Abstract
Background
Whether associations between circulating metabolites and prostate cancer are causal is unknown. We report on the largest study of metabolites and prostate cancer (2,291 cases and 2,661 controls) and appraise causality for a subset of the prostate cancer-metabolite associations using two-sample Mendelian randomization (MR).
Materials and Methods
The case-control portion of the study was conducted in nine UK centres with men aged 50–69 years who underwent prostate-specific antigen (PSA) screening for prostate cancer within the Prostate testing for cancer and Treatment (ProtecT) trial. Two data sources were used to appraise causality: a genome-wide association study (GWAS) of metabolites in 24,925 participants and a GWAS of prostate cancer in 44,825 cases and 27,904 controls within the Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium.
Results
Thirty-five metabolites were strongly associated with prostate cancer (p <0.0014, multiple-testing threshold). These fell into four classes: i) lipids and lipoprotein subclass characteristics (total cholesterol and ratios, cholesterol esters and ratios, free cholesterol and ratios, phospholipids and ratios, and triglyceride ratios); ii) fatty acids and ratios; iii) amino acids; iv) and fluid balance. Fourteen top metabolites were proxied by genetic variables, but MR indicated these were not causal.
Conclusions
We identified 35 circulating metabolites associated with prostate cancer presence, but found no evidence of causality for those 14 testable with MR. Thus, the 14 MR-tested metabolites are unlikely to be mechanistically important in prostate cancer risk.
Impact
The metabolome provides a promising set of biomarkers that may aid prostate cancer classification.
Introduction
Prostate cancer is the most frequently diagnosed malignancy among men worldwide.1 Despite huge geographical variation in incidence and mortality (suggestive of environmental causes), the only established risk factors are age, race, and family history,2 while adiposity is associated with advanced prostate cancer.3 Metabolic dysregulation is a hallmark of carcinogenesis4, and various circulating metabolites have been associated with both prostate cancer presence and aggressiveness.2,5–15 However, there are limitations with the existing evidence-base. First, the metabolites detected as being associated with prostate cancer have varied between studies, with no clear pattern of associations emerging. Second, most previous studies have been small, having fewer than 100 cases (Supplementary Table 1 in the Supplement)5 and hence liable to the play of chance, yielding both false-positive and false-negative findings. Third, the nature of any observed associations must be interpreted cautiously, because epidemiological studies are highly susceptible to various biases that preclude causal inference.16,17 For instance, metabolites may be associated with other factors that affect prostate cancer development (confounding), or the presence of prostate cancer may alter metabolites (reverse causation). Mendelian randomization (MR) is a method for appraising causality that uses genetic instrumental variables to proxy for exposures that can be otherwise confounded or subject to reverse causation. Such an approach may be used to distinguish causally relevant intervention targets from biomarkers that are non-causally associated with prostate cancer. The latter may nevertheless be of potential value in risk prediction (e.g. if the biomarker acts as a strong proxy for other factors leading to disease) or disease detection (e.g. if early disease leads to alterations in the circulating metabolome).
Methods
Observational study
Study Population
We undertook a case-control study within the Prostate testing for cancer and Treatment (ProtecT) trial (ISRCTN20141297).18–20 During recruitment to ProtecT, 228,966 men aged 50–69 years at 337 general practices in nine UK centres (Birmingham, Bristol, Cambridge, Cardiff, Edinburgh, Leeds, Leicester, Newcastle, Sheffield) were invited between 2001 and 2009 to attend a clinic for consideration of a prostate-specific antigen (PSA) test. Overall, 100,444 men attended the clinic where a PSA test was offered to 82,429 men deemed eligible to enroll into the ProtecT treatment trial, after a fully-informed, 30-minute discussion with a nurse. Men with PSA levels ≥3 ng/mL were offered a 10-core diagnostic biopsy. Tumours were histologically confirmed, assigned a Gleason score by uropathologists, and clinically staged using the TNM classification.21 Men with a PSA <3 ng/mL or a raised PSA (≥3 ng/mL) but a negative biopsy without development of prostate cancer during the follow-up protocol were eligible as controls. Controls were randomly selected from the same five-year age-band (age at PSA test) and GP/family practice, as cases.22
In the current analysis, in concert with the guidelines of the National Institute for Health and Care Excellence (NICE)23, men with stage T3 or T4 tumours (‘localized advanced’), Gleason score ≥8, or with a PSA level at diagnosis >20 ng/ml, were classified as ‘high-risk’. Men with stage T1 or T2 tumours, Gleason score <8, or with a PSA level at diagnosis ≤20 ng/ml were classified as ‘low-risk’. Participants in the present analysis consisted of those with self-reported (White) European ancestry.
Ethics
All men provided written informed consent prior to inclusion into ProtecT. The Trent Multicentre Research Ethics Committee (MREC) approved ProtecT (MREC/01/4/025) and the linked ProMPT study, which collected biological material (MREC/01/4/061), including serum used for the present study of metabolites.
