Abstract
Introduction: Autosomal dominant tubulointerstitial kidney disease (ADTKD) is a rare genetic cause of renal impairment resulting from mutations in the MUC1, UMOD, HNF1B, REN, and SEC61A1 genes. Neither the national or global prevalence of these diseases has been determined. We aimed to establish a database of patients with ADTKD in Ireland and report the clinical and genetic characteristics of these families.
Methods: We identified patients via the Irish Kidney Gene Project and referral to the national renal genetics clinic in Beaumont Hospital who met the clinical criteria for ADTKD (chronic kidney disease, bland urinary sediment, and autosomal dominant inheritance). Eligible patients were then invited to undergo genetic testing by a variety of methods including panel-based testing, whole exome sequencing and, in five families who met the criteria for diagnosis of ADTKD but were negative for causal genetic mutations, we analyzed urinary cell smears for the presence of MUC1fs protein.
Results: We studied 54 individuals from 16 families. We identified mutations in the MUC1 gene in three families, UMOD in five families, HNF1beta in two families, and the presence of abnormal MUC1 protein in urine smears in three families (one of which was previously known to carry the genetic mutation). We were unable to identify a mutation in 4 families (3 of whom also tested negative for urinary MUC1fs).
Conclusions: There are 4443 people with ESRD in Ireland, 24 of whom are members of the cohort described herein. We observe that ADTKD represents at least 0.54% of Irish ESRD patients.
Keywords: ADTKD, genetic, MUC-1, UMOD, chronic kidney disease, HNF-1B, urinary smear, frameshift
Introduction
Autosomal dominant tubulointerstitial kidney disease (ADTKD) is a rare genetic cause of progressive chronic kidney disease (CKD) and end-stage renal disease (ESRD) [1]. Causative mutations have been identified in the MUC1 gene encoding mucin-1, the UMOD gene encoding uromodulin (previously Tamm-Horsfall protein), the REN gene encoding renin, the HNF1B gene encoding hepatocyte nuclear factor 1beta, and the SEC61A1 gene encoding translocon subunit SEC61A [1–4]. The cardinal features include gradual loss of kidney function, histological evidence of tubular atrophy and interstitial fibrosis and an autosomal dominant inheritance pattern. Urinary sediment is typically bland. The mean age of ESRD is approximately 45 years but this is variable [5,6]. KDIGO consensus guidelines recommend criteria for diagnosis of confirmed or suspected ADTKD [7].
Genetic testing for UMOD, REN, and HNF1B mutations is reasonably straightforward. For ADTKD-MUC1, identification of causative mutations was recalcitrant to whole exome and whole genome sequencing and ultimately required cloning, standard Sanger sequencing and de novo assembly of MUC1 genomic sequence. The identified mutations were cytosine duplications in one of the heptanucleotide cytosine tracts that is present in each of the 20–125 canonical 60 base pair variable number of tandem repeat (VNTR) units of MUC1 [8]. Duplication of a cytosine results in the mistake, a frameshift, in protein translation and the production of a unique and structurally abnormal mucin protein, MUC1fs (MUC1 frameshift protein). It is thought that MUC1fs deposition results in accelerated apoptosis of renal tubular cells, tubulo-interstitial fibrosis, and progressive chronic kidney disease [1]. Other mutations (e.g., an adenosine insertion, guanosine insertion, deletion of two cytosine residues, or duplication of 16 base pairs) causing the same frameshift can also cause ADTKD-MUC1 [9,10]. Due to the high guanosine-cytosine content and the number of repeats, the MUC1 gene cannot be sequenced by traditional methods.
At present, genetic analysis for the most prevalent mutations 27dupC and 28dupA can be obtained by the Clinical Laboratory Improvement Amendments of 1988 (CLIA) certified mass-spectrometry-based genotyping assay provided at the Broad Institute [10,11]. Identification of other unique frameshift mutations is possible by Illumina sequencing and specific bioinformatic analysis of the VNTR region of MUC1 [10]. However, not all mutations resulting in ADTKD-MUC1 can be determined in this manner [12,13]. A recent development in the diagnosis of ADTKD-MUC1 is the detection of abnormal MUC1fs protein in urine samples from individuals with suspected ADTKD. This technique relies on immunohistochemical intracellular detection of the unique disease-causing frameshift protein on urinary cell smears [10].
