Abstract
The dopamine transporter (DAT) is a plasma membrane protein that mediates the reuptake of extracellular dopamine (DA) and controls the spatiotemporal dynamics of dopaminergic neurotransmission. The transporter is subject to fine control that tailors clearance of transmitter to physiological demands, and dysregulation of reuptake induced by psychostimulant drugs, transporter polymorphisms, and signaling defects may impact transmitter tone in disease states. We previously demonstrated that DAT undergoes complex regulation by palmitoylation, with acute inhibition of the modification leading to rapid reduction of transport activity, and sustained inhibition of the modification leading to transporter degradation and reduced expression. Here, to examine mechanisms and outcomes related to increased modification, we co-expressed DAT with palmitoyl acyltransferases (PATs), also known as DHHC enzymes, which catalyze palmitate addition to proteins. Of twelve PATs tested, DAT palmitoylation was stimulated by DHHC2, DHHC3, DHHC8, DHHC15, and DHHC17, with others having no effect. Increased modification was localized to previously identified palmitoylation site Cys580 and resulted in upregulation of transport kinetics and elevated transporter expression mediated by reduced degradation. These findings confirm palmitoylation as a regulator of multiple DAT properties crucial for appropriate DA homeostasis and identify several potential PAT pathways linked to these effects. Defects in palmitoylation processes thus represent possible mechanisms of transport imbalances in DA disorders.
Keywords: post translational modification, palmitoyl acyl transferase, protein palmitoyl thioesterase, acyl protein thioesterase, protein degradation, [35S]-methionine labeling
In the central nervous system several processes including reward, motor function, and cognition are controlled by the neurotransmitter dopamine (DA)1, and dysregulation of DA homeostasis has been implicated in neurological and mood disorders such as Parkinson disease, schizophrenia, bipolar disorder, attention deficit hyperactivity disorder, and drug addiction2. Extracellular levels of DA are controlled primarily by the dopamine transporter (DAT), a plasma membrane protein that drives transmitter reuptake into the presynaptic neuron following vesicular release3–5. DAT dysfunction has been postulated to result in imbalanced transmitter levels in DA disorders, and the transporter is a target for psychostimulant and therapeutic drugs that increase extracellular DA by inhibiting reuptake or inducing efflux6–9.
DAT function is tightly regulated by signaling pathways that serve to coordinate transmitter clearance with physiological demands. Mechanisms include kinetic control of forward and reverse transport, and modulation of surface copy numbers through recruitment or endocytosis. While much remains to be understood about these processes, uptake and efflux kinetics have been linked to N-terminal phosphorylation, and transporter trafficking has been linked to N-terminal ubiquitylation and C-terminal binding partner interactions1, 10–15. Many of these processes are perturbed in disease states associated with psychostimulant drugs, transporter polymorphisms, and signaling pathway defects, highlighting their importance in DA control16.
We recently discovered that DAT is modified by S-palmitoylation, the thioesterification of a 16-carbon fatty acid to the sulfydryl group of cysteine17. A major fraction of palmitoylation on DAT occurs on Cys580 located at the membrane-cytoplasm interface of transmembrane domain (TM) 12, with the remainder occurring on one or more unknown sites18. S-palmitoylation of proteins is reversible and dynamic, allowing for regulated control, and is catalyzed by palmitoyl acyltransferases (PATs), also known as DHHC enzymes based on the conserved Asp-His-His-Cys catalytic motif17. Palmitoylation exerts pleiotropic effects on membrane proteins, regulating functions such as activity, trafficking, targeting, protein-protein interactions, and membrane raft partitioning18–21. The mechanisms of DAT regulation by palmitoylation are still under investigation, but pharmacological or mutational inhibition of the modification leads to reduced transport velocity, enhancement of protein kinase C (PKC)-dependent down regulation, and reduced transporter protein levels18.
Human, mouse, and rat genomes contain 23 highly homologous PATs, many of which are expressed in the nervous system22–27. Those utilizing DAT as a substrate are unknown, and to investigate this issue, we co-transfected DAT-expressing cells with a panel of PAT enzymes expressed in mouse substantia nigra. Our findings identify a subset of PATs that enhance DAT palmitoylation and demonstrate that the modification increases transporter protein levels and stimulates DA uptake. Direct measurement of DAT turnover by [35S]-methionine pulse-chase analysis supports a role for palmitoylation in promotion of transporter stability that likely underlies expression increases. However, although palmitoylation enhances total transporter levels, plasma membrane levels do not show concomitant changes, indicating that increased transport results from kinetic upregulation of surface transporters. These results demonstrate that palmitoylation represents a mechanism capable of bidirectional control of both short- and long-term DAT properties crucial for DA neurotransmission and may be dysregulated in disease conditions.
RESULTS
Identification of DAT palmitoyl acyl transferases
Because there are few pharmacological agents available for specific control of DHHC enzymes28, we utilized co-expression strategies to identify those capable of modifying and regulating DAT. We performed many initial studies in stably transfected rDAT-LLCPK1 cells, which we previously used for palmitoylation analyses18, and in some later studies moved to Neuro-2a (N2a) cells transiently transfected with WT or C580A DATs. Characterization of rDAT-N2a cells for palmitoylation properties (Supplemental Fig. S1), indicates that they may provide a superior model system for these studies, as they support DAT palmitoylation and uptake responses to the global PAT inhibitor 2-bromopalmitate (2BP) with time frames (30–120 min) that are considerably more similar to those we previously demonstrated18 in rat striatal synaptosomes (10–60 min) than those obtained in rDAT-LLCPK1 cells (12–18h).
For these initial studies we did not test all PATs, but focused on a subset with reported mRNA expression in mouse substantia nigra (DHHCs 2, 3, 5, 7, 8, 9, 11, 15 17, 20, 21, and 22)29. Vectors for HA-tagged mouse DHHC proteins and an HA-GST control were the generous gift of Dr. M. Fukata23, and we used the DHHC2 template to generate a catalytically inactive DHHA2 construct by mutation of active site Cys156 to Ala30. Immunoblotting with anti-HA verified expression of all forms and comparable expression of DHHC2 and DHHA2 (Supplemental Fig. S2).
For palmitoylation analyses, cells were transfected in parallel with control or DHHC vectors, and DAT modification was assessed and normalized to total transporter protein in each sample. For most experiments, DAT palmitoylation was analyzed by the acyl-biotinyl exchange (ABE) method18, 31, in which endogenous palmitate moieties are cleaved by hydroxylamine (NH2OH) and the liberated side chains are modified by a sulfhydryl – specific biotinylated reagent. Samples are chromatographed on NeutrAvidin® resin to capture biotinylated proteins and eluates immunoblotted for DAT. The specificity of the immunoblot signal for S-palmitoylation is determined in every experiment by parallel assessment of Tris controls that do not undergo acyl group exchange, with background signals typically <5% of total.
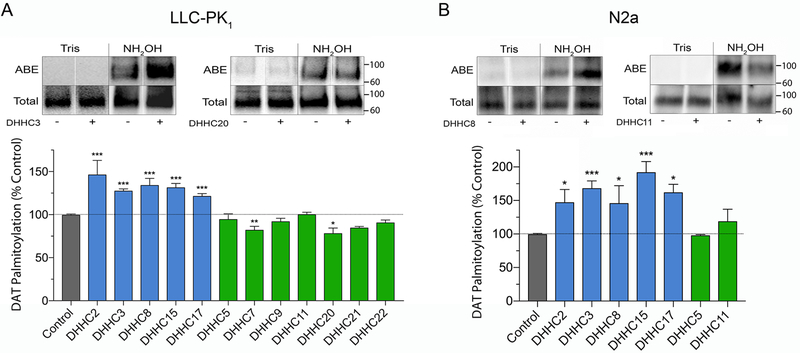
Figure 1 summarizes the findings from rDAT-LLCPK1 cells (Fig. 1A) and rDAT-N2a cells (Fig. 1B). The blots show representative examples of enzymes that increase (DHHC3 and DHHC8) or have no effect (DHHC11 and DHHC20) on DAT palmitoylation levels, and the histograms summarize findings from all tested PATs. In LLCPK1 cells, DAT palmitoylation was enhanced relative to control by co-expression of DHHC2 (146 ± 17%), DHHC3 (128 ± 2%), DHHC8 (134 ± 8%), DHHC15 (132 ± 5%), and DHHC17 (122 ± 3%) (all p<0.001), whereas expression of DHHCs 5, 7, 9, 11, 20, 21, or 22 did not increase (p>0.05) and in some cases, decreased, palmitoylation (DHHCs 7 and 20; p<0.05). In N2a cells we did not analyze the entire DHHC panel but focused on the forms that were positive in LLCPK1 cells. For those enzymes tested, the pattern of responses was essentially identical (Fig. 2B), with enhanced palmitoylation driven by DHHC2 (147 ± 19%), DHHC3 (173 ± 6%), DHHC8 (146 ± 26%), DHHC 15 (192 ± 16%) and DHHC 17 (162 ± 12%) (all p<0.05–0.001), but not by DHHC5 or DHHC11 (p>0.05).
