Abstract
The microbiome of the female reproductive tract has implications for women’s reproductive health. We examined the vaginal microbiome in two cohorts of women who experienced normal term births: a cross-sectionally sampled cohort of 613 pregnant and 1,969 non-pregnant women, focusing on 300 pregnant and 300 non-pregnant women of African, Hispanic or European ancestry case-matched for race, gestational age and household income; and a longitudinally sampled cohort of 90 pregnant women of African or non-African ancestry. In these women, the vaginal microbiome shifted during pregnancy toward Lactobacillus-dominated profiles at the expense of taxa often associated with vaginal dysbiosis. The shifts occurred early in pregnancy, followed predictable patterns, were associated with simplification of the metabolic capacity of the microbiome and were significant only in women of African or Hispanic ancestry. Both genomic and environmental factors are likely contributors to these trends, with socioeconomic status as a likely environmental influence.
Growing evidence suggests that the impact of the vaginal microbiome extends to the health of pregnant women and their neonates in utero and beyond. The vaginal microbiome consists of a finite number of discrete microbial communities dominated by different bacterial taxa or combinations thereof1,2. A vaginal microbiome with microbial communities dominated by species of Lactobacillus has been associated with adverse conditions of health of the female reproductive tract, whereas a microbiome dominated by complex microbial communities of Gardnerella, Atopobium, Dialister, Peptoniphilus, Lachnospiraceae members (bacterial vaginosis (BV)-associated bacterium 1 (BVAB1)) and other anaerobes3-5 has been associated with a higher risk. A complex vaginal microbiome is associated with BV, the most common gynecological condition of women of reproductive age6, as well as a higher risk of sexually transmitted infection, pelvic inflammatory disease and adverse pregnancy outcomes including preterm birth (PTB)3,5,7.
More than 10% of neonates in the United States are delivered preterm (<37 weeks’ gestation), and certain racial and ethnic groups have even higher rates8-10. Women of African ancestry in the United States are significantly more likely than women of European ancestry to have a premature birth or experience very preterm delivery (<32 weeks’ gestation). This health disparity could be due to differences in the vaginal microbiomes of these women as the population attributable risk of BV for spontaneous PTB at <32 weeks’ gestation has been estimated to be ~40%11. Although environmental factors, including socioeconomic status (for example, household income, access to care and so on) are known to contribute to these differences, genetic factors also play a role12-14. Recent studies1,2,15-23 show that the vaginal microbiomes of women of African ancestry are less likely to be dominated by species of Lactobacillus, and more likely to comprise primarily Gardnerella vaginalis, Atopobium vaginae, Sneathia amnii, BVAB1 and other anaerobes. Independent of the underlying cause, this difference is a probable contributor to the observed disparities in risk of PTB.
Here, we report an analysis of data from a collaborative effort under the umbrella of the National Institutes of Health Human Microbiome Project phase 1 (HMP1)24 and the integrative HMP (HMP2)25. In our HMP1 study, the Vaginal Human Microbiome Project (VaHMP), we collected more than 40,000 vaginal, cervical, introital and buccal swab samples cross-sectionally from a racially diverse cohort of 4,851 women visiting VCU Medical Center women’s clinics (Fig. 1a). In our HMP2 study, the Multi Omic Microbiome Study: Pregnancy Initiative (MOMS-PI), we collected more than 200,000 vaginal, buccal, rectal, birth product and other samples longitudinally from a racially diverse cohort of 1,572 women and neonates during and after their pregnancies from VCU Medical Center Clinics and clinics associated with the Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) in Seattle (Fig. 1b). We present our findings from women in these two cohorts who experienced uncomplicated term birth (>37 weeks’ gestation). The samples were analyzed by 16S ribosomal RNA taxonomic profiling, metagenomic and metatranscriptomic sequencing, and metabolic pathway analysis to track dynamics of the vaginal microbiome through pregnancy. Our results show that the vaginal microbiome changes during normal pregnancy, becoming more Lactobacillus-dominated at the expense of G. vaginalis and other anaerobes. These changes are most significant in women of African ancestry, occur early in pregnancy, and seem to be the result of stabilization of Lactobacillus colonization and destabilization of colonization by other taxa. Pathway profiles generated from our metagenomic and metatranscriptomic data confirmed a transition early in pregnancy. Together, these observations suggest that the compositions of the vaginal microbiomes of women of African and non-African ancestry respond differently during pregnancy due to complex interactions between human and microbial physiology and environmental influences.
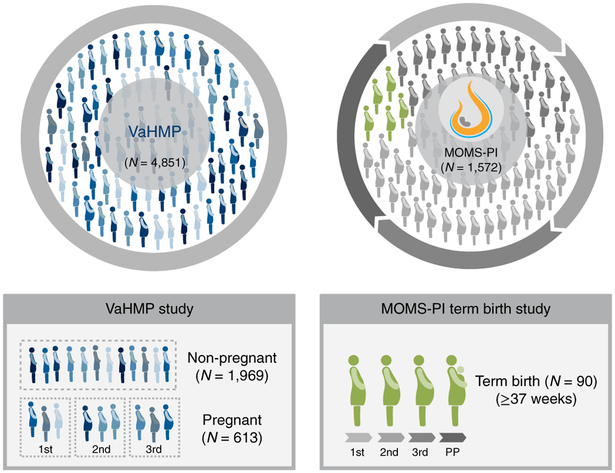
Fig. 1 ∣. Overview of the VaHMP study and the MOMS-PI Term Birth study.
a, Of the 4,851 women enrolled in the VaHMP study, 613 pregnant and 1,969 non-pregnant women who reported no health complaints were analyzed in this project. A subset of 600 of these women (that is, 300 pregnant and 300 non-pregnant women, case-matched for self-reported race, gestational age at sampling and household income) was selected for analysis. Of these, there were 156 case-matched pairs of women with African ancestry, 61 case-matched pairs of women with European ancestry, and 83 case-matched pairs of women with Hispanic ancestry. These women were sampled at regular visits to VCU women’s health clinics. b, The 90 women (49 of African ancestry, 41 of European ancestry) forming the MOMS-PI Term Birth cohort were selected from a Phase 1 cohort (dark shade, pregnant women, N = 627) of women enrolled from women’s clinics at VCU. These participants were sampled longitudinally throughout pregnancy. 1st, 2nd and 3rd refer to the trimesters of pregnancy; PP, postpartum.
Results
Pregnancy alters vaginal microbiome profiles.
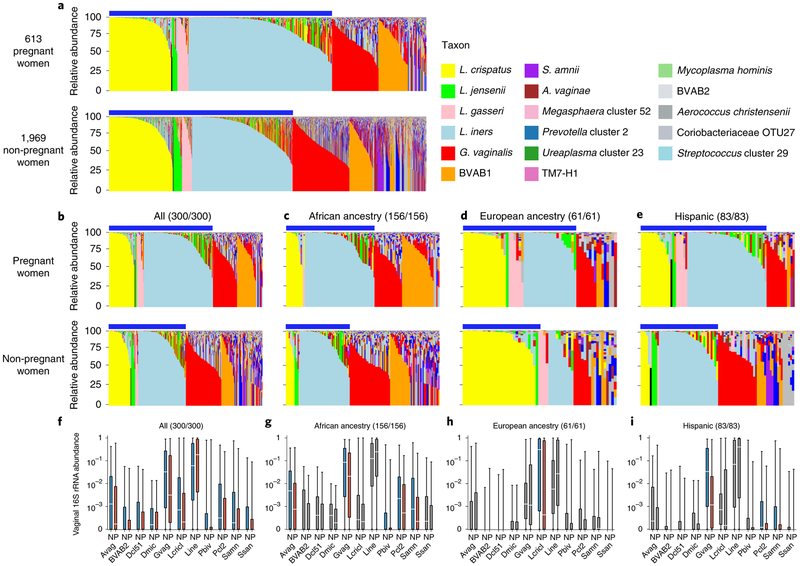
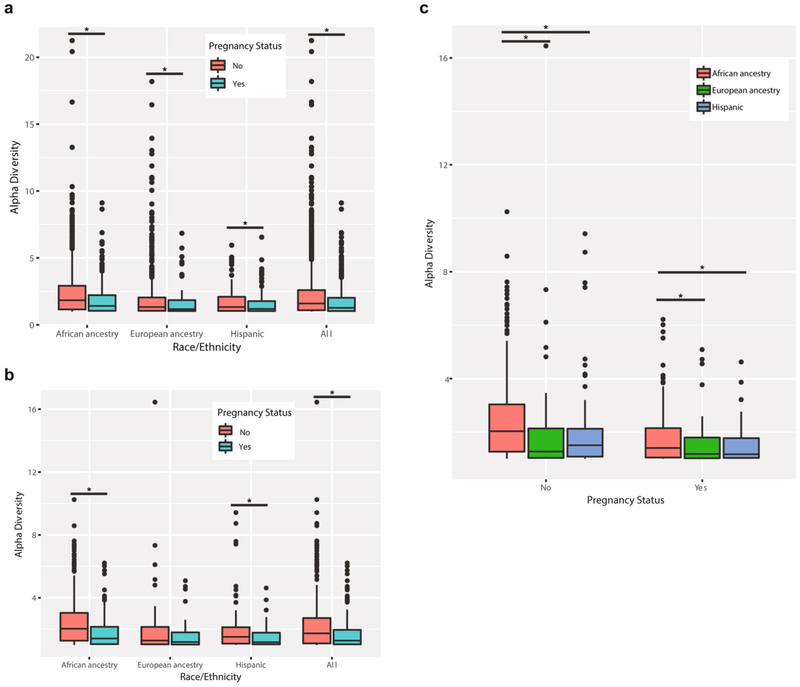
We generated vaginal microbiome profiles from 613 pregnant and 1,969 non-pregnant apparently healthy women from the VaHMP study as previously described15,26,27 (Supplementary Table 1a and Fig. 2a). The results suggested that pregnant women have a significantly higher (P < 0.01) prevalence of the four most common Lactobacillus vagitypes (L. crispatus, L. iners, L. gasseri and L. jensenii) and a commensurately lower prevalence of vagitypes dominated by other taxa. The primary driver of these differences was a higher (P < 0.01) prevalence of the L. iners-dominated vagitype in pregnancy at the expense of G. vaginalis and more complex vagitypes. Interestingly, incidence of BVAB1 vagitypes, a taxon associated with risk of BV and PTB7,28,29, did not decrease in pregnant women. The alpha diversity of the profiles of these pregnant women was also lower than that of non-pregnant women (P < 0.01, Extended Data Fig. 1a). These observations suggest that a pregnancy-related physiological or environmental influence change in the vagina is conducive to a less complex microbiota. To further explore these observations, we compared the vaginal microbiome profiles of 300 pregnant and 300 non-pregnant case-matched women (Supplementary Table 1b and Fig. 2b). As expected from the above-noted results, pregnant women were significantly (P < 0.01) more likely to have vagitypes of lactobacilli, with a higher predominance (P < 0.01) of the L. iners vagitype, and a commensurately lower prevalence of the G. vaginalis vagitype (P < 0.05) and other microbiome profiles. A taxon-specific analysis of these profiles confirmed a significantly higher abundance of L. iners and lower abundance of L. crispatus, G. vaginalis, A. vaginae, S. amnii, Prevotella cluster 2, Prevotella bivia and so on (Fig. 2f and Supplementary Table 2). Again, BVAB1 vagitypes were not affected by pregnancy. The alpha diversity of the profiles of pregnant women in this cohort was significantly lower than that of non-pregnant women (P < 0.01, Extended Data Fig. 1b).
Fig. 2 ∣. Pregnant and non-pregnant women of different ancestry exhibit different vaginal microbiome profiles.
a, Microbiome profiles of 613 pregnant and 1,969 non-pregnant women, 1 sample each, of all racial backgrounds taken from the VaHMP data set. Profiles were generated by 16S rRNA taxonomic classification and sorted into vagitypes representing the most dominant taxon present in the profile as previously described15,26,27. The legend is shown for a-e. Blue bars above the charts (a-e) indicate samples with Lactobacillus-dominated vagitypes. b, Microbiome profiles of a subset of 300 pregnant and 300 non-pregnant women from the VaHMP, case-matched for self-identified race, age and socioeconomic status. c, Microbiome profiles of 156 pregnant and 156 non-pregnant women of African ancestry from the VaHMP and case-matched for age and socioeconomic status. d, Microbiome profiles of 61 pregnant and 61 non-pregnant women of European ancestry from the VaHMP and case-matched for age and socioeconomic status. e, Microbiome profiles of 83 pregnant and 83 non-pregnant women of Hispanic ancestry from the VaHMP and case-matched for age and socioeconomic status. f-h, Taxon abundance differences in pregnant and non-pregnant women from b-e. Colored boxes (red and blue) indicate significant (q < 0.05 in two-sided Mann-Whitney U test after correction for false discovery rate (FDR)) differences in the abundance of that taxon between samples in women who are pregnant or not, respectively. Sample sizes for pregnant/non-pregnant cohorts are listed above each plot. Gray boxes indicate differences that are not significant. Boxes show median and interquartille range. The whiskers show the minimum and maximum values. Avag, A. vaginae; BVAB2, BV-associated bacterium 2; Dcl51, Dialister cluster 51; Dmic, Dialister micraerophilus; Gvag, G. vaginalis; Lcricl, L. crispatus cluster; Lini, L. iners; Pbiv, P. bivia; Pcl2, Prevotella cluster 2 (including P. timonensis and P. buccalis); Samn, S. amnii; Ssan, Sneathia sanguinis.
Shifts in the vaginal microbiome in pregnancy are most pronounced in women of African ancestry.
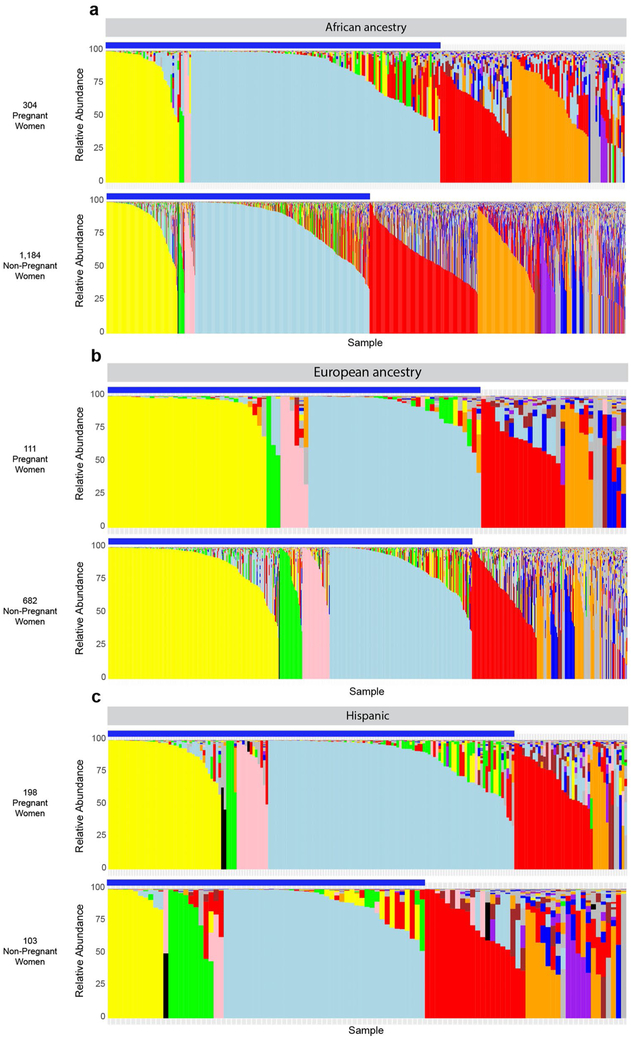
Owing to the overlap between the taxa of higher prevalence in women of African ancestry and taxa that are decreased in abundance in pregnancy, we examined the vaginal microbiome profiles of pregnant and non-pregnant women of different ancestries (Extended Data Fig. 2). Stratification of the profiles of our non-case-matched cohort according to self-reported ancestry revealed minimal differences in the microbiome profiles of pregnant and non-pregnant women of European ancestry (Extended Data Fig. 2b). However, pregnant women of African and Hispanic ancestry showed a higher (P < 0.01) prevalence of vagitypes dominated by the four most prevalent Lactobacillus species than their non-pregnant counterparts (Extended Data Fig. 2a,c). Similarly, no vagitype exhibited a significant difference in pregnant and non-pregnant women of European ancestry (Extended Data Fig. 2b), and the higher prevalence of the L. iners vagitype identified in the overall cohort (Fig. 2b) was driven by the higher prevalence (P < 0.01) of this vagitype in women of African ancestry. This difference was coupled with a lower prevalence (P < 0.01) of the G. vaginalis vagitype specific to this racial group (Extended Data Fig. 2a). Pregnant Hispanic women also exhibited an increased, but not quite significant, prevalence of the four Lactobacillus vagitypes. Notably, despite the lack of significant differences in the prevalence of specific vagitypes in women of European and Hispanic ancestry, the alpha diversities of the vaginal microbiomes of pregnant women in all groups were lower (P < 0.01) than those of their non-pregnant counterparts (Extended Data Fig. 1a). Interestingly, the alpha diversity of women of African ancestry was consistently greater than that of women of other ancestries (Extended Data Fig. 1c), independent of pregnancy status. These observations are consistent with the concept that pregnancy both favors taxa of Lactobacillus at the expense of other taxa and presents an environment conducive to a vaginal microbiome of reduced complexity.
Analysis of the case-matched cohorts of African, European and Hispanic ancestry confirmed that the differences in the profiles of the overall cohort were driven by significantly higher (P < 0.01) prevalence of Lactobacillus vagitypes with commensurately lower prevalence of other vagitypes in women of African and Hispanic ancestry (Fig. 2c,e). Women of European ancestry exhibited no significant alteration of vagitype associated with pregnancy (Fig. 2d), despite a trend toward a higher prevalence of L. iners at the expense of L. crispatus. The trends toward a higher prevalence of L. iners vagitypes and a lower prevalence of G. vaginalis vagitypes observed in the non-racially stratified group above were also observed in pregnant women of African and Hispanic ancestry, although these changes did not reach significance. All of the groups exhibited an enhanced prevalence of the L. iners vagitype in pregnancy.
A taxon-level analysis of the microbiome profiles of all women (Fig. 2f) or racially stratified groups of case-matched women demonstrated that, in women of African ancestry, G. vaginalis, A. vaginae, Prevotella cluster 2, P. bivia and S. amnii were significantly (P < 0.05) less abundant during pregnancy (Fig. 2g and Supplementary Table 2). Pregnant women of Hispanic ancestry exhibited a lower abundance of G. vaginalis, S. amnii and Prevotella cluster 2 (P < 0.05), and a lower, but not significant, abundance of A. vaginae and P. bivia (Fig. 2i and Supplementary Table 2). Pregnant women of European ancestry showed only a lower abundance of L. crispatus (P < 0.05) complemented by a higher abundance of L. iners (Fig. 2h and Supplementary Table 2). Again, BVAB1 showed no significant change in abundance during pregnancy in these groups. The alpha diversity of the vaginal microbiomes of pregnant women in the overall case-matched cohort was significantly lower (P < 0.01, Extended Data Fig. 1b). The alpha diversities of the microbiome profiles of pregnant women of African and Hispanic ancestry were also significantly lower (P < 0.05), but pregnant women of European ancestry showed a non-significant difference.
Vaginal microbiomes of women of African ancestry shift early in pregnancy.
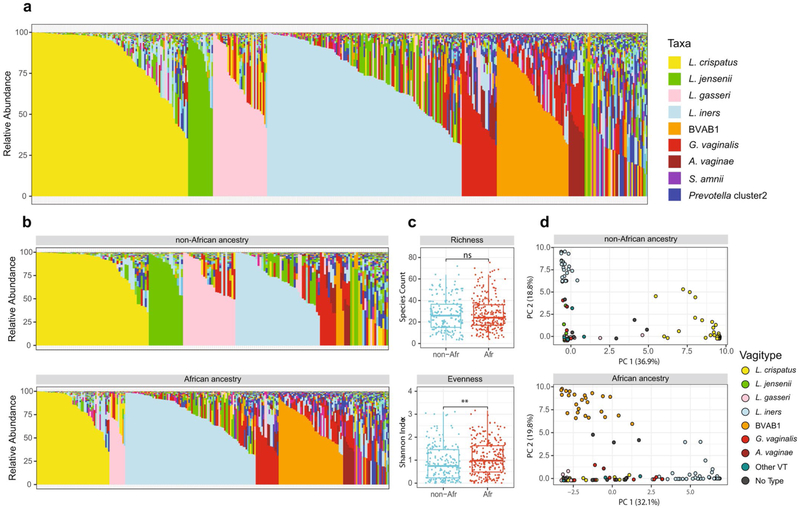
We characterized the vaginal microbiomes in 90 pregnant women from the MOMS-PI study, 49 of African ancestry and 41 of European ancestry, who delivered at term (>37 weeks’ gestation, Supplementary Table 1c). Overall, these women displayed an array of vagitypes reminiscent of those observed in the VaHMP study (Extended Data Fig. 3a), and the combined profiles of samples from women of African and non-African ancestry (Extended Data Fig. 3b) showed differences reflecting those described above (Fig. 2). Thus, pregnant women of African ancestry displayed significantly (P < 0.01) lower representation of the L. crispatus, L. gasseri and L. jensenii vagitypes, and higher (P < 0.01) representation of L. iners and BVAB1 vagitypes. Variability in women of African ancestry was driven by BVAB1 and L. iners, whereas variability in women of non-African ancestry was driven by L. crispatus and L. iners (Extended Data Fig. 3d). Again, pregnancy had no significant effect on prevalence of the BVAB1 vagitype. The differences between women of African and non-African ancestry were also clearly reflected in the significantly (P < 0.01) higher alpha diversity of samples from women of African ancestry (Extended Data Fig. 3c).
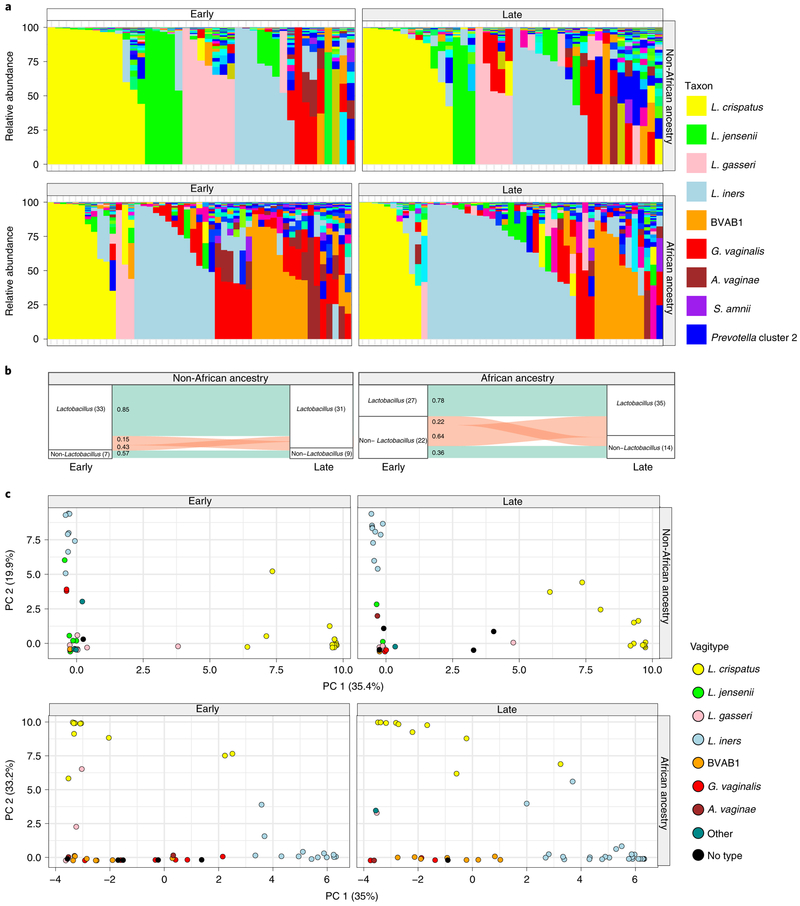
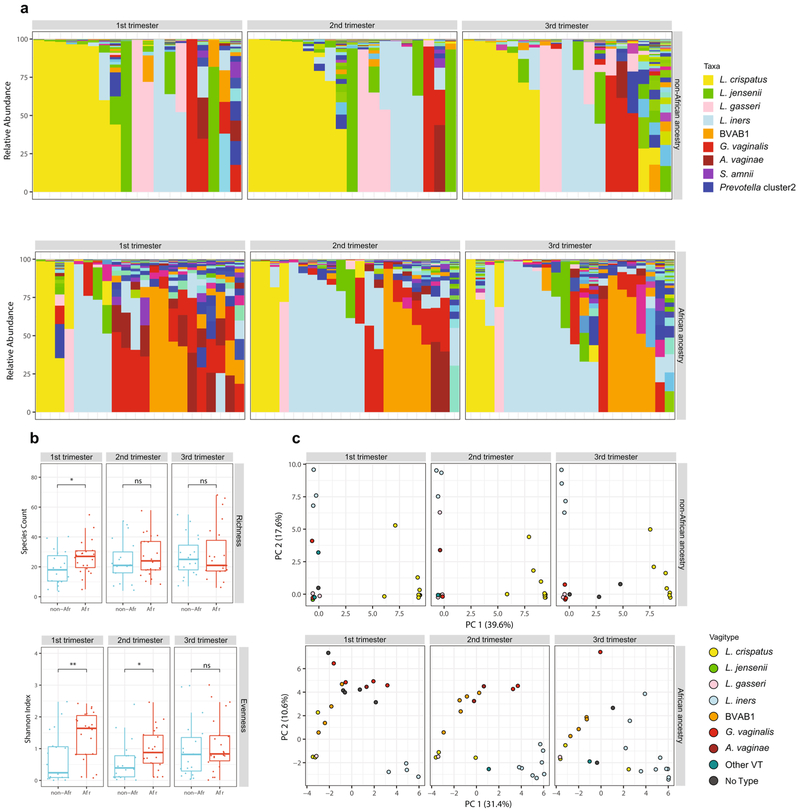
Samples collected longitudinally across these pregnancies suggested that pregnant women of African ancestry transition to Lactobacillus-dominated vagitypes, often to a L. iners vagitype (Fig. 3a). Prevalence of Lactobacillus-dominated profiles in women of African ancestry was lower in the first than in later trimesters, whereas women of European ancestry had a higher prevalence of Lactobacillus vagitypes throughout pregnancy. Women of African ancestry showed significantly (P < 0.05) increased prevalence of the L. iners vagitype, and a decrease of G. vaginalis and other vagitypes often associated with dysbiotic conditions. Women of non-African ancestry, who were over 75% dominated by Lactobacillus vagitypes early in pregnancy, showed no significant changes in vagitypes across pregnancy.
Fig. 3 ∣. Vaginal microbiome profiles of women of African ancestry change early in pregnancy.
a, Vaginal microbiome profiles of pregnant women, 49 of African ancestry and 41 of non-African ancestry, collected early (the first sample before 23 weeks of gestation) and late (last sample collected after 32 weeks of gestation) during pregnancy. b, An alluvial diagram showing trajectories of transitions from Lactobacillus-dominated and non-Lactobacillus-dominated profiles of women of non-African and African ancestry across pregnancy. The number of participants in each group is indicated in brackets, with the fraction of participants transitioning indicated on the stratum. c, L1-norm principal component analysis (PCA) of samples from a. An L1-norm PCA (see Methods) is a method for ordination that replaces the traditional sum-of-squared errors criterion with the outlier-insensitive L1 norm54. L1-norm PCA methods capture baseline behavior in the presence of outliers when traditional PCA and principal coordinate analysis can be adversely affected. See Supplementary Table 5 for sequence read statistics for data presented in this figure.
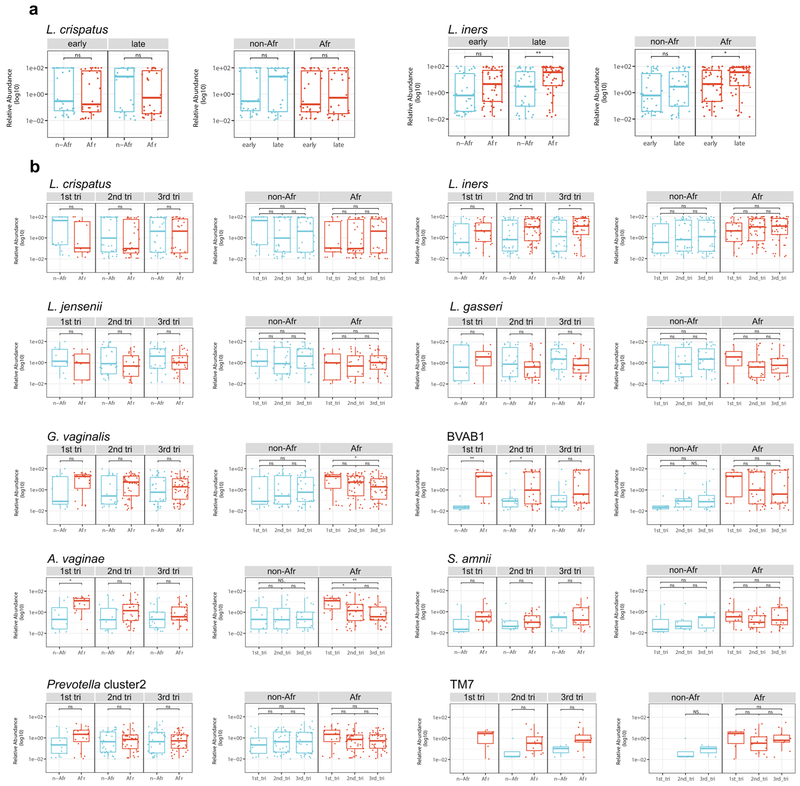
By the second trimester, the composition of the vaginal microbiomes of pregnant women of African ancestry shows an increased prevalence of taxa of Lactobacillus driven by an increase in predominance of the L. iners vagitype (Fig. 3b,c and Extended Data Fig. 4). In the first trimester, the alpha diversity of the vaginal microbiomes of women of African ancestry was greater (P < 0.05) than that of other women, but conversion to a more Lactobacillus-dominated profile as pregnancy progressed was accompanied by a significant (P < 0.05) decrease in the alpha diversity and the number of taxa present that no longer differed significantly from that of women of non-African ancestry (Extended Data Fig. 4b). In contrast, the alpha diversity of women of non-African ancestry was low and showed no significant variation throughout pregnancy (not shown). Taxon-specific analysis confirmed that, as for the cross-sectional cohort, L. iners was more abundant (P < 0.05, Extended Data Fig. 5), and G. vaginalis and A. vaginae were significantly (P < 0.05 and P < 0.01, Extended Data Fig. 5a) less abundant late in pregnancy in women of African ancestry. Other taxa (for example, Prevotella cluster 2, S. amnii and TM7), following the significant trends of the more numerous participants in the cross-sectional analyses described above, were likewise less abundant late in pregnancy in women of African ancestry, although the difference did not reach significance in this smaller cohort (Extended Data Fig. 5b).
Thus, the differences in the vaginal microbiomes of pregnant women are driven by changes that occur early in pregnancy in women of African ancestry. Many of the taxa decreased in pregnancy (that is, G. vaginalis, A. vaginae, Prevotella cluster 2, S. amnii and TM7) are associated with BV, pelvic inflammatory disease, sexually transmitted infections, risk of preterm birth and other adverse conditions of the female reproductive tract3,5-7.
Microbiome transitions during pregnancy follow predictable patterns.
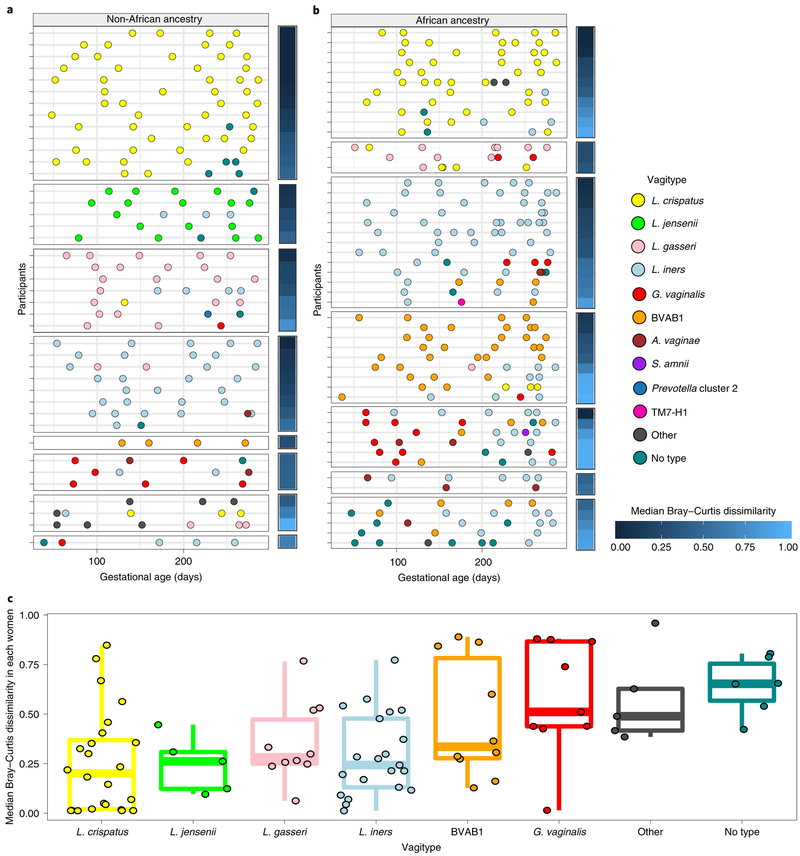
Prevalence of a vagitype is a product of the stability of that community, the lack of stability of other vagitypes and the probabilities of switching among different dominant communities. Figure 4 shows that vagitypes dominated by lactobacilli, in particular L. crispatus, were quite stable across pregnancy. Other vagitypes (for example, G. vaginalis) exhibited less stability. Figure 4c illustrates longitudinal beta diversity measures of the samples within each participant across pregnancy. For example, samples from women with microbiomes dominated by L. crispatus at first visit are highly similar to subsequent samples collected across pregnancy, but higher instability was observed in other vagitypes (Kruskal–Wallis; P < 0.05). Stability of vagitypes examined only in women of African ancestry or in women of non-African ancestry differed but did not reach significance (not shown).
Fig. 4 ∣. Temporal dynamics of vagitype transitions during pregnancy.
a, Transitions for 41 women of non-African ancestry. b, Transitions for 49 women of African ancestry. Each row represents all of the samples from a single participant, with vagitypes assigned based on 16S rRNA taxonomic profiles, shown as different color circles (see legend for code) across their pregnancies. The median Bray–Curtis dissimilarity within samples collected from the same participant is shown as a heat map to the right of each primary panel. Participants are grouped according to the vagitype of the first sample, and further sorted by decreasing median Bray–Curtis dissimilarities. c, The distribution of median Bray–Curtis dissimilarities between longitudinal samples of each women (90 women; 421 samples), grouped based on the vagitype of the first sample, is shown (see Methods). Box plots were generated in R using standard approaches. The bar represents the median and the boxes indicate interquartile ranges. The whiskers show the minimum and maximum values.
Tracking of the microbiome profiles of individual women through pregnancy (Fig. 4a,b and Extended Data Fig. 6a,b) shows that transitions occur more frequently from vagitypes often associated with dysbiotic conditions, but transitions to a vagitype dominated by a taxon poorly represented in the previous profile occurs infrequently. Thus, vagitype transitions follow predictable patterns. Most transitions away from a L. crispatus vagitype changed to the ‘no type’ or L. iners vagitype, and subsequent changes were often back to L. crispatus. The L. crispatus vagitype was stable and generally persistent in both women of African and non-African ancestry. In contrast, vagitypes dominated by L. iners, G. vaginalis and other vagitypes often associated with dysbiotic conditions were less persistent. Thus, G. vaginalis vagitypes transitioned frequently but only infrequently reverted to a G. vaginalis vagitype.
Table 1 fits a Markov chain model to these transitions, providing the probability that a vagitype will remain constant or transition over a 90-d window. Our initial effort modeled five vagitype groups, one consisting of L. crispatus, L. gasseri and L. jensenii as a ‘Lacto’ group, and four others; that is L. iners, G. vaginalis, BVAB1 and ‘other’ vagitypes often associated with vaginal dysbiosis. In this model, the Lacto group had an ~75% probability of not transitioning. The L. iners vagitype was also stable (~71%), and its most common transition (~19%) was to the ‘other’ category. BVAB1-dominated vagitypes had lower stability (~58%), most often changed to L. iners vagitypes (~25%) and rarely (4%) changed to the Lacto group vagitypes. G. vaginalis vagitypes were unstable; nearly 80% underwent transition, usually (~74%) to L. iners, BVAB1 or the ‘other’ vagitypes and only infrequently (~5%) to the Lacto group. The ‘other’ vagitypes usually (84%) remained vagitypes often associated with dysbiosis; only ~16% transitioned to Lacto group vagitypes.
Table 1 ∣.
Transition probabilities of vagitypes during pregnancy over a 90-d window using a simplified Markov model
| All samplesa | Lacto group (%) |
L. iners (%) |
G. vaginalis (%) |
BVAB1 (%) |
Other (%)a |
|---|---|---|---|---|---|
| Lacto group | 75 | 7 | 2 | 1 | 15 |
| L. iners | 5 | 71 | 3 | 2 | 19 |
| G. vaginalis | 5 | 32 | 21 | 20 | 22 |
| BVAB1 | 4 | 25 | 1 | 58 | 12 |
| Other | 16 | 31 | 8 | 9 | 36 |
| All samplesb | Lacto group | L. iners | Otherb | ||
| Lacto group | 75 | 9 | 16 | ||
| L. iners | 7 | 71 | 22 | ||
| Other | 13 | 34 | 53 | ||
| Non-Africanb | Lacto group | L. iners | Otherb | ||
| Lacto group | 83 | 2 | 15 | ||
| L. iners | 9 | 83 | 8 | ||
| Other | 27 | 14 | 59 | ||
| Africanb | Lacto group | L. iners | Otherb | ||
| Lacto group | 76 | 12 | 12 | ||
| L. iners | 1 | 76 | 23 | ||
| Other | 4 | 34 | 62 | ||
Probabilities over a 90-d window are shown as percentages. The initial vagitype is shown in the first column, and the transition frequency (%) to the subsequent groups is shown for each of the vagitype groups. In brief, the R package msm was used to fit a continuous-time Markov chain model for vagitype transitions. The model takes as input the subject, state, and gestational age in days for each sample. The states were Lacto, L. iners, BVAB1, G. vaginalis, and ‘other’ in the initial analysisa, or Lacto, L. iners, and ‘other’ in the analysis examining ancestry differencesb. In each analysis, the Lacto group included the more protective Lactobacillus vagitypes (that is, L. crispatus, L. gasseri, and L. jensenii). Samples from women of African and non-African ancestry were included, and ancestry was modeled as a covariate. 95% confidence intervals for transition probabilities were used to determine whether there were statistically significant differences between ancestry groups. Ttransitions that exhibited large differences between the women of African and non-African ancestry are highlighted in bold.
Model examining transition of all 90 women across five vagitype groupings.
Model examining transitions in all women in the cohort across three vagitype groups. The same groupings are used to examine transitions in the 49 women of African or the 41 non-African ancestry.
Owing to the lower numbers of women in the African and non-African groups, we collapsed the vagitypes into three groups (Lacto, L. iners and ‘other’), rationalizing that G. vaginalis and BVAB1 could group with ‘other’ taxa as they are all often associated with dysbiosis. In this model, the Lacto and L. iners vagitypes were quite stable (~75% and ~71%, respectively), but the ‘other’ group tended to remain in this higher diversity category (53%) or transition to the L. iners vagitype (34%), and only infrequently transitioned to the more protective Lacto group vagitypes (~13%). These trends held in both groups of women (Table 1c,d), but women of African ancestry with the Lacto vagitypes were sixfold more likely to transition to L. iners than women of non-African ancestry. Women of African ancestry with an L. iners or ‘other’ vagitype were also less likely (about ninefold and about sevenfold, respectively) to transition to the Lacto vagitypes. Conversely, women of African ancestry with L. iners vagitypes were threefold more likely to transition to an ‘other’ vagitype. Although these transitions failed to reach significance due to relatively low numbers, women of African ancestry underwent significantly (P < 0.05) more transitions over pregnancy than other women. Together, these observations may explain the prevalence of dysbiosis-linked vagitypes in women of African ancestry; that is, women of African ancestry seem to be less likely to transition from dysbiosis-linked vagitypes to ‘Lacto’ vagitypes and more likely to transition from ‘Lacto’ vagitypes to vagitypes more often associated with dysbiosis.
The functional potential of the vaginal microbiome changes early in pregnancy.
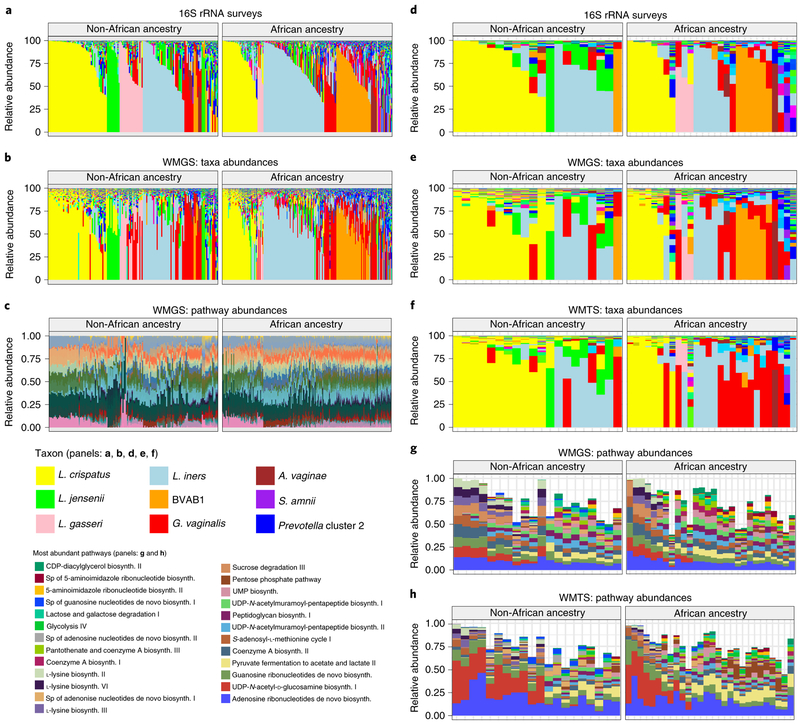
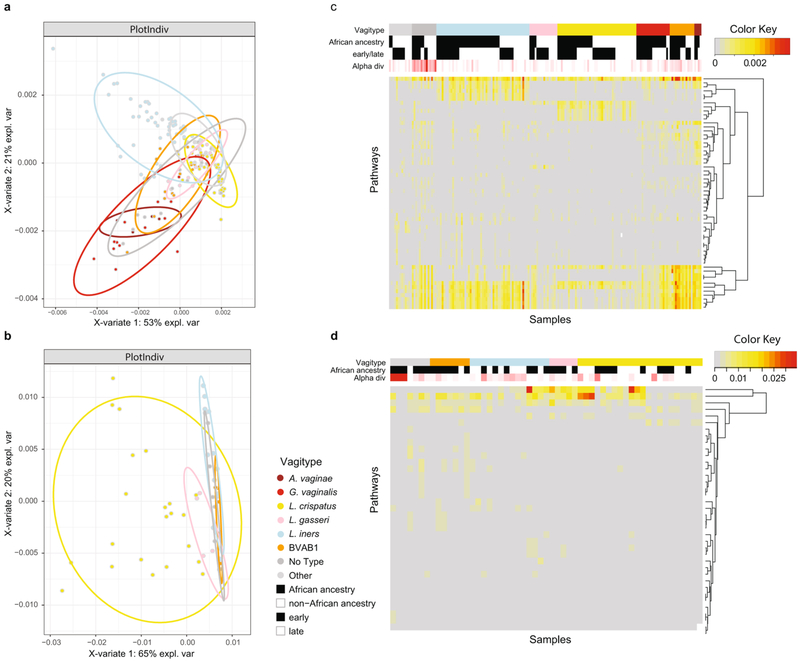
Metagenomic and metatranscriptomic taxonomic profiles of the vaginal microbiomes of women in this study largely recapitulated the vagitypes (Fig. 5a,b,d-f). However, G. vaginalis transcripts unexpectedly seemed to predominate over transcripts from BVAB1 and other taxa (Fig. 5f), suggesting that G. vaginalis is more transcriptionally active. Analysis of pathways predicted from the metagenomic and metatranscriptomic sequence data (Fig. 5c,g,h) and analysis by sparse partial least squares discriminant analysis (PLS-DA; Extended Data Fig. 7a,b) largely reflected the vagitypes in these samples. Whereas L. crispatus and L. iners showed discrete clusters in both metagenomic and metatranscriptomic pathway data, other vagitypes exhibited less discrete overlapping groups. Heat maps of the pathway analysis also resolved the vagitypes (Extended Data Fig. 7c,d). A pathway centric analysis of the metagenomic data (Supplementary Table 3) identified only ten largely unrelated pathways that significantly (P < 0.05) discriminated ancestry, likely due to the substantial partitioning of the vaginal microbiomes of women of both African and non-African ancestry into relatively discrete community states or vagitypes. However, analysis of the metatranscriptomic data identified eight pathways associated with nucleotide metabolism that were significantly (P < 0.05) less prevalent in the more complex microbial communities in women of African ancestry.
Fig. 5 ∣. Metagenomic, metatranscriptomic and pathway analyses of vaginal microbiome samples support metabolic differences among vagitypes of pregnant women of African and non-African ancestry.
Longitudinal samples from 90 women, 49 of African and 41 of non-African ancestry were compared. a, 16S rRNA taxonomic assignments of samples from all three trimesters from women of non-African and African ancestry. b, Taxonomic profiles from metagenomic sequence data of samples from all three trimesters from women of non-African and African ancestry. c, Relative abundances of metabolic pathways estimated using HUMAnN255 from metagenomic sequence data of samples from all three trimesters from women of non-African and African ancestry. d, 16S rRNA taxonomic assignments of samples from the second or early third trimester (one sample per pregnancy) from women of non-African and African ancestry. e, Taxonomic profiles from metagenomic sequence data from samples as in d. f, Taxonomic profiles from metatranscriptomic sequence from samples as in d. g, Relative abundances of 25 highly abundant metabolic pathways estimated using HUMAnN2 from metagenomic sequence data from samples as in d. h, Relative abundances of 25 highly abundant metabolic pathways estimated using HUMAnN2 from metatranscriptomic sequence from samples as in d. See Supplementary Table 6 for mapping statistics for b, e and f. ‘sp’ in the pathways key means ‘superpathway’.
Analysis of metabolic capacity of the microbiomes across trimesters showed 17 pathways significantly (P < 0.05) reduced, and 90 of 94 abundant pathways reduced in prevalence between trimesters 1 and 2. Only two pathways were significantly (P < 0.05) more reduced in prevalence between trimesters 2 and 3. Thus, the most prevalent changes in the functional microbiome occur by the first trimester. Of the 17 pathways significantly less prevalent in pregnancy, 5 were associated with protein synthesis, 5 with carbohydrate metabolism and cell wall synthesis, 4 with nucleotide metabolism, and 3 with coenzyme A and other cofactor biochemistry. Thus, pregnancy is associated with a simplification of the metabolic capacity of the resident vaginal microbiome, consistent with a transition toward a less complex Lactobacillus-dominated microbiome.
Noting a discrete pattern of pathways in L. crispatus- dominated vagitypes (Fig. 5c,g,h), we directly compared the pathways of L. crispatus vagitypes to those of other major vagitypes (Supplementary Table 3). The L. iners vagitype showed the fewest significant differences (P < 0.05), mostly in pathways associated with cell wall/ membrane biochemistry. G. vaginalis, BVAB1, ‘no type’ and ‘all other’ vagitypes differed significantly (P < 0.05) in many pathways (for example, carbohydrate metabolism, cell wall/membrane biochemistry, nucleotide metabolism) from the L. crispatus vagitype. Many of the pathways (for example, carbohydrate metabolism, cell wall/envelope biochemistry, protein synthesis and nucleotide metabolism) that differed significantly (P < 0.05) between the L. crispatus vagitypes and the G. vaginalis, BVAB1, ‘no type’ and ‘all other’ vagitypes, were common. The majority of the pathways, except those associated with l-lysine biosynthesis, UDP-N-acetyl-d-glucosamine biosynthesis and acetylene degradation, were less prevalent in L. crispatus vagitypes than in other microbiome profiles. These observations are consistent with the sparse PLS-DA analysis of pathways (Extended Data Fig. 7a,b), which showed a significant overlap of the G. vaginalis, BVAB1, ‘no type’ and ‘all other’ vagitypes.
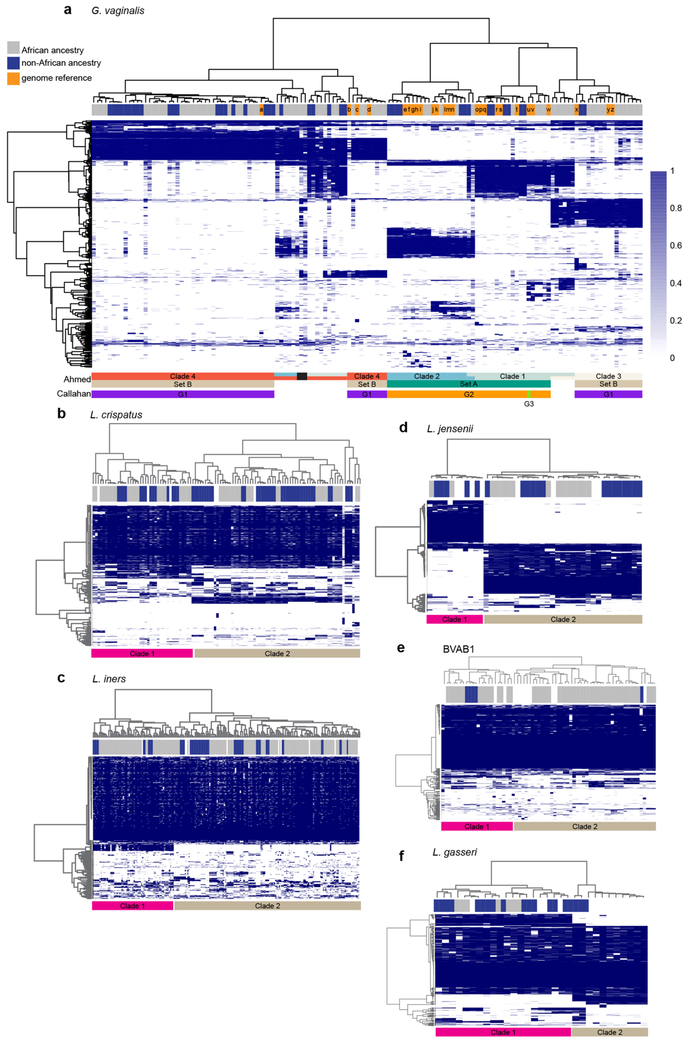
Strains of G. vaginalis show racial and taxonomic proclivities.
Bacterial strains of the same species often exhibit significant sequence heterogeneity. G. vaginalis is now subclassified into at least four clades exhibiting nucleotide sequence divergence of ~4 to 20%30-33. Examination of our metagenomic data using PanPhlAn34 readily discriminated the four major clades of Gardnerella (Extended Data Fig. 8 and Supplementary Table 4). G. vaginalis clade 4 was most common, but clade 3 was tightly associated with BVAB1 (P < 0.01, see Methods), and was detected only in vaginal microbiomes with BVAB1 concurrently or at a previous or subsequent sampling. Although BVAB1 is found primarily in women of African ancestry, the correlation between G. vaginalis clade 3 and African ancestry was not significant, perhaps because there are many women of this ancestry whose microbiomes lack BVAB1. However, 10 of 11 women with clade 3 were of African ancestry, suggesting a linkage between this clade and ancestry. No other G. vaginalis clade showed a strong linkage to any other taxon or racial group. Since both BVAB1 and G. vaginalis are associated with higher risk of adverse reproductive health effects7,28,35-37 and both are prevalent in women of African ancestry, the apparent linkage between BVAB1 and G. vaginalis clade 3 may be relevant to racioethnic disparities in women’s reproductive health.
These analyses also revealed discrete but previously uncharacterized clades of L. crispatus, L. jensenii, L. gasseri, L. iners and BVAB1 (Extended Data Fig. 8). We observed no relationship between these clades and any other parameter, although, strain-level discrimination is likely relevant to understanding the impact of these microbial communities on human health.
Discussion
Our VaHMP and MOMS-PI cohorts of women of European, African and Hispanic ancestry exhibit different vaginal microbiome compositions and dynamics during pregnancy. The cause of these differences remains unclear, although both genetic and environmental influences are likely contributors. Potential confounding factors that our analyses have yet to resolve include socioeconomic status and age. Thus, in our cohorts, women of African and Hispanic ancestry are significantly younger and have lower household income than women of European ancestry (Supplementary Table 1). Our previous analysis15 of non-pregnant women indicated that race has a stronger association with diversity of the vaginal microbiome than socioeconomic status. Thus, although we were unable to completely decouple race from socioeconomic status herein, we believe both have an impact.
Our longitudinal analysis argues that the transitions of the vaginal microbiomes occur early in pregnancy. Stout et al.38 reported a decrease in richness and diversity early in pregnancy in the vaginal microbiomes of predominantly African American women who would later deliver preterm. However, in that study of only 149 longitudinal samples from 77 women, no change was observed in women who would deliver at term. DiGiulio et al.39 reported a stable pregnancy microbiome of the gut, saliva and mouth, and in contrast to results reported herein, the vagina. The cohort of women in that project was predominantly (22 of 34) self-reported as ‘White’ with only 1 woman self-reported as ‘Black’. Goltsman et al.40, following ten white pregnant women (six who delivered term and four who delivered preterm), presented data that suggest stability of the vaginal microbiome in pregnancy, although four of the women with vaginal microbiomes initially dominated by L. iners showed an increase in diversity with gestation. Together, these results are largely consistent with our observation that vaginal microbiome changes are most significant in women of African ancestry.
It appears that changes occurring during pregnancy may render the reproductive tracts of women of all racial backgrounds more hospitable to taxa of Lactobacillus and less favorable for G. vaginalis and other taxa associated with BV and dysbiosis. Since the changes are generally toward a microbiome composition resembling that of non-pregnant women of European ancestry, it is not surprising that the shifts are more striking in women of African and Hispanic ancestry. As we and others have speculated7,16,17,41,42, hormonal and other physiological changes in pregnancy may promote a vaginal environment conducive to a less dysbiotic vaginal microbiome. We previously demonstrated43 that non-pregnant women taking combined oral contraceptives have vaginal microbiome profiles with a higher predominance of Lactobacillus taxa, and several studies by others44-47 have shown that post-menopausal women receiving hormone replacement therapy tend to have a more Lactobacillus-dominated microbiome. Elevated estrogens in pregnancy enhance glycogen synthesis in the vaginal epithelium. Although most lactobacilli lack alpha-amylase, human enzymes and other bacterial pathways process glycogen into compounds (for example, maltose, maltotriose, maltopentaose and maltodextrins) that are fermented by these bacteria48. Thus, Lactobacillus are apparently selectively enriched, to the detriment of other microbial taxa, in pregnant women. Interestingly, BVAB1, which has been associated with dysbiotic vaginal conditions and risk of PTB7,28,29, and which is present as a major vagitype largely in women of African ancestry, is not noticeably decreased in prevalence in pregnancy. Thus, BVAB1, for reasons yet to be determined, is apparently resistant to factors sculpting the microbiome in pregnant women, possibly explaining in part the enhanced risk for PTB experienced by women of African ancestry.
We and others1,2,15-23 have previously shown that vaginal microbiomes of non-pregnant women of European ancestry are less complex than those of non-pregnant women of African ancestry. However, the differential impact of pregnancy on the vaginal microbiomes of women of diverse ancestry has not been comprehensively explored. An early cross-sectional study by Aagaard et al.49 probed the microbiomes of 24 pregnant and 60 non-pregnant women and found that, in pregnancy, the vaginal microbiome is reduced in complexity and enriched in species of Lactobacillus. Walther-Antonio et al.41 and Romero et al.42 followed 8 pregnant women and 22 pregnant/32 non-pregnant women, respectively, and also concluded that lactobacilli dominate the vaginal microbiome during pregnancy. MacIntyre et al.16, in a longitudinal study of a British cohort of 42 pregnant women who experienced term birth, and Freitas et al.50 in a cross-sectional study of 182 pregnant and 310 non-pregnant Canadian women, reported reduced diversity and higher prevalence of lactobacilli in vaginal microbiomes during pregnancy. Although these studies are consistent with the results reported herein, numbers of participants were quite modest, non-pregnant case-matched controls were often not available, taxon-specific changes other than Lactobacillus species were generally not addressed, and variables associated with race and ethnicity were usually not explored.
In our study, taxa that are decreased in prevalence in pregnancy include G. vaginalis, A. vaginae, S. amnii, Prevotella cluster 2, P. bivia and others. Many of these taxa have been associated with adverse health conditions, including BV, susceptibility to sexually transmitted infections and human immunodeficiency virus, and adverse pregnancy outcomes including preterm birth7,28,35-37. Thus, our results are suggestive of a relationship between physiological changes occurring during pregnancy and the development of a less dysbiotic vaginal microbiota (that is, a more disease-resistant state).
Our examination of the probability of transitions among vagitypes provides clues to the reasons that women of African ancestry generally have more complex vaginal microbiome profiles. Thus, the more favorable Lactobacillus vagitypes (that is, the L. crispatus, L. jensenii and L. gasseri vagitypes) are more stable, and transition to those vagitypes is less common in women of African ancestry. In contrast, vagitypes associated with dysbiosis (BVAB1, G. vaginalis, A. vaginae, S. amnii and other vagitypes) are less stable and transition to other vagitypes at a higher rate. Moreover, transitions from dysbiotic vagitypes to favorable vagitypes are more frequent in women of non-African ancestry. As a result, women of African ancestry are more likely to have a dysbiotic vagitype than women of non-African ancestry. These observations are reminiscent of Gajer et al.17, who in a study of 32 non-pregnant reproductive age women reported that vaginal communities dominated by Lactobacillus species were quite stable in contrast to more complex vaginal microbiomes, although that report did not focus on differences in transition rates associated with ancestry or pregnancy. The fact that the directions of these microbiome transitions are similar in pregnant and non-pregnant women suggests that the forces driving them may be related.
Metagenomic analysis illustrated minimal differences in metabolic potential associated with the overall vaginal microbiomes of women of African and non-African ancestry. In contrast, our metatranscriptome analysis identified pathways associated with nucleotide metabolism that are less prevalent in women of African ancestry. This result is consistent with a hypothesis suggesting that a more complex microbiome may be associated with greater damage to the vaginal epithelium, which may release metabolic precursors (for example, nucleotides and nucleotide precursors) that could be scavenged by the vaginal microbiota. Interestingly, nucleotide biosynthesis has been identified as a critical capability for bacterial proliferation in the human bloodstream51. It would not be surprising if this were also true in other niches of the human body.
Metagenomic and metatranscriptomic data permitted subspecies classifications. Thus, four previously identified clades of G. vaginalis30-33 and two new clades each of L. crispatus, L. jensenii, L. gasseri, L. iners and BVAB1 were discriminated. Clade 3 of G. vaginalis was tightly associated with BVAB1, suggesting a synergistic or symbiotic relationship. Although BVAB1 is highly correlated with women of African ancestry, the relationship between clade 3 of G. vaginalis and race was not significant, probably because many women of African ancestry have microbiome profiles dominated by other taxa. Recently, Janulaitiene et al.52, studying a population of 109 non-pregnant women with and without BV, found, as did we, that clade 4 was most common, followed by clade 3, and identified a correlation between clades 1 and 2 and a high Nugent score indicative of BV. Callahan et al.53, studying a cohort of 39 pregnancies, 9 of which ended prematurely (<37 weeks’ gestation), showed data suggesting an association between a G. vaginalis group, termed G3, that includes isolates from both clades 1 and 2, and preterm birth. We have not yet detected a correlation between clades 1, 2 or 4 and any other taxon or clinical condition. However, these observations of close linkage between specific microbial taxa and clinical and demographic phenotypes are suggestive of interdependencies that remain to be elucidated.
The fact that some groups frequently have vaginal microbiome profiles that are clearly associated with dysbiosis, BV and risk of adverse outcomes in pregnancy, including but not limited to preterm birth, underscores the compelling need to further explore these phenomena. That the vaginal microbiomes known to confer higher risk of poor health and adverse outcomes of pregnancy are more highly associated with women of African and Hispanic ancestry, but that pregnancy tends to drive these microbiomes toward more favorable microbiota, suggests that an external intervention that favors this trend might be beneficial for these populations. Characterization of the composition of the vaginal microbiome is now readily achieved. What remains is to verify the most favorable microbiome and the most effective strategy for intervention.
Methods
Participant enrollment, informed consent and health history collection.
Participants in this study were enrolled under the VaHMP or the MOMS-PI, as part of the National Institutes of Health HMP phases 1 and 224,25 (https://commonfund.nih.gov/hmp). All participants in this study were enrolled from visitors to women’s clinics in Virginia. All study procedures involving human subjects were reviewed and approved by the Virginia Commonwealth University institutional review board (IRB nos. HM12169 or HM15527). The study was performed in compliance with all relevant ethical regulations. Written informed consent was obtained from all participants and parental permission and assent was obtained for participating minors at least 15 years of age. Women were informed that they could withdraw from the study at any time and that providing samples was at their sole discretion. Women who were incapable of understanding the informed consent or assent forms, or incarcerated were excluded from the study. Comprehensive demographic, health history and dietary assessment surveys were administered, and relevant clinical data (for example, gestational age, height, weight, blood pressure, vaginal pH, diagnosis and so on) were recorded. Relevant clinical information was also obtained from neonates at birth and discharge. At subsequent prenatal visits, triage, in labor and delivery, and at discharge, additional surveys were administered, relevant clinical data were recorded and samples were collected. Vaginal samples were not collected at labor and delivery or at discharge. Women with any of the following conditions were excluded from sampling at a given visit: incapable of self-sampling due to mental, emotional or physical limitations; more than minimal vaginal bleeding as judged by the clinician; ruptured membranes before 37 weeks’ gestation; active herpes lesions in the vulvovaginal region. The 2,582 VaHMP cohort was selected according to the following: participants with abnormal discharge, bacterial vaginosis, trichomoniasis, yeast infection, gonorheae, herpes or warts as indicated by the physician at the time of the vist were excluded; in addition, participants with missing clinical data and missing ethinicity information were also excluded.
Participant/sample selection and case-control design.
We analyzed samples from 613 pregnant and 1,969 non-pregnant women with no known health issues who were sampled cross-sectionally in the VaHMP study. A subset of 300 pregnant and 300 non-pregnant women, case-matched for age, race and socioeconomic status, was selected from this group for more detailed analysis. Case-matching was performed blinded to all other study data. From the MOMS-PI study, we selected 90 women, 41 of European ancestry and 49 of African ancestry, who experienced full-term birth at >37 weeks’ gestation, reported no known adverse health conditions, and who had samples collected at least one visit per trimester during their pregnancies. These cohorts are briefly described in Supplementary Table 1.
Sample collection.
Samples were collected from consented women at their first visit, either in the VaHMP or MOMS-PI studies, and again longitudinally at each prenatal visit, at triage, and at labor and delivery for women in the MOMS-PI study. Samples were collected with BD BBL CultureSwab EZ swabs. Vaginal samples were collected either by a clinician during a pelvic exam or by self-sampling. Research coordinators instructed the participants on self-sampling procedures, provided a self-sampling instructional brochure, and provided the participant a private room for self-sampling. Self-sampling has been shown to provide results equivalent to those collected by a trained clinician56.
Samples were collected as follows: a single- or double-tipped CultureSwab EZ swab was placed carefully on the vaginal sidewall about halfway between the introitus and the cervix, pressed firmly into the sidewall to a depth of roughly the diameter of the swab, rolled dorsally–ventrally back and forth four times to coat the swab, and removed. Vaginal pH was collected using commercial applicators with pH paper. Briefly, the applicators were inserted ~1.5–2 inches into the vagina, applied gently to the vaginal wall and withdrawn. The research coordinator compared the color of the pH indicator to a color chart and recorded vaginal pH.
Sample processing.
After collection, swabs were immediately immersed in transfer buffer depending on the objectives. Swabs for DNA isolation were immersed in MoBio PowerSoil-htp Bead Solution; swabs for RNA purification were immersed in RNAlater (Qiagen). Swabs were either processed immediately or stored until processing at −80 °C. Swabs for DNA purification were processed using the MoBio PowerSoil DNA Isolation Kit, as described by the manufacturer. Swabs for RNA purification were processed using the MoBio PowerMicrobiome RNA Isolation Kit as described by the manufacturer. Total RNA was depleted of human and microbial rRNA using the Epicentre/Illumina Ribo-Zero Magnetic Gold Kit (Epidemiology) as described by the manufacturer. DNA and RNA samples were stored at −80 °C.
16S rRNA taxonomic surveys of the vaginal microbiome.
DNA in each sample was amplified with barcoded primers targeting the V1–V3 region of the 16S rRNA and validated for vaginal taxa essentially as previously reported7,26. The samples were randomized at the PCR stage and again at the sequencing stage. Samples from the VaHMP study were amplified and sequenced on the 454 Titanium Sequencer (Roche) and analyzed as previously described4. Samples from the MOMS-PI study were multiplexed (384 samples per run) and sequenced using 2 × 300 base (b) PE technology on our Illumina MiSeq sequencer to generate a depth of coverage of at least 50,000 reads per sample5. Briefly, the raw paired-end sequence data were demultiplexed into sample-specific paired-end fastq files based on unique barcode sequences using custom python scripts. The merging of overlapping pairs and quality filtering was performed using the MeFiT57 pipeline, with a maximum expected error cutoff of 1.0 (Supplementary Table 5). Each high-quality amplicon sequence was taxonomically classified to the species level using STIRRUPS26, by alignment to a custom reference database of vaginally relevant species. Samples were filtered based on the sequencing depth—samples having fewer than 1,000 high-quality amplicons were removed from further analysis. Taxa assigned below-threshold by STIRRUPS and less than 0.01% in abundance were also filtered out, and relative proportions were re-computed.
Vagitype assignment and analyses of diversity and richness.
Using 16S taxonomic profiles, samples were assigned to vagitypes based on the taxon with the largest proportion of reads and at least 30% representation, essentially as previously reported26. Samples with no taxon with over 30% representation were assigned to the ‘no type’ vagitype. This dataset-independent method of categorizing microbiome profiles has been shown to be in concordance with clustering-based methods58.
Fisher’s exact test was used to assess the differences in proportions of Lactobacillus and non-Lactobacillus vagitypes among samples from pregnant and non-pregnant subjects in Fig. 2a (n = 2,582, odds ratio = 0.5722). The proportions of L. iners,G. vaginalis, L jensenii, L. gasseri and BVAB1 in pregnant and non-pregnant subjects were also tested using Fisher’s exact test with a FDR adjustment using the function p.adjust() in R. The difference in mean diversity between pregnant and non-pregnant subjects was tested using a two-sample t-test (df = 1,758.4, t = 8.1927, n = 2,582). The same tests were used for the case-matched cohorts of 300 pregnant and 300 non-pregnant subjects in Fig. 2b (for the two-sample t-test (df = 433.40, t = 5.6252). The Fisher’s exact tests and two-sample t-test were applied to the two sets of subjects after stratification by race/ethnicity (for the complete cohort/African/Fisher’s: n = 1,488, odds ratio = 0.5454; complete cohort/European/Fisher’s: n = 793, odds ratio = 0.9144, P = 0.7374; complete cohort/Hispanic/Fisher’s: n = 301, odds ratio = 0.4382; complete cohort/African/t-test: df = 698.79, t = 5.616, n = 1,488; complete cohort/European/t-test: df = 250.20, t = 3.2696, n = 793; complete cohort/Hispanic/t-test: df = 165.34, t = 2.011, n = 301; case-match/African/Fisher’s: n = 312, odds ratio = 0.5249; case-match/European/Fisher’s: n = 122, odds ratio = 1.0, P = 1.0; case-match/Hispanic/Fisher’s: n = 166, odds ratio = 0.2281; case-match/African/t-test: df = 245.13, t = 4.8336, n = 312; case-match/European/t-test: df = 78.589, t = 1.678, n = 122, P = 0.09732; case-match/Hispanic/t-test: df = 110.01, t = 2.8475, n = 166) (Fig. 2c-e).
Within-sample richness and evenness (beta and alpha diversity) of microbial communities were determined by computing the species count and Shannon index respectively, using vegan in the R statistical package (Extended Data Figs. 3c and 4b). Compositional dissimilarity between samples from each participant was quantified using the Bray–Curtis index (Fig. 4). To visualize this dissimilarity between samples, we employed L1-norm PCA54 ordination on square-root-transformed relative abundances using the function sharpel1pcal in the R package pcaL1 (rdrr.io/cran/pcaL1/man/pcaL1-package.html), which is less sensitive to outliers (Fig. 3c).
Univariate analysis to identify taxa significantly different in abundance in different cohorts (Fig. 2f-I and Supplementary Table 2).
We analyzed vaginal 16S rRNA profiles from pregnant or non-pregnant women of African, Hispanic and European/Non-African ancestry from the VaHMP and MOMS-PI projects to identify taxa significantly enriched in each group. First, from the joint cohorts (n = 600) we removed taxa of low abundance. We used two abundance criteria: we retained taxa for which either 5% of the profiles exhibited an abundance of at least 0.01 (1%), or at least 15% of the profiles exhibited an abundance of at least 0.001 (0.1%). Taxa that failed to meet both criteria were removed. For the remaining taxa, sample abundance values below 0.0001 (0.01%) were rounded to zero. Next, in each ancestry cohort, we performed a Mann–Whitney U test (two-sided) for each taxon to identify significant differences in abundance between pregnant and non-pregnant subjects. Taxa abundance was considered significantly different between cohorts if the q-value was less than a FDR of 5% after correction via the Benjamini–Hochberg59,60 procedure. For each taxon we also calculated the median and 75th percentile in each of the relevant cohorts. For each box plot, the box indicates the interquartile range and the median is indicated. The whiskers extend to the data point that is at most 1.5 times the interquartile range from the box. In Fig. 1f-I, outlier points are not plotted.
Metagenomic and metatranscriptomic sequencing.
Metagenomic and metatranscriptomic sequencing were performed as previously described7. Briefly, DNA libraries were prepared using the KAPA Biosystems HyperPlus Library Kit and sequenced on our Illumina HiSeq 4000 (2 × 150 b PE), with 24 samples per lane to yield ~4 Gb per sample. The rRNA-depleted messenger RNA was prepared for sequencing by constructing complementary DNA libraries using the KAPA Biosystems RNA HyperPrep Kit. Indexed cDNA libraries were pooled in equimolar amounts and sequenced on the Illumina HiSeq 4000 (2 × 75 b and 2 × 150 b PE) instrument with 8 samples per lane to yield an average of 10 Gb per sample, sufficient to provide >200× coverage of the expression profiles of the most abundant 15–20 taxa in each sample (Supplementary Table 6). Raw sequence data were demultiplexed into sample-specific fastq files using bcl2fastq conversion software from Illumina. The pre-processing of raw paired-end sequence data was performed using Trimmomatic61, by removing Illumina specific adapters from the reads and trimming the 5′ end for quality using a sliding-window approach (average Q20 over a window of 4 b). Reads shorter than 70 b after trimming were discarded. This was followed by removal of contaminant reads of vector and human origin by aligning against the UniVec database (https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec/) and the hg19 build of the human genome using bwa62. Samples with fewer than 100,000 high-quality reads were removed from further downstream analysis.
Taxonomic classification and functional analysis of metagenomic and metatranscriptomic sequence data.
Taxonomic classification of the metagenomics and metatranscriptomic sequence data was performed on high-quality non-human reads using two tools: MetaPhlAn234 and by alignment to a custom database of bacterial genomes using CLARK-S63, a discriminative space k-mer-based approach. Bacterial genomes were downloaded from the NCBI Reference Sequence Database, accessed in March 2017, supplemented with our in-house genome assembly of Lachnospiraceae_BVAB17. Species abundances were estimated by normalizing the assigned read counts to species genome size.
Strain classification of metagenomic and metatranscriptomic data.
Pangenome-based Phylogenomic Analysis (PanPhlAn) version 1.2.2.455 was used to identify strain-specific gene repertoires of a given species using metagenomic data compared to reference genomes for different bacterial strains using default parameters of -min_coverage 1. The G. vaginalis genome abbreviation (strain name/GenBank accession number) used in the analysis shown in Extended Data Fig. 7: a, (409-05/NC_013721.1), b, (6420LIT (B475)/ADEO01000003.1); c, (AMD/NZ_ADAM01000095.1); d, (6420B (B476)/NZ_ADEP01000014.1); e, (JCP8151B/NZ_KE347039.1); f, (JCP8151A/NZ_KE347187.1); g, (JCP8070/NZ_KE347476.1); h, (JCP8066/NZ_KE347620.1); i, (JCP8522/NZ_ KE346595.1); j, (JCP8017B/NZ_KE347778.1); k, (JCP8017A/NZ_KE347941.1); l, (JCP7659/NZ_KE348406.1); m, (JCP7719/NZ_KE348086.1); n, (00703C2Mash (B482)/NZ_ADEU01000022.1); o, (ATCC14019/NC_014644.1); p, (ATCC 14018 (JCM 11026)/NZ_AP012332.1); q, (315-A/NZ_AFDI01000013.1); r, (284V (B472)/NZ_ADEL01000014.1); s, (75712 (B473)/NZ_ADEM01000003.1); t, (3549624/NZ_LFWD01000001.1); u, (1400E (B478)/NZ_ADER01000028.1); v, (JCP8108/NZ_KE347338.1); w, (41V/NZ_AEJE01000002.1); x, (1500E (B479)/NZ_ADES01000027.1); y, (101/NZ_AEJD01000002.1); z, (00703DMash (B483)/NZ_ ADEV01000011.1). The metabolic potential and activity of the microbial community were profiled from the metagenomic and metatranscriptomic dataset using HUMAnN264, which estimates the presence/absence and abundance of microbial pathways against the MetaCyc Metabolic Pathway Database.
Pathway analysis.
For metagenomic and metatranscriptomic data, normalized relative abundance from output generated by HUMAnN264 was used for each pathway. Unintegrated and unmapped sequences were excluded from the analyses. Pathways were screened by taking the sum of the normalized relative abundances across samples. Those with a value of 0.01 or greater were retained for downstream analysis. Heat maps were generated using the heatmap.3 function in R installed from https://raw.githubusercontent.com/obigriffith/biostar-tutorials/master/Heatmaps/heatmap.3.R, accessed on July 17, 2018. A significance level of 0.05 for all adjusted P values was employed to assess significance.
For analysis of longitudinal pathway data, we examined results from subjects who had samples in each of the three trimesters of pregnancy. To assess whether the pathway measurements could discriminate vagitypes derived by 16S rRNA taxonomic analysis, sparse PLS-DA was applied using the function splsda() in the R package mixomics (mixomics.org). The method was constrained to select up to 15 taxa to contribute to the resulting loading vectors.
The effects on pathway abundance of vagitype, African ancestry and trimester were assessed by fitting a linear mixed-effects model with a subject random effect to account for multiple samples from subjects. Models were fitted for each pathway using the function lme() from the R package nlme()65. The P values for all of the coefficients across the models were adjusted using a FDR adjustment implemented in the R function p.adjust(). Differences between trimesters for each pathway were assessed using a post hoc test for estimated marginal means. The function emmeans() in the R package (https://cran.r-project.org/web/packages/emmeans/emmeans.pdf) was used to test for differences in pathway abundances for each pair of trimesters. The P values from the post hoc tests across all of the pathways were adjusted using a FDR adjustment implemented in the R function p.adjust().
For metatranscriptomic data, linear mixed-effects models were fitted for each pathway to assess the effect of African ancestry and vagitype. P values were adjusted as for the metagenomics data.
Strain association analysis.
An analysis was conducted to detect associations between strain groups identified by PanPhlan and environmental factors including ethnic background, trimester and vagitype. The associations were restricted to use only the strain groups as antecedents. Rules were generated with minimum support of 10% (at least 10% of the samples satisfied the rule) and minimum confidence of 60% (confidence: among all subjects with the environmental characteristics, the percentage having a particular strain). An FDR correction for the Fisher’s exact test was applied to the generated rules. To confirm the associations and avoid bias from repeated measures, a mixed-effects logistic regression model was fitted with the strain as the response, the environmental variable as a predictor, and a subject random effect.
The R package arules66 was used to generate and analyze rules. The function glmer() in the R package lme465 was used to fit mixed-effects logistic regression models.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
16S rRNA sequence data and metadata for each sample have been deposited in the HMP DACC (https://portal.hmpdacc.org/). Data that are of controlled access (that is, metagenomic and metatranscriptomic sequence data, which can include some sensitive human sequence and subject metadata) have been deposited at NCBI’s controlled-access dbGaP (study accession IDs phs001523 and phs000256) and Sequence Read Archive (SRA; BioProject IDs PRJNA326441, PRJNA430481, PRJNA430482, PRJNA74947, PRJNA51443 and PRJNA46877). Additional metadata have been deposited in, and are available through, the RAMS Registry (https://ramsregistry.vcu.edu). Project information is also available at the project website (http://vmc.vcu.edu).
Code availability
Custom code is available at https://github.com/Vaginal-Microbiome-Consortium/TBS. Open-source software is described in the text.
Extended Data
Extended Data Fig. 1 ∣. Differences in microbiome diversity in pregnant and non-pregnant women of different ancestry.
a, Differences in alpha diversities of the vaginal microbiomes in 613 pregnant and 1,969 non-pregnant women of different racial descendance due to pregnancy. b, Differences in alpha diversities of the vaginal microbiomes of 300 pregnant and 300 non-pregnant women of different racial descendance case-matched for race, age and socioeconomic status due to pregnancy. c, Differences in alpha diversities of the vaginal microbiomes of 300 pregnant and 300 non-pregnant women of different racial descendance case-matched for race, age and socioeconomic status. Box plots were generated in R using standard approaches. The bar represents the median and the boxes indicate interquartile ranges. Significant differences are indicated (*P < 0.05).
Extended Data Fig. 2 ∣. Effects of pregnancy on the vaginal microbiome in different racial backgrounds.
a, Microbiome profiles of 304 pregnant women (upper panel) and 1,184 non-pregnant women of African ancestry. b, Microbiome profiles of 111 pregnant women of European ancestry and 682 non-pregnant women of European ancestry. c, Microbiome profiles of 198 pregnant women of Hispanic ancestry and 103 non-pregnant women of Hispanic ancestry. Legend is as shown for Fig. 2. The blue bars denote the Lactobacillus taxa (L. crispatus, L. jensenii, L. gasseri and L. iners).
Extended Data Fig. 3 ∣. Vaginal microbiome profiles of 90 women, 49 of African and 41 of non-African ancestry.
a, Microbiome profiles of all samples (421 total, 175 from women of non-African ancestry and 246 from women of African ancestry) from each of these 90 women. Taxa are color-coded as indicated. b, Microbiome profiles of these same samples from women of non-African (top) and African ancestry (bottom). Taxa are color-coded as in a. c, Alpha diversity measures of richness (species counts) and evenness (Shannon index) of these samples (described in a) from women of non-African (n-Afr) and African (Afr) ancestry, measured using the vegan package. Alpha diversities and statistical analysis were calculated as indicated in the Methods. Box plots were generated in R using standard approaches. The bar represents the median and the boxes indicate interquartile ranges. d, L1-Norm PCA analysis of the same samples (see Methods). Legend of vagitypes is as indicated. See Supplementary Table 5 for sequence read statistics for data presented in this figure.
Extended Data Fig. 4 ∣. Longitudinal changes in microbiome profiles across trimesters during pregnancy.
a, Vaginal microbiome profiles of 41 pregnant women of African (n = 22) or non-African (n = 19) ancestry who provided at least 1 sample from each of 3 trimesters. b, Alpha diversity measures of richness (species counts) and evenness (Shannon index) of samples from a. Diversity measures calculated using the vegan package (see Methods). Box plots were generated in R using standard approaches. The bar represents the median and the boxes indicate interquartile ranges. Asterisks indicate statistical significance (*P < 0.05; **P < 0.01). c, L1-Norm PCA analysis (see Methods) of samples from a. Legends are indicated. n-Afr, women of non-African ancestry; Afr: women of African ancestry. See Supplementary Table 5 for sequence read statistics for data presented in this figure.
Extended Data Fig. 5 ∣. Changes in abundance of taxa across pregnancy.
a, Relative abundances of L. crispatus and L. iners in 1 early and 1 late sample from each of 90 participants, 41 of non-African (n-Afr) and 49 of African (Afr) ancestry. b, Longitudinal differences in relative abundance of select taxa—L. crispatus, L. iner, L. jensenii, L. gasseri, G. vaginalis, BVAB1, A. vaginae, S. amnii, Prevotella cluster 2 and TM7_OTU-H1, from 1 sample collected in each trimester from 90 participants, 41 of non-African (n-Afr) and 49 of African (Afr) ancestry. For both a and b, the medians for each group were compared using a two-sided Wilcoxon test, with FDR adjustments for multiple comparisons where applicable (ns, not significant; *P < 0.05; **P < 0.01).
Extended Data Fig. 6 ∣. Stability of vagitypes in pregnancy showing the variation of the microbiomes of each woman across all samples collected during that pregnancy.
a, Vaginal microbiome profiles from 41 women of non-African ancestry. Each facet represents the data from a single participant across all vaginal samples collected during her pregnancy. The samples, within each facet, are ordered from left to right based on their gestational age at sampling; same as Fig. 3a,b. The bars below each stacked bar indicate the strain of L. crispatus (1 or 2), L. jensenii (1 or 2), L. gasseri (1 or 2), L. iners (1 or 2), BVAB1 (1 or 2) or G. vaginalis (1, 2, 3 or 4). b, Vaginal microbiome profiles from 49 women of African ancestry. As for Extended Data Fig. 7, each facet represents the data from a single participant across all vaginal samples collected during her pregnancy. The samples, within each facet, are ordered from left to right based on their gestational age at sampling; same as Fig. 3a,b. The bars below each stacked bar indicate the strain of L. crispatus (1 or 2), L. jensenii (1 or 2), L. gasseri (1 or 2), L. iners (1 or 2), BVAB1 (1 or 2) or G. vaginalis (1, 2, 3 or 4).
Extended Data Fig. 7 ∣. Functional metabolic potential and transcriptional activity in vaginal microbiomes cluster according to vagitype.
a, Sparse partial least squares discriminant analysis (PLS-DA) of pathways derived from metagenomic sequence analysis of all 373 samples (147 samples from the 41 women of non-African ancestry, and 226 samples from the 49 women of African ancestry) from the 90 women in this study. Samples are color-coded according to vagitype (see legend). b, Sparse PLS-DA of pathways derived from metatranscriptomic sequence analysis of 1 sample from each pregnancy taken in the second or early third trimester (20 samples from the women of non-African ancestry and 28 from the women of African ancestry). c, Heat map of pathways from metagenomic analysis of samples as for a. Samples are sorted according to major vagitype (see legend). Samples from women of African ancestry (African) and from prior to 26 weeks’ gestation (early) are indicated. Alpha diversity is shown. d, Heat map of pathways from metatranscriptomic analysis of samples as for b. Samples are sorted as in c. Abundance and alpha diversity value scales are indicated. Sparse PLS-DA is a technique for fitting classification models that simultaneously selects features (via an L1 norm penalty term) that best describe group separation. The resulting model is sparse so that only a small subset of bacteria is included; the discriminant functions allow for visualization of the classification rule.
Extended Data Fig. 8 ∣. Association of G. vaginalis, L. crispatus, L. jensenii, L. gasseri, L. iners and Lachnospiracea BVAB1 strains with ancestry and other taxa.
Samples with these taxa were analysed in parallel with known reference strains using PanPhlan software to discriminate strain designations using default parameters of -min_coverage 1 (see Methods). a, G. vaginalis. Using these parameters, 121 samples provided sufficient numbers of G. vaginalis reads to provide accurate strain designations. Strain designations, which were previously reported by Ahmed et al.33 or Callahan et al.53, are indicated by the colored bars below the heat map. Note that G1 of Callahan et al. is within Set B of Ahmed et al., which also overlaps clades 3 and 4, and G2 of Callahan et al. includes Set A of Ahmed et al., which is also subdivided into clades 1 and 2. G3 of Callahan et al. classifies in clade 1 of Ahmed et al. The ancestry of each participant is indicated in the bar above the heat map, where blue indicates non-African and gray indicates African ancestry, and orange indicates a reference strain genome. Note: several samples contained multiple strains of different lineage. The black bar indicates two samples that contained three strains from clades 2, 3 and 4. b-f, L. crispatus, L. jensenii, L. gasseri, L. iners, and Lachnospiracea BVAB1. Analyses similar to that done for G. vaginalis above were performed with samples containing sufficient presence of these taxa (see above, and Methods). The ancestry of each participant is indicated in the bar above the heat map, where blue indicates non-African and gray indicates African ancestry, and white indicates a reference strain genome. Clades are differentiated by pink and light brown bars under each heat map.
Supplementary Material
Acknowledgements
The study team would like to gratefully acknowledge the participants who contributed specimens and data to the study. The authors would also like to acknowledge other members of the Vaginal Microbiome Consortium whose contributions made the study possible, including the team of research coordinators, the team of sample processors and the team of clinicians and nurses who assisted with sample collection. This study was funded by NIH grants UH3AI083263 and U54HD080784 to G.A.B., K.K.J. and J.F.S. We would also like to thank the Common Fund, the National Center for Complementary and Integrative Health, the Office of Research on Women’s Health, the Eunice Kenedy Shriver National Institute of Child Heatlh and Human Development, and the National Institute of Allergy and Infectious Disease at NIH for their generous support of this project. Other grants that provided partial support include a GAPPS BMGF PPB grant to G.A.B. and J.M.F. and NIH grant R21HD092965 to J.M.F. and E. P. Wickham and 1R01HD092415 to G.A.B. and T.J.A. N.R.J. was supported by grant R25GM090084 for the VCU Initiative For Maximizing Student Development (IMSD) programme. All sequence analysis reported herein was performed in the Nucleic Acids Research Facilities at VCU, and all informatics analysis was performed in servers provided by the Center for High Performance Computing at VCU.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41591-019-0465-8.
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41591-019-0465-8.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-019-0465-8.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ravel J et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108, 4680–4687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravel J et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younes JA et al. Women and their microbes: the unexpected friendship. Trends Microbiol. 26, 16–32 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan S et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PloS One 5, e10197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrova MI, Lievens E, Malik S, Imholz N & Lebeer S Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol 6, 81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobel JD Bacterial vaginosis. Annu. Rev. Med 51, 349–356 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Fettweis JM et al. The vaginal microbiome and preterm birth. Nat. Med 10.1038/s41591-019-0450-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah R et al. Incidence and risk factors of preterm birth in a rural Bangladeshi cohort. BMC Pediatr. 14, 112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tielsch JM Global incidence of preterm birth. Nestle Nutr. Inst. Workshop Ser 81, 9–15 (2015). [DOI] [PubMed] [Google Scholar]
- 10.WHO. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity Bull. World Health Org; 88, 31–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldenberg RL et al. The preterm prediction study: the value of new vs standard risk factors in predicting early and all spontaneous preterm births. NICHD MFMU Network. Am. J. Public Health 88, 233–238 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.York TP, Eaves LJ, Neale MC & Strauss JF The contribution of genetic and environmental factors to the duration of pregnancy. Am. J. Obstet. Gynecol 210, 398–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barcelona de Mendoza V et al. A systematic review of DNA methylation and preterm birth in African American women. Biol. Res. Nurs 19, 308–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modi BP et al. Mutations in fetal genes involved in innate immunity and host defense against microbes increase risk of preterm premature rupture of membranes (PPROM). Mol. Genet. Genom. Med 5, 720–729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fettweis JM et al. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiolology 160, 2272–2282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacIntyre DA et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep 5, 8988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gajer P et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med 4, 132ra52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyman RW et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod. Sci 21, 32–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma B, Forney LJ & Ravel J The vaginal microbiome: rethinking health and diseases. Annu. Rev. Microbiol 66, 371–389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey RJ, Zhou X, Pierson JD, Ravel J & Forney LJ Understanding vaginal microbiome complexity from an ecological perspective. Transl. Res. J. Lab. Clin. Med 160, 267–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin DH & Marrazzo JM The vaginal microbiome: current understanding and future directions. J. Infect. Dis 214, S36–S41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1, 121–133 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Beamer MA et al. Bacterial species colonizing the vagina of healthy women are not associated with race. Anaerobe 45, 40–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson J et al. The NIH Human Microbiome Project. Genome Res. 19, 2317–2323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Integrative Human Microbiome Project. Dynamic analysis of microbiome–host omics profiles during periods of human health and disease. Cell Host Microbe 16, 276–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fettweis JM et al. Species-level classification of the vaginal microbiome. BMC Genom. 13, S17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks JP et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 15, 66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson DB et al. Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery. Paediatr. Perinat. Epidemiol 28, 88–96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB & Marrazzo JM Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J. Clin. Microbiol 45, 3270–3276 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopes dos Santos Santiago G et al. Gardnerella vaginalis comprises three distinct genotypes of which only two produce sialidase. Am. J. Obstet. Gynecol 204, 450.e1–7 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Piot P et al. Biotypes of Gardnerella vaginalis. J. Clin. Microbiol 20, 677–679 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingianni A, Petruzzelli S, Morandotti G & Pompei R Genotypic differentiation of Gardnerella vaginalis by amplified ribosomal DNA restriction analysis (ARDRA). FEMS Immunol. Med. Microbiol 18, 61–66 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Ahmed A et al. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J. Bacteriol 194, 3922–3937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segata N et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onderdonk AB, Delaney ML & Fichorova RN The human microbiome during bacterial vaginosis. Clin. Microbiol. Rev 29, 223–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menard JP et al. High vaginal concentrations of Atopobium vaginae and Gardnerella vaginalis in women undergoing preterm labor. Obstet. Gynecol 115, 134–140 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Eastment MC & McClelland RS Vaginal microbiota and susceptibility to HIV. AIDS Lond. Engl 32, 687–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stout MJ et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am. J. Obstet. Gynecol 208, 226.e1–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiGiulio DB et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl Acad. Sci. USA 112, 11060–11065 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goltsman DSA et al. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 28, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walther-António MRS et al. Pregnancy’s stronghold on the vaginal microbiome. PloS One 9, e98514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero R et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks JP et al. Effects of combined oral contraceptives, depot medroxyprogesterone acetate and the levonorgestrel-releasing intrauterine system on the vaginal microbiome. Contraception 95, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muhleisen AL & Herbst-Kralovetz MM Menopause and the vaginal microbiome. Maturitas 91, 42–50 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Cauci S et al. Prevalence of bacterial vaginosis and vaginal flora changes in peri- and postmenopausal women. J. Clin. Microbiol 40, 2147–2152 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pabich WL et al. Prevalence and determinants of vaginal flora alterations in postmenopausal women. J. Infect. Dis 188, 1054–1058 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Hillier SL & Lau RJ Vaginal microflora in postmenopausal women who have not received estrogen replacement therapy. Clin. Infect. Dis 25, S123–126 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Spear GT et al. Human α-amylase present in lower-genital-tract mucosal fluid processes glycogen to support vaginal colonization by. Lact. J. Infect. Dis 210, 1019–1028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aagaard K et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PloS One 7, e36466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freitas AC et al. The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci. Rep 7, 9212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samant S et al. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLOS Pathog. 4, e37 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janulaitiene M et al. Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis. BMC Infect. Dis 17, 394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callahan BJ et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc. Natl Acad. Sci. USA 114, 9966–9971 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks JP, Dulá JH & Boone EL A pure L1-norm principal component analysis. Comput. Stat. Data Anal 61, 83–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forney LJ et al. Comparison of self-collected and physician-collected vaginal swabs for microbiome analysis. J. Clin. Microbiol 48, 1741–1748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parikh HI, Koparde VN, Bradley SP, Buck GA & Sheth NU MeFiT: merging and filtering tool for Illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinform. 17, 491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brooks JP, Dulá JH & Pakyz AL Identifying hospital antimicrobial resistance targets via robust ranking. IISE Trans. Healthc. Syst. Eng 7, 121–128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 60.Benjamini Y, Drai D, Elmer G, Kafkafi N & Golani I Controlling the false discovery rate in behavior genetics research. Behav. Brain Res 125, 279–284 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H & Durbin R Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ounit R, Wanamaker S, Close TJ & Lonardi S CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genom. 16, 236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abubucker S et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol 8, e1002358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bates D, Mächler M, Bolker B & Walker S Fitting linear mixed-effects models using lme4. J. Stat. Softw 67, 1–48 (2015). [Google Scholar]
- 66.Hahsler M, Chelluboina S, Hornik K & Buchta C The arules R-package ecosystem: analyzing interesting patterns from large transaction data sets. J. Mach. Learn. Res 12, 2021–2025 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA sequence data and metadata for each sample have been deposited in the HMP DACC (https://portal.hmpdacc.org/). Data that are of controlled access (that is, metagenomic and metatranscriptomic sequence data, which can include some sensitive human sequence and subject metadata) have been deposited at NCBI’s controlled-access dbGaP (study accession IDs phs001523 and phs000256) and Sequence Read Archive (SRA; BioProject IDs PRJNA326441, PRJNA430481, PRJNA430482, PRJNA74947, PRJNA51443 and PRJNA46877). Additional metadata have been deposited in, and are available through, the RAMS Registry (https://ramsregistry.vcu.edu). Project information is also available at the project website (http://vmc.vcu.edu).