In Ireland, 11.8% of the population has CKD [14] and more than 4400 people, 928 per million population, have ESRD [15]. Genetic causes of CKD e.g., Autosomal dominant polycystic kidney disease and Alport syndrome have been described in the Irish population previously [16]. Recently, the Irish Kidney Gene Project (IKGP) indicated that 34% of patients attending Irish nephrology units have a relevant family history [17]. Patients with unspecified tubulointerstitial kidney disease were significantly more likely to have a family history of kidney disease than those with other causes of CKD. Whole exome sequencing of 114 Irish families with CKD identified a monogenic cause of CKD in 37% [18]. The IKGP and subsequent work enabled identification of individuals in Ireland who might suffer from ADTKD.
We set out to identify individuals recruited through the IKGP who had clinical characteristics, family history, and renal biopsy results (where available) in keeping with ADTKD and to identify causative mutations in these patients. Of 1840 individuals included in the IKGP study, 29 had a history of unspecified tubulointerstitial kidney disease and two-thirds of these reported a relevant family history [17].
The goals of this work were to identify a cohort of Irish patients with ADTKD and to describe clinical characteristics and causative mutations in our population. We aimed to provide an estimate of prevalence within the Irish population. Furthermore, establishing a database of Irish patients with ADTKD will allow us to contribute to global progress in the understanding of this disease. This could include future enrollment of Irish patients with ADTKD in clinical trials of disease modifying agents.
Methods
The IKGP was a multi-centre cross-sectional study of adult patients with CKD attending Irish nephrology units. Ethical approval for patient enrollment, collection of clinical data, and DNA collection was granted by each local medical ethics committee. Patients provided written consent at enrollment. The full IKGP study protocol has been described elsewhere [17].
Nineteen individuals were identified via the IKGP study as meeting criteria for suspected ADTKD. An additional 23 relatives were recruited to give a total of 42 individuals from 11 families. Patients referred to the Renal Genetics Clinic in Beaumont Hospital were also considered, yielding a further 12 individuals from five families.
We then performed a review of electronic medical records, biochemical and histologic (kidney biopsy) results to compile a database of clinical characteristics. Some data fields (e.g., proteinuria quantification at diagnosis) were not available for all patients and in these cases was recorded as missing.
We included only patients who met KDIGO diagnostic criteria for a definite ADTKD diagnosis (See Table 1). Inclusion criteria were as follows: (1) A clinical history compatible with ADTKD and either (2) A genetic diagnosis in the patient or affected family member OR (3) A family history suggestive of autosomal dominant inheritance of kidney disease AND a kidney biopsy compatible with a diagnosis of ADTKD in the patient or an affected family member. Exclusion criteria were as follows: (1) Clinical characteristics suggestive of an alternative diagnosis (including hematuria, drug exposure accounting for renal pathology, suggestion of an alternative diagnosis on renal biopsy); (2) Non-availability of samples for genetic testing in at least one member of an affected family.
Table 1.
KDIGO criteria for suspected and definite diagnosis of ADTKD which were used to define inclusion in our study.
| Suspected diagnosis | Confirmed diagnosis |
|---|---|
| Family history of autosomal dominant inheritance of CKD with clinical characteristics in keeping with ADTKD | Compatible family history (minimum of 1 first degree relative) and compatible kidney biopsy in one affected individual |
| Without a compatible family history – compatible histology on a kidney biopsy of compatible extra-renal mutations e.g., Gout or Diabetes | Demonstrated mutation in one of the 4 known genes (MUC1, UMOD, HNF1B, and REN) |
ADTKD: autosomal dominant tubulointerstitial kidney disease; CKD: chronic kidney disease.
Collection and bio-banking of DNA samples was carried out by a Rare Kidney Disease Registry and Biobank in Trinity Kidney Health Centre and the Royal College of Surgeons in Ireland.
We initially performed targeted genetic testing for pathogenic mutations in the MUC1 and UMOD genes (facilitated through Anthony Bleyer, Wake Forest School of Medicine, Winston-Salem, NC USA or the Broad Institute). Genetic testing for MUC1 mutations was carried out at the Broad Institute of Harvard University and the Massachusetts Institute of Technology, Cambridge, MA as previously described [11]. This analysis identified individuals with previously described cytosine duplication (27dupC) or adenine duplication (28dupA) in the VNTR of the MUC1 gene [10]. Genetic testing for UMOD mutations was carried out by the Rare Inherited Kidney Disease Team, Wake Forest School of Medicine, Winston-Salem, USA using full sequence analysis of the UMOD gene [19].