Laboratory Analyses
Two hundred and twenty-seven quantified metabolic traits (henceforth “metabolites”) were obtained per sample of serum using a proton nuclear magnetic resonance (NMR) spectroscopy-based metabolomics platform (Nightingale Health, Helsinki, Finland). Details of the methodology have been described elsewhere.24 Briefly, 100 μL serum was mixed with sodium phosphate buffer and transferred to NMR tubes using an eight-channel, Varispan Janus liquid handling robot (PerkinElmer). Two 1D NMR spectra were acquired using a 500 MHz Bruker Avance III HD spectrometer and analysed bioinformatically for absolute quantification of lipoprotein subclasses, their particle concentrations and composition, lipoprotein particle size, apolipoprotein A-I and B, multiple cholesterol and triglyceride measures, albumin, various fatty acids, as well as numerous low-molecular-weight metabolites covering amino acids (including branched-chained and aromatic), glycolysis-related measures, and ketone bodies. The method has been widely used in epidemiological research and recently reviewed.24,25
Statistical Analysis
All analyses were performed using R (version 3.4.1). Two thousand two hundred and ninety-one men with screen-detected prostate cancer (348 high-risk; 1939 low-risk) and 2,661 controls had NMR metabolites measured in ProtecT. The distribution of baseline characteristics in cases versus controls was compared using Wilcoxon rank sum tests for continuous variables and a Chi-squared statistic for categorical variables. Multiple imputation using the ‘mice’ R package, and based on a subset of 78 metabolites chosen at random (given imputation constraints), was used to impute family history of prostate cancer, unknown for 11% of participants. Family history and age were selected as covariates in multi-variable models of prostate cancer risk, as those factors are strongly associated with prostate cancer and are potential confounders of the exposure-outcome relationship. We also adjusted for the primary-care centre where patients were registered. Metabolite trait concentrations/ratios were log-transformed and then scaled to standard-deviation scores to allow direct comparison of the magnitude of the effect of traits with different units on prostate cancer. A dictionary of metabolic traits with units before standardization is available in Supplementary Table 2 in the Supplement.
Multivariable logistic regression was performed to compare the odds of total prostate cancer (versus controls) per log-transformed, then standard-deviation-scaled metabolite concentration, such that each metabolite has a standard deviation of one. As a sensitivity analysis, we also examined the odds of prostate cancer by high-and low-risk case status and performed tests of the differences between odds ratios (took the absolute difference between the odds ratios (δ); calculated the standard error (SE) for δ using the SEs from each comparison set, such that and refer respectively to the SEs of the first comparison and second comparison sets,; calculated z scores, ; and calculated p-values for the z scores) for the following comparisons: differences in odds ratios for high-risk results and total results, the low-risk results and total results, and high-risk and low-risk results. In addition (also as a sensitivity analysis) we examined the correlation between the metabolites and PSA, given that our population of participants was screen-(PSA) detected.
To account for multiple testing and the correlation between the metabolic measures, principal component analysis was carried out on z-scored metabolic trait data.26 We calculated that the first 37 principal components explained >99% of the variance in the data and set our statistical threshold top < 0.05/37 (=0.0014), equivalent to p<0.05 after adjusting for multiple testing.
Causal Analysis
To assess causality, we used MR, a causal analysis method which exploits the random assortment of alleles in an instrumental variable (IV) framework to address confounding and reverse causation that preclude causal inference in observational studies.27,28 Germline genetic variants associated with each metabolite of interest can serve as proxies (IVs) for those metabolites in models examining the causal effects of metabolic traits on prostate cancer, if a number of assumptions are met: i) the IVs (genetic variants) are robustly associated with metabolites; ii) the IVs are independent of confounders of the metabolites and prostate cancer; and iii) the IVs are not pleiotropically associated with the prostate cancer; i.e. they are associated with prostate cancer only through the metabolites they are instrumenting and not associated with prostate cancer through other exposures.29
From the literature, we know there are strong associations between single-nucleotide polymorphisms (SNPs) and metabolite levels;30–33 therefore, these SNPs can serve as instruments in Mendelian randomization analyses.34–36 For instance, the median proportion of variance explained for metabolite associations in Kettunen et al. (2016) was 5% and ranged from 0.2% for acetoacetate to 12.5% for glycine.33 To implement MR, we identified independent (those not in linkage disequilibrium; r2 <0.01) SNPs that were robustly associated at genome-wide signficance (i.e. p-value < 5×10–8) with metabolites in the Kettunen et al. (2016) genome-wide association study (GWAS) of 123 circulating metabolites in 24,925 participants from 14 European cohorts.33 These SNPs were chosen as IVs for our metabolites. We could not instrument 113 of the 227 NMR-quantified metabolic traits; sixty five of these traits were ratio measures not included in the GWAS and 48 were other types of traits that had no genetic proxy.
