Abstract
Tumors comprise cancer stem cells (CSCs) and their heterogeneous progeny within a stromal micro-environment. In response to transforming growth factor–β (TGF-β), epithelial and carcinoma cells undergo a partial or complete epithelial-mesenchymal transition (EMT), which contributes to cancer progression. This process is seen as reversible, because cells revert to an epithelial phenotype upon TGF-β removal. However, we found that prolonged TGF-β exposure, mimicking the state of in vivo carcinomas, promotes stable EMT in mammary epithelial and carcinoma cells, in contrast to the reversible EMT induced by a shorter exposure. The stabilized EMT was accompanied by stably enhanced stem cell generation and anticancer-drug resistance. Furthermore, prolonged TGF-β exposure enhanced mammalian target of rapamycin (mTOR) signaling. A bitopic mTOR inhibitor repressed CSC generation, anchorage-independence, cell survival, and chemoresistance, and efficiently inhibited tumorigenesis in mice. These results reveal a role for mTOR in the stabilization of stemness and drug resistance of breast cancer cells and position mTOR inhibition as a treatment strategy to target CSCs.
Introduction
The cell heterogeneity of tumors is a major cause of problems in therapeutically interfering with cancer progression. Epithelial tumors, or carcinomas, comprise heterogeneous cancer cell populations, including cancer stem cells (CSCs), differentiated cancer cells, stromal cancer-associated fibroblasts, immune cells and endothelial cells. CSCs are a small population of self-renewing cells with the ability to initiate tumor formation. In contrast to a linear model of CSC differentiation, epithelial cancer cells are now seen to have substantial differentiation plasticity (1, 2). This plasticity allows a dynamic balance between dedifferentiated CSCs and differentiated cancer cells.
In carcinomas, dedifferentiation of cancer cells and generation of CSCs correlate with epithelial plasticity through a process called epithelial-mesenchymal transition (EMT) (3–5). As epithelial cells progress through EMT, they lose epithelial cell-cell contacts and apical-basal polarity, reorganize their cytoskeleton and reprogram gene expression to enable, among many changes, increased deposition of extracellular matrix components and matrix metalloproteases (6). EMT is essential in development, and is repurposed in cancer progression to enable cancer cell invasion, contribute to cancer stroma formation, generate CSCs and decrease sensitivity to anticancer drugs (7, 8). EMT is thought of as a reversible process, whereby cancer cells that acquired mesenchymal properties can revert to an epithelial state through mesenchymal-epithelial transition, which has been correlated with CSC differentiation. The epithelial plasticity is controlled by signals from the cancer microenvironment.
Among the many signals in the cancer microenvironment, transforming growth factor-β (TGF-β) signaling, which is commonly upregulated in carcinomas, often initiates and drives EMT of carcinoma cells (9). Associated with EMT, and perhaps best illustrated with breast carcinomas, TGF-β potently induces carcinoma cell invasion and CSC generation (10). TGF-β signaling is initiated upon ligand binding to a cell surface complex of two TGF-β type II receptors (TβRII) and two TGF-β type I receptors (TβRI), which then activates the signaling effectors Smad2 and Smad3 through C-terminal phosphorylation (11). The activated Smad proteins form complexes with Smad4 and regulate target gene expression through association with high-affinity DNA-binding transcription factors at regulatory sequences (11, 12). TGF-β-induced, Smad3/4-mediated gene expression drives the gene reprogramming that characterizes the EMT process, starting with activation of expression of EMT master transcription factors, such as Snail, ZEB1 and ZEB2, and Twist, and cooperation of Smad3/4 complexes with these transcription factors in driving EMT (6). In addition to Smad signaling, TGF-β also activates phosphoinositide 3-kinase (PI3K)–AKT, extracellular signal-regulated kinase (ERK)-mitogen-activated protein kinase (MAPK), p38 MAPK, and Rho-guanosine triphosphatase (GTPase) pathways (11, 13). Among these, TGF-β-induced signaling through the PI3K-AKT-mammalian target of rapamycin (mTOR) pathway is required for progression through EMT (14, 15). Cell culture studies enable the dissection of the TGF-β-induced EMT program, and documented its reversible nature upon TGF-β withdrawal (16).
In breast cancer progression, the exposure of carcinoma cells to increased TGF-β signaling from either the carcinoma cells themselves or the stromal cells is not likely to be limited to a few days that would mimic the cell culture conditions used by most researchers. Because there is no evidence for dramatic TGF-β level changes within the tumor, it is logical to assume that the carcinoma cells are exposed to TGF-β for longer times (17, 18). This raises the question whether prolonged exposure to TGF-β, rather than short-term exposure, as routinely done in cell culture, allows the carcinoma cells to maintain the reversible character of EMT, and may result in additional changes of relevance for cancer progression. In this study, we addressed this question using an established human mammary epithelial cell population and a derivative, H-Ras-transformed carcinoma cell population that have been previously studied (3, 19–21). We found that prolonged TGF-β exposure stabilized the mesenchymal phenotype, and enhanced the stemness and resistance to anticancer drugs, in contrast to and beyond what is seen in reversible EMT following short-term TGF-β exposure. Reversible EMT and stabilized EMT contributed differently to tumorigenesis and dissemination in vivo. Stabilized EMT is suggested to contribute more to tumor latency and persistence, and less to cancer dissemination, which is strongly enhanced by reversible EMT. We also found that the carcinoma cells after prolonged exposure to TGF-β have activated mTOR complex 1 and complex 2 (mTORC1 and mTORC2) signaling. The stem cell characteristics and increased cancer drug resistance accompanying stabilized EMT could no longer be efficiently inhibited by blocking Smad signaling, yet were fully reversed by mTOR inhibition using a novel bitopic mTOR inhibitor, which efficiently inhibited tumor growth in mice. These results illustrate the transition from reversible EMT into a stabilized EMT phenotype, which may add more roles of TGF-β-induced EMT in cancer progression than we previously understood. They also reveal a role of enhanced mTOR signaling in the stabilization of stemness and drug resistance of breast cancer cells, thereby suggesting a basis for therapeutic interference with mTOR signaling.
Results
Prolonged exposure to TGF-β stabilizes a mesenchymal state in mammary epithelial cells.
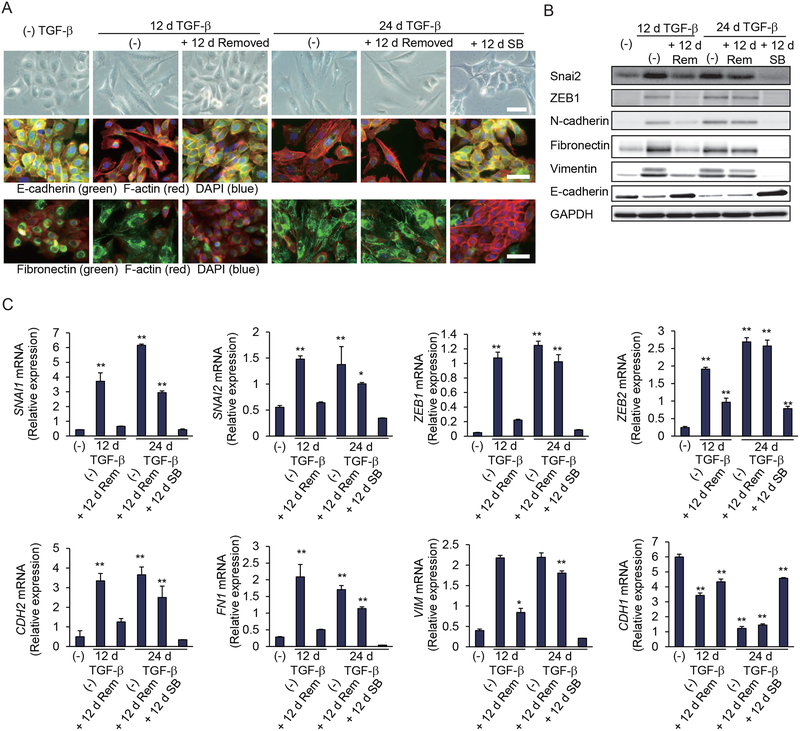
To characterize the effects of prolonged TGF-β exposure on epithelial plasticity, we used human mammary epithelial HMLE cells, an immortalized cell population derived from primary cells that is well studied to correlate stem cell properties with EMT (3, 19, 20). In these cells, TGF-β induced EMT over a period of 9–12 days, which was apparent from the change in cell shape, actin stress fiber formation, loss of E-cadherin at cell-cell contacts, and increased fibronectin deposition. These cells readily reverted to the epithelial phenotype when TGF-β was subsequently removed (Fig. 1A, left). Treatment with TGF-β for 24 days stabilized the EMT phenotype; thus, cells retained their mesenchymal state after subsequent removal of TGF-β for 12 days (Fig. 1A, right). They maintained their changed morphology, the increased expression of EMT transcription factors Snai1, Snai2, ZEB1 and ZEB2, as well as mesenchymal N-cadherin, fibronectin and vimentin, and the decreased expression of E-cadherin (Fig. 1, B and C, and fig. S1A). A partial reversion was, however, apparent when the cells were treated for 12 days with the TβRI kinase inhibitor SB431542 (Fig. 1, A to C, and fig. S1A). Exposure of the cells to a neutralizing anti-TGF-β monoclonal antibody for 12 days after they were treated with TGF-β for 24 days also partially reversed the mesenchymal phenotype (fig. S1, B and C), albeit less efficiently than SB431542, suggesting autocrine TGF-β signaling in close association or perhaps even inside the cells.
Figure 1. Prolonged TGF-β treatment stabilizes a mesenchymal state in HMLE cells.
(A) HMLE cells were treated with TGF-β for 12 or 24 days (d) followed by its removal, and then cultured with or without SB431542 for 12 days. The cells were observed by phase-contrast microscopy or subjected to staining for E-cadherin, fibronectin, F-actin or DAPI. Scale bars, 50 μm. Images are representative of three independent experiments. (B and C) HMLE cells were treated with TGF-β, followed by its removal as in (A), and were then cultured without TGF-β (Rem) or with SB431542 (SB) for 12 days. The expression of epithelial and mesenchymal markers was analyzed by immunoblotting (B) or quantified by qRT-PCR and normalized to rPL19 mRNA (C). Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 by a Dunnet’s test.
Prolonged TGF-β exposure also stabilized the EMT of mouse mammary epithelial NMuMG cells, a non-transformed, immortalized cell line that is commonly used to study TGF-β-induced EMT and the underlying signaling mechanisms (16). Similar to HMLE cells, TGF-β induced reversible EMT in NMuMG cells, albeit over a shorter period of 2 days, whereas treatment of the cells with TGF-β for 8 days stabilized the EMT (fig. S1, D to F). These data illustrate that long-term treatment of mammary epithelial cells with TGF-β confers a mesenchymal phenotype that no longer depends on externally added TGF-β.
Prolonged TGF-β exposure stabilizes a stem cell-like state.