Families without an identified pathogenic mutation in either UMOD or MUC1 underwent genetic testing by either whole exome sequencing (SeqCap EZ Exome v3.0) or gene panel testing (individuals from five families) on a custom gene panel of 227 genes (Supplementary Table 1). Figure 1 shows the sequential use of genetic testing strategies. Whole exome sequencing was performed in Boston Children’s Hospital as previously described [18,20].
Figure 1.
Flow chart demonstrating recruitment to this cohort and methods of genetic testing for ADTKD. Sixteen families have been identified with a total solve rate of 12/16. Individuals with tubulointerstitial kidney disease and a relevant family history identified from the Irish Kidney Gene Project (IKGP) were further screened via review of biopsy reports and clinical records. Five individuals were excluded from this report because they did not meet KDIGO criteria for a definite diagnosis of ADTKD, 3 because they did not have an affected first degree relative (or confirmed mutation) and 2 because they did not have a compatible biopsy report (or confirmed mutation). The Renal Genetics Clinic (RGC) was an additional source of recruitment. *Two families recruited via the RGC went directly to gene panel testing without prior targeted testing for MUC1 and UMOD mutations. †One individual had both gene panel and whole exome sequencing and was found to have a pathogenic mutation in the UMOD gene on both methods.
Gene panel testing was performed in the Royal College of Surgeon’s Ireland. Library preparation was conducted using standard library preparation methods (Roche Diagnostics). Sequencing was conducted using Illumina MiSeq and NextSeq sequencers. Sequencing data were analyzed using an in-house bioinformatic pipeline. Sequencing reads were aligned to the reference genome using Burrows-Wheeler Aligner [21], and the Genome Analysis Toolkit (GATK) pipeline [22] was used for base quality score recalibration, indel realignment, and duplicate removal. Variant identification was conducted using GATK standard hard filtering parameters [23]. ANNOVAR [24] was used for variant annotation, and variants were assessed for pathogenicity following guidelines of the American College of Medical Genetics [25].
As discussed above, the MUC1fs protein can be detected by immunohistochemical staining of urinary cell smears. In six individuals without a previously identified genetic diagnosis, we collected urinary samples for immunohistochemical analysis to identify MUC1fs protein. This was performed by Dr. Živná in Research Unit for Rare Diseases in the First Faculty of Medicine, Charles University, Prague, as previously described [10].
Following the above diagnostic methods, 5 individuals remained in the ADTKD-NOS group.
In accordance with the 2015 KDIGO guidelines (2015), patients were categorized as ADTKD-MUC1, ADTKD-UMOD, ADTKD-HNF1ß, or ADTKD-NOS. Diagnostic criteria are shown in Table 1.
Descriptive statistical analysis and statistical testing for differences between were performed using Minitab 18® (Minitab, State College, PA) statistical software. Differences in mean values between groups were compared using one-way ANOVA testing and differences in proportion by group were compared using Chi-Square testing. Differences in mean values for those with and without an identified mutation were compared using two-sample t-testing. A p value of <0.05 was deemed statistically significant.
Results
Overall cohort
This case series describes 16 families comprised of 54 individuals that meet KDIGO guideline definitions for ‘Definite ADTKD’. Five families have ADTKD-MUC1 (in two of which diagnosis was based on urinary intracellular detection of MUC1fs), five ADTKD-UMOD, and two ADTKD-HNF1B (Figure 2). Following the above diagnostic methods, five individuals (from four families) remained in the ADTKD-NOS group. Although we were unable to diagnose a pathogenic mutation, these individuals had both a compatible biopsy (in at least one family member) and an autosomal dominant family history of kidney disease. Therefore, they meet criteria for confirmed ADTKD by KDIGO guidelines [7].
Figure 2.
Frequency of each genetic subtype of ADTKD within this Irish cohort. MUC1: ADTKD-MUC1; UMOD: ADTKD-UMOD; HNF1B: ADTKD-HNF1B; NOS: not otherwise specified.