To leverage power from large samples, we performed two-sample MR,27,37,38 whereby we obtained summary data on the effects of the SNPs that acted as genetic instruments for each metabolite on prostate cancer from a separate data source, the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium. This consortium involved 52 prostate cancer case-control studies on which genome-wide association studies (GWAS) had been conducted.39–44 The current MR analysis was based on 44,825 prostate cancer cases and 27,904 controls from within 48 of the 52 PRACTICAL cohorts of men with European ancestry.
To implement MR, we undertook the following process for each of the SNPs identified as a proxy for each metabolite: we extracted the effect and non-effect allele, and the log odds ratio (OR) and standard error per copy of the effect allele from the PRACTICAL GWAS. We used these data to construct our genetic instruments for our circulating metabolic traits and calculated the log OR for prostate cancer per standard deviation (SD) increase in metabolic measure levels using the two-sample MR Maximum likelihood estimator.45 (Supplementary Table 3 in the Supplement contains the characteristics of the genetic variants associated with metabolites that were selected as instruments.) In computing the effect estimates based on MR, the effect estimates for each SNP-prostate cancer association were meta-analysed.
Two-sample MR analyses were performed in R using the TwoSampleMR package46.
Results
Characteristics of men in the ProtecT study
Table 1 compares the distribution of selected characteristics in cases versus controls. The median age (63 years) and body mass index (BMI, 27 kg/m2) were the same, but more cases than controls had a family history of prostate cancer (8% versus 5%; p <0.001).
Table 1.
Baseline characteristics (medians and interquartile ranges, or percents) for ProtecT cases and controls
| Characteristic | Case (n=2291) | Control (n=2661) |
P-value1 |
|---|---|---|---|
| Age | 63 (59–67) | 63 (59–66) | 0.709 |
| Family history of prostate cancer (%)2 | 170 (8) | 128 (5) | <0.001 |
| BMI (kg/m2) | 27 (24–29) | 27 (24–29) | 0.872 |
ProtecT=Prostate Testing for Cancer and Treatment trial.
P-value based on Chi-squared tests (for categorical variables) and Wilcoxon rank sum tests (for continuous variables).
Family history data available on only 90% of these subjects.
BMI data available on only 64% of these subjects.
Observational associations of metabolic traits on prostate cancer (ProtecT)
Thirty-five metabolites were associated with odds of prostate cancer at p <0.0014 (Table 2, Figure 1, and Supplementary Table 4 in the Supplement). The following increased the odds of prostate cancer: i) Lipids and lipoprotein particle concentration, specifically, total lipids (TL) in small high-density lipoprotein (HDL) and concentration of small HDL particles; ii) Total cholesterol (TC) and TC compositional ratios, namely, TC in HDL3, TC in medium low-density lipoprotein (LDL), small HDL, and small LDL; and TC:TL ratios in medium LDL, small HDL, small LDL, and small very low-density lipoprotein (VLDL); iii) Cholesterol esters (CE) and CE compositional ratios, specifically: CE in medium LDL, small HDL, and small LDL; CE-to-TL ratios in medium LDL, CE:TL ratios in small HDL and small LDL; iv) Free cholesterol (FC) and a FC compositional ratios, namely, FC in IDL, large LDL, and medium HDL, and FC-to-TL ratio in medium HDL; v) Phospholipids (PL) and PL compositional ratios, including: PL in intermediate-density lipoprotein (IDL) and very small VLDL, and PL:TL ratios in medium VLDL and very small VLDL; vi) and the protein albumin; vii) the ratio of omega-6 fatty acids (FA) to total FA.
Table 2.