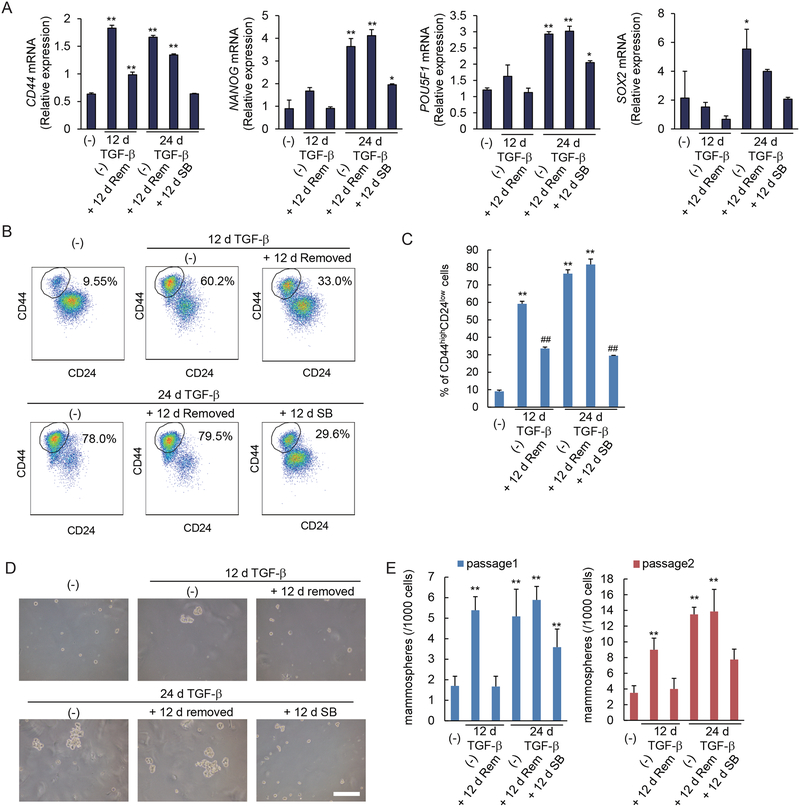
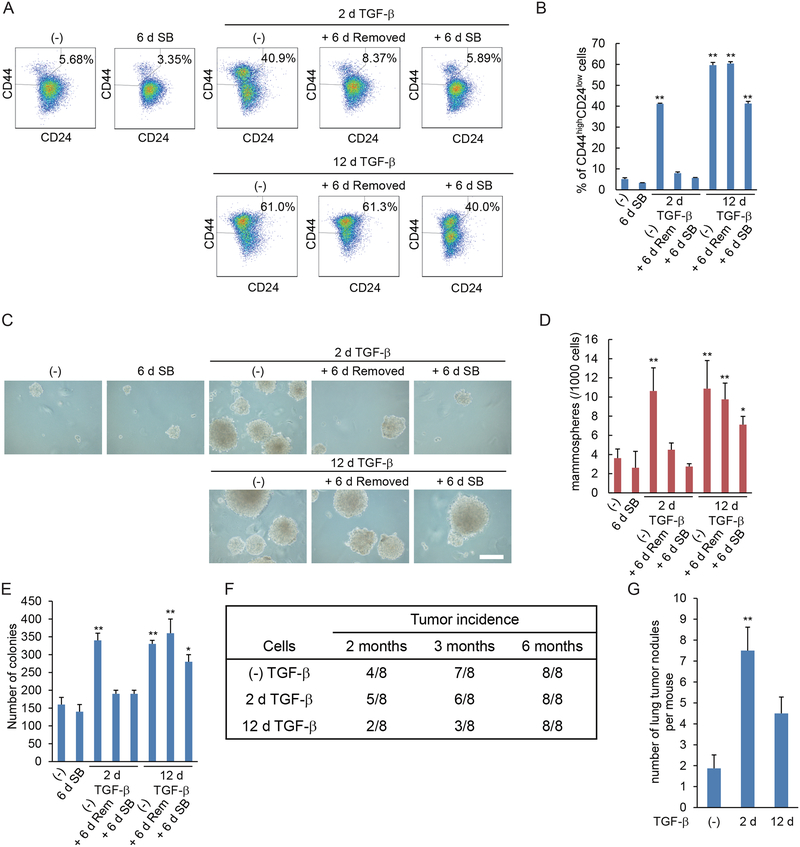
EMT and acquisition of mesenchymal characteristics have been shown to correlate with the generation of epithelial and carcinoma stem cells (3, 5). We therefore evaluated in HMLE cells the effect of prolonged TGF-β exposure on the generation of stem cells in relation to EMT. TGF-β treatment for 12 days, which induces reversible EMT (Fig. 1, A to C), induced the expression of the mammary epithelial stem cell marker gene CD44 (Fig. 2A), and increased the relative abundance of the stem-cell-enriched CD44highCD24low cell population (Fig. 2, B and C). Subsequent removal of TGF-β for 12 days resulted in decreased CD44 expression and a decrease in the CD44highCD24low cell population, consistent with a reversible induction. TGF-β treatment for 24 days, however, stabilized the increased CD44 expression and relative size of the CD44highCD24low stem cell population, since they did not decrease after removal of TGF-β for 12 days (Fig. 2, A to C). Furthermore, the stabilization resulted in a larger CD44highCD24low stem cell population than what was seen in response to the 12-day TGF-β treatment (Fig. 2, B and C). Stabilization of stem cell characteristics was also apparent in the maintained induction of NANOG and POU5F1 (encoding the transcription factor Oct4), as well as SOX2 expression, which were expressed at substantially higher amounts at 24 days of TGF-β than in cells treated for 12 days. While removal of added TGF-β had little if any effect, SB431542 treatment for 12 days attenuated the stem cell marker expression and percentage of CD44highCD24low cells following long-term treatment with TGF-β (Fig. 2, A to C).
Figure 2. Prolonged TGF-β treatment stabilizes a stem cell-like state.
(A) HMLE cells were treated with TGF-β for 12 or 24 days (d). TGF-β was removed and cells were cultured without TGF-β (Rem) or in the presence of SB431542 (SB) for 12 days. The expression of the CD44 stem cell marker gene and the stemness-associated NANOG, POU5F1 and SOX2 genes was assessed by qRT-PCR and normalized to rPL19 mRNA. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 by a Dunnet’s test. (B and C) HMLE cells were treated with TGF-β for 12 or 24 days, followed by removal of TGF-β, and the cells were cultured with or without SB431542 for 12 days. The expression of the cell surface markers CD44 and CD24 was analyzed by flow cytometry (B). The graph (C) shows the percentages of CD24lowCD44high stem cells in the entire cell populations. Data are mean ± S.E. from N=3 independent experiments. **P< 0.01 vs. untreated cells, and ##P< 0.01 vs. cells treated with TGF-β for 12 d or 24 days, by a Tukey’s test. (D and E) HMLE cells were treated with TGF-β for 12 or 24 days. TGF-β was removed and cells were cultured in the presence (SB) or absence (Rem) of SB431542 for 12 days. Cells were assessed for primary and secondary mammosphere formation. The mammospheres were observed by phase-contrast microscopy (D) and counted (E). Scale bar, 100 μm. Data are mean ± S.E. from N=3 independent experiments. **P< 0.01 by a Dunnet’s test.
To further assess their self-renewal and differentiation capacities, we evaluated the short-term and long-term TGF-β-treated HMLE cells in mammosphere formation assays. These assays score growth in suspension, starting from a single cell, and correlate with the presence of mammary epithelial stem cells (22). Short-term (12 days) TGF-β treatment induced a reversible increase in primary and secondary mammosphere formation (Fig. 2, D and E). Cells treated long-term (24 days) with TGF-β also showed increased mammosphere formation, which, however, remained unchanged when subsequently cultured without TGF-β for 12 days prior to the assay. SB431542 treatment after long-term TGF-β treatment partially reduced the generation of mammospheres (Fig. 2, D and E).
Because TGF-β has been shown to induce the expression of TGF-β1 and TGF-β2 (23), we evaluated whether the stabilization of EMT and stem cell phenotype resulted from increased TGF-β expression. Transcript expression for TGF-β1 and TGF-β2 was transiently increased upon TGF-β treatment but decreased after TGF-β removal, whereas that of TGF-β3 remained largely unchanged (fig. S2). These data indicate that increased TGF-β expression does not explain the persistence of EMT and stem cell characteristics after removal of TGF-β.
Together, these data show that prolonged TGF-β treatment enhanced the population of cells with stem cell-like properties to an extent above that seen with short-term treatment, and stabilized the stem cell phenotype to become independent of external TGF-β.
Prolonged TGF-β exposure stabilizes the mesenchymal phenotype of breast cancer cells.
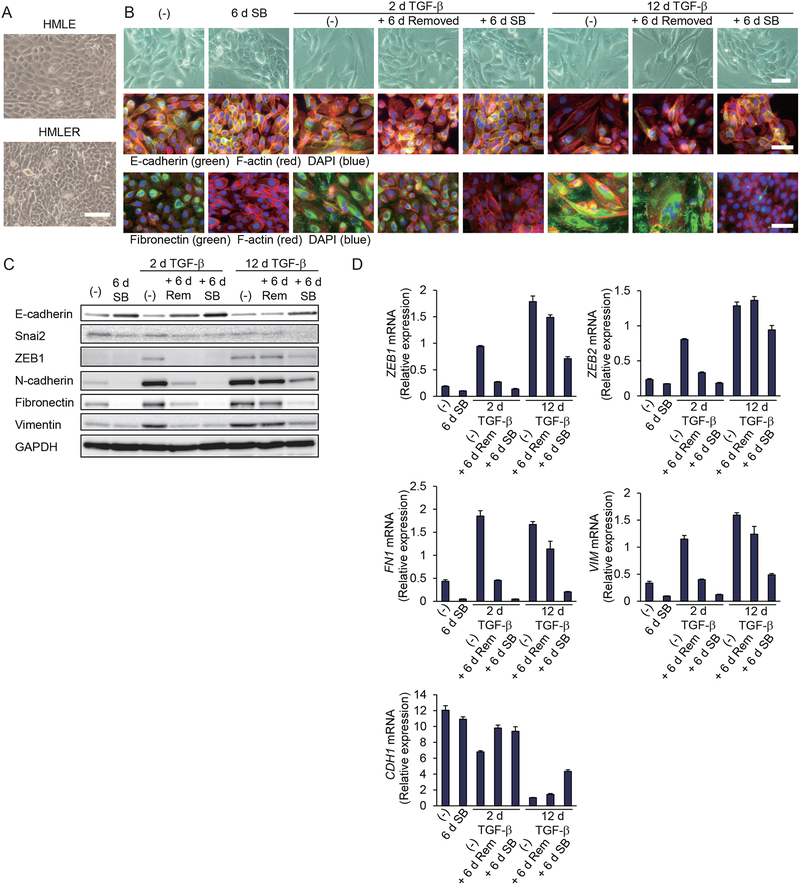
To examine whether our findings on EMT stabilization in non-transformed epithelial cells could be extended to breast carcinoma cells, we transformed HMLE cells with oncogenic H-Ras (G12V), thus generating HMLER cells (21). When grown to confluence, HMLE cells formed a monolayer with contact inhibition. In contrast, HMLER cells showed impaired contact inhibition, and cells piled up intermittently (Fig. 3A). Without adding TGF-β, HMLER cells showed some mesenchymal characteristics, apparent from their cell morphology and the expression of both epithelial and mesenchymal markers (Fig. 3, B to D, and fig. S3A). SB431542 treatment for 6 days resulted in repression of the mesenchymal characteristics and conferred pronounced epithelial characteristics in these cells (Fig. 3, B to D, and fig. S3A), indicating that Ras-induced EMT depends on autocrine TGF-β signaling.