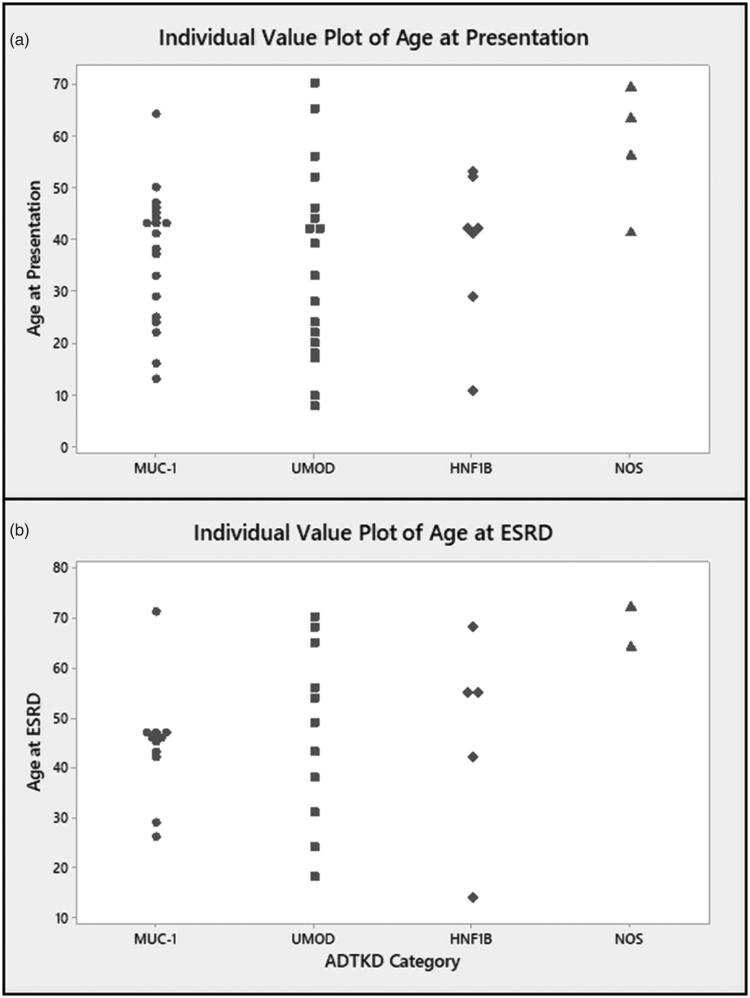
In the overall cohort 28/54 (52%) individuals were male, the mean age at presentation is 38.3 ± 16.0 years and the mean creatinine at presentation is 210.6 ± 163.5 micromoles/litre (µmol/L). ESRD was reached by 32/54 individuals (57%) at a mean age of 47.4 years (Table 2). Figure 3 demonstrates the range of ages at presentation (a) and ESRD (b) by genetic category.
Table 2.
Clinical characteristics of individuals in this cohort by ADTKD-subgroup.
| ADTKD (All) n = 54 | ADTKD-MUC1n = 23 | ADTKD-UMOD n = 19 | ADTKD-HNF1B n = 7 | ADTKD-NOS n = 5 | p Value | |
|---|---|---|---|---|---|---|
| Confirmed mutation n (%) | 28 (52%) | 15*/23 (65%) | 9 (48%) | 7 (100%) | 0 (0%) | NA |
| Number of families | 16 | 5 | 5 | 2 | 4 | NA |
| Mean age at presentation, years (SD) | 38 (16.0) | 37.0 (12.8) | 35.3 (18.3) | 38.6 (14.5) | 57.25 (12.0) | 0.090** |
| Mean creatinine at presentation, µmol/l (SD) | 210.6 (163.5) | 136.4 (45.6) | 272 (217.2) | 171 (20.4) | 372 (214.0) | 0.030** |
| Number reached ESRD (%) | 32 (59%) | 12 (52%) | 12 (63%) | 5 (71%) | 3 (56%) | NA |
| Mean age at ESRD, years | 47.4 ( 16.0) | 44.5 (11.47) | 46.9 (17.8) | 46.8 (20.5) | 68.0 (5.7) | 0.303** |
| Gout (%) | 15/54 (27%) | 0% (0/19) | 68% (13/19) | 14% (1/7) | 20% (1/5) | <0.001*** |
ADTKD: autosomal dominant tubulointerstitial kidney disease; ESRD: end stage renal disease; n: Number; NA: non-applicable; SD: standard deviation. *For two individuals in this group, diagnosis was based on urinary detection of MUC1fs protein in urinary smears; **p Values by one-way ANOVA testing comparing ADTKD sub-categories; ***p Values by chi-square testing comparing ADTKD sub-categories.
Figure 3.
(a) Age at presentation and (b) Age at end stage renal disease (ESRD) by genetic category of ADTKD. MUC1: ADTKD-MUC1; UMOD: ADTKD-UMOD; HNF1B: ADTKD-HNF1B; NOS: not otherwise specified.
Twenty-four individuals included in this report are currently living with ESRD. The most recent national renal office figures indicate that 4443 patients are living with ESRD in Ireland [15]. We therefore estimate that ADTKD accounts for at least 0.54% of ESRD cