresses to AML. Likewise, one needs to determine which preleukemic mutations represent the highest risk for being positively selected by chemotherapy to provide a reservoir for disease relapse.
Limitations of current AML mouse models
Apart from the obvious species-specific differences a major drawback of currently used mouse models is that the engineered genetic lesions only partially mimic those found in the leukemic blasts of a given patient, as even in the presence of leading cytogenetic lesions AML blasts often contain one or several potentially cooperating mutations.2 The current tools generally limit expression to 2 to 3 putative oncogenic drivers in the same cell, either by viral cotransduction or by breeding-in different (conditional) transgenes. Improved genome editing strategies, for example, by using the CRISPR/Cas9 system in mouse zygotes will most likely facilitate the generation of more complex mouse models.92 Another important limitation is the inability to ensure that a single cell at a particular stage of hematopoietic differentiation will undergo the initiating oncogenic mutation. In most models, even when conditionally regulated, the mutation occurs in a large population of genetically identical cells, failing to appropriately model the process of clonal selection starting from a single cell. Most in vivo human AML studies are based on PDX of patients into immune-deficient mouse strains. A major drawback of these models is the difference in the crosstalk between human hematopoietic cells and the murine stroma that are pivotal for a permissive microenvironment.93 To overcome these limitations, newer mouse strains are continuously being established that carry multiple transgenes expressing critical human hematopoietic factors, which have improved expansion of AML cells in mice.94,95
In addition, many mouse AML models are based on BM transplants into irradiated recipients. It was recently shown that total body irradiation of recipient mice permanently damages the BM stroma interfering with efficient engraftment and significantly affecting innate immunity.96 However, there is increasing evidence that the immune system plays an essential role in expansion of leukemic clones in the BM. Using conditional transgenic mouse models, researchers showed that the absence of an intact immune system resulted in a 10- to 1000-fold reduction in the rate, extent, and duration of regression of mouse T-cell ALL upon inactivation of a potent driver oncogene.97 The transplantation of rMLL-AF9 cells into an immune-competent mouse model revealed the potential role of the cytotoxic T-cell response in recognizing immunogenic antigens expressed on AML cells.98 Importantly, it was reported that numerous empty niches exist in the mouse BM in steady-state hematopoiesis that allow low-level donor HSPC engraftment in nonconditioned recipients.99 Several strategies are currently being explored to improve the transfer of hematopoietic cells without irradiation such as the application of blocking or toxin-conjugated antibodies targeting c-Kit or CD45, respectively, that selectively reduce the number HSC in the niche.100,101 It remains open for debate whether and how such strategies may affect the long-term innate and adaptive immunity of the host and clonal expansion of AML cells.
In contrast to the majority of the patients, putative leukemogenic mutations are often expressed in the BM of young mice that most likely do not carry any additional mutations that define clonal hematopoiesis or a preleukemic state. Therefore, novel mouse strains are needed in which the animals carry 1, 2, or more mutated alleles to appropriately phenocopy this situation. These mice will be particularly useful to determine the potency of a given mutation for progression toward AML. In addition, it is of great importance to model how these mutations will allow the cells to escape immune surveillance, and to ascertain whether boosting immune recognition might facilitate the elimination of relapsing clones. It is thus essential to functionally dissect which alterations are “seed” mutations (capable of inducing a preleukemic state and ultimately lead to AML) from those that provide “soil” for other events supporting leukemogenesis. Conditional activation of mutant alleles will be essential to control the appropriate sequence of events, as some of the most prevalent mutations such as FLT3-ITD seem to occur rather late during leukemogenesis, whereas others such as DNMT3A, IDH1/2, or TET2 mutations may primarily act as inducers of a preleukemic state. In fact, several studies have shown that expression of mutations associated with preleukemic states such as NPM1, TET2, or DNMT3A in HSPC carrying the FLT3-ITD mutation induce an AML phenotype in mice.102–107 However, whether conditional FLT3-ITD expression in preleukemic cells with DNMT3A or TET2 mutations will result in the same phenotype, remains to be shown. Recent studies suggest that the FLT3 protein is predominantly expressed in multipotent myeloid progenitor cells but only in a very small fraction of HSC.108 It is also not clear how to appropriately model AML originating from HSC as this cell compartment seems to be rather heterogeneous.109 In addition, expression of FLT3-ITD mutations may actively deplete the normal HSC reservoir suggesting that cooperating FLT3-ITD mutations arise in a more differentiated GMP-like cells rather than in the HSC compartment harboring preleukemic mutations.108 Several studies have shown that the nature of a given AML-associated mutation necessitates careful choice of approach in order to appropriately model its effect in different levels of the hematopoietic hierarchy. Inactivation of a gene that is recurrently targeted by loss-of-function mutations in AML will not necessarily result in an identical phenotype as conditional activation of mutant knock-in allele driven by its native promoter, as illustrated by the divergent phenotypes of different approaches to model the biology of AML-associated ASXL1 (additional sex combs like 1) gene mutations: while Asxl1 deletion resulted in MDS, expression of a mutated knock-in allele resulted in increased susceptibility for leukemic transformation of the HSC compartment in mice.110–112
What can one learn from mouse AML models?
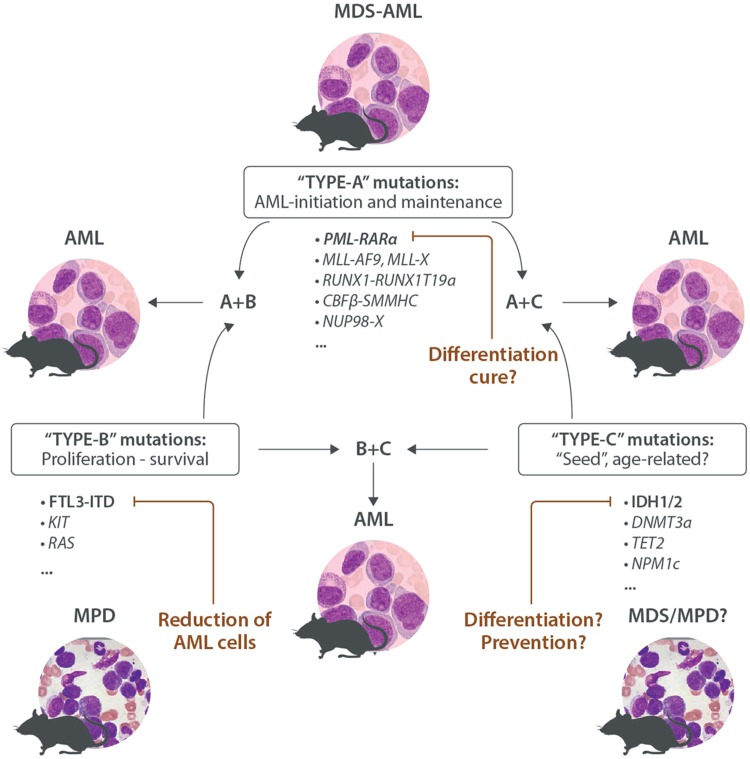
Despite the inherent limitations, mouse models have proven to be important pillars for a better understanding of AML biology and for searching for and validating novel therapeutic strategies. As outlined above, AML is generally modeled by expression of distinct leukemia-associated mutations in particular cells of the hematopoietic system. Hereby, expression of AML-associated fusion genes involving transcription factors or other epigenetic regulators may result in a leukemic disease after a long latency (eg, some MLL fusions) or is not sufficient to induce any disease (eg, CBF or RARA fusions) (Fig. 4). However, functional studies have shown that these alterations are generally necessary for the maintenance of the transformed phenotype and might therefore be rational therapeutic targets. The PML-RARA fusion in APL is the archetype of this type of mutations (hereby called “TYPE-A mutations”): targeted degradation of the fusion protein by treatment with pharmacologic doses of all-trans retinoid acid (ATRA) and/or arsenic trioxide (As2O3) is able to successfully treat patients without traditional chemotherapy.113
Figure 4.
Mouse models functionally classify AML-associated mutations. “TYPE-A mutations” initiate and/or maintain a leukemic phenotype in the mouse. Targeted inactivation might have the potential to cure the disease, as illustrated by successful therapy of PML-RARA-driven APL by ATRA and arsenic. “TYPE-B mutations” support proliferation and survival of HSPCs; they induce a myeloproliferative disease (MPD) in mice. Targeting TYPE-B mutations has antileukemic potential but is probably insufficient to eradicate the disease as illustrated by the clinical efficacy of small molecule FLT3 inhibitors. “TYPE-C mutations” (often affecting epigenetic regulators of DNA methylation) are often found in older individuals with clonal hematopoiesis and patients with MDS, MPN, and AML. Mouse models suggest that they may affect differentiation and self-renewal, as well as proliferation, leading to myeloproliferation and/or dysplasia, but they seem not sufficient to induce AML. TYPE-C mutations also seem to be suitable therapeutic targets as illustrated by the potent clinical activity recently reported for small molecules that selectively target AML-associated IDH2 mutations. Mouse models have demonstrated that the combination of A + B, A + C, and B + C TYPE mutations is sufficient to rapidly induce an AML-like disease. AML = acute myeloid leukemia, APL = acute promyelocytic leukemia, ATRA = all-trans retinoid acid, HSPCs = hematopoietic stem and progenitor cells, MDS = myelodysplasia, MPN = myeloproliferative neoplasm.
Alterations that constitutively activate kinases, such as fusions or mutants involving ABL, PDGFR, KIT, FLT3, or JAK2 or related signaling mediators (such as signaling mediators of the RAS-MAPK pathway) primarily support proliferation/survival rather than affecting differentiation of HSPC. Transgenic expression of such mutations (hereby called “TYPE-B mutations”) in the hematopoietic system of the mouse generally leads to the development of a lethal MPD. Mouse models of tyrosine kinase-driven MPD helped to preclinically explore the in vivo efficacy of several highly potent and selective small molecules such as inhibitors blocking JAK2 or FLT3.24,114–116 Based on the coexistence of TYPE-A and TYPE-B mutations in some AML or CML blast crisis patients, a large number of mouse models were consequentially generated and used to show that coexpression of TYPE-A and TYPE-B mutations did indeed cooperate to rapidly induce AML-like phenotypes often after a short latency.62,117–121
The third type of genetic alterations classically characterizes clonal hematopoiesis and preleukemic states including point mutations in IDH1/IDH2, DNMT3A, TET2, NPM1c, and others. Based on their potential we call them “seed mutations” or “TYPE-C mutations.” Several studies have shown that expression of these lesions in the hematopoietic system of the mouse induce a wide spectrum of diseases including MPD, MDS, or both but are generally not sufficient to induce AML.105,122–125 Several recent studies demonstrated that TYPE-C mutations functionally collaborate with TYPE-A or TYPE-B mutations resulting in AML with high penetrance in mice.106,107,126–129
All 3 types of mutations (A-B-C) are potential therapeutic targets. Targeting of TYPE-A mutations is most likely the best path to take to cure a fraction of AML in which the blasts remain fully addicted to the activity of the respective potent driver oncogene. Several critical protein-protein interactions have been identified and an increasing number of specific and potent small molecules are currently being tested that impair the transforming activity of these driver lesions. Following the success of the APL-associated PML-RARA fusion, most likely only targeted degradation below a yet-to-be-defined biological threshold will be able to efficiently eliminate the leukemic clones and ultimately cure the disease. Although targeting of TYPE-B mutations (eg, by small molecule inhibitors) clearly has clinical antileukemic activity it seems to be insufficient to eliminate the disease and therefore needs to be combined with traditional chemotherapy.130 Recently compounds have been developed that selectively block particular TYPE-C mutations such as mutant IDH1 and IDH2. Emerging mouse model studies have demonstrated that these compounds are able to differentiate AML blasts with potent preclinical and promising clinical activity leading to fast FDA approval as antileukemic therapeutics.131–133
Collectively, mouse models represent the cornerstones to study AML biology and provide platforms to explore novel therapeutic strategies. Nardella and coworkers proposed that mouse models could be integrated more closely with clinical trials, the so-called “coclinical trial,”134 where trials in mice would be operated contemporaneously with phase I/II clinical trials. Identification of key genetic and/or molecular factors that could affect patient outcome might be identified in appropriate mouse models allowing for better stratification of patient cohorts as well as the possibility of testing drug combinations to overcome acquired resistance.
Footnotes
Citation: Fisher JN, Kalleda N, Stavropoulou V, Schwaller J. The Impact of the Cellular Origin in Acute Myeloid Leukemia: Learning From Mouse Models. HemaSphere, 2018;00:00 http://dx.doi.org/10.1097/HS9.0000000000000152.
Funding/support: Our research is supported by grants from the Swiss National Science Foundation (SNF-31003A_149714/1), Swiss Cancer League (KFS-3019-08-2012), the Novartis Research Foundation (14B058), and the Gertrude von Meissner Foundation, Basel.
Disclosure: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–494. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Dohner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med. 2016;375:900–901. [DOI] [PubMed] [Google Scholar]
- 3.Fialkow PJ, Gartler SM, Yoshida A. Clonal origin of chronic myelocytic leukemia in man. Proc Natl Acad Sci USA. 1967;58:1468–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fialkow PJ, Singer JW, Adamson JW, et al. Acute nonlymphocytic leukemia: heterogeneity of stem cell origin. Blood. 1981;57:1068–1073. [PubMed] [Google Scholar]
- 5.Mehrotra B, George TI, Kavanau K, et al. Cytogenetically aberrant cells in the stem cell compartment (CD34+lin-) in acute myeloid leukemia. Blood. 1995;86:1139–1147. [PubMed] [Google Scholar]
- 6.Haase D, Feuring-Buske M, Konemann S, et al. Evidence for malignant transformation in acute myeloid leukemia at the level of early hematopoietic stem cells by cytogenetic analysis of CD34+ subpopulations. Blood. 1995;86:2906–2912. [PubMed] [Google Scholar]
- 7.Rowley JD. The role of chromosome translocations in leukemogenesis. Semin Hematol. 1999;36 suppl 7:59–72. [PubMed] [Google Scholar]
- 8.Early E, Moore MA, Kakizuka A, et al. Transgenic expression of PML/RARalpha impairs myelopoiesis. Proc Natl Acad Sci USA. 1996;93:7900–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grisolano JL, Wesselschmidt RL, Pelicci PG, et al. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 10.He LZ, Tribioli C, Rivi R, et al. Acute leukemia with promyelocytic features in PML/RARalpha transgenic mice. Proc Natl Acad Sci USA. 1997;94:5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David G, Terris B, Marchio A, et al. The acute promyelocytic leukemia PML-RAR alpha protein induces hepatic preneoplastic and neoplastic lesions in transgenic mice. Oncogene. 1997;14:1547–1554. [DOI] [PubMed] [Google Scholar]
- 12.Corral J, Lavenir I, Impey H, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. [DOI] [PubMed] [Google Scholar]
- 13.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. [DOI] [PubMed] [Google Scholar]
- 14.Grundler R, Miething C, Thiede C, et al. FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model. Blood. 2005;105:4792–4799. [DOI] [PubMed] [Google Scholar]
- 15.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 16.McCormack E, Bruserud O, Gjertsen BT. Animal models of acute myelogenous leukaemia—development, application and future perspectives. Leukemia. 2005;19:687–706. [DOI] [PubMed] [Google Scholar]
- 17.Fortier JM, Graubert TA. Murine models of human acute myeloid leukemia. Cancer Treat Res. 2010;145:183–196. [DOI] [PubMed] [Google Scholar]
- 18.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. [DOI] [PubMed] [Google Scholar]
- 19.So CW, Karsunky H, Wong P, et al. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. [DOI] [PubMed] [Google Scholar]
- 20.Cozzio A, Passegue E, Ayton PM, et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiMartino JF, Ayton PM, Chen EH, et al. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. [DOI] [PubMed] [Google Scholar]
- 22.Lavau C, Luo RT, Du C, et al. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci USA. 2000;97:10984–10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavau C, Szilvassy SJ, Slany R, et al. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly LM, Yu JC, Boulton CL, et al. CT53518, a novel selective FLT3 antagonist for the treatment of acute myelogenous leukemia (AML). Cancer Cell. 2002;1:421–432. [DOI] [PubMed] [Google Scholar]
- 25.Milne TA. Mouse models of MLL leukemia: recapitulating the human disease. Blood. 2017;129:2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.So CW, Karsunky H, Passegue E, et al. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–171. [DOI] [PubMed] [Google Scholar]
- 27.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. [DOI] [PubMed] [Google Scholar]
- 28.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. [DOI] [PubMed] [Google Scholar]
- 29.Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. [DOI] [PubMed] [Google Scholar]
- 30.Krivtsov AV, Figueroa ME, Sinha AU, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27:852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren R. Modeling the dosage effect of oncogenes in leukemogenesis. Curr Opin Hematol. 2004;11:25–34. [DOI] [PubMed] [Google Scholar]
- 32.Haviernik P, Zhang Y, Bunting KD. Retroviral transduction of murine hematopoietic stem cells. Methods Mol Biol. 2008;430:229–241. [DOI] [PubMed] [Google Scholar]
- 33.von Laer D, Thomsen S, Vogt B, et al. Entry of amphotropic and 10A1 pseudotyped murine retroviruses is restricted in hematopoietic stem cell lines. J Virol. 1998;72:1424–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orlic D, Girard LJ, Jordan CT, et al. The level of mRNA encoding the amphotropic retrovirus receptor in mouse and human hematopoietic stem cells is low and correlates with the efficiency of retrovirus transduction. Proc Natl Acad Sci USA. 1996;93:11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosticardo M, Ghosh A, Du Y, et al. Self-inactivating retroviral vector-mediated gene transfer induces oncogene activation and immortalization of primary murine bone marrow cells. Mol Ther. 2009;17:1910–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W, Kumar AR, Hudson WA, et al. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindels EM, Havermans M, Lugthart S, et al. EVI1 is critical for the pathogenesis of a subset of MLL-AF9-rearranged AMLs. Blood. 2012;119:5838–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stavropoulou V, Kaspar S, Brault L, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30:43–58. [DOI] [PubMed] [Google Scholar]
- 40.Horton SJ, Walf-Vorderwulbecke V, Chatters SJ, et al. Acute myeloid leukemia induced by MLL-ENL is cured by oncogene ablation despite acquisition of complex genetic abnormalities. Blood. 2009;113:4922–4929. [DOI] [PubMed] [Google Scholar]
- 41.Schwieger M, Schuler A, Forster M, et al. Homing and invasiveness of MLL/ENL leukemic cells is regulated by MEF2C. Blood. 2009;114:2476–2488. [DOI] [PubMed] [Google Scholar]
- 42.Liu T, Jankovic D, Brault L, et al. Functional characterization of high levels of meningioma 1 as collaborating oncogene in acute leukemia. Leukemia. 2010;24:601–612. [DOI] [PubMed] [Google Scholar]
- 43.Takacova S, Slany R, Bartkova J, et al. DNA damage response and inflammatory signaling limit the MLL-ENL-induced leukemogenesis in vivo. Cancer Cell. 2012;21:517–531. [DOI] [PubMed] [Google Scholar]
- 44.Ugale A, Norddahl GL, Wahlestedt M, et al. Hematopoietic stem cells are intrinsically protected against MLL-ENL-mediated transformation. Cell Rep. 2014;9:1246–1255. [DOI] [PubMed] [Google Scholar]
- 45.Stavropoulou V, Almosailleakh M, Royo H, et al. A novel inducible mouse model of MLL-ENL-driven mixed lineage acute leukemia. HemaSphere. 2018;2:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang K, Volk AG, Haug JS, et al. Therapeutic targeting of MLL degradation pathways in MLL-rearranged leukemia. Cell. 2017;168:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barabe F, Kennedy JA, Hope KJ, et al. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. [DOI] [PubMed] [Google Scholar]
- 48.Castilla LH, Wijmenga C, Wang Q, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. [DOI] [PubMed] [Google Scholar]
- 49.Yergeau DA, Hetherington CJ, Wang Q, et al. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. [DOI] [PubMed] [Google Scholar]
- 50.Okuda T, Cai Z, Yang S, et al. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 51.Rhoades KL, Hetherington CJ, Harakawa N, et al. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000;96:2108–2115. [PubMed] [Google Scholar]
- 52.Higuchi M, O’Brien D, Kumaravelu P, et al. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. [DOI] [PubMed] [Google Scholar]
- 53.Kuo YH, Landrette SF, Heilman SA, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9:57–68. [DOI] [PubMed] [Google Scholar]
- 54.de Guzman CG, Warren AJ, Zhang Z, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol. 2002;22:5506–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fenske TS, Pengue G, Mathews V, et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA. 2004;101:15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabezas-Wallscheid N, Eichwald V, de Graaf J, et al. Instruction of haematopoietic lineage choices, evolution of transcriptional landscapes and cancer stem cell hierarchies derived from an AML1-ETO mouse model. EMBO Mol Med. 2013;5:1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heidenreich O, Krauter J, Riehle H, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157–3163. [DOI] [PubMed] [Google Scholar]
- 58.Hatlen MA, Wang L, Nimer SD. AML1-ETO driven acute leukemia: insights into pathogenesis and potential therapeutic approaches. Front Med. 2012;6:248–262. [DOI] [PubMed] [Google Scholar]
- 59.Gillard EF, Solomon E. Acute promyelocytic leukaemia and the t(15;17) translocation. Semin Cancer Biol. 1993;4:359–367. [PubMed] [Google Scholar]
- 60.Brown D, Kogan S, Lagasse E, et al. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:2551–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westervelt P, Lane AA, Pollock JL, et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARalpha expression. Blood. 2003;102:1857–1865. [DOI] [PubMed] [Google Scholar]
- 62.Kelly LM, Kutok JL, Williams IR, et al. PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model. Proc Natl Acad Sci USA. 2002;99:8283–8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanyam D, Belair CD, Barry-Holson KQ, et al. PML-RAR{alpha} and Dnmt3a1 cooperate in vivo to promote acute promyelocytic leukemia. Cancer Res. 2010;70:8792–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agrawal-Singh S, Koschmieder S, Gelsing S, et al. Pim2 cooperates with PML-RARalpha to induce acute myeloid leukemia in a bone marrow transplantation model. Blood. 2010;115:4507–4516. [DOI] [PubMed] [Google Scholar]
- 65.Chapiro E, Delabesse E, Asnafi V, et al. Expression of T-lineage-affiliated transcripts and TCR rearrangements in acute promyelocytic leukemia: implications for the cellular target of t(15;17). Blood. 2006;108:3484–3493. [DOI] [PubMed] [Google Scholar]
- 66.Lin P, Hao S, Medeiros LJ, et al. Expression of CD2 in acute promyelocytic leukemia correlates with short form of PML-RARalpha transcripts and poorer prognosis. Am J Clin Pathol. 2004;121:402–407. [DOI] [PubMed] [Google Scholar]
- 67.Albano F, Mestice A, Pannunzio A, et al. The biological characteristics of CD34+ CD2+ adult acute promyelocytic leukemia and the CD34 CD2 hypergranular (M3) and microgranular (M3v) phenotypes. Haematologica. 2006;91:311–316. [PubMed] [Google Scholar]
- 68.Welch JS, Yuan W, Ley TJ. PML-RARA can increase hematopoietic self-renewal without causing a myeloproliferative disease in mice. J Clin Invest. 2011;121:1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heuser M, Beutel G, Krauter J, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–3905. [DOI] [PubMed] [Google Scholar]
- 70.Heuser M, Yun H, Berg T, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer Cell. 2011;20:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamel-Reid S, Dick JE. Engraftment of immune-deficient mice with human hematopoietic stem cells. Science. 1988;242:1706–1709. [DOI] [PubMed] [Google Scholar]
- 72.Kamel-Reid S, Letarte M, Sirard C, et al. A model of human acute lymphoblastic leukemia in immune-deficient SCID mice. Science. 1989;246:1597–1600. [DOI] [PubMed] [Google Scholar]
- 73.Sawyers CL, Gishizky ML, Quan S, et al. Propagation of human blastic myeloid leukemias in the SCID mouse. Blood. 1992;79:2089–2098. [PubMed] [Google Scholar]
- 74.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. [DOI] [PubMed] [Google Scholar]
- 75.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. [DOI] [PubMed] [Google Scholar]
- 76.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci USA. 2000;97:7521–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goardon N, Marchi E, Atzberger A, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. [DOI] [PubMed] [Google Scholar]
- 78.Quek L, Otto GW, Garnett C, et al. Genetically distinct leukemic stem cells in human CD34- acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213:1513–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ley TJ, Miller C, et al. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKerrell T, Park N, Moreno T, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559:400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Desai P, Mencia-Trinchant N, Savenkov O, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24:1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shlush LI, Minden MD. Preleukemia: the normal side of cancer. Curr Opin Hematol. 2015;22:77–84. [DOI] [PubMed] [Google Scholar]
- 88.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4:149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Corces-Zimmerman MR, Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 2014;28:2276–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shlush LI, Mitchell A, Heisler L, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547:104–108. [DOI] [PubMed] [Google Scholar]
- 92.Sakurai T, Kamiyoshi A, Kawate H, et al. A non-inheritable maternal Cas9-based multiple-gene editing system in mice. Sci Rep. 2016;6:20011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rivera-Cruz CM, Shearer JJ, Figueiredo Neto M, et al. The immunomodulatory effects of mesenchymal stem cell polarization within the tumor microenvironment niche. Stem Cells Int. 2017;2017:4015039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ellegast JM, Rauch PJ, Kovtonyuk LV, et al. inv(16) and NPM1mut AMLs engraft human cytokine knock-in mice. Blood. 2016;128:2130–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rongvaux A, Willinger T, Martinek J, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abbuehl JP, Tatarova Z, Held W, et al. Long-term engraftment of primary bone marrow stromal cells repairs niche damage and improves hematopoietic stem cell transplantation. Cell Stem Cell. 2017;21:241–255. [DOI] [PubMed] [Google Scholar]
- 97.Rakhra K, Bachireddy P, Zabuawala T, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell. 2010;18:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hasegawa K, Tanaka S, Fujiki F, et al. An immunocompetent mouse model for MLL/AF9 leukemia reveals the potential of spontaneous cytotoxic T-cell response to an antigen expressed in leukemia cells. PLoS ONE. 2015;10:e0144594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shimoto M, Sugiyama T, Nagasawa T. Numerous niches for hematopoietic stem cells remain empty during homeostasis. Blood. 2017;129:2124–2131. [DOI] [PubMed] [Google Scholar]
- 100.Xue X, Pech NK, Shelley WC, et al. Antibody targeting KIT as pretransplantation conditioning in immunocompetent mice. Blood. 2010;116:5419–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palchaudhuri R, Saez B, Hoggatt J, et al. Non-genotoxic conditioning for hematopoietic stem cell transplantation using a hematopoietic-cell-specific internalizing immunotoxin. Nat Biotechnol. 2016;34:738–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mallardo M, Caronno A, Pruneri G, et al. NPMc+ and FLT3_ITD mutations cooperate in inducing acute leukaemia in a novel mouse model. Leukemia. 2013;27:2248–2251. [DOI] [PubMed] [Google Scholar]
- 103.Mupo A, Celani L, Dovey O, et al. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia. 2013;27:1917–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rau R, Magoon D, Greenblatt S, et al. NPMc+ cooperates with Flt3/ITD mutations to cause acute leukemia recapitulating human disease. Exp Hematol. 2014;42:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rasmussen KD, Jia G, Johansen JV, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meyer SE, Qin T, Muench DE, et al. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Discov. 2016;6:501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mead AJ, Neo WH, Barkas N, et al. Niche-mediated depletion of the normal hematopoietic stem cell reservoir by Flt3-ITD-induced myeloproliferation. J Exp Med. 2017;214:2005–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eaves CJ. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125:2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abdel-Wahab O, Gao J, Adli M, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang J, Li Z, He Y, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nagase R, Inoue D, Pastore A, et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018;215:1729–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lo-Coco F, Avvisati G, Vignetti M, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369:111–121. [DOI] [PubMed] [Google Scholar]
- 114.Armstrong SA, Kung AL, Mabon ME, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. [DOI] [PubMed] [Google Scholar]
- 115.Geron I, Abrahamsson AE, Barroga CF, et al. Selective inhibition of JAK2-driven erythroid differentiation of polycythemia vera progenitors. Cancer Cell. 2008;13:321–330. [DOI] [PubMed] [Google Scholar]
- 116.Meyer SC, Keller MD, Chiu S, et al. CHZ868, a type II JAK2 inhibitor, reverses type I JAK inhibitor persistence and demonstrates efficacy in myeloproliferative neoplasms. Cancer Cell. 2015;28:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dash AB, Williams IR, Kutok JL, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci USA. 2002;99:7622–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schessl C, Rawat VP, Cusan M, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest. 2005;115:2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stubbs MC, Kim YM, Krivtsov AV, et al. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao L, Melenhorst JJ, Alemu L, et al. KIT with D816 mutations cooperates with CBFB-MYH11 for leukemogenesis in mice. Blood. 2012;119:1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reckzeh K, Bereshchenko O, Mead A, et al. Molecular and cellular effects of oncogene cooperation in a genetically accurate AML mouse model. Leukemia. 2012;26:1527–1536. [DOI] [PubMed] [Google Scholar]
- 122.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Inoue S, Li WY, Tseng A, et al. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30:337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mayle A, Yang L, Rodriguez B, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Quivoron C, Couronne L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. [DOI] [PubMed] [Google Scholar]
- 126.Shih AH, Meydan C, Shank K, et al. Combination targeted therapy to disrupt aberrant oncogenic signaling and reverse epigenetic dysfunction in IDH2- and TET2-mutant acute myeloid leukemia. Cancer Discov. 2017;7:494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kunimoto H, Meydan C, Nazir A, et al. Cooperative epigenetic remodeling by TET2 loss and NRAS mutation drives myeloid transformation and MEK inhibitor sensitivity. Cancer Cell. 2018;33:44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Palam LR, Mali RS, Ramdas B, et al. Loss of epigenetic regulator TET2 and oncogenic KIT regulate myeloid cell transformation via PI3K pathway. JCI Insight. 2018;3:94679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang X, Su J, Jeong M, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48:1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377:454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yen K, Travins J, Wang F, et al. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 2017;7:478–493. [DOI] [PubMed] [Google Scholar]
- 132.Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant-IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Amatangelo MD, Quek L, Shih A, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 2017;130:732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nardella C, Lunardi A, Patnaik A, et al. The APL paradigm and the “co-clinical trial” project. Cancer Discov. 2011;1:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]