Figure 3. Prolonged TGF-β treatment stabilizes a mesenchymal state in breast cancer cells.
(A) HMLE cells and Ras-transformed HMLER cells were grown in a monolayer culture to confluence, and the cells were observed by phase-contrast microscopy. Scale bar, 100 μm. Images are representative of three independent cultures. (B) HMLER cells were treated with TGF-β for 2 or 12 days (d), followed by its removal, and then cultured with or without SB431542 for 6 days. The cells were observed by phase-contrast microscopy or subjected to staining for E-cadherin, fibronectin, F-actin or DAPI. Scale bars, 50 μm. Images are representative of three independent experiments. (C and D) HMLER cells were treated with TGF-β, followed by its removal, as in (B), and were then cultured without added TGF-β (Rem) or with SB431542 (SB) for 6 days. Epithelial and mesenchymal gene expression was analyzed at the protein level by immunoblot (C) or quantified at the transcript level by qRT-PCR normalized to rPL19 mRNA (D). Data are mean ± S.E. from N=3 independent experiments.
In these cancer cells, addition of TGF-β for two days was sufficient to induce EMT, which was reversed toward a mild epithelial phenotype when TGF-β was removed for 6 days, and a more pronounced epithelial phenotype after SB431542 treatment for 6 days (Fig. 3, B to D, and fig. S3A). Twelve days of TGF-β treatment stabilized the mesenchymal state, and this phenotype was maintained after removing TGF-β for 6 days (Fig. 3, B to D, and fig. S3A). Consistent with the EMT phenotype, short-term TGF-β treatment increased the migration and invasion of HMLER cells, which were reduced when TGF-β was removed for 6 days (fig. S3, B and C). Long-term TGF-β treatment further enhanced the cell migration capacity and maintained the invasion at a similar level as that in short-term treated cells (fig. S3, B and C). In contrast to short-term TGF-β-treated cells, the cell migration and invasion of long-term TGF-β-treated cells were maintained after removal of TGF-β (fig. S3, B and C). Treatment of these stabilized cells with SB431542 for 6 days mildly repressed the mesenchymal phenotype and promoted differentiation toward epithelial characteristics (Fig. 3, B to D, and fig. S3A). We conclude from these data that Ras-mediated transformation confers a partial EMT, dependent upon TGF-β signaling, and that prolonged exposure of these transformed cells to TGF-β resulted in enhanced EMT that no longer depended on exogenous TGF-β, as seen with non-transformed epithelial cells. Our observation is consistent with the reported cooperation of Ras and TGF-β in modulating plasticity (24–26).
Prolonged TGF-β exposure promotes anticancer drug resistance
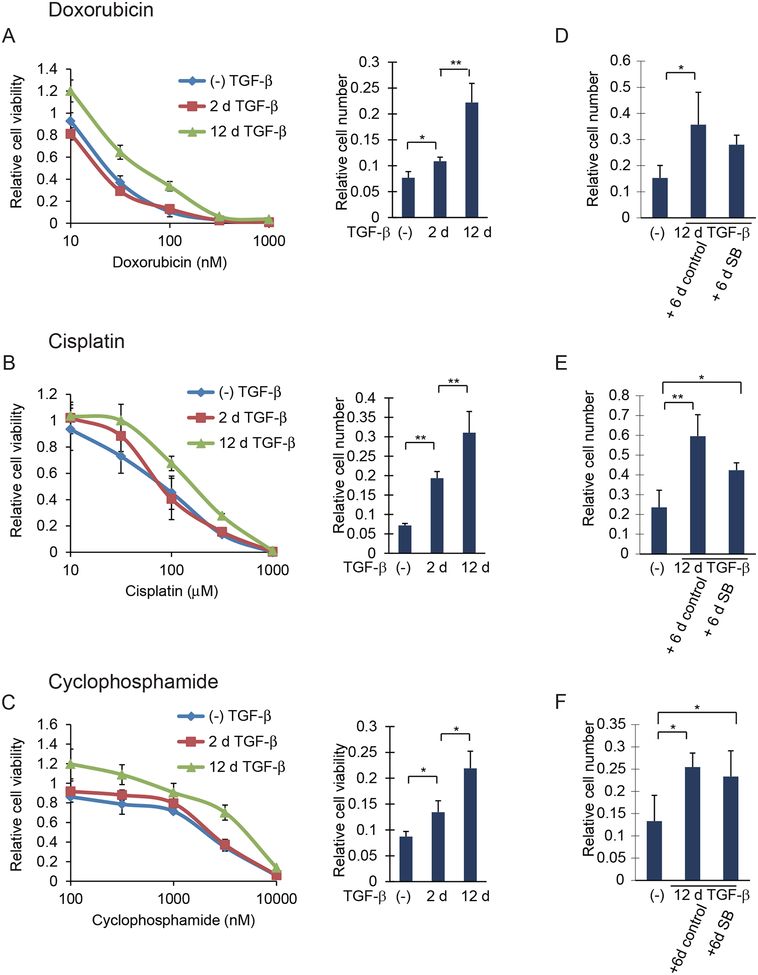
Because EMT has been correlated with increased resistance to some anticancer drugs (8), we evaluated the anticancer drug sensitivity of HMLER cells that had been exposed to TGF-β for 2 or 12 days. Cells were treated with increasing amounts of doxorubicin, cisplatin or cyclophosphamide, which resulted in reduced cell viability (Fig. 4, A to C). HMLER cells that were treated with TGF-β for 2 days were slightly less sensitive to these drugs than cells that were not treated with TGF-β. In contrast, long-term TGF-β treatment substantially decreased the sensitivity of the cells to doxorubicin, cisplatin or cyclophosphamide, resulting in increased cell viability in the presence of the drugs (Fig. 4, A to C). Long-term TGF-β treatment also increased the resistance to 5-fluoruracil and paclitaxel, although the differences between short-term versus long-term treatment with TGF-β were less striking (fig. S4, A and B). The increased survival of cells with stabilized EMT in these assays did not result from increased cell proliferation. In fact, these cells showed a lower proliferation rate, both in the presence or absence of TGF-β, than the two other cell populations (fig. S4C). We therefore conclude that EMT stabilization is accompanied by resistance to the tested anticancer drugs to an extent that far exceeds the resistance induced by short-term TGF-β treatment. In contrast to the partial inhibition of EMT marker expression by SB431542 (Fig. 3, A to C), blocking TGF-β signaling using SB431542 only slightly reduced the resistance of cells with stabilized EMT to doxorubicin, cisplatin or cyclophosphamide (Fig. 4, D to F).
Figure 4. Prolonged TGF-β treatment, resulting in stabilized EMT, increases drug resistance in breast cancer cells.
(A to C) HMLER cells treated with TGF-β for 0, 2 or 12 days (d), were trypsinized and reseeded. The cells were then incubated with doxorubicin (A, left), cisplatin (B, left), cyclophosphamide (C, left) for 48 hours at the concentrations indicated, and the cell viability was quantified and shown relative to the cells not treated with anticancer drugs. For the bar graphs to the right, the cells were cultured for 48 h with 250 nM doxorubicin (A, right), 250 μM cisplatin (B, right), or 10 μM cyclophosphamide (C, right) and the cell number was quantified and shown relative to the cells not treated with anticancer drugs. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 by a Tukey’s test. (D to F) HMLER cells were treated with or without TGF-β for 12 days and then cultured for 6 days with SB431542 or DMSO solvent as a control. The cells were then cultured for 48 hours with 250 nM doxorubicin (D), 250 μM cisplatin (E), or 10 μM cyclophosphamide (F), and the cell number was quantified and shown relative to the cells not treated with anticancer drugs. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 by a Tukey’s test.
Stabilization of a mesenchymal phenotype promotes stem cell generation in breast cancer cells.
Stabilization of a mesenchymal state also correlated with an increased and stabilized stem cell-like phenotype in HMLER cells. TGF-β treatment for two days increased the population of CD44highCD24low cells, but this increase was reversed upon withdrawal of TGF-β for 6 days. Longer treatment (12 days), however, resulted in greater and stabilized expression of stem cell markers (Fig. 5, A and B). Short-term TGF-β treatment also induced an increase in mammosphere formation that depended on added TGF-β, and thus decreased after removal of TGF-β (Fig. 5, C and D). In contrast, long-term TGF-β-treatment increased the mammosphere formation, and this ability was maintained when the cells were cultured without TGF-β. SB431542 treatment for 6 days after long-term TGF-β-treatment only partially reduced the mammosphere formation (Fig. 5, C and D), perhaps similarly to its partial effect on increased anticancer drug resistance (Fig. 4). Thus, stabilization of EMT in the Ras-transformed cells enhanced the fraction of cells with stem cell characteristics and stabilized the stem cell-like phenotype so that it no longer requires exogenous TGF-β.
Figure 5. Prolonged TGF-β treatment stabilizes a stem cell state and induces latency in breast cancer cells.
(A and B) HMLER cells were treated with TGF-β for 2 or 12 days (d), followed by its removal, as indicated, and then cultured with (SB) or without (Rem) SB431542 for 6 days. The expression of cell surface markers CD44 and CD24 was analyzed by flow cytometry (A). The graph (B) shows the percentages of CD24lowCD44high stem cells in the entire cell population. Data are mean ± S.E. from N=3 independent experiments. **P< 0.01 by a Dunnet’s test. (C and D) HMLER cells were treated as in (A) and were assessed for mammosphere formation. The mammospheres were observed by phase-contrast microscopy (C) and counted (D). Scale bar, 100 μm. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 by a Dunnet’s test. (E) HMLER cells were treated as in (A) and assessed for colony formation in soft agar. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 by a Dunnet’s test. (F) HMLER cells were treated with TGF-β for 0, 2 or 12 days then suspended in PBS with 50% Matrigel, and 10,000 cells were injected orthotopically into a mammary fat pad of NSG mice. The number of mice with a palpable tumor is indicated in the table. (G) HMLER cells were treated with TGF-β for 0, 2 or 12 days then suspended in PBS, and 500,000 cells were injected into tail vein of NSG mice. Six weeks after injection, the mice were sacrificed, and lungs were harvested. Cancer dissemination into the lungs was measured by counting the tumor nodules in the lungs. Data are mean ± S.E. from N=3 independent experiments with N=7 mice. **P< 0.01 by a Dunnet’s test.
The TGF-β-induced mesenchymal state controls tumorigenesis.