Figure 1. Effect of DHHC enzymes on DAT palmitoylation.
A, rDAT-LLCPK1 or B, rDAT-N2a cells were transfected with indicated DHHC plasmids and equal amounts of DAT protein were assessed for palmitoylation. Blots show representative ABE (palmitoylated) and total DAT samples and histograms show quantification of DAT palmitoylation (% Control, means ± S.E), *p<0.05, **p<0.01, ***p<0.001 vs Control (ANOVA with Dunnett’s posttest, n=3–4). Shading indicates DAT palmitoylation values for vector control (gray), DHHC enzymes that increased DAT palmitoylation (blue), and DHHC enzymes that did not increase DAT palmitoylation (green). Vertical white dividing lines indicate the rearrangement of lane images from the same blot. Mr markers for all gels are shown at right.
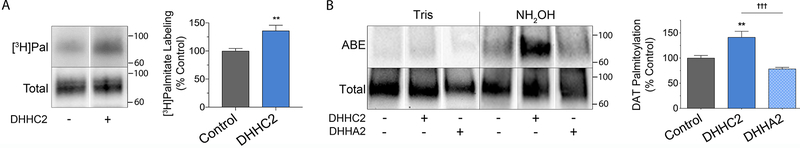
Figure 2. DAT palmitoylation specificity controls.
A, rDAT-LLCPK1 cells transfected with control or DHHC2 plasmids were labeled for 18h with [3H]palmitic acid. Equal amounts of DAT were immunoprecipitated and subjected to SDS-PAGE/autofluorography. Left, representative autoradiogram and matching immunoblot. Right, quantification of [3H]palmitate labeling (% Control, means ± S.E.). **p<0.01 DHHC2 vs control (Student’s t-test, n=3). B, rDAT-LLCPK1 cells were transfected with indicated plasmids and equal amounts of DAT analyzed for palmitoylation. Left, representative ABE and total DAT blots. Right, quantification of palmitoylation (% Control, means ± S.E). **p<0.01, DHHC2 vs Control; †††p<0.001 DHHA2 vs DHHC2 (ANOVA with Tukey’s posttest, n=4). Shading indicates DAT palmitoylation responses for vector control (gray), DHHC2 (blue) and catalytically inactive DHHA2 (stippled blue). Vertical white dividing lines indicate rearrangement of lane images from the same immunoblot or autoradiogram.
To verify that the enhanced signals detected in vitro by ABE represent metabolically-generated palmitoylation, we performed labeling of DAT with [3H]palmitic acid (Fig. 2A). In control conditions, DAT shows a constitutive level of [3H]palmitate labeling, with co-expression of DHHC2 increasing the labeling to 138 ± 10% of control (p<0.01). Experiments to verify that increased palmitoylation derives from enzymatic activity of the transfected PATs were performed using DHHA2, the catalytically inactive form of DHHC2. Fig. 2B shows that in parallel transfections, DAT palmitoylation was increased relative to control by DHHC2 (122 ± 7% of control, p<0.05), but not by DHHA2 (79 ± 4% of control) (p>0.05 vs control, p<0.001 vs DHHC2). DHHA2 was thus used as an additional negative control in several subsequent experiments.
Palmitoylation enhances total DAT levels
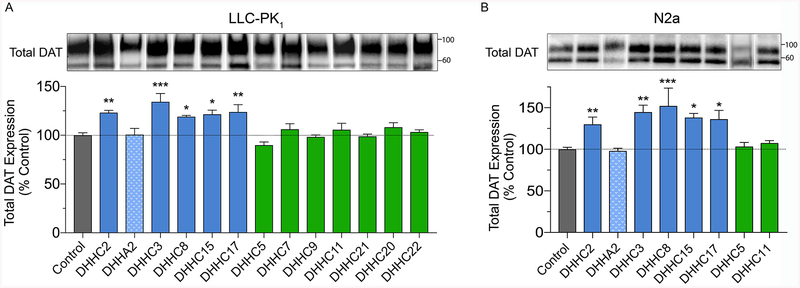
We previously found in both cells and rat striatal synaptosomes that DAT protein levels were reduced by sustained suppression of palmitoylation18, suggesting that the modification functions to maintain or enhance transporter levels. To investigate this, rDAT-LLCPK1 or rDAT-N2a cells were transfected with DHHC enzymes and equal amounts of protein were immunoblotted for DAT (Fig. 3). Our findings show that DAT protein levels paralleled palmitoylation status, with expression in rDAT-LLCPK1 cells increased by DHHC2 (123 ± 2%), DHHC3 (134 ± 9%), DHHC8 (119 ± 1%), DHHC15 (121 ± 4%), and DHHC17 (124 ± 8%) (all p<0.05–0.001), but not changed by DHHA2 or DHHCs 5, 7, 9, 11, 20, 21, or 22 (all p>0.05) (Fig. 3A). Similar results were obtained in rDAT-N2a cells (Fig. 3B), with DAT expression increased by DHHC2 (130 ± 9%), DHHC3 (145 ± 8%), DHHC8 (152 ± 21%), DHHC15 (138 ± 5%), and DHHC17 (136 ± 10%) (all p<0.05–0.01), but not changed by DHHA2, DHHC5, or DHHC11 (all p>0.05).
Figure 3. Effect of DHHC enzymes on DAT expression.
A, rDAT-LLCPK1 or B, rDAT-N2a cells were transfected with indicated DHHC plasmids and equal amounts of protein were immunoblotted for DAT. Top panels show representative blots (lanes ordered as in graphs), and histograms show quantification of band densities (% Control, means ± S.E.) *p<0.05, **p<0.01, ***p<0.001 vs Control (ANOVA with Dunnett’s posttest, n=4). Shading indicates DAT expression values for vector control (gray), DHHC enzymes that increased DAT palmitoylation (blue), DHHC enzymes that did not increase DAT palmitoylation (green), and catalytically inactive DHHA2 (stippled blue). Vertical white dividing lines indicate rearrangement of lane images from the same blot.
Palmitoylation increases DAT transport capacity via kinetic upregulation
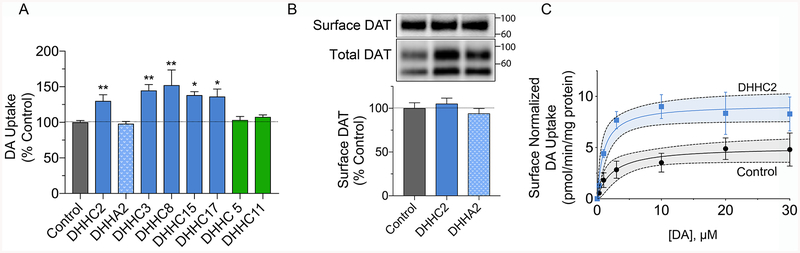
We also investigated the effects of enhanced DAT palmitoylation on [3H]DA transport, with uptake activity normalized to total cellular protein levels in each sample (pmol/min/mg). The results showed that uptake was increased in parallel with palmitoylation status, with transport levels significantly increased by DHHC2 (130 ± 9%), DHHC3 (145 ± 8%), DHHC8 (152 ± 21%), DHHC15 (138 ± 5%), and DHHC17 (136 ± 10%), (all p<0.05–0.01), but not changed by DHHA2, DHHC5, or DHHC11 (all p>0.05) (Fig. 4A).
Figure 4. Effect of DHHC enzymes on DA transport.
A, rDAT-N2a cells transfected with the indicated DHHC plasmids were assayed for [3H]DA uptake, and transport values were normalized to total protein and expressed as % control, means ± S.E. *p<0.05, **p<0.01 vs Control (ANOVA with a Dunnett’s post-hoc test, n=3–5). Shading indicates DA uptake values for cells transfected with vector control (gray), DHHC enzymes that increased DAT palmitoylation (blue), DHHC enzymes that did not increase DAT palmitoylation (green), and catalytically inactive DHHA2 (stippled blue). B, Surface biotinylation analysis of rDAT-N2a cells transfected with Control, DHHC2, or DHHA2 plasmids. Upper and lower panels show representative blots of surface or total DATs from 100 μg or 25 μg protein, respectively, and histogram shows quantification of surface band densities (% Control, means ± S.E.), all samples p>0.05 vs control (ANOVA with Dunnett’s posttest, n=5–8). C, Transport saturation analysis of rDAT-N2a cells transfected with Control or DHHC2 plasmids. Each point represents means ± S.E. of three independent experiments, normalized to surface DAT, and results were fit to Michaelis-Menten kinetics. Gray and blue shading indicates 95% confidence intervals for each curve.
To determine if these changes in uptake capacity were driven by increased surface expression following from increased total transporter levels, we performed cell surface biotinylation studies. For these experiments we analyzed the effects of DHHC2 on DAT expression and surface levels, using DHHA2 as a negative control (Fig. 4B). The findings show that DAT expression normalized to total cellular protein is enhanced by DHHC2 but not DHHA2 (lower panel), as independently demonstrated in Fig. 3. Surface biotinylation of these samples, however, shows that, despite the increased levels of DAT induced by DHHC2, transporter plasma membrane levels (upper panel) were unchanged in all conditions (DHHC2, 105 ± 6% of control; DHHA2, 94 ± 6% of control, both p>0.05) (Fig. 4B). These results indicate that the increased DA transport velocity induced by DHHC2 is not driven by enhancement of transporter surface levels and thus occurs via kinetic upregulation of surface transporters. Unchanged surface levels of DAT were also found in cells transfected with DHHC3 and DHHC8, which upregulate DA transport activity, as well as DHHC11, which does not affect DAT function (Supplemental Fig. S3), supporting similar mechanistic outcomes from other PATs. The experiments in Fig. S3 also demonstrate reduced surface expression of DAT with amphetamine treatment, which induces transporter endocytosis, verifying the responsiveness of the cells to known trafficking signals.
Quantification of the results in Fig. 4B also demonstrates that the ratio of surface to total DAT is reduced in DHHC2 conditions (Control 0.70 ± 0.02; DHHC2 0.58 ± 0.03; p<0.05 vs control; DHHA2 0.65 ± 0.02; p>0.05 vs control). In conjunction with the unchanged surface levels in DHHC2 conditions, these findings suggest that palmitoylation per se does not constitute a signal for DAT plasma membrane recruitment, and rather, indicate that palmitoylated transporters accumulate in one or more internal membrane compartments.
Saturation analysis of DHHC2-increased transport normalized for surface expression (Fig. 4C), showed that the effect was due to enhanced Vmax (DHHC2, 9.2 ± 0.9 pmol/min/mg vs control, 5.1 ± 0.7 pmol/min/mg, p<0.05), with no effect on Km (DHHC2, 1.2 ± 0.4 μM vs control, 2.3 ± 1.0 μM, p = 0.4).
DAT palmitoylation effects are mediated through Cys580
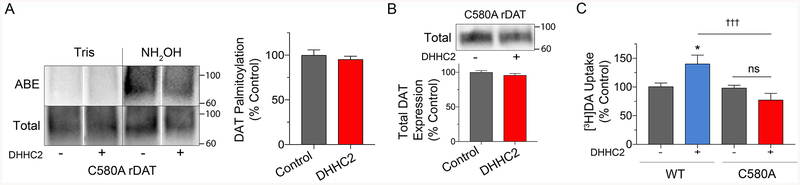
In our previous studies, we identified Cys580 as one of the major sites for DAT palmitoylation, with mutation of the residue to Ala reducing transporter modification by ~50%18. To determine if PAT-driven palmitoylation and associated functions are mediated through this site, we analyzed responses of C580A DAT to DHHC2. In contrast to its ability to modify and regulate WT DAT, DHHC2 did not stimulate C580A DAT palmitoylation (95% ± 3% of control p>0.05) (Fig. 5A) or increase C580A DAT levels (96 ± 2% of control, p>0.05) (Fig 5B). For examination of uptake responses to DHHC2 we performed parallel analyses of WT and C580A DAT. In these experiments, the WT protein showed the expected increase in transport activity (141 ± 15 %; p<0.05 vs control), whereas activity of C580A DAT was not increased relative to its own control (78 ± 11%, p>0.05), or relative to the fold stimulation obtained for WT DAT, p<0.001) (Fig. 5C). These results thus support Cys580 as the site of enhanced palmitoylation in the WT protein and indicate that the functional effects of DHHC2 and likely other PATs, follows directly from modification of the transporter rather than indirectly through effects on other proteins. It is possible that lack of C580A DAT responsiveness in these experiments could arise from a ceiling effect of endogenous PATs in these cells. However, C580A DATs have both lower expression and transport activity than WT DAT32 arguing against this possibility.
Figure 5. Cys580 mediates DHHC2 effects.
A, LLCPK1 cells, or B and C, N2a cells expressing WT or C580A DAT as indicated, were transfected with control or DHHC2 plasmids and assessed for palmitoylation (A), expression (B), or uptake (C). A, Left, representative ABE and total C580A DAT blots; Right, quantification of palmitoylation (% Control, means ± S.E). p>0.05, Student’s t-test (n=4). B, Representative immunoblot and quantification of band densities for C580A DAT expression (% Control, mean ± S.E., n=3). C, [3H]DA uptake in WT- or C580A-DAT cells. (% Control for each form, means ± S.E.) *p<0.05, WT/DHHC2 vs WT Control; †††p<0.001 C580A/DHHC2 vs WT/DHHC2; ns, no significant difference (Two-way ANOVA with Tukey’s post-test; C580A: F(1, 29) = 15.30; DHHC: F(1, 29) = 1.257; interaction: F(1, 29) = 13.13; p < 0.001; n=3–5). Gray shading indicates responses of WT or C580 DAT to control conditions, red shading indicates C580A DAT responses to DHHC2, and blue shading indicates WT DAT responses to DHHC2. Vertical white dividing lines indicate rearrangement of lane images from the same blot.
Reduced palmitoylation increases DAT turnover
Our previous studies showed that steady-state expression of C580A DAT was lower than that of WT DAT, and that pharmacological inhibition of DAT palmitoylation induced acute loss of transporter protein and production of proteolytic fragments18, suggesting that the modification functions to oppose transporter turnover. To directly test the role of palmitoylation in regulation of DAT turnover, we performed [35S]methionine ([35S]Met) pulse-chase analysis of WT and C580A DATs (Fig. 6). Cells were given a 30 min pulse of [35S]Met and harvested at various intervals afterward. DAT levels in all samples were determined by immunoblotting, and equal amounts of DAT were immunoprecipitated for determination of labeling. Protein synthesis inhibitors were not used, and total DAT levels in each sample remained relatively constant throughout the analysis. In addition, cells were 80–90% confluent at the beginning of the analysis, and we saw no major increases in total cellular protein or DAT levels due to cell division.
Figure 6. Cys580 palmitoylation regulates DAT turnover.
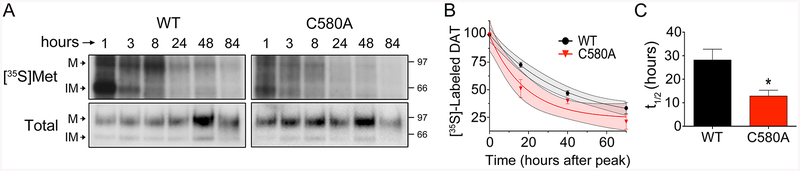
WT and C580A rDAT-LLCPK1 cells were labeled with [35S]Met for 30 min followed by chase with unlabeled medium, and samples were collected at the indicated times post-pulse. DAT levels were determined by immunoblotting and equal amounts immunoprecipitated for analysis of [35S]Met labeling. A, Representative autoradiograms of [35S]Met-labeled DATs with matching immunoblots. M, mature form; IM, immature form. B, Quantification of [35S]Met labeling in 90kDa (M) bands of WT and C580A DAT forms, normalized to peak levels for each form at 8h post-pulse. Curves were fit to One Phase Decay, (goodness of fit WT DAT r2 = 0.97; C580A DAT, r2 = 0.91; Two-way ANOVA; C580A: F(1, 4) = 4.6; Time: F(1.348, 4.942) = 181.5; interaction: F(3, 11) = 3.8; p<0.05). Gray and red shading indicates 95% confidence intervals for WT and C580A decay curves, respectively. C, Half-life of WT and C580A DAT proteins obtained from decay curves (means ± S.E., three independent experiments). * p<0.05 C580A vs WT (Student’s t-test, n=3).