Among ProtecT cases and controls, odds of prostate cancer for top metabolites1
| Name | OR | LCI | UCI | P-value2 |
|---|---|---|---|---|
| Lipids and Lipoprotein Subclass Characteristics | ||||
| Small HDL (particle concentration) | 1.102 | 1.042 | 1.167 | 0.00070 |
| VLDL (mean particle diameter) | 0.906 | 0.856 | 0.958 | 0.00056 |
| Cholesterol Esters (CE) | ||||
| CE in medium LDL | 1.105 | 1.044 | 1.170 | 0.00058 |
| CE to total lipids ratio in medium LDL | 1.108 | 1.044 | 1.180 | 0.00062 |
| CE in small HDL | 1.135 | 1.071 | 1.205 | 0.00002 |
| CE to total lipids ratio in small HDL | 1.100 | 1.038 | 1.167 | 0.00111 |
| CE in small LDL | 1.099 | 1.039 | 1.165 | 0.00107 |
| CE to total lipids ratio in small LDL | 1.100 | 1.037 | 1.169 | 0.00139 |
| Free (FC) & Total Cholesterol (TC) | ||||
| FC in IDL | 1.105 | 1.044 | 1.170 | 0.00057 |
| FC in large LDL | 1.101 | 1.041 | 1.166 | 0.00080 |
| FC in medium HDL | 1.109 | 1.045 | 1.179 | 0.00060 |
| FC to total lipids ratio in medium HDL | 1.109 | 1.045 | 1.179 | 0.00055 |
| TC in HDL3 | 1.098 | 1.038 | 1.162 | 0.00109 |
| TC in medium LDL | 1.100 | 1.039 | 1.164 | 0.00095 |
| TC to total lipids ratio in medium LDL | 1.100 | 1.039 | 1.167 | 0.00105 |
| TC in small HDL | 1.144 | 1.080 | 1.213 | <0.00001 |
| TC to total lipids ratio in small HDL | 1.099 | 1.039 | 1.165 | 0.00106 |
| TC in small LDL | 1.097 | 1.037 | 1.161 | 0.00132 |
| TC to total lipids ratio in small LDL | 1.100 | 1.039 | 1.166 | 0.00107 |
| TC to total lipids ratio in small VLDL | 1.099 | 1.038 | 1.163 | 0.00105 |
| Phospholipids (PL) & Total Lipids (TL) | ||||
| PL in IDL | 1.100 | 1.040 | 1.164 | 0.00092 |
| PL to total lipids ratio in medium LDL | 0.904 | 0.853 | 0.957 | 0.00046 |
| PL to total lipids ratio in medium VLDL | 1.145 | 1.082 | 1.211 | <0.00001 |
| PL in very small VLDL | 1.099 | 1.039 | 1.163 | 0.00103 |
| PL to total lipids ratio in very small VLDL | 1.120 | 1.056 | 1.190 | 0.00013 |
| TL in small HDL | 1.108 | 1.048 | 1.173 | 0.00035 |
| Triglycerides (TG) | ||||
| TG to total lipids ratio in medium VLDL | 0.907 | 0.857 | 0.959 | 0.00064 |
| TG to total lipids ratio in small VLDL | 0.906 | 0.856 | 0.958 | 0.00055 |
| Fatty Acids (FA) | ||||
| Ratio of omega-6 FA to total FA | 1.102 | 1.041 | 1.166 | 0.00080 |
| Ratio of saturated FA to total FA | 0.890 | 0.841 | 0.942 | 0.00006 |
| Amino Acids | ||||
| Isoleucine | 0.893 | 0.844 | 0.944 | 0.00008 |
| Leucine | 0.901 | 0.851 | 0.953 | 0.00027 |
| Tyrosine | 0.886 | 0.837 | 0.937 | 0.00003 |
| Valine | 0.913 | 0.863 | 0.965 | 0.00139 |
| Fluid Balance | ||||
| Albumin | 1.104 | 1.043 | 1.168 | 0.00065 |
ProtecT=Prostate Testing for Cancer and Treatment trial; OR=odds ratio; LCI=lower limit of 95% confidence interval; UCI=upper limit of 95% confidence interval; HDL= high-density lipoprotein; LDL=low-density lipoprotein; VLDL=very low-density lipoprotein; IDL=intermediate-density lipoprotein.
Models adjusted for age, centre, and imputed family history of prostate cancer (imputed because family history was only available for 90% of subjects).
P-value threshold corrected for multiple testing (P<0.05/37=0.0014).
Figure 1.
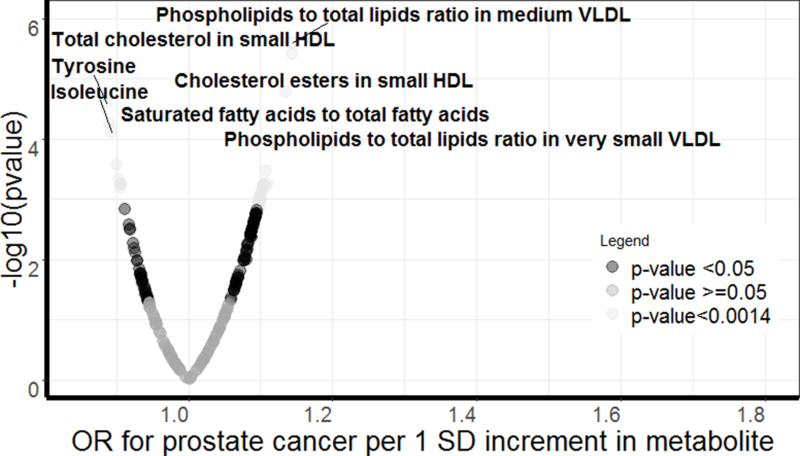
Volcano plot of the odds of prostate cancer in ProtecT.
Figure 1 displays a volcano plot of the odds of prostate cancer in ProtecT (Prostate Testing for Cancer and Treatment trial). OR=odds ratio. Labelled metabolites are Bonferroni significant (<0.05/227). Light-grey dots indicate p-value <0.0014; dark-grey dots indicate p-value <0.05; and medium-grey dots indicate p-value>=0.05.