We next evaluated the effects of short-term versus long-term TGF-β treatment, resulting in reversible versus stabilized EMT, respectively, on the tumorigenic potential of HMLER cells. We first evaluated their colony formation ability in soft agar, which generally correlates with tumorigenesis (27). Short-term TGF-β treatment enhanced the colony formation and this increase was reversible (Fig. 5E). Long-term TGF-β treatment enhanced the colony formation to a similar extent as that seen in cells treated with TGF-β for only two days. However, in contrast to the short-term treated cells, the increased colony forming ability was stabilized, meaning it did not revert following removal of TGF-β, and was only mildly decreased when cells were treated with SB431542. This increase in colony formation after long-term TGF-β treatment did not result from increased proliferation, as the cells showed decreased proliferation (fig. S4C). Rather, as has been established, colony formation in soft agar reflects increased anchorage-independence and anchorage-independent survival, and predicts in vivo tumorigenesis (28).
To evaluate and compare the tumorigenesis of short-term or long-term TGF-β–treated cells, we performed limiting dilution experiments with a small number of cancer cells orthotopically injected into mammary fat pads of NOD/SCID/IL2rγnull (NSG) mice. At 3 months after orthotopic injection, cells subjected to long-term TGF-β treatment showed a decreased ability to seed tumors compared with untreated or short-term TGF-β–treated cells (Fig. 5F). However, they formed tumors at 6 months after orthotopic injection, thus reflecting an increased latency period (Fig. 5F). Tumors established from untreated, short-term TGF-β-treated, and long-term TGF-β-treated cells showed similar morphology with heterogeneous carcinoma cells, when examined histologically by hematoxylin and eosin (H&E) staining (fig. S5A).
To assess the dissemination and colonization of the HMLER cells, we injected cells that were untreated or treated with TGF-β for 2 or 12 days into the tail vein of NSG mice. HMLER cells treated with TGF-β for 2 days showed strongly increased lung colonization, assessed by numbers of lung tumor nodules (Fig. 5G and fig. S5B). Stabilization of EMT by prolonged TGF-β exposure resulted in a lower ability to colonize compared to cells with a reversible mesenchymal state after 2 days of TGF-β treatment. These differences suggest that induction of a reversible EMT phenotype facilitates the dissemination and seeding to give rise to lung nodules. The abundance of Ki67-expressing, proliferating cells was similar in tumors derived from untreated and short-term treated HMLER cells and seemed to be lower in tumors derived from cells after long-term TGF-β treatment (fig. S5C). The tumors that originated from untreated, short-term and long-term treated HMLER cells all contained heterogeneous tumor cells, with some of them expressing E-cadherin and some expressing vimentin (fig. S5C). These results suggest that cells with stabilized EMT may need to undergo MET to colonize the lung and establish tumors. Hence, as suggested by our data, the lower ability of cells with stabilized EMT to colonize, compared to cells with reversible EMT, may derive from an impaired ability to undergo MET.
Prolonged TGF-β treatment does not stabilize Smad3 activation in breast cancer cells.
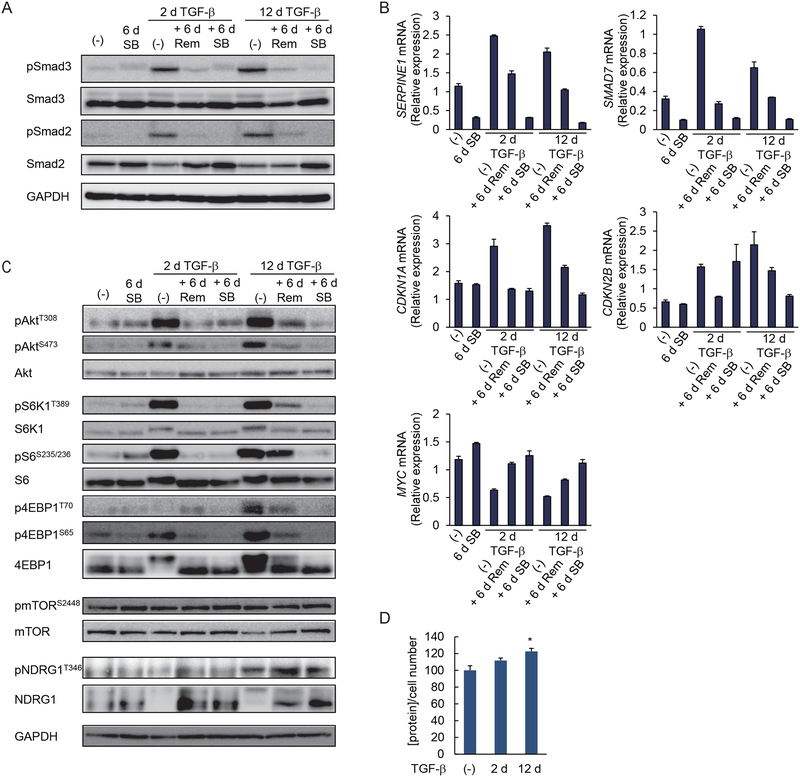
We next addressed changes in TGF-β signaling that accompany stabilization of the mesenchymal state and stemness, and the increased cancer drug resistance, to find out why the stabilized phenotype is not fully inhibited when TGF-β/Smad signaling is blocked. Because SB431542 attenuated the stabilized mesenchymal state toward an epithelial state, we first assessed the activation of TGF-β signaling through Smad2 and Smad3. C-terminal phosphorylation, and thus activation, of Smad2 and Smad3 was enhanced in HMLER cells treated with TGF-β for 2 or 12 days. With either treatment, removal of TGF-β for 6 days decreased the Smad2 and Smad3 activation to an amount comparable to autocrine TGF-β stimulation in control cells, and that amount was further decreased by SB431542 (Fig. 6A and fig. S6A). Consistent with the reversibility of Smad3 activation, short-term and long-term TGF-β treatment activated the expression of direct TGF-β/Smad3 target genes encoding plasminogen activator inhibitor-1 (PAI1) or Smad7, and their upregulation was not stabilized (Fig. 6B). The activation of the genes encoding the cyclin-dependent kinase (CDK) inhibitors CDKN1A/p21Cip1 and CDKN2B/p15Ink4B, and repression of that encoding the transcription factor c-Myc, which are also direct TGF-β/Smad3 target genes and control cell proliferation, was similarly reversible (Fig. 6B). Expression of transcripts encoding TGF-β1 and TGF-β2 was transiently increased with TGF-β treatment and decreased after TGF-β removal, whereas expression of the transcript encoding TGF-β3 did not change (fig. S6B). These results suggest that the stabilization of the mesenchymal and stem cell phenotypes did not arise from differences in Smad activation and did not merely depend on continuously enhanced autocrine TGF-β signaling through Smad activation.
Figure 6. Prolonged TGF-β treatment enhances and stabilizes AKT-mTOR signaling.
(A) HMLER cells were treated with TGF-β for 2 or 12 days (d), followed by its removal, as indicated, and were cultured in the media without added TGF-β (Rem) or with SB431542 (SB) for 6 days. C-terminal phosphorylation of Smad3 and Smad2 was analyzed by immunoblotting. Blots are representative of N=3 independent experiments. (B) HMLER cells were treated as in (A), and the expression of TGF-β target genes encoding PAI1, Smad7, p21Cip1, p15Ink4B or c-Myc was assessed by qRT-PCR and normalized to rPL19 mRNA. Data are mean ± S.E. from N=3 independent experiments. (C) HMLER cells were treated as in (A) and phosphorylation of AKT, S6K1, S6, 4EBP1, mTOR and NDRG1 was analyzed by immunoblotting. Blots are representative of N=3 independent experiments. (D) HMLER cells were treated with TGF-β for 0, 2 or 12 days. The cells were trypsinized and counted. The total protein content normalized for cell number is shown relative to untreated cells. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 by a Dunnet’s test.
To evaluate whether the cells treated with TGF-β for 2 or 12 days still responded to TGF-β, we starved the treated cells for 4 hours in the absence of TGF-β, and then treated them with TGF-β for 30 min up to 4 hours (fig. S6C). Prolonged TGF-β treatment did not change the basal amount of Smad3 activation following starvation and did not impair the TGF-β-induced Smad3 activation (fig. S6C) nor the TGF-β-induced expression of transcript encoding PAI1 and Smad7 (fig. S6D), suggesting that the prolonged exposure to TGF-β did not render the cells refractory to TGF-β signaling.
Prolonged TGF-β treatment results in enhanced mTOR signaling of breast cancer cells.
We previously reported that TGF-β-induced activation of mTORC1 and mTORC2 is required for full transition from epithelial to mesenchymal state (14, 15). Because additionally AKT and mTOR signaling are often upregulated in breast cancers and enhance tumor cell survival (29–31), we evaluated the activation of AKT and mTOR signaling in HMLER cells treated with TGF-β for 2 or 12 days. Both cell populations showed increased AKT phosphorylation at Thr308 as well as Ser473, when compared with cells that were not treated with TGF-β (Fig. 6C and fig. S6E). When cells treated with TGF-β for 2 days were cultured in medium lacking TGF-β or containing SB431542, thus reversing the mesenchymal state, AKT activation was decreased to the basal abundance seen in unstimulated cells. In contrast, in cells with a stabilized mesenchymal phenotype, removal of TGF-β after 12 days of TGF-β treatment did not decrease AKT activation to the basal abundance but conferred only a partial reduction of AKT phosphorylation at Thr308 and Ser473 (Fig. 6C and fig. S6E); however, treatment with SB431542 after 12 days of TGF-β treatment further reduced AKT activation (Fig. 6C and fig. S6E). The cells with a stabilized mesenchymal state also showed increased phosphorylation of the mTORC1 target p70 ribosomal protein S6 kinase 1 (S6K1) and the S6K1 target S6, and a strong increase in the phosphorylation of the mTORC1 target eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4EBP1), and maintained this activation following removal of TGF-β (Fig. 6C and fig. S6E). The phosphorylation of mTOR, however, was not or was minimally increased. Prolonged TGF-β treatment also enhanced mTORC2 activation, as apparent from enhanced phosphorylation of AKT at Ser473, and enhanced phosphorylation of N-myc downstream-regulated gene 1 (NDRG1), a substrate of the mTORC2 target serum- and glucocorticoid-induced protein kinase 1 (SGK1) (Fig. 6C and fig. S6E). As with the mTORC1 activation and other parameters of stabilized EMT, withdrawal of TGF-β for 6 days resulted in only a mild if any attenuation of mTORC2 activity. Prolonged SB431542 treatment inhibited the activation of mTORC1 and mTORC2 targets more strongly (Fig. 6C and fig. S6E). Finally, consistent with our previous report that activation of AKT-mTOR signaling in TGF-β-induced EMT results in increased protein content and cell size (15), long-term TGF-β-treated HMLER cells showed increased total protein abundance (Fig. 6D). Together, our data strongly suggest that the stabilized mesenchymal phenotype of HMLER cells induced by prolonged TGF-β stimulation not only increases Smad3 activation that is reversed on TGF-β withdrawal, but, more strikingly, increases activation of AKT, mTORC1 and mTORC2 that is resilient to removal of TGF-β.
Inhibition of mTOR signaling in cells with stabilized EMT suppresses the CSC phenotype.
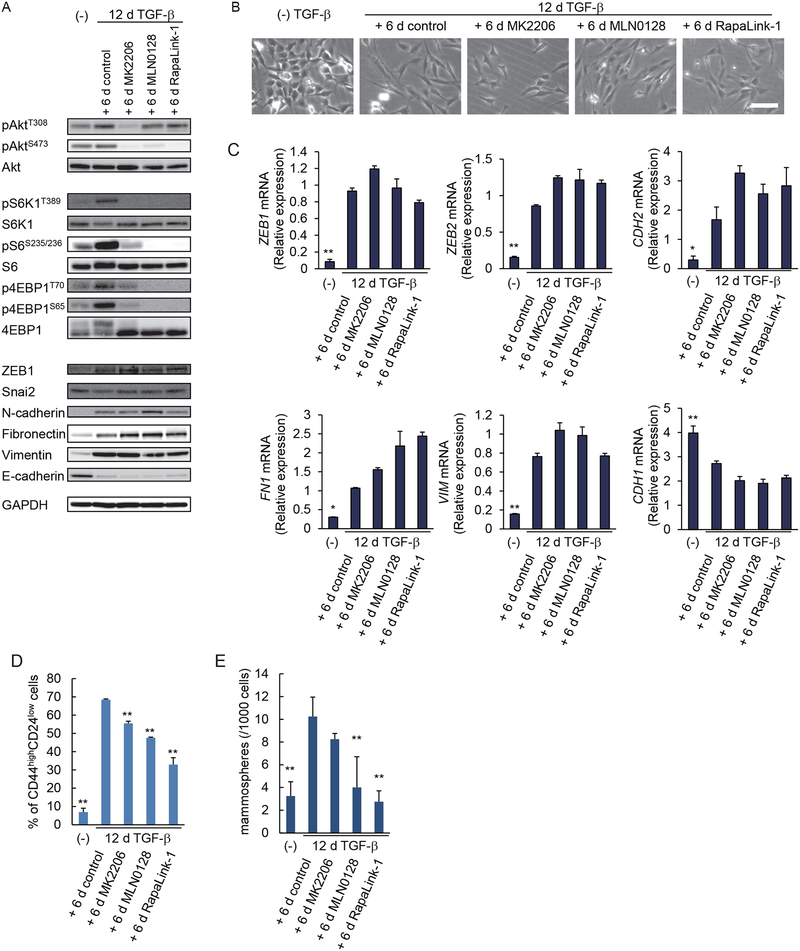
Because cells with stabilized mesenchymal and stem cell phenotypes showed upregulated AKT-mTOR signaling, and inhibition of Smad3 activation only moderately reversed these cells to an epithelial phenotype, we next focused on the role of AKT-mTOR signaling in the stabilization of mesenchymal and CSC-like states. We treated HMLER cells with TGF-β for 12 days, and then with pharmacological inhibitors for 6 days, and assessed their effects on the mesenchymal and stem cell characteristics. AKT signaling was inhibited with the AKT kinase inhibitor MK2206, whereas mTOR signaling was inhibited using MLN0128, an ATP-competitive mTOR kinase inhibitor, or RapaLink-1, a novel bitopic mTOR inhibitor that links MLN0128 to rapamycin and is more potent than either rapamycin or MLN0128 alone (32). As shown above (Fig. 6C), withdrawal of TGF-β for 6 days in the absence of inhibitor did not decrease the increased AKT and mTOR target activation (Fig. 7A and fig. S7A). MK2206 inhibited AKT phosphorylation at Thr308 and Ser473, and reduced the phosphorylation of S6K1, S6 and 4EBP1, which are downstream targets of the AKT-mTORC1 pathway (Fig. 7A and fig. S7A). MLN0128 and RapaLink-1 did not inhibit AKT phosphorylation at Thr308, but strongly inhibited the phosphorylation of AKT-Ser473, and S6K1, S6 and 4EBP1 (Fig. 7A and fig. S7A), consistent with their ability to inhibit both mTORC1 and mTORC2 (32). Inhibition of AKT-mTOR signaling with either inhibitor did not change the morphology of the long-term TGF-β-treated cells (Fig. 7B), consistent with our observations that mTORC1 or mTORC2 inhibition does not affect the mesenchymal morphology of cells undergoing EMT (14). In addition, these three inhibitors did not attenuate the EMT-associated changes in expression of the epithelial and mesenchymal markers and EMT transcription factors that were tested (Fig. 7, A and C, and fig. S7B).
Figure 7. Inhibition of mTOR signaling suppresses the CSC phenotype.
(A) HMLER cells were treated with or without TGF-β for 12 days (d), and then cultured for 6 days with MK-2206 (500 nM), MLN0128 (100 nM), RapaLink-1 (5 nM) or DMSO solvent as a control. Activation of the AKT-mTOR signaling pathway and expression of epithelial and mesenchymal markers at the protein level were analyzed by immunoblotting. Blots are representative of N=3 independent experiments. (B) HMLER cells were treated as in (A), and the cells were observed by phase-contrast microscopy. Images are representative of N=3 independent experiments. Scale bar, 100 μm. (C) HMLER cells were treated as in (A), and the expression of epithelial and mesenchymal markers was analyzed by qRT-PCR and normalized to rPL19 mRNA. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 vs the treated+6d-control by a Dunnet’s test. (D) HMLER cells were treated as in (A), and the percentage of CD24lowCD44high stem cells in the entire cell population was determined by flow cytometry. Data are mean ± S.E. from N=3 independent experiments. **P< 0.01 vs the treated+6d-control by a Dunnet’s test. (E) HMLER cells were treated as in (A), and then assessed for formation of secondary mammospheres. Data are mean ± S.E. from N=3 independent experiments. **P< 0.01 vs the treated+6d-control by a Dunnet’s test.
We next studied the effects of these inhibitors on the stabilization of stem cell state. The mTOR inhibitors MLN0128 and RapaLink-1 reduced the CD24lowCD44high stem cell population of HMLER cells treated with TGF-β for 12 days, whereas the AKT inhibitor MK2206 was less effective (Fig. 7D). We also examined the effect of these inhibitors on mammosphere formation of long-term TGF-β-treated cells. MLN0128 and RapaLink-1 efficiently reduced the mammosphere formation to an amount similar to those of HMLER cells that were not treated with TGF-β (Fig. 7E). MK2206, however, showed a much milder inhibition, suggesting that inhibition of AKT signaling alone is not sufficient to inhibit the stem cell-like phenotype of long-term TGF-β treated cells (Fig. 7E). Considering the target specificities of these inhibitors, these results show that the increased mTOR signaling is required for the maintenance of the stabilized CSC phenotype, and that inhibition of mTOR is much more efficient than inhibition of AKT in repressing the stem cell phenotype.
mTOR inhibition reduces cell viability and cancer drug resistance of cells with stabilized EMT.
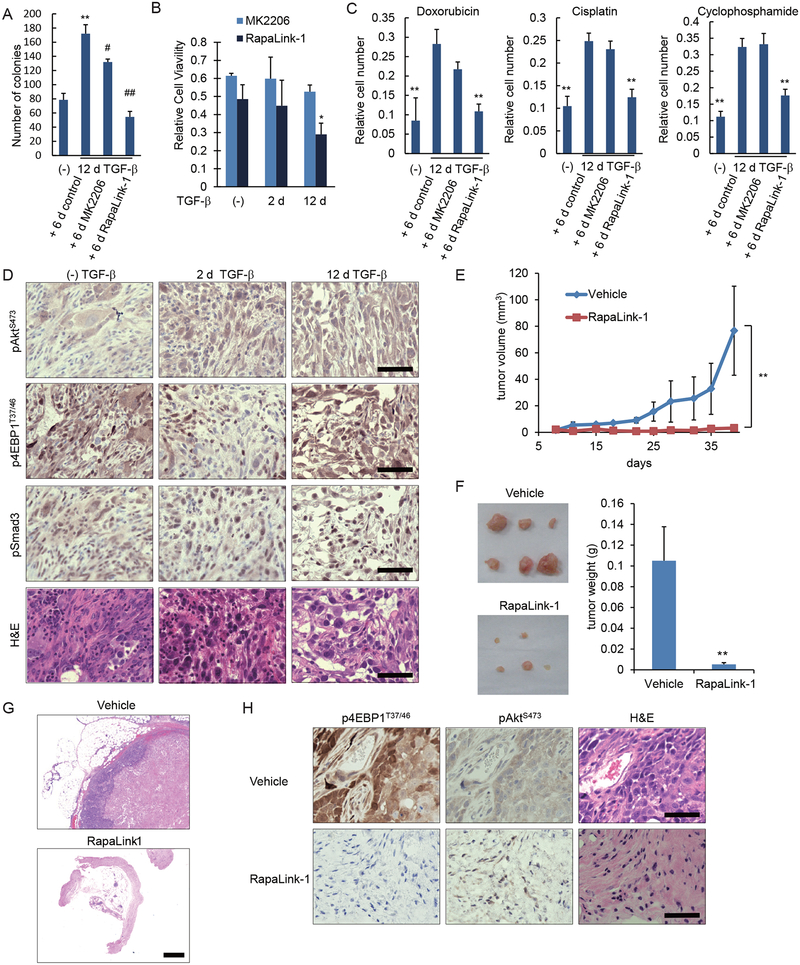
Since stabilized EMT is characterized by enhanced mTOR activation and stem cell characteristics, and mTOR inhibition repressed the CSC phenotype, we tested whether blocking mTOR also inhibits tumor cell survival. We compared the mTOR inhibitor RapaLink-1, which potently inhibits mammosphere formation, and the AKT inhibitor MK2206. RapaLink-1 inhibited the anchorage-independent colony formation of HMLER cells with a stabilized mesenchymal phenotype to a level that was comparable to that of cells that were not treated with TGF-β. In contrast, MK2206 had only a low ability to inhibit anchorage-independent colony growth (Fig. 8A). The relative effects of these two inhibitors resembled those in the mammosphere assay (Fig. 7E). RapaLink-1 moderately and partially decreased the colony formation of untreated cells and short-term TGF-β−treated cells (fig. S8).
Figure 8. Inhibition of mTOR signaling suppresses cancer drug resistance and tumorigenesis.
(A) HMLER cells were treated with or without TGF-β for 12 days (d) and then cultured for 6 days with MK2206 (500 nM), RapaLink-1 (5 nM) or DMSO solvent as a control. The cells were assessed for colony formation in soft-agar. Data are mean ± S.E. from N=3 independent experiments. **P< 0.01 vs (−) TGF-β; #P<0.05 and ##P< 0.01 vs. control, by a Tukey’s test. (B) HMLER cells were treated with TGF-β for 0, 2 or 12 days then reseeded and cultured with MK2206 (500 nM) or RapaLink-1 (5 nM) for 2 days. The live cells were counted, and numbers of viable cells relative to cells cultured without the inhibitors were shown as a graph. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 vs. (−)TGF-β+RapaLink-1, by a Dunnet’s test. (C) HMLER cells were treated with TGF-β for 12 days, and subsequently for 48 hours with doxorubicin (250 nM), cisplatin (250 μM) or cyclophosphamide (10 μM) in the presence of MK2206 (500 nM), RapaLink-1 (5 nM) or DMSO solvent. The live cells were counted, and their numbers were normalized to the initial total cell number. Data are mean ± S.E. from N=3 independent experiments. *P< 0.05 and **P< 0.01 vs. 12d TGF-β + control, by a Dunnet’s test. (D) HMLER cells were treated with TGF-β for 0, 2 or 12 days then suspended in 1:1 PBS/Matrigel, and 10,000 cells were injected orthotopically into mammary fat pad. Primary tumors were stained with antibodies to pSmad3, AKT-pSer473, or 4EBP1-pThr37/46, or with H&E. Images are each representative of 8 independent tumors. Scale bars, 50 μm. (E to H) HMLER cells were treated with TGF-β for 12 days, and 2 × 106 cells were injected orthotopically into a mammary fat pad. Mice were treated by intraperitoneal injection of vehicle or RapaLink-1 (1.5 mg/kg) every 5 or 7 days starting at day 8. Tumor sizes were measured by caliper twice per week (E). (F) Mice were sacrificed at 6 weeks after injection and tumors or residual stroma at the site of injection were harvested, photographed (F), and sections were stained by H&E (G; scale bar, 500 μm) or with antibodies to 4EBP1-pThr37/46 and AKT-pSer473 (H; scale bars, 50 μm). Images are representative of the cohort (in G and H, the Rapa-link sections were from normal tissue and remaining stroma at the site of injection), and data are mean ± S.E. from N=6 mice. **P< 0.01 vs. vehicle by a Student’s t test.
RapaLink-1 was also more efficient than MK2206 in reducing the viability of HMLER cells. Using cells that were not treated with TGF-β or treated with TGF-β for two days, MK2206 mildly reduced the proliferation of HMLER cells, whereas RapaLink-1 was slightly more effective than MK2206 (Fig. 8B). In contrast, treatment of HMLER cells with TGF-β for 12 days made the cells more sensitive to RapaLink-1, reducing their viability to about 30%, whereas the effect of MK2206 was less (Fig. 8B). These results suggest that the increased mTOR pathway activation seen in cells with a stabilized mesenchymal phenotype makes the cells more sensitive to mTOR inhibition by RapaLink-1. Therefore, mTOR inhibition may be used to target the breast cancer cells with stabilized EMT that are enriched in stem cells and show increased cancer latency.
Given that long-term TGF-β treatment increased the resistance to anticancer drugs (Fig. 4, A to C, and fig. S4, A and B), we also tested the effects of MK2206 or RapaLink-1 on the increased resistance of HMLER cells with stabilized EMT to anticancer drugs. Following treatment with TGF-β for 12 days, MK2206 had little or no effect on the resistance of the cells to doxorubicin, cisplatin or cyclophosphamide (Fig. 8C). In contrast, RapaLink-1 strongly increased the sensitivity to these anticancer drugs (Fig. 8C). These data strongly suggest that the increased drug resistance of HMLER cells following long-term TGF-β treatment results from increased mTOR activity and illustrate that inhibiting mTOR activity represses the resistance to a level that is comparable to that of cells that were not treated with TGF-β.
Inhibition of mTOR signaling reduces the tumorigenicity of cells with stabilized EMT.
To examine the effect of mTOR inhibition on tumor formation in vivo, we used the orthotopic implantation model of HMLER cells that we used above (Fig. 5F) but with a greater number of injected cells to enhance tumorigenesis. Immunostaining of tumors derived from HMLER cells that were treated with TGF-β for 12 days showed activation of mTOR, apparent through staining for phospho-AKT-Ser473 and phospho-4EBP1, and Smad3 activation, assessed by phospho-Smad3 staining (Fig. 8D). Tumors derived from untreated HMLER cells or HMLER cells treated with TGF-β for only 2 days also showed some mTOR and Smad3 activation, suggesting TGF-β activity in the tumor (Fig. 8D).
To test the effect of mTOR inhibition in vivo, we administered RapaLink-1 to mice orthotopically injected with long-term TGF-β-treated cells, starting at day 8 after injection of the tumor cells. We compared the tumor size progression in the inhibitor- and vehicle-treated mice. RapaLink-1 treatment resulted in almost complete inhibition of tumor formation (Fig. 8, E and F). Histological examination of H&E-stained samples revealed no detectable tumor cells at the site of injection in 5 out of 6 mice treated with RapaLink-1 (Fig. 8G). Furthermore, immunohistochemistry revealed that the mTORC1-mediated 4EBP1 phosphorylation was substantially reduced in the residual tumor stroma of RapaLink-1-treated mice, when compared to untreated tumors (Fig. 8H), which is consistent with the potent inhibition of mTORC1 by RapaLink-1. AKT phosphorylation at Ser473 was mildly reduced in RapaLink-1-treated mice (Fig. 8H), consistent with the less effective mTORC2 inhibition by RapaLink-1 in vivo (33). We therefore conclude that the upregulation of AKT-mTOR signaling in cells with stabilized mesenchymal and stem cell phenotypes renders the cells sensitive to mTOR inhibition, enabling RapaLink-1 to greatly impact tumor progression.
Discussion
To interfere with cancer progression, it is essential to understand the heterogeneity of cancer cells, their interactions with stromal cells, and the signaling networks that control their differentiation plasticity. At the basis of the heterogeneity among carcinoma cells is the epithelial plasticity that allows the cells to lose epithelial characteristics and reprogram their organization and gene expression, and thus partially or substantially resemble mesenchymal cells. That EMT is reversible has been concluded based on cell culture and in vivo observations, and is apparent by the reversion of TGF-β-induced EMT toward the epithelial phenotype when TGF-β is removed or TGF-β/Smad signaling is blocked after a few days (16). Given that, in the context of breast cancer, carcinoma cells are continuously exposed to increased levels of activated TGF-β, we investigated whether exposure to TGF-β for longer times than a few days would maintain the reversible nature of EMT. Using mammary epithelial and carcinoma cells as models, we found that prolonged TGF-β treatment stabilized EMT, rendering the phenotype independent of added TGF-β.
Very few studies have examined EMT resulting from sustained TGF-β treatment (34, 35). In one study, TGF-β treatment of non-transformed NMuMG cells for 2–3 weeks gave rise, after substantial cell death, to a cell population with a mesenchymal phenotype and suppressed Smad activation that correlated with resistance to TGF-β-induced growth inhibition. These changes were reversed after removal of TGF-β (34, 35). Another study found that prolonged exposure of NMuMG cells to TGF-β leads to an attenuated mesenchymal morphology that is suggestive of autophagy (36). These observations differ from our results using NMuMG cells and did not address how changes in EMT phenotype induced by prolonged TGF-β exposure contribute to the stem cell phenotype, anticancer drug resistance and cancer progression. Here, we found that stabilized EMT confers a stable and enhanced stem cell-like state of breast cancer cells and increases their resistance to anticancer drugs. Our findings support the existence of multiple EMT states that differentially contribute to cancer progression.
In contrast to reversible EMT, SB431542 or neutralizing anti-TGF-β antibody only partially reversed the stabilized EMT phenotype, suggesting changes in underlying signaling. Specifically, blocking TGF-β signaling was not effective at suppressing the cancer stem cell characteristics and cancer drug resistance. EMT is accompanied by epigenetic changes, and stabilization of the mesenchymal phenotype may result from such changes induced by TGF-β signaling. Indeed, epigenetic regulation of initiation or maintenance of EMT has been documented (37). The methyltransferases G9a, EZH2, SUV39H1 and SETDB1 are required for and epigenetically control TGF-β-induced EMT of mammary epithelial and carcinoma cells (38–41). Furthermore, the maintenance of the mesenchymal phenotype requires autocrine TGF-β signaling to drive sustained ZEB expression, and DNA methylation of the miR-200 locus that regulates ZEB expression (42). Our observations raise the questions how reversible and stabilized EMT differ epigenetically, and how TGF-β-mediated EMT itself is required for stem cell formation and drug resistance. HMLEs represent a pool of cells with epithelial and mesenchymal traits (3); further analyses of different subpopulations may help address these questions.
Notably, our data reveal that reversible EMT and stabilized EMT confer different cancer cell properties. In carcinomas, EMT presents itself with intermediate states, rarely reaching full mesenchymal differentiation (7). Partial or complete EMT allows for cancer cell migration and invasion, enabling cancer dissemination (43), and is linked to CSC generation, and decreased anticancer drug sensitivity (5, 8). Whether EMT facilitates metastatic colonization is subject of discussion, which may relate to a simplistic view of the metastasis process after cell dissemination, and a perception of EMT that does not take into account the diversity of partial and transitional EMT stages.
We found that in both non-transformed and transformed epithelial cells, the reversible EMT transitioned with continued TGF-β exposure into a stabilized EMT that no longer depends on added TGF-β. Additionally, Ras-mediated transformation facilitated the EMT response to TGF-β, resulting in a more rapid response, which may reflect the cooperation of Ras-ERK MAPK signaling with TGF-β/Smad signaling. It should be noted that the plasticity response resulting from enhanced Ras activity is reversed by SB431542, and thus depends on TGF-β/Smad signaling.
Reversible EMT and stabilized EMT endow the cells with different properties. The changes in phenotype and gene expression that classically define EMT are similar in cells with reversible EMT and those with stabilized EMT. In contrast, stabilization of EMT in the Ras-transformed cells increased the resistance to anticancer drugs to a much higher level than what was seen with reversible EMT and enhanced and stabilized the stem cell-like properties. Additionally, reversible EMT was accompanied with enhanced lung colonization, whereas sustained TGF-β exposure resulted in a lower increase in lung colonization, when compared to reversible EMT. These observations suggest that EMT stabilization prolongs latency and persistence of cancer cells in vivo, which correlates with increased stemness and drug resistance. These results are reminiscent of the lack of metastatic outgrowth from cells with mesenchymal traits (44, 45), and consistent with the chemoresistance associated with EMT in studies that found a limited contribution of EMT to experimental metastasis (46, 47). Our observations suggest that reversible EMT and stabilized EMT have different roles in cancer progression. Stabilized EMT may contribute to tumor latency and persistence, which correlates with strongly enhanced CSC generation and anticancer drug resistance, while reversible EMT strongly enhances dissemination and metastasis. These findings add new insights for understanding the roles of EMT in cancer progression and metastatic colonization.
Our study further reveals that mTOR’s role in stabilized EMT provides an opportunity to effectively target epithelial plasticity in cancer progression. We have previously reported that TGF-β-induced activation of mTORC1 is required for EMT-associated cell migration and invasion (15) and that mTORC2 activation is required to complete EMT (14). Stabilized EMT was accompanied by increased AKT, mTORC1 and mTORC2 activation, at higher levels than those seen in the context of reversible EMT. As in cells with reversible EMT, SB431542 fully repressed the activation of Smad3 and TGF-β/Smad3 target genes in cells with stabilized EMT. Furthermore, SB431542 inhibited the EMT-associated cell shape change and marker gene expression completely in cells with reversible EMT and only partially in cells with stabilized EMT. In contrast, the acquisition of stem cell properties and cancer drug resistance, which are enhanced with EMT stabilization were less susceptible to inhibition of TGF-β/Smad signaling. Conversely, mTOR inhibition in cells with stabilized EMT did not reverse the mesenchymal cells to the epithelial morphology but repressed the increased stem cell generation and cancer drug resistance that associate with stabilized EMT. SB431542 repressed mTORC1 and mTORC2 activation only partially, consistent with its inability to block TGF-β-induced AKT activation in a cancer cell line (48). These findings reinforce the proposed cooperation of AKT-mTOR signaling with TGF-β/Smad signaling in EMT (14, 15), and highlight the role of upregulated mTOR signaling in increasing and stabilizing the CSC phenotype and resistance to anticancer drugs. Our observations also illustrate the different roles of Smad and mTOR signaling downstream from TGF-β in stabilizing EMT and stemness, and suggest that cancer drug resistance and stemness are not merely associated with EMT but require additional signaling and/or epigenetic changes.
The established notion of reversible EMT in response to TGF-β has invited studies of TβRI kinase inhibitors to inhibit epithelial plasticity in cancer progression (49). Many studies have been unsuccessful, likely due to poor pharmacokinetics, requiring impractically frequent dosing, and difficulties to stably repress TGF-β-induced Smad activation in vivo. Moreover, the pleiotropic nature of TGF-β/Smad signaling and concerns of anticipated tumor suppression and promotion effects raise the desire to inhibit an alternative pathway. Our findings that long-term exposure to TGF-β stabilizes EMT and CSC properties, that inhibition of TGF-β/Smad signaling only partially inhibits the stabilized EMT phenotype, and that upregulated mTOR activity helps stabilize the CSC properties, invited an evaluation of the effects of mTOR inhibition. Inhibition of mTOR suppressed the anchorage independence, viability and anticancer drug resistance of cancer cells with stabilized EMT. Consistent with previous observations (32, 33), RapaLink-1 was more effective than MLN0128. Thus, in contrast to cells with reversible EMT, the increased mTOR activation that associates with stabilized EMT and stemness renders the cells more sensitive to mTOR inhibition by RapaLink-1. However, inhibition of AKT had only a minimal effect on anchorage independence, which correlates with tumorigenesis and is consistent with the minimal effects of blockade of PI3K or AKT on mTOR activity in some cancers (50). Consistent with our results that mTOR inhibition reduces chemoresistance, mTOR activation was shown to correlate with increased resistance to inhibitors of BRAF-MEK-ERK MAPK and PI3K signaling, as well as CDK inhibitors (51). In gliomas, mTORC2 activation was correlated with resistance to cisplatin and temozolomide (52, 53).
Consistent with its effects on stem cell characteristics and cell viability, mTOR inhibition fully suppressed the tumor formation of HMLER cells with stabilized EMT. Among many processes driven by mTOR, phosphorylation of 4EBP1 that leads to translational initiation has been suggested to be critical in tumorigenesis (31, 54, 55). HMLER cells with stabilized EMT showed increased 4EBP1 phosphorylation, and RapaLink-1, which blocked 4EBP1 phosphorylation, prevented tumor formation, suggesting key roles of mTOR and 4EBP1 phosphorylation in this stable phenotype.
Together, our results highlight the heterogeneity of the differentiation plasticity of carcinoma cells in response to signals provided by the tumor microenvironment. They also highlight the enhanced stem cell properties and resistance to anticancer drugs that accompany stabilized EMT, and not reversible EMT, and may be much more relevant in cancer progression than the mere loss of epithelial characteristics and acquisition of mesenchymal traits. The substantial contribution of mTOR signaling to the stabilized EMT phenotype invites approaches to inhibit mTOR signaling in combination with anticancer drugs as a treatment strategy to target breast cancer stem cells and cancer progression.
Materials and Methods
Cell culture and reagents.
NMuMG cells were cultured in DMEM with 10% FBS (Sigma-Aldrich Co. LLC.), 10 μg/ml insulin (Sigma-Aldrich Co. LLC), 100 U/ml penicillin, and 100 μg/ml streptomycin. Immortalized human mammary epithelial (HMLE) cells were from Jing Yang (UCSD) and maintained in MGEM (Lonza), as described (20). To induce EMT, NMuMG cells were treated with 1 ng/ml TGF-β (R&D), and HMLE cells were treated with 2 ng/ml TGF-β (R&D), as indicated. SB431542 was from Sigma-Aldrich and was used at 5 μM. MK2206 and MLN0128 were from Selleck Chemicals and were used at 500 nM and 100 nM respectively. RapaLink-1 was synthesized as described (32), and was used in cell culture at 5 nM. Doxorubicin and 4-hydroperoxycyclophosphamide was from Toronto Research Chemicals. Cisplatin and 5-fluorouracil were from Wako. Paclitaxel was from Bristol-Myers.
Immunofluorescence and cell imaging.
NMuMG cells and HMLE cells, cultured on glass slides, were fixed with 4% paraformaldehyde for 15 min, permeabilized in 0.2% Triton X-100 for 10 min, and blocked with 3% BSA in PBS for 2 h at room temperature. The slides were incubated overnight at 4°C with anti–E-cadherin (BD Biosciences, Cat# 610181), or anti-fibronectin (Sigma-Aldrich, Cat# F3648) diluted 1:400 in PBS and 3% BSA, washed with PBS at room temperature, and incubated for 1 h with Alexa Fluor-conjugated secondary antibody (1:1,000; Thermo Fisher Scientific) or rhodamine-conjugated phalloidin (1:1,000; Thermo Fisher Scientific, Cat# R415) to visualize actin filaments. The slides were mounted with ProLong™ Gold Antifade Mountant with DAPI (Thermo Fisher Scientific) to visualize nuclei. Immunofluorescence images were obtained with an inverted light microscope (DMI5000, Leica Microsystems), a laser scanning confocal microscope (SP5, Leica Microsystems) or All-in-One Fluorescence Microscope (BZ-700, Keyence).
Immunoblot analyses.
Cells were washed once with ice-cold PBS and lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholate, and protease inhibitor cocktail). Cell debris was removed by centrifugation at 15,000 rpm for 10 min. Immunoblotting was performed as described (56), using the indicated antibodies, and the bound antibodies were visualized with horseradish peroxidase-conjugated antibodies against rabbit or mouse IgG. The following antibodies were used: anti-ZEB1 (Novus Biologicals, Cat# NBP1–05987), anti-Slug (Cell Signaling, Cat# 9589), anti-N-cadherin (BD Biosciences, Cat# 610920), anti-fibronectin (BD Biosciences, Cat# 610077), anti-vimentin (Cell Signaling, Cat# 5741), anti-E-cadherin (BD Biosciences, Cat# 610181), anti-Smad3 (Cell Signaling, Cat# 9523), anti-phospho-Smad3-Ser423/425 (Abcam Cat# ab52903 ), anti-mTOR (Cell Signaling, Cat# 2983), anti-phospho-mTOR-Ser2448 (Cell Signaling, Cat# 2971), anti-phospho-AKT-Thr308 (Cell Signaling, Cat# 2965), anti-phospho-AKT-Ser473 (Cell Signaling, Cat# 9271), anti-AKT (Cell Signaling, Cat# 9272), anti-phospho-S6K1-Thr389 (Cell Signaling, Cat# 9234), anti-S6K1 (Cell Signaling, Cat# 9202), anti-phospho-S6-Ser235/236 (Cell Signaling, Cat# 2211), anti-S6 (Cell Signaling, Cat# 2217), anti-phospho-4EBP1-Thr70 (Cell Signaling, Cat# 9455), anti-phospho-4EBP1-Ser65 (Cell Signaling, Cat# 9451), anti-4EBP1 (Cell Signaling, Cat#), anti-phospho-NDRG1-Thr346 (Cell Signaling, Cat# 3217), anti-NDRG1 (Cell Signaling, Cat# 9408) and anti-GAPDH (Sigma Aldrich, Cat# G8795).
Quantitative real-time PCR measurements of mRNA.
Total RNA was extracted using RNAeasy extraction kit (Qiagen). cDNAs were generated using the one-step iScript cDNA synthesis kit (Biorad) or PrimeScript2 reverse transcriptase (TaKaRaBio). Quantitative real-time PCR was performed with cDNA samples using iQ SYBR green supermix (Biorad) or Fast Start Universal SYBR Green Master Mix with ROX (Roche Diagnostics), and the signal was detected by CFX96 real-time PCR system (Biorad) or StepOnePlus Real-Time PCR system (Thermo Fisher Scientific). Sequences of the primers used are listed in table S1.
Flow cytometry.
Antibodies for the human CD24 (Clone ML5) and human CD44 (Clone G44–26) were purchased from BD Biosciences. Cells were dissociated into single cells and stained with antibodies to CD24 and CD44 in PBS. Flow cytometry analysis was done using an LSRII (BD) with BD FACSDiva Software or Gallios Flow Cytometer (Beckman Coulter).
Mammosphere formation assay.
Mammosphere formation assay was performed as described (22). Cells were seeded in 12-well ultra-low attachment plates at a density of 5000 cells/well in MEGM medium (Lonza) supplemented with B27 supplement (Thermo Fisher Scientific), 10 ng/ml bFGF, 20 ng/ml EGF and 1% methylcellulose. After incubating the cells for 6 days, the mammospheres were observed by phase-contrast microscopy and quantified. For secondary sphere formation, primary spheres were collected by centrifugation at 800 g for 5 min, dissociated into single cells by trypsinization, and replated in ultra-low attachment plates at 2000 cells/well.
Cell survival and proliferation assays.
HMLER cells were seeded at 2,000 cells per well in 96-well plates. 24 hours later, the cells were treated for 48 hours with doxorubicin, cisplatin, cyclophosphamide, 5-fluoruracil or paclitaxel at the indicated concentrations. Cell viability was measured using Cell Count Reagent SF (Nacalai Tesque) according to the manufacturer’s instruction. The reagent was added to the cells and the cells were incubated at 37°C for 4 hours. Absorbance at 450 nm was measured with a Model 680 Microplate Reader (Bio-Rad), followed by subtraction of absorbance at 595 nm. For the cell survival and proliferation assay, HMLER cells were seeded at 10,000 cells per well in 24 well plates. 24 hours later, the cells were incubated with anticancer drugs, TGF-β, or AKT-mTOR inhibitors for 48 hours. The cells were trypsinized and the number of viable cells was counted.
Soft-agar colony formation assay.
Colony formation assay in soft agar was performed as described (56). Cells were seeded at 2 × 104 cells per well in 0.3% agar and cultured for 2 weeks. The colonies were observed by phase-contrast microscopy and quantified.
In vivo tumorigenesis assays in mice.
All mouse experiments were performed in accordance with protocols approved by the UCSF Institutional Animal Care and Use Committee or the Animal Care and Use Committee of the Graduate School of Medicine, The University of Tokyo. For experimental metastasis experiments, 5×105 HMLER cells were injected laterally into the tail vein of 7- to 12-week-old female NOD/SCID/IL2rγnull (NSG) mice. Mice were sacrificed at 8 weeks and the lungs were harvested. Tissues were fixed overnight in 4% paraformaldehyde and embedded in paraffin. Sections were stained with hematoxylin and eosin. Lung sections were analyzed using bright field microscopy, and the number of tumor nodules in the lungs was determined based on visual inspection.
For orthotopic xenograft experiments, 1×104 HMLER cells in 1:1 PBS/Matrigel were injected orthotopically into the fourth mammary fat pad. Primary tumor size was measured twice a week. Mice were sacrificed and the tumors were dissected when their sizes reached 1 cm in diameter. For orthotopic injections and treatment studies, 2×106 HMLER cells treated with TGF-β for 12 days were suspended in 1:1 PBS/Matrigel and injected into the fourth mammary fat pad of 7- to 12-week-old female NSG mice. Mice were treated with i.p. injection of vehicle (20% DMSO, 40% PEG-300, and 40% PBS [v/v]) or RapaLink-1 (1.5 mg/kg in 20% DMSO, 40% PEG-300, and 40% PBS [v/v]), starting at day 8 after tumor implantation, and every 5 days for 20 days, then once a week for 2 weeks. Primary tumor size was measured twice a week. All the mice were sacrificed at 6 weeks after tumor implantation, and the primary tumors were harvested, fixed in 4% paraformaldehyde, and embedded in paraffin. Sections were stained with hematoxylin and eosin or were subjected to immunohistochemistry.
Protein content measurements.
To measure protein content, cells were trypsinized, and the cell number was determined. The cells were lysed in RIPA buffer with protease inhibitors, and the protein concentrations were quantified using BCA protein assay (ThermoFisher Scientific). The protein content was normalized to cell number.
Immunohistochemistry of mouse tissues.
Formalin-fixed, paraffin-embedded sections were deparaffinized in xylene and rehydrated in graded alcohol. Antigen retrieval was performed using autoclave treatment (121°C, 15 min) in 10 mM sodium citrate (pH 6.0). Staining was performed using the Vectastain Elite ABC kit and DAB Liquid System. The primary antibodies used in this study included rabbit anti-human phospho-AKT-Ser473 antibody (Cell Signaling, # 4060), rabbit anti-human phospho-4EBP1-Thr37/46 antibody (Cell Signaling # 2855), and rabbit anti-human phospho-Smad3-Ser423/425 antibody (Rockland Immunochemicals #600-401-920).
Statistical analyses.
Quantitative data are presented as mean ± standard error of mean (S.E.M) from at least three independent experiments. Differences between groups were analyzed using one-way analysis of variance (Tukey’s test or Dunnet’s test). Unpaired Student’s t-tests were used to compare the two groups. Values of p <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank J. Yang (UCSD) for providing the HMLE cells, Y. Morishita (Univ Tokyo) and the UCSF HDFCCC Preclinical Therapeutics Core for expert technical assistance, and the Derynck laboratory and Miyazono laboratory members for valuable suggestions.
Funding
This research was supported by grants RO1-CA63101 and RO1-CA136690 from the National Cancer Institute to R.D., grants R21CA164772, 5P30 CA082103 and RO1CA210561 from the National Cancer Institute to R.J.A, KAKENHI, grants-in-aid for Scientific Research (S) (15H05774) from the Japan Society for the Promotion of Science (JSPS) to K.M., NIH P50 AA017072, the Stand Up 2 Cancer Lung Cancer Dream Team, the Samuel Waxman Cancer Research Foundation and the Howard Hughes Medical Institute to K.M.S., a postdoctoral fellowship and subsequent grant-in-aid for Young Scientists (B) (16K20974) from JSPS to Y.K., and a postdoctoral fellowships from the Swiss National Science Foundation to D.S.M..
Footnotes
Competing interests
R.D. is a Founder of Pliant Therapeutics, and a consultant to Genentech Inc. and N.G.M. Biopharmaceuticals Inc. K.M.S. is a co-inventor of a patent covering Rapalink-1, owned by UCSF and licensed to Revolution Medicines; a Founder, stock holder, and consultant of Revolution Medicines; a co-inventor of patents related to MLN-128 owned by UCSF and licensed to Takeda Pharmaceuticals; and a consultant to Takeda Pharmaceuticals. R.J.A. has sponsored research agreements with Pfizer Corp. and Plexxikon and is a co-inventor of a pending patent application on the use of anti-TGF-β antibodies that is co-owned by UCSF and Xoma. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
Supplemental Materials
Fig. S1. Neutralizing anti-TGF-β monoclonal antibody and SB431542 partially reverse the stabilized mesenchymal phenotype.
Fig. S2. Prolonged TGF-β treatment does not maintain the expression of TGF-β ligand.
Fig. S3. Prolonged TGF-β treatment stabilizes cell migration and invasion.
Fig. S4. Prolonged TGF-β treatment increases the resistance of breast cancer cells against 5-fluorouracil and paclitaxel.
Fig. S5. Histological evaluation of tumors from mice injected with HMLER cells.
Fig. S6. HMLER cells treated with TGF-β for 2 or 12 days maintain their TGF-β responsiveness.
Fig. S7. Quantification of the phosphorylation of mTOR targets and EMT marker expression.
Fig. S8. RapaLink-1 partially reduces the colony formation of untreated or short-term TGF-β–treated HMLER cells.
Table S1: Primer sequences used for quantitative real-time PCR.
References and Notes
- 1.Visvader JE, Lindeman GJ, Cancer stem cells: current status and evolving complexities. Cell Stem Cell 10, 717–728 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Pattabiraman DR, Weinberg RA, Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov 13, 497–512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA, The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A, Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 3, e2888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheel C, Weinberg RA, Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol 22, 396–403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamouille S, Xu J, Derynck R, Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieto MA, Huang RY, Jackson RA, Thiery JP, EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Shibue T, Weinberg RA, EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14, 611–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morikawa M, Derynck R, Miyazono K, TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol 8, a021873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsuno Y, Lamouille S, Derynck R, TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 25, 76–84 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Derynck R, Budi EH, Specificity, versatility and control of TGF-β family signaling. Sci. Signal 11, eaav5183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng XH, Derynck R, Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21, 659–693 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Zhang YE, Non-Smad signaling pathways of the TGF-β family. Cold Spring Harb Perspect Biol 9, a022129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R, TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci 125, 1259–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamouille S, Derynck R, Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol 178, 437–451 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miettinen PJ, Ebner R, Lopez AR, Derynck R, TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol 127, 2021–2036 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M, Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: a tissue microarray study. Cancer Res 62, 497–505 (2002). [PubMed] [Google Scholar]
- 18.Massagué J, TGFβ in cancer. Cell 134, 215–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA, Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 15, 50–65 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA, Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L, Yang J, Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell 19, 372–386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS, In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17, 1253–1270 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bascom CC, Wolfshohl JR, Coffey RJ Jr., Madisen L, Webb NR, Purchio AR, Derynck R, Moses HL, Complex regulation of transforming growth factor β1, β2, and β3 mRNA expression in mouse fibroblasts and keratinocytes by transforming growth factors β1 and β2. Mol Cell Biol 9, 5508–5515 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E, TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev 10, 2462–2477 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Oft M, Akhurst RJ, Balmain A, Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol 4, 487–494 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S, Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol 156, 299–313 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamburger AW, Salmon SE, Primary bioassay of human tumor stem cells. Science 197, 461–463 (1977). [DOI] [PubMed] [Google Scholar]
- 28.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li S, Long YO, Thach C, Liu Y, Zarieh A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P, Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589 e517 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Engelman JA, Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 9, 550–562 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Guerrero-Zotano A, Mayer IA, Arteaga CL, PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev 35, 515–524 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Saxton RA, Sabatini DM, mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, Barratt DG, Cosulich S, Klinowska T, Rosen N, Shokat KM, Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 534, 272–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Q, Aksoy O, Wong RA, Ilkhanizadeh S, Novotny CJ, Gustafson WC, Truong AY, Cayanan G, Simonds EF, Haas-Kogan D, Phillips JJ, Nicolaides T, Okaniwa M, Shokat KM, Weiss WA, A kinase inhibitor targeted to mTORC1 drives regression in glioblastoma. Cancer Cell 31, 424–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caulin C, Scholl FG, Frontelo P, Gamallo C, Quintanilla M, Chronic exposure of cultured transformed mouse epidermal cells to transforming growth factor-β1 induces an epithelial-mesenchymal transdifferentiation and a spindle tumoral phenotype. Cell Growth Differ 6, 1027–1035 (1995). [PubMed] [Google Scholar]
- 35.Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A, Sustained TGFβ exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene 27, 1218–1230 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Jiang Y, Woosley AN, Sivalingam N, Natarajan S, Howe PH, Cathepsin-B-mediated cleavage of Disabled-2 regulates TGF-β-induced autophagy. Nat Cell Biol 18, 851–863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam WL, Weinberg RA, The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 19, 1438–1449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP, G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 122, 1469–1486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong C, Wu Y, Wang Y, Wang C, Kang T, Rychahou PG, Chi YI, Evers BM, Zhou BP, Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene 32, 1351–1362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schubeler D, van Nimwegen E, Christofori G, Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Du D, Katsuno Y, Meyer D, Budi EH, Chen SH, Koeppen H, Wang H, Akhurst RJ, Derynck R, Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls Snail1 expression and epithelial-mesenchymal transition. EMBO Rep 19, 135–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, Lindeman GJ, Shannon MF, Drew PA, Khew-Goodall Y, Goodall GJ, An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell 22, 1686–1698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert AW, Pattabiraman DR, Weinberg RA, Emerging biological principles of metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J, Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ocaña OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA, Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D, Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R, Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamidi A, Song J, Thakur N, Itoh S, Marcusson A, Bergh A, Heldin CH, Landstrom M, TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci Signal 10, aal4186 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Akhurst RJ, Targeting TGF-β signaling for therapeutic gain. Cold Spring Harb Perspect Biol 9, a022301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan QW, Cheng C, Knight ZA, Haas-Kogan D, Stokoe D, James CD, McCormick F, Shokat KM, Weiss WA, EGFR signals to mTOR through PKC and independently of Akt in glioma. Sci Signal 2, ra4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guri Y, Hall MN, mTOR signaling confers resistance to targeted cancer drugs. Trends Cancer 2, 688–697 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, Dang J, Zhu S, Yang H, De Jesus J, Amzajerdi AN, Zhang Y, Dibble CC, Dan H, Rinkenbaugh A, Yong WH, Vinters HV, Gera JF, Cavenee WK, Cloughesy TF, Manning BD, Baldwin AS, Mischel PS, Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov 1, 524–538 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiler M, Blaes J, Pusch S, Sahm F, Czabanka M, Luger S, Bunse L, Solecki G, Eichwald V, Jugold M, Hodecker S, Osswald M, Meisner C, Hielscher T, Rubmann P, Pfenning PN, Ronellenfitsch M, Kempf T, Schnolzer M, Abdollahi A, Lang F, Bendszus M, von Deimling A, Winkler F, Weller M, Vajkoczy P, Platten M, Wick W, mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci U S A 111, 409–414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, Baselga J, Ramon y Cajal S, 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res 67, 7551–7555 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D, The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katsuno Y, Ehata S, Yashiro M, Yanagihara K, Hirakawa K, Miyazono K, Coordinated expression of REG4 and aldehyde dehydrogenase 1 regulating tumourigenic capacity of diffuse-type gastric carcinoma-initiating cells is inhibited by TGF-β. J Pathol 228, 391–404 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.