Fig. 6A shows that [35S]Met was rapidly incorporated into the immature (IM) 60 kDa form of DAT. Processing of labeled transporters into the mature (M) glycosylated 90 kDa form was seen by 3h, with peak appearance of label in the mature form occurring about 8h after the pulse. Maturation time frames did not seem overtly different between WT and C580A DATs, although we did not pursue this aspect in detail. However, even though equal amounts of DAT protein determined by immunoblotting were analyzed in each form, [35S]Met labeling of C580A DAT consistently appeared lower than that of WT DAT, indicative of elevated turnover.
For quantification of transporter degradation, we monitored [35S]Met levels in the 90 kDa band of each form at 0h, 16h, 40h, and 70h after the peak protein labeling at 8h post-pulse. [35S]Met signals for each form were normalized for DAT protein and expressed as % of the 0h value. In three independent experiments we found that [35S]Met signals decayed more rapidly in C580A DATs than in WT DATs. Fig. 6A shows autoradiographs and blots from a representative experiment, and Fig. 6B shows the quantification of labeling for all experiments. The accelerated loss of [35S]Met from C580A DAT is evident, especially at the earlier time points. Turnover rates obtained from these curves indicate mean half-lifes of 28 ± 5h for WT DAT and 13 ± 2h for C580A DAT (p<0.05) (Fig. 6C), directly demonstrating that Cys580 palmitoylation functions to oppose DAT metabolic degradation.
Model of DAT palmitoylation functions
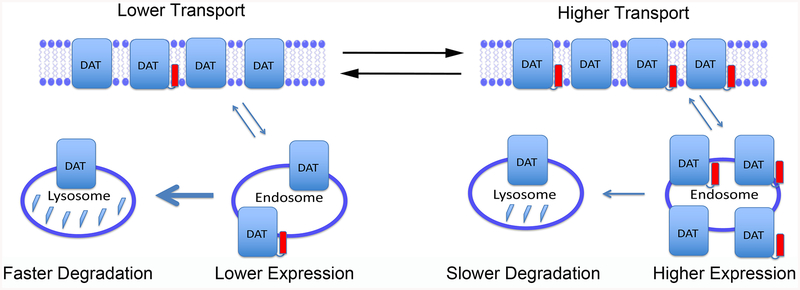
A model summarizing these findings is shown in Figure 7, which depicts DAT populations with lower (left) or higher (right) palmitoylation stoichiometries. Overall expression correlates with palmitoylation (lower on left, higher on right), with equal numbers of transporters at the cell surface and expression differences residing in internal endosome or vesicular pools. Transport velocities correlate with palmitoylation (lower on left, higher on right), indicating kinetic regulation of surface transporters, and internal pools of transporters undergo degradation at different rates (faster at left, slower at right) that likely underlie the expression differences.
Figure 7. Model of DAT palmitoylation functions.
Schematic representation of DAT populations with lower (left) or higher (right) stoichiometries of Cys580 palmitoylation (red rectangles). Total transporter expression is indicated by number of DAT symbols, with equal numbers of transporters at the surface in both conditions, and higher numbers of transporters in internal endosome or vesicular compartments on the right. Relative rates of transport activity are indicated at top, and large and small arrows leading from vesicles to lysosomes represent relative rates of transporter degradation.
DISCUSSION
The findings presented here demonstrate that enhancement of DAT palmitoylation functions to increase DA uptake capacity via control of transport kinetics and to enhance total levels of transporter copy numbers. In conjunction with our previous studies using palmitoylation inhibitors, these findings establish the ability of palmitoylation to bidirectionally regulate multiple DAT functions that could profoundly impact DA clearance and DA neurotransmission.
Although the overexpression approaches used in this study cannot probe rapid time dependency of functional outcomes related to palmitoylation changes, our previous 2BP studies showed that acute inhibition of DAT palmitoylation led to rapid changes in uptake capacity, providing a mechanism for short-term responsiveness of uptake to momentary physiological demands. Palmitoylation also exerts longer-term changes to DAT via impacts on expression. Control of DAT levels and activity is crucial to ensure adequate clearance during neurotransmission33, and losses or compensatory changes of DAT protein levels are observed in numerous DA disorders, including drug abuse, withdrawal, and Parkinson disease34–37. Palmitoylation may thus represent a contributing mechanism in these conditions.
Palmitoylation is known to exert pleiotropic effects on proteins, and the regulation of both uptake velocity and transporter degradation by palmitoylation suggests influences on DAT via multiple mechanisms. Kinetic control of uptake indicates an effect on the transporter alternating access cycle, in which extracellular DA binds to an outwardly facing form of DAT, conformational changes convert the transporter to an inwardly facing form that releases transmitter to the cell interior, and the empty protein reorients back to the outward form for another round of transport4, 38, 39. These events are driven by conformational changes of core TM helices that form the permeation pathway, coupled to concerted opening and closing of extracellular and intracellular gates that control pathway access and stabilize specific transporter conformations. At present the specifics of these processes are incompletely understood, and how Cys580 modification could accelerate events is not known. However, effects are likely to be mediated indirectly, as TM12 is located outside the substrate permeation pathway.
Structurally, the addition of palmitate to Cys580 will increase the hydrophobicity of the intracellular end of TM12, which will likely affect its hydrophobic matching with the lipid bilayer and affect tilt or orientation. These conformational differences could then be propagated to adjacent domains to indirectly affect events occurring during transport. In a similar vein, TM12 may serve as a dimer interface40 that could be affected by palmitoylation to regulate DAT oligomerization and associated functions41–45. Another possibility is that control of transport may follow from TM12 impacts on intracellular gating. Crystal structures of Drosophila DAT and the serotonin transporter revealed the presence of a short helix at the cytoplasmic end of TM12 that interacts with internal residues to stabilize the inwardly-closed form of the transporter46. The homologous sequence in mammalian DAT lies just downstream of Cys580, suggesting that palmitoylation effects on TM12 conformation may propagate to this domain and impact its ability to participate in these interactions. Finally, palmitoylation-induced transport upregulation may follow from its reciprocal relationship with transport down-regulation mediated by N-terminal phosphorylation32, although the mechanisms underlying the integration of these modifications is not known.
Regulation of transport kinetics by palmitoylation could also follow from effects on subcellular targeting and/or transporter interactome. For many proteins, palmitoylation directs raft-nonraft partitioning, and numerous DAT properties including uptake, efflux, and phosphorylation are impacted by raft localization47–52, which could be mediated by transporter interactions with cholesterol or regulatory binding partners46–48, 52–61. In addition, many DAT binding partners such as syntaxin 1A, flotillin 1, and DA receptors are also palmitoylated, which may further influence their raft partitioning and/or transporter interactions62–65.
Possible mechanisms for control of DAT turnover include regulation of transporter targeting to lysosomes or the necessity for depalmitoylation to occur as part of the degradation process. Post-endocytic trafficking of DAT is complex, with different stimuli directing transporter sorting to recycling endosomes for plasma membrane return or to late endosome/lysosome pathways for degradation66. Lysosomal targeting and degradation of DAT are stimulated by PKC67, which we previously showed inhibits DAT palmitoylation32, providing a potential mechanistic link between this modification and lysosomal targeting/degradation, and palmitoylation may also serve as a mechanism for regulating protein entry into retromer vs degradative pathways68, 69. In addition, once at the lysosome, palmitate moieties on proteins must be removed as part of the degradation process17. Terminal depalmitoylation is mediated by the lysosomal enzyme protein palmitoyl thioesterase 1 (PPT1), a process that is distinct from regulatory depalmitoylation catalyzed by acyl protein thioesterase 1 (APT1)17. Slower degradation rates of modified vs unmodified transporters may thus follow from the necessity for this additional step.
The PATs that act on DAT endogenously in the brain are not known, and although we have not yet examined all PAT enzymes for effects on DAT, our current findings have identified enzymes DHHC2, DHHC3, DHHC8, DHHC15 and DHHC17 as enhancing DAT palmitoylation. In situ hybridization findings indicate expression of these enzymes in mouse brain and/or substantia nigra70, 71, suggesting them as possibilities for catalyzing DAT modification in vivo, and we have performed immunoblotting studies that support endogenous expression of DHHC2 in rat striatum and N2a cells (Supplemental Fig. S4). The PAT enzymes we identify in this study as affecting DAT play major roles in synaptic physiology via palmitoylation of key components including vesicular fusion proteins, ligand- and voltage-gated ion channels, and neurotransmitter and growth factor receptors. Similar to DAT, these and many other neuronal substrates are palmitoylated by multiple PATs22, 23, 25, 27. This redundancy is not understood, but may function to ensure appropriate palmitoylation in different conditions or subcellular compartments25. The possibility should also be considered that the endogenous palmitoylation occurring in our cells systems may be masking effects of transfected enzymes, requiring the use of knock-down strategies to confirm outcomes.