The following decreased the odds of total prostate cancer: VLDL particle size, PL-to-TL ratios in medium LDL; triglycerides (TG)-to-total lipid ratios in small and medium VLDL; amino acids (AA), specifically, the branched-chain AA, isoleucine, leucine, and valine, and the aromatic AA tyrosine; and saturated FA-to-total FA.
In the sensitivity analysis of the effect of metabolic traits on high-risk prostate cancer versus controls, albumin was associated with high-risk case status (OR 1.12; 95% CI 1.08–1.36; p<0.0014); 138 (61%) had ORs reversed from those in the combined (total, case-control) analysis; and, though 53 ORs were statistically different from those in the total analysis (p-value threshold <0.05), none of the differences survived multiple comparisons (p<0.05/227; 0.0002). (Supplementary Tables 5 and 6 in the Supplement). Consistent with these results, in the comparison of high-versus low-risk ORs, 78 metabolites had ORs that were statistically different at the <0.05 threshold and two at the multiple-testing threshold (p-value <0.0002: TC:TL in small HDL and PL:TL in small HDL); 63% of metabolites had directionally reversed ORs. Notably, among the 35 top metabolites in the total analysis, eight were included in the set of those with statistically different ORs in the high-versus low-risk comparison (p-value for multiple testing set to 0.05/35=0.0014) (Supplementary Tables 7 and 8 in the Supplement). Conversely, the sensitivity analysis for the effect of metabolites on low-risk prostate cancer (versus controls) revealed patterns of association that mirrored the magnitude and direction of effects observed for the total analysis versus controls; only four (0.02%) metabolites in the low-risk analysis had ORs directionally reversed from those in the total analysis; and none of the ORs were statistically different from those in the total analysis (p-value <0.05) (Supplementary Tables 9 and 10 in the Supplement>).
None of the metabolite-PSA correlations exceeded |0.06| (Supplementary Table 11 in the Supplement).
Mendelian randomization causal analysis (PRACTICAL)
Fourteen of the top 35 metabolites observationally associated with total prostate cancer were analysable using MR. Of the 14 metabolites that were instrumental, none appear causal for prostate cancer risk (Figure 2, Supplementary Table 12 and 13 in the Supplement).
Figure 2.
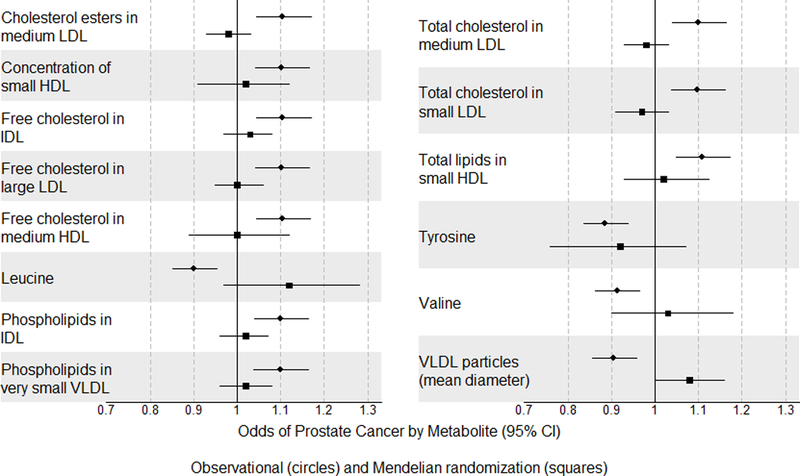
Forest plot showing odds of prostate cancer by metabolite for top observational findings in ProtecT along with causal estimates from Mendelian randomization.
Figure 2 is a forest plot of the odds of prostate cancer by metabolite for top observational findings in the Prostate Testing for Cancer and Treatment (ProtecT) trial with models adjusted for age, centre, and imputed family history of prostate cancer. Summary data for the effects of metabolite loci on prostate cancer for the Mendelian randomization analysis was obtained from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium. The squares and lines indicate odds ratios and 95% confidence intervals for top findings in ProtecT. The circle dots and lines indicate the causal estimates for the effects of the metabolites on prostate cancer in PRACTICAL.
Discussion
Main findings
We identified 35 potential biomarkers for prostate cancer. The majority of these were cholesterols, followed by glycerides and phospholipids. Steroid hormones, including androgens that drive prostate cancer, are derived from cholesterol,47 and high levels of cholesterol are required by rapidly proliferating cells.48 Hence, it is possible that our findings point to the underlying relationship between prostate cancer and androgens. Moreover, the observed effects appear to be driven solely by the low-risk cases, which were more abundant in our screen-detected cohort. The weak correlations between the metabolites and PSA suggest that our findings are not a bi-product of screening.
Fifteen of the top non-instrumented metabolites were ratios, which means that we were able to test the causal effects for the majority (70%) of our top metabolites that were not ratios (14/20).
Comparison with previous literature
A few recent studies have explored the relationship between serum metabolites and prostate cancer using metabolites detected from chromatography-mass spectrometry.2,14,15 In a pilot study, Mondul et al. (2014) compared 420 metabolic compounds in fasting serum collected prospectively from 74 clinically detected prostate cancer cases and 74 matched controls within the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) cohort. In their study, circulating 1-stearoylglycerol (1-SG) was inversely associated with prostate cancer (OR 0.34, 95% CI 0.20–0.58).15 In the present study, we did not quantify 1-SG. In their replication study, also within the ATBC cohort, Mondul et al. (2015) analysed fasting serum collected prospectively for 626 metabolic compounds in 200 clinically detected cases and 200 matched controls.14 Notably, there was no overlap between the findings of the present study and those of Mondul et al. (2015).14
Similarly, Huang et al. (2016) undertook an investigation of prostate cancer within the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO), for which they prospectively examined 695 known serum metabolites in 380 screen-detected cases and 380 matched controls. Their set of top metabolites differed from both from ours and the set observed in the ATBC (clinically detected) studies.2
The present study differed from both the two ATBC and the PLCO metabolome studies—studies perhaps the most comparable to ours— in that, instead of using chromatography-mass spectrometry to detect agnostically any measurable serum metabolites, we used a quantitative high-throughput NMR metabolomics platform with a pre-chosen set of metabolites that cover metabolic pathways for lipoprotein lipids and subclasses, fatty acids, amino acids, and glycolysis precursors. As such, some of the deviation between our findings and theirs are explained by this—we examined different sets of metabolites. Another difference is that the ATBC and PLCO studies were prospective and the observational portion of the present study is cross-sectional.
We observed a family history of prostate cancer in 8% of cases, likely reflecting that they were identified in a screening versus a clinical setting.
Strengths
Our analysis uses MR to interrogate whether some of our top findings (those with genetic instruments) appear causal. It is the first study of circulating metabolic traits and prostate cancer to do so. Moreover, it is the largest (more than 4x larger than the largest previous study2) examination of the role of circulating metabolites in prostate cancer, and it yielded novel and promising associations with metabolic traits that may be useful clinically as biomarkers to better distinguish presence of disease and disease severity.
Limitations
Our study has a few limitations. As the blood samples were collected at diagnosis for cases, we were unable to determine the direction of causality in our observational analyses. Likewise, there is potential, due to the way we selected our controls [men with PSA <3 ng/mL or a raised PSA (≥3 ng/mL) and negative biopsy] for there to be some misclassification of case status. Use of MR, at least for the instrumentable metabolites, allowed us, nonetheless, to appraise causality for a subset of our top findings, and we had at least 80% power to detect effect estimates within the range of those observed in our observational analysis for most metabolites. Another limitation is that there was a lack of specificity for many of the available genetic instruments, potentially biasing our causal analysis towards the null. Given this, while our MR found no evidence for causality, future MR analyses containing a larger number of specific genetic instruments for the metabolites are needed to strengthen causal assessment of the role of the metabolites we have detected as marking the presence of prostate cancer.
Conclusion
We identified 35 circulating metabolites associated with prostate cancer presence, but found no evidence of causality for those 14 testable with MR. Thus, the 14 metabolites tested with MR are unlikely to be mechanistically important in prostate cancer risk. We cannot speculate about the causality for those not tested with MR.
Supplementary Material
Acknowledgments
Funding:
This work was supported by Cancer Research UK (CRUK) (C18281/A19169) and the Medical Research Council Integrative Epidemiology Unit at the University of Bristol, which is supported by the Medical Research Council (MRC) and the University of Bristol (MC_UU_12013/2) (author C.D.A). R.R. is supported by CRUK (C18281/A19169) and MRC (MC_UU_12013/2); W.S is supported by MRC (MC_UU_12013/1) and a Wellcome Trust studentship (108902/15/Z); V.T. is supported by CRUK (C18281/A19169); P.W. is funded by the Academy of Finland (grant numbers: 312476 and 312477); D.N. is funded by (C11043/A4286, C18281/A8145 and C18281/A11326); J.A.L is supported by CRUK (C18281/A19169); C.R is supported by MRC (MC_UU_12013/2) and CRUK (C18281/A19169); R.A.E. is supported by CRUK (C5047/A17528); Z.K-J. is supported by CRUK (C5047/A17528); K-T.K with EPIC Norfolk is supported by grants from the MRC (MR/N003284/1; G1000143) and CRUK (14136); R.T. is supported by CRUK (C8221/A19170), CRUK (14136 for EPIC-Norfolk and C570/A16491 for EPIC-Oxford), and the MRC (1000143 for EPIC-Norfolk and MR/M012190/1 for EPIC-Oxford; R.M.M is supported by CRUK (C18281/A19169).
The National Institute for Health Research Biomedical Research Centre supported R.M.M, C.R., G.D.S. Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) [U19 CA 148537 for ELucidating Loci Involved in Prostate cancer SuscEptibility (ELLIPSE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract number HHSN268201200008I]. Additional analytic support was provided by NIH NCI U01 CA188392 (PI: Schumacher). The PRACTICAL consortium was supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692, C16913/A6135, European Commission’s Seventh Framework Programme grant agreement n° 223175 (HEALTH-F2-2009-223175), and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148537-01 (the GAME-ON initiative). We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust.
Footnotes
Information for the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium can be found at http://practical.icr.ac.uk/.
Conflict-of-Interest Disclosures: Peter Wurtz is an employee and shareholder of Nightingale Health Ltd, a company offering NMR-based metabolic profiling. Paul A. Townsend was a speaker at MedLabs in 2017/18 and was compensated (<$10,000).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Mondul AM, Weinstein SJ, et al. Serum metabolomic profiling of prostate cancer risk in the prostate, lung, colorectal, and ovarian cancer screening trial. Br J Cancer. 2016;115(9):1087–1095. doi: 10.1038/bjc.2016.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markozannes G, Tzoulaki I, Karli D, et al. Diet, body size, physical activity and risk of prostate cancer: an umbrella review of the evidence. Eur J Cancer. 2016;69:61–69. doi: 10.1016/j.ejca.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly RS, Heiden MGV, Giovannucci E, Mucci LA. Metabolomic biomarkers of prostate cancer: Prediction, diagnosis, progression, prognosis, and recurrence. Cancer Epidemiol Biomarkers Prev. 2016;25(6):887–906. doi: 10.1158/1055-9965.EPI-15-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osl M, Dreiseitl S, Pfeifer B, et al. A new rule-based algorithm for identifying metabolic markers in prostate cancer using tandem mass spectrometry. Bioinformatics. 2008;24(24):2908–2914. doi: 10.1093/bioinformatics/btn506. [DOI] [PubMed] [Google Scholar]
- 7.Lokhov PG, Dashtiev MI, Moshkovskii SA, Archakov AI. Metabolite profiling of blood plasma of patients with prostate cancer. Metabolomics. 2010;6(1):156–163. doi: 10.1007/s11306-009-0187-x. [DOI] [Google Scholar]
- 8.Fan Y, Murphy TB, Byrne JC, Brennan L, Fitzpatrick JM, Watson RWG. Applying random forests to identify biomarker panels in serum 2D-DIGE data for the detection and staging of prostate cancer. J Proteome Res. 2011;10(3):1361–1373. doi: 10.1021/pr1011069. [DOI] [PubMed] [Google Scholar]
- 9.Miyagi Y, Higashiyama M, Gochi A, et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS One. 2011;6(9):1–12. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saylor P Prospective study of changes in the metabolomic profiles of men during their first three months of androgen deprivation therapy for prostate cancer. Clin Cancer Res. 2012;18(13):3677–3685. doi: 10.1158/1078-0432.CCR-11-3209.Prospective. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Mao J, Ai J, et al. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7(11):1–11. doi: 10.1371/journal.pone.0048889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang G, Liu X, Jiao L, et al. Metabolomic evaluation of the response to endocrine therapy in patients with prostate cancer. Eur J Pharmacol. 2014;729(1):132–137. doi: 10.1016/j.ejphar.2014.01.048. [DOI] [PubMed] [Google Scholar]
- 13.Zang X, Jones CM, Long TQ, et al. Feasibility of detecting prostate cancer by ultra performance liquid chromatography-mass spectrometry serum metabolomics. J Proteome Res. 2014;3(13(7)):3444–3454. [DOI] [PubMed] [Google Scholar]
- 14.Mondul AM, Moore SC, Weinstein SJ, Karoly ED, Sampson JN, Albanes D. Metabolomic analysis of prostate cancer risk in a prospective cohort: the alpha-tocolpherol, beta-carotene cancer prevention (ATBC) study. Int J Cancer. 2015;137(9):2124–2132. doi: 10.1002/ijc.29576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mondul AM, Moore SC, Weinstein SJ, Männistö S, Sampson JN, Albanes D. 1-Stearoylglycerol is associated with risk of prostate cancer: results from a serum metabolomic profiling analysis. Metabolomics. 2014:1–6. doi: 10.1007/s11306-014-0643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fewell Z, Davey Smith G, Sterne JAC. The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am J Epidemiol. 2007;166(6):646–655. doi: 10.1093/aje/kwm165. [DOI] [PubMed] [Google Scholar]
- 17.Davey Smith G, Ebrahim S. Data dredging, bias, or confounding. BMJ. 2002;325(7378):1437–1438. doi: 10.1136/bmj.325.7378.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane JA, Hamdy FC, Martin RM, Turner EL, Neal DE, Donovan JL. Latest results from the UK trials evaluating prostate cancer screening and treatment: the CAP and ProtecT studies. Eur J Cancer. 2010;46(17):3095–3101. doi: 10.1016/j.ejca.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Hamdy FC. The Prostate Testing for Cancer and Treatment (ProtecT) study: what have we learnt? BJU Int. 2016;118(6):843. doi: 10.1111/bju.13699. [DOI] [PubMed] [Google Scholar]
- 20.Hamdy FC, Donovan JL, Lane JA, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med. 2016;375(15):1415–1424. doi: 10.1056/NEJMoa1606220. [DOI] [PubMed] [Google Scholar]
- 21.Lane JA, Donovan JL, Davis M, et al. Active monitoring, radical prostatectomy, or radiotherapy for localised prostate cancer: study design and diagnostic and baseline results of the ProtecT randomised phase 3 trial. Lancet Oncol. 2014;15(10):1109–1118. doi: 10.1016/S1470-2045(14)70361-4. [DOI] [PubMed] [Google Scholar]
- 22.Bonilla C, Lewis SJ, Martin RM, et al. Pubertal development and prostate cancer risk: Mendelian randomization study in a population-based cohort. BMC Med. 2016;14(1):66. doi: 10.1186/s12916-016-0602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Institute For Health and Care Excellence. Prostate cancer: diagnosis and management. https://www.nice.org.uk/guidance/cg175. Published on January 2014. Accessed on October 10, 2017. doi: 10.1016/j.juro.2007.01.121. [DOI]
- 24.Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative Serum NMR Metabolomics in Large-Scale Epidemiology: a Primer on -Omic Technology. Am J Epidemiol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Würtz P, Havulinna AS, Soininen P, et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131(9):774–785. doi: 10.1016/j.bbamem.2015.02.010.Cationic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32(4):361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 27.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. doi: 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davey Smith G, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 29.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16(4):309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 30.Joshi PK, Esko T, Mattsson H, et al. Directional dominance on stature and cognition in diverse human populations. Nature. 2015. doi: 10.1038/nature14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen AK, Zeilinger S, Kastenmuller G, et al. Epigenetics meets metabolomics: an epigenome-wide association study with blood serum metabolic traits. Hum Mol Genet. 2014;23(2):534–545. doi: 10.1093/hmg/ddt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gieger C, Geistlinger L, Altmaier E, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4(11):e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kettunen J, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:1–9. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurtz P, Wang Q, Kangas AJ, et al. Metabolic Signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med. 2014;11(12):e1001765. doi: 10.1371/journal.pmed.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lotta LA, Scott RA, Sharp SJ, et al. Genetic predisposition to an impaired metabolism of the branched-Chain Amino Acids and risk of Type 2 diabetes: a Mendelian randomisation analysis. PLoS Med. 2016;13(11):1–22. doi: 10.1371/journal.pmed.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varbo A, Benn M, Tybjærg-Hansen A, Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG. Remnant cholesterol as a causal risk factor for ischemic heart disease. J Am Coll Cardiol. 2013;61(4):427–436. doi: 10.1016/j.jacc.2012.08.1026. [DOI] [PubMed] [Google Scholar]
- 37.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce BL, Burgess S. Efficient design for mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178(7):1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kote-jarai Z, Easton DF, Stanford JL, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL consortium. Cancer Epidemiol Biomarkers Prev. 2008;17(8):2052–2061. doi: 10.1158/1055-9965.EPI-08-0317.Multiple. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eeles RA, Kote-jarai Z, Amin A, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2010;41(10):1116–1121. doi: 10.1038/ng.450.Identification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kote-Jarai Z, Olama AAA, Giles GG, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43(8):785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al Olama AA, Kote-Jarai Z, Schumacher FR, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22(2):408–415. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eeles, Rosalind A et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45(9):1–22. doi: 10.1523/JNEUROSCI.5473-10.2011.Loss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao F, Xu H, Torrey N, Road P, Jolla L. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46(10):1103–1109. doi: 10.1126/scisignal.274pe36.Insulin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemani G, Zheng J, Wade KH, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 2016. doi: 10.1101/078972 [DOI] [Google Scholar]
- 47.Berg JM, Tymoczko JL SL. Section 26.4, Important Derivatives of Cholesterol Include Bile Salts and Steroid Hormones In: Biochemisty. 5th ed. New York: W H Freeman; 2002. https://www.ncbi.nlm.nih.gov/books/NBK22339/. Accessed October 10, 2017. [Google Scholar]
- 48.Harshman LC, Wang X, Nakabayashi M, et al. Statin use at the time of initiation of androgen deprivation therapy and time to progression in patients with hormone-sensitive prostate cancer. JAMA Oncol. 2015;1(4):495. doi: 10.1001/jamaoncol.2015.0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