Palmitoylation and depalmitoylation inputs may link to signaling pathways connected to DAT and contribute to functional dysregulation associated with imbalanced DA levels in disease states. A variety of neuropsychiatric diseases including schizophrenia, X-linked intellectual disability, Alzheimer disease, Huntington disease, and Infantile Neuronal Ceroid Lipofuscinosis are associated with DHHC, PPT1 and APT mutations that lead to palmitoylation defects25, 72–78. This suggests the potential for DA clearance imbalances to follow from dysregulated DAT palmitoylation, through enzyme defects or from transporter-based processes that prevent appropriate modification ensuing from polymorphisms or drug use.
EXPERIMENTAL PROCEDURES
Materials
[7,8-3H]DA (45 Ci/mmol) was from Perkin Elmer; [9,10-3H]palmitic acid (73.4 Ci/mmol) was from Moravek; [35S]-methionine was from MP Biomedical; DA was from Research Biochemicals International; DAT polyclonal antibody 16 (poly 16) and monoclonal antibody 16 (MAb 16) have been previously authenticated79, 80; Anti-HA antibody was from Covance. Anti-DHHC2 antibody was from ThermoFisher. X-tremeGENE HP transfection reagent was from Roche Applied Bioscience; lipofectamine 2000 was from life technologies; methyl methanethiosulfonate (MMTS), sulfhydryl-reactive (N-(6(biotinamido)hexyl)-3`-(2`-pyridyldithio)-pro-pionamide (HPDP-biotin), sulfo-NHS-SS-biotin, high capacity NeutrAvidin® agarose resin, protease inhibitor tablets and bicinchoninic acid (BCA) protein assay reagent were from Thermo Scientific; (–)-Cocaine and other fine chemicals were from Sigma-Aldrich. HA-tagged human DHHC cDNA in pEF-BOS-HA vectors were the generous gift of Dr. Masaki Fukata23. Construct DHHA2 was generated from the DHHC2 cDNA template by mutating Cys156 to alanine using the Stratagene QuickChange kit, with codon substation verified by sequencing (Eurofins MWG Operon, Huntsville, Al ).
Cell culture, transient transfection, and mutagenesis
Lilly laboratory cell-porcine kidney (LLCPK1) cells stably expressing the rat dopamine transporter (rDAT)81 were maintained with α-minimum essential medium (α-MEM) supplemented with 5% fetal bovine serum, 2 mM L-glutamine, 100 μg/ml penicillin/streptomycin, and 200 μg/ml G418 at 37°C in a 5% CO2 incubator. Neuro-2A cells (N2a) were maintained with α-minimum essential medium (α-MEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 μg/ml penicillin/streptomycin at 37°C in a 5% CO2 incubator. For transient transfection, LLCPK1 or N2a cells were grown to ~70 % confluence in 24-well plates and transfected using X-tremeGENE HD (Roche Applied Bioscience) or lipofectamine 2000 (Life Technologies) transfection reagent. N2a cells were transfected with 1 μg of WT or C580A DAT and 2 μg of PAT DHHC, and cells were used after 18–20 hours.
Cell membrane isolation
rDAT-LLCPK1 cells were grown in 100 mm plates to 90% confluency. Cells were washed twice and resuspended in 0.25 M sucrose, 10 mM triethanolamine, 10 mM acetic acid, (pH 7.8) at 4°C, and pelleted at 700 x g for 8 min. Cells were then resuspended in buffer C (0.25 M sucrose, 10 mM triethanolamine, 10 mM acetic acid, 1 mM EDTA, pH 7.8) and homogenized in a Dounce homogenizer. Homogenate was centrifuged at 700 x g for 10 min to remove nuclei and debris, the post-nuclear supernatant was further centrifuged at 16,000 x g for 12 min, and the resulting membrane pellet was resuspended in SP buffer ~ 1 mg/ml (10 mM Na2HPO4, 0.32 M sucrose, pH 7.4).
Acyl-biotinyl exchange
DAT palmitoylation was assessed by ABE using a method modified from Wan et al31. Cell membranes (200 μg protein) were pelleted by centrifugation (17,000 × g for 12 min at 4°C) and pellets were solubilized in lysis buffer (50 mM HEPES, pH 7.0, 2% SDS, 1 mM EDTA) containing protease inhibitor and 20 mM methanethiosulfonate (MMTS) to block free thiols. Each sample was divided into two equal portions that were treated for 2 h at 27 ˚C with 50 mM Tris-HCl, pH 7.4 (control) or 0.7 M NH2OH, pH 7.4 to cleave endogenous palmitoyl thioester linkages and liberate the Cys SH side group. NH2OH was removed with three sequential acetone precipitations followed by resuspension of pellets in 4SB (4% SDS, 50 mM Tris, 5 mM EDTA, pH 7.4). Samples were diluted with 50 mM Tris containing 0.4 mM sulfhydryl-reactive (N-(6(biotinamido)hexyl)-3`-(2`-pyridyldithio)-pro-pionamide (HPDP) to biotinylate the NH2OH-liberated SH groups that were the original sites of endogenous palmitoylation. Samples were incubated for 1 h at 27 ˚C, unreacted HPDP was removed by three sequential acetone precipitations, and the final pellet resuspended in lysis buffer. Equal amounts of DAT protein determined by immunoblotting were chromatographed on NeutrAvidin® resin to capture biotinylated protein, and eluates were immunoblotted for DAT and quantified where indicated as previously described18.
[3H]Palmitate metabolic labeling
rDAT-LLCPK1 cells were metabolically labeled with [9,10-3H] palmitic acid (0.5 mCi/ml) for 18 hours at 37°C in α-MEM media. Cells were lysed in radioimmunoprecipitation assay buffer (RIPA: 10 mM sodium phosphate, 150 mM NaCl, 2 mM EDTA, 50 mM sodium fluoride, 1% Triton X-100, 0.1% SDS, pH 7.2) and aliquots were immunoblotted to determine DAT levels. Equal amounts of DAT were immunoprecipitated with polyclonal Ab16 and resolved on 4–20% SDS-polyacrylamide gels. Gels were soaked in Fluro-Hance (Research Products International) fluorographic reagent for 30 min, dried, and exposed to pre-flashed X-ray film for 30–90 days. Fluorographic band intensities were quantified using Quantity One software (Bio-Rad), normalized to total DAT protein, and expressed as % control.
Surface biotinylation
N2a cells transiently co-transfected with WT rDAT and the indicated DHHC plasmids, were washed three times with ice cold Hank’s balanced salt solution (HBSS) Mg-Ca (HBSS, 1mM MgSO4, 0.1 mM CaCl2, pH 7.4), incubated twice with 0.5 mg/mL of membrane-impermeable sulfo-NHS-SS-biotin for 25 min at 4°C. The biotinylation reagent was removed and the reaction was quenched by two sequential incubations with 100 mM glycine in HBSS Mg-Ca for 20 min at 4°C. Cells were washed with HBSS Mg-Ca and lysed with RIPA containing protease inhibitor. Equal amounts of cell protein (100 μg) were immunoblotted for DAT, and equal amounts of DAT were chromatographed on NeutrAvidin® agarose with incubation overnight at 4°C. The beads were washed three times with RIPA buffer and the bound protein was eluted with 32 μl of sample buffer. Eluted proteins were immunoblotted for DAT with MAb16. Specificity of biotinylating reagent for surface protein reactivity was verified by immunoblotting for the cytosolic enzyme protein phosphatase 1α (PP1α), which was only detected in non-biotinylated fractions. The binding capacity of NeutrAvidin® agarose for DAT was verified to be within the linear range as previously described82.
[3H]DA uptake assay
N2a cells were grown to 70–80% confluence and transiently transfected with WT rDAT and the indicated DHHC coding plasmids. After 18–20 h, the cells were washed twice with Krebs-Ringer HEPES (KRH) buffer (25 mM HEPES, 125 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.3 mM CaCl2, 1.2 mM MgSO4, 5.6 mM glucose, pH 7.4). DA uptake was initiated by addition of 10 μl of a 50× DA stock solution to 500 μl KRH bringing the final concentration of [3H]DA to 10 nM and total DA to 3 μM. Nonspecific uptake was determined in the presence of 100 μM (−)–cocaine and subtracted from total uptake values. Uptake assays were performed at 37°C for 8 min and terminated by rapidly washing the cells two times with ice-cold KRH buffer. Cells were solubilized in RIPA containing protease inhibitors, and radioactivity in lysates measured by liquid scintillation counting. Lysates were assessed for total protein content and DA uptake values (pmol/min/mg protein) from saturation analyses were normalized to DAT surface expression by dividing uptake values by relative surface abundance for each where the control surface abundance was set to 1 and the DHHC treated was either less than or greater than 1 as determined by cell surface biotinylation with equal amounts of total protein for each sample loaded on the high capacity NeutrAvidin® resin. Vmax and Km values were determined by nonlinear regression analysis of the normalized saturation analysis uptake values.
[35S]methionine pulse chase assay
WT and C580A rDAT-LLCPK1 cells were grown in 35 mm dishes to ~80–90% confluence. Cells were rinsed twice with HBSS and once with methionine/cysteine free media and incubated with methionine/cysteine free media at 37 °C for 30 min. Cells were labeled with 0.5 mCi/ml [35S]methionine for 30 min at 37 °C, followed by replacement with complete medium and harvested at times 0, 1, 8, 24, 48 and 84 h post-chase. Cells were pelleted by centrifugation at 2,000 × g for 5 min at 4 °C and lysed with lysis buffer (10 mM triethanolamine acetate pH 7.8, 150 mM NaCl, 0.1% Triton X-100, 15% sucrose, 100 mM DTT, and protease inhibitors). Lysates were centrifuged at 4,000 × g for 2 min, supernatants adjusted to contain 0.5% SDS, and centrifuged at 20,000 × g for 30 min to remove insoluble material. DAT levels were determined by immunoblotting, and equal amounts of DAT were immunoprecipitated with poly 16 Ab followed by SDS-PAGE/autoradiography, and parallel aliquots were immunoblotted with Mab 16 to detect total DAT protein. t½ values were calculated from decay curves for each experiment by non-linear curve fitting analysis with best fit to a one phase decay model (GraphPad Prism).
Supplementary Material
Acknowledgments:
This work was supported by grants R15 DA031991 (JDF), R01 DA13147 (RAV), ND EPSCOR Doctoral Dissertation Fellowship (DEB); P20 GM104360 (to UND) from the COBRE program and P20 GM103442 (to UND) from the INBRE program of the National Institute of General Medical Sciences. We thank Dr. Masaki Fukata, National Institute for Physiological Sciences, Japan, for the generous gift of DHHC coding plasmids and Dr. Keith Henry for providing the molecular model that we modified in the graphic for the Abstract .
The abbreviations used are:
- LLCPK1
Lilly lab cell-porcine kidney cells
- BME
β-mercaptoethanol
- 2BP
2-bromopalmitate
- ABE
acyl-biotinyl exchange
- APTs
acyl protein thioesterases
- BCA
bicinchoninic acid
- DA
dopamine
- DAT
dopamine transporter
- KRH
Krebs-Ringer/HEPES buffer
- MMTS
methyl methanethiosulfonate
- MAb 16
monoclonal antibody 16
- PAT
palmitoyl acyl transferase
- Poly 16
polyclonal antibody 16
- PKC
protein kinase C
- PPTs
protein palmitoyl thioesterases
- APTs
acyl protein thioesterase
- HPDP biotin
sulfhydryl-reactive (N-(6-(biotinamido) hexyl)-3′-(2′-pyridyldithio)-propionamide
- RIPA
radioimmunoprecipitation assay
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- NH2OH
hydroxylamine
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the content of this article.
REFERENCES
- (1).Torres GE, Gainetdinov RR, and Caron MG (2003) Plasma membrane monoamine transporters: structure, regulation and function, Nat Rev Neurosci 4, 13–25. [DOI] [PubMed] [Google Scholar]
- (2).Iversen SD, and Iversen LL (2007) Dopamine: 50 years in perspective, Trends Neurosci 30, 188–193. [DOI] [PubMed] [Google Scholar]
- (3).Eriksen J, Jorgensen TN, and Gether U (2010) Regulation of dopamine transporter function by protein-protein interactions: new discoveries and methodological challenges, J Neurochem 113, 27–41. [DOI] [PubMed] [Google Scholar]
- (4).Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, and Gether U (2011) SLC6 neurotransmitter transporters: structure, function, and regulation, Pharmacol Rev 63, 585–640. [DOI] [PubMed] [Google Scholar]
- (5).Rudnick G, Kramer R, Blakely RD, Murphy DL, and Verrey F (2014) The SLC6 transporters: perspectives on structure, functions, regulation, and models for transporter dysfunction, Pflugers Arch 466, 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, and Loland CJ (2008) The binding sites for cocaine and dopamine in the dopamine transporter overlap, Nat Neurosci 11, 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cervinski MA, Foster JD, and Vaughan RA (2005) Psychoactive substrates stimulate dopamine transporter phosphorylation and down-regulation by cocaine-sensitive and protein kinase C-dependent mechanisms, J Biol Chem 280, 40442–40449. [DOI] [PubMed] [Google Scholar]
- (8).Kuhar MJ, Ritz MC, and Boja JW (1991) The dopamine hypothesis of the reinforcing properties of cocaine, Trends Neurosci 14, 299–302. [DOI] [PubMed] [Google Scholar]
- (9).Mazei-Robison MS, Bowton E, Holy M, Schmudermaier M, Freissmuth M, Sitte HH, Galli A, and Blakely RD (2008) Anomalous dopamine release associated with a human dopamine transporter coding variant, J Neurosci 28, 7040–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Foster JD, Cervinski MA, Gorentla BK, and Vaughan RA (2006) Regulation of the dopamine transporter by phosphorylation, Handb Exp Pharmacol, 197–214. [DOI] [PubMed] [Google Scholar]
- (11).Foster JD, Pananusorn B, Cervinski MA, Holden HE, and Vaughan RA (2003) Dopamine transporters are dephosphorylated in striatal homogenates and in vitro by protein phosphatase 1, Brain Res Mol Brain Res 110, 100–108. [DOI] [PubMed] [Google Scholar]
- (12).Moritz AE, Foster JD, Gorentla BK, Mazei-Robison MS, Yang JW, Sitte HH, Blakely RD, and Vaughan RA (2013) Phosphorylation of dopamine transporter serine 7 modulates cocaine analog binding, J Biol Chem 288, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Miranda M, Dionne KR, Sorkina T, and Sorkin A (2007) Three ubiquitin conjugation sites in the amino terminus of the dopamine transporter mediate protein kinase C-dependent endocytosis of the transporter, Mol Biol Cell 18, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Miranda M, Wu CC, Sorkina T, Korstjens DR, and Sorkin A (2005) Enhanced ubiquitylation and accelerated degradation of the dopamine transporter mediated by protein kinase C, J Biol Chem 280, 35617–35624. [DOI] [PubMed] [Google Scholar]
- (15).Vaughan RA, Huff RA, Uhl GR, and Kuhar MJ (1997) Protein kinase C-mediated phosphorylation and functional regulation of dopamine transporters in striatal synaptosomes, J Biol Chem 272, 15541–15546. [DOI] [PubMed] [Google Scholar]
- (16).Vaughan RA, and Foster JD (2013) Mechanisms of dopamine transporter regulation in normal and disease states, Trends Pharm Sci 34, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Korycka J, Lach A, Heger E, Boguslawska DM, Wolny M, Toporkiewicz M, Augoff K, Korzeniewski J, and Sikorski AF (2012) Human DHHC proteins: a spotlight on the hidden player of palmitoylation, Eur J Cell Biol 91, 107–117. [DOI] [PubMed] [Google Scholar]
- (18).Foster JD, and Vaughan RA (2011) Palmitoylation controls dopamine transporter kinetics, degradation, and protein kinase C-dependent regulation, J Biol Chem 286, 5175–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Linder ME, and Deschenes RJ (2003) New insights into the mechanisms of protein palmitoylation, Biochemistry 42, 4311–4320. [DOI] [PubMed] [Google Scholar]
- (20).Adams MN, Christensen ME, He Y, Waterhouse NJ, and Hooper JD (2011) The role of palmitoylation in signalling, cellular trafficking and plasma membrane localization of protease-activated receptor-2, PLoS One 6, e28018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Iwanaga T, Tsutsumi R, Noritake J, Fukata Y, and Fukata M (2009) Dynamic protein palmitoylation in cellular signaling, Prog Lipid Res 48, 117–127. [DOI] [PubMed] [Google Scholar]
- (22).Fang C, Deng L, Keller CA, Fukata M, Fukata Y, Chen G, and Luscher B (2006) GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses, J Neurosci 26, 12758–12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fukata M, Fukata Y, Adesnik H, Nicoll RA, and Bredt DS (2004) Identification of PSD-95 palmitoylating enzymes, Neuron 44, 987–996. [DOI] [PubMed] [Google Scholar]
- (24).Fukata Y, Iwanaga T, and Fukata M (2006) Systematic screening for palmitoyl transferase activity of the DHHC protein family in mammalian cells, Methods 40, 177–182. [DOI] [PubMed] [Google Scholar]
- (25).Greaves J, and Chamberlain LH (2011) DHHC palmitoyl transferases: substrate interactions and (patho)physiology, Trends Biochem Sci 36, 245–253. [DOI] [PubMed] [Google Scholar]
- (26).Ohno Y, Kihara A, Sano T, and Igarashi Y (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins, Biochim Biophys Acta 1761, 474–483. [DOI] [PubMed] [Google Scholar]
- (27).Tsutsumi R, Fukata Y, and Fukata M (2008) Discovery of protein-palmitoylating enzymes, Pflugers Arch 456, 1199–1206. [DOI] [PubMed] [Google Scholar]
- (28).Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, and Linder ME (2009) 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro, J Lipid Res 50, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Xu M, St Clair D, and He L (2010) Testing for genetic association between the ZDHHC8 gene locus and susceptibility to schizophrenia: An integrated analysis of multiple datasets, Am J Med Genet B Neuropsychiatr Genet 153B, 1266–1275. [DOI] [PubMed] [Google Scholar]
- (30).Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, Tani H, Iwanari H, Mochizuki Y, Kodama T, Matsuura Y, Bredt DS, Hamakubo T, and Fukata M (2009) Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95, J Cell Biol 186, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wan J, Roth AF, Bailey AO, and Davis NG (2007) Palmitoylated proteins: purification and identification, Nat Protoc 2, 1573–1584. [DOI] [PubMed] [Google Scholar]
- (32).Moritz AE, Rastedt DE, Stanislowski DJ, Shetty M, Smith MA, Vaughan RA, and Foster JD (2015) Reciprocal Phosphorylation and Palmitoylation Control Dopamine Transporter Kinetics, J Biol Chem 290, 29095–29105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Giros B, Jaber M, Jones SR, Wightman RM, and Caron MG (1996) Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter, Nature 379, 606–612. [DOI] [PubMed] [Google Scholar]
- (34).Masoud ST, Vecchio LM, Bergeron Y, Hossain MM, Nguyen LT, Bermejo MK, Kile B, Sotnikova TD, Siesser WB, Gainetdinov RR, Wightman RM, Caron MG, Richardson JR, Miller GW, Ramsey AJ, Cyr M, and Salahpour A (2015) Increased expression of the dopamine transporter leads to loss of dopamine neurons, oxidative stress and l-DOPA reversible motor deficits, Neurobiol Dis 74, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lohr KM, Masoud ST, Salahpour A, and Miller GW (2017) Membrane transporters as mediators of synaptic dopamine dynamics: implications for disease, Eur J Neurosci 45, 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wu C, Garamszegi SP, Xie X, and Mash DC (2017) Altered Dopamine Synaptic Markers in Postmortem Brain of Obese Subjects, Front Hum Neurosci 11, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Papapetropoulos S, Basel M, and Mash DC (2007) Dopaminergic innervation of the human striatum in Parkinson’s disease, Mov Disord 22, 286–288. [DOI] [PubMed] [Google Scholar]
- (38).Forrest LR, and Rudnick G (2009) The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters, Physiology (Bethesda) 24, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, and Rudnick G (2008) Mechanism for alternating access in neurotransmitter transporters, Proc Natl Acad Sci U S A 105, 10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yamashita A, Singh SK, Kawate T, Jin Y, and Gouaux E (2005) Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters, Nature 437, 215–223. [DOI] [PubMed] [Google Scholar]
- (41).Seidel S, Singer EA, Just H, Farhan H, Scholze P, Kudlacek O, Holy M, Koppatz K, Krivanek P, Freissmuth M, and Sitte HH (2005) Amphetamines take two to tango: an oligomer-based counter-transport model of neurotransmitter transport explores the amphetamine action, Mol Pharmacol 67, 140–151. [DOI] [PubMed] [Google Scholar]
- (42).Kobe F, Renner U, Wlodarczyk J, Papusheva E, Bao G, Zeug A, Richter DW, Neher E, and Ponimaskin E (2008) Stimulation- and palmitoylation-dependent changes in oligomeric conformation of serotonin 5-HT1A receptors, Biochim Biophys Acta 1783, 1503–1516. [DOI] [PubMed] [Google Scholar]
- (43).Zhen J, Antonio T, Cheng SY, Ali S, Jones KT, and Reith ME (2015) Dopamine transporter oligomerization: impact of combining protomers with differential cocaine analog binding affinities, J Neurochem 133, 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sitte HH, Farhan H, and Javitch JA (2004) Sodium-dependent neurotransmitter transporters: oligomerization as a determinant of transporter function and trafficking, Mol Interv 4, 38–47. [DOI] [PubMed] [Google Scholar]
- (45).Torres GE, Carneiro A, Seamans K, Fiorentini C, Sweeney A, Yao WD, and Caron MG (2003) Oligomerization and trafficking of the human dopamine transporter. Mutational analysis identifies critical domains important for the functional expression of the transporter, J Biol Chem 278, 2731–2739. [DOI] [PubMed] [Google Scholar]
- (46).Penmatsa A, Wang KH, and Gouaux E (2013) X-ray structure of dopamine transporter elucidates antidepressant mechanism, Nature 503, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Adkins EM, Samuvel DJ, Fog JU, Eriksen J, Jayanthi LD, Vaegter CB, Ramamoorthy S, and Gether U (2007) Membrane mobility and microdomain association of the dopamine transporter studied with fluorescence correlation spectroscopy and fluorescence recovery after photobleaching, Biochemistry 46, 10484–10497. [DOI] [PubMed] [Google Scholar]
- (48).Foster JD, Adkins SD, Lever JR, and Vaughan RA (2008) Phorbol ester induced trafficking-independent regulation and enhanced phosphorylation of the dopamine transporter associated with membrane rafts and cholesterol, J Neurochem 105, 1683–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Jayanthi LD, Annamalai B, Samuvel DJ, Gether U, and Ramamoorthy S (2006) Phosphorylation of the norepinephrine transporter at threonine 258 and serine 259 is linked to protein kinase C-mediated transporter internalization, J Biol Chem 281, 23326–23340. [DOI] [PubMed] [Google Scholar]
- (50).Jayanthi LD, Samuvel DJ, and Ramamoorthy S (2004) Regulated internalization and phosphorylation of the native norepinephrine transporter in response to phorbol esters. Evidence for localization in lipid rafts and lipid raft-mediated internalization, J Biol Chem 279, 19315–19326. [DOI] [PubMed] [Google Scholar]
- (51).Gabriel LR, Wu S, Kearney P, Bellve KD, Standley C, Fogarty KE, and Melikian HE (2013) Dopamine transporter endocytic trafficking in striatal dopaminergic neurons: differential dependence on dynamin and the actin cytoskeleton, J Neurosci 33, 17836–17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jones KT, Zhen J, and Reith ME (2012) Importance of cholesterol in dopamine transporter function, J Neurochem 123, 700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Epand RM (2006) Cholesterol and the interaction of proteins with membrane domains, Prog Lipid Res 45, 279–294. [DOI] [PubMed] [Google Scholar]
- (54).Hong WC, and Amara SG (2010) Membrane cholesterol modulates the outward facing conformation of the dopamine transporter and alters cocaine binding, J Biol Chem 285, 32616–32626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Torres GE (2006) The dopamine transporter proteome, J Neurochem 97 Suppl 1, 3–10. [DOI] [PubMed] [Google Scholar]
- (56).Garcia-Olivares J, Torres-Salazar D, Owens WA, Baust T, Siderovski DP, Amara SG, Zhu J, Daws LC, and Torres GE (2013) Inhibition of Dopamine Transporter Activity by G Protein betagamma Subunits, PLoS One 8, e59788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Binda F, Dipace C, Bowton E, Robertson SD, Lute BJ, Fog JU, Zhang M, Sen N, Colbran RJ, Gnegy ME, Gether U, Javitch JA, Erreger K, and Galli A (2008) Syntaxin 1A interaction with the dopamine transporter promotes amphetamine-induced dopamine efflux, Mol Pharmacol 74, 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Carvelli L, Blakely RD, and DeFelice LJ (2008) Dopamine transporter/syntaxin 1A interactions regulate transporter channel activity and dopaminergic synaptic transmission, Proc Natl Acad Sci U S A 105, 14192–14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Cervinski MA, Foster JD, and Vaughan RA (2010) Syntaxin 1A regulates dopamine transporter activity, phosphorylation and surface expression, Neuroscience 170, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Cremona ML, Matthies HJ, Pau K, Bowton E, Speed N, Lute BJ, Anderson M, Sen N, Robertson SD, Vaughan RA, Rothman JE, Galli A, Javitch JA, and Yamamoto A (2011) Flotillin-1 is essential for PKC-triggered endocytosis and membrane microdomain localization of DAT, Nature Neurosci 14, 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Navaroli DM, Stevens ZH, Uzelac Z, Gabriel L, King MJ, Lifshitz LM, Sitte HH, and Melikian HE (2011) The plasma membrane-associated GTPase Rin interacts with the dopamine transporter and is required for protein kinase C-regulated dopamine transporter trafficking, J Neurosci 31, 13758–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Blaskovic S, Blanc M, and van der Goot FG (2013) What does S-palmitoylation do to membrane proteins?, FEBS J 280, 2766–2774. [DOI] [PubMed] [Google Scholar]
- (63).Charollais J, and Van Der Goot FG (2009) Palmitoylation of membrane proteins (Review), Mol Membr Biol 26, 55–66. [DOI] [PubMed] [Google Scholar]
- (64).Abrami L, Kunz B, Deuquet J, Bafico A, Davidson G, and van der Goot FG (2008) Functional interactions between anthrax toxin receptors and the WNT signalling protein LRP6, Cell Microbiol 10, 2509–2519. [DOI] [PubMed] [Google Scholar]
- (65).Joseph M, and Nagaraj R (1995) Interaction of peptides corresponding to fatty acylation sites in proteins with model membranes, J Biol Chem 270, 16749–16755. [DOI] [PubMed] [Google Scholar]
- (66).Hong WC, and Amara SG (2013) Differential targeting of the dopamine transporter to recycling or degradative pathways during amphetamine- or PKC-regulated endocytosis in dopamine neurons, FASEB J 8, 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Daniels GM, and Amara SG (1999) Regulated trafficking of the human dopamine transporter. Clathrin-mediated internalization and lysosomal degradation in response to phorbol esters, J Biol Chem 274, 35794–35801. [DOI] [PubMed] [Google Scholar]
- (68).McCormick PJ, Dumaresq-Doiron K, Pluviose AS, Pichette V, Tosato G, and Lefrancois S (2008) Palmitoylation controls recycling in lysosomal sorting and trafficking, Traffic 9, 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Modica G, Skorobogata O, Sauvageau E, Vissa A, Yip CM, Kim PK, Wurtele H, and Lefrancois S (2017) Rab7 palmitoylation is required for efficient endosome-to-TGN trafficking, J Cell Sci 130, 2579–2590. [DOI] [PubMed] [Google Scholar]
- (70).Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, David Daly B, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, and Jones AR (2012) An anatomically comprehensive atlas of the adult human brain transcriptome, Nature 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Lein ES, and Hawrylycz MJ, and Ao N, and Ayres M, and Bensinger A, and Bernard A, and Boe AF, and Boguski MS, and Brockway KS, and Byrnes EJ, and Chen L, and Chen L, and Chen TM, and Chin MC, and Chong J, and Crook BE, and Czaplinska A, and Dang CN, and Datta S, and Dee NR, and Desaki AL, and Desta T, and Diep E, and Dolbeare TA, and Donelan MJ, and Dong HW, and Dougherty JG, and Duncan BJ, and Ebbert AJ, and Eichele G, and Estin LK, and Faber C, and Facer BA, and Fields R, and Fischer SR, and Fliss TP, and Frensley C, and Gates SN, and Glattfelder KJ, and Halverson KR, and Hart MR, and Hohmann JG, and Howell MP, and Jeung DP, and Johnson RA, and Karr PT, and Kawal R, and Kidney JM, and Knapik RH, and Kuan CL, and Lake JH, and Laramee AR, and Larsen KD, and Lau C, and Lemon TA, and Liang AJ, and Liu Y, and Luong LT, and Michaels J, and Morgan JJ, and Morgan RJ, and Mortrud MT, and Mosqueda NF, and Ng LL, and Ng R, and Orta GJ, and Overly CC, and Pak TH, and Parry SE, and Pathak SD, and Pearson OC, and Puchalski RB, and Riley ZL, and Rockett HR, and Rowland SA, and Royall JJ, and Ruiz MJ, and Sarno NR, and Schaffnit K, and Shapovalova NV, and Sivisay T, and Slaughterbeck CR, and Smith SC, and Smith KA, and Smith BI, and Sodt AJ, and Stewart NN, and Stumpf KR, and Sunkin SM, and Sutram M, and Tam A, and Teemer CD, and Thaller C, and Thompson CL, and Varnam LR, and Visel A, and Whitlock RM, and Wohnoutka PE, and Wolkey CK, and Wong VY, and Wood M, and Yaylaoglu MB, and Young RC, and Youngstrom BL, and Yuan XF, and Zhang B, and Zwingman TA, and Jones AR (2007) Genome-wide atlas of gene expression in the adult mouse brain, Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- (72).Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, Karayiorgou M, and Gogos JA (2008) Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion, Nat Neurosci 11, 1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Mukai J, Liu H, Burt RA, Swor DE, Lai WS, Karayiorgou M, and Gogos JA (2004) Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia, Nat Genet 36, 725–731. [DOI] [PubMed] [Google Scholar]
- (74).Maynard TM, Meechan DW, Dudevoir ML, Gopalakrishna D, Peters AZ, Heindel CC, Sugimoto TJ, Wu Y, Lieberman JA, and Lamantia AS (2008) Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes, Mol Cell Neurosci 39, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Raymond FL, Tarpey PS, Edkins S, Tofts C, O’Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, Cole J, Dicks E, Gray K, Halliday K, Hills K, Hinton J, Jones D, Menzies A, Perry J, Raine K, Shepherd R, Small A, Varian J, Widaa S, Mallya U, Moon J, Luo Y, Shaw M, Boyle J, Kerr B, Turner G, Quarrell O, Cole T, Easton DF, Wooster R, Bobrow M, Schwartz CE, Gecz J, Stratton MR, and Futreal PA (2007) Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus, Am J Hum Genet 80, 982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Shin HD, Park BL, Bae JS, Park TJ, Chun JY, Park CS, Sohn JW, Kim BJ, Kang YH, Kim JW, Kim KH, Shin TM, and Woo SI (2010) Association of ZDHHC8 polymorphisms with smooth pursuit eye movement abnormality, Am J Med Genet B Neuropsychiatr Genet 153B, 1167–1172. [DOI] [PubMed] [Google Scholar]
- (77).Young FB, Butland SL, Sanders SS, Sutton LM, and Hayden MR (2012) Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases, Prog Neurobiol 97, 220–238. [DOI] [PubMed] [Google Scholar]
- (78).Gupta P, Soyombo AA, Atashband A, Wisniewski KE, Shelton JM, Richardson JA, Hammer RE, and Hofmann SL (2001) Disruption of PPT1 or PPT2 causes neuronal ceroid lipofuscinosis in knockout mice, Proc Natl Acad Sci U S A 98, 13566–13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Gaffaney JD, and Vaughan RA (2004) Uptake inhibitors but not substrates induce protease resistance in extracellular loop two of the dopamine transporter, Mol Pharmacol 65, 692–701. [DOI] [PubMed] [Google Scholar]
- (80).Vaughan RA (1995) Photoaffinity-labeled ligand binding domains on dopamine transporters identified by peptide mapping, Mol Pharmacol 47, 956–964. [PubMed] [Google Scholar]
- (81).Gu H, Wall SC, and Rudnick G (1994) Stable expression of biogenic amine transporters reveals differences in inhibitor sensitivity, kinetics, and ion dependence, J Biol Chem 269, 7124–7130. [PubMed] [Google Scholar]
- (82).Yang JW, Larson G, Konrad L, Shetty M, Holy M, Jantsch K, Kastein M, Heo S, Erdem FA, Lubec G, Vaughan RA, Sitte HH, and Foster JD (2018) Dephosphorylation of human dopamine transporter at threonine 48 by protein phosphatase PP½A upregulates transport velocity, J Biol Chem 294, 3419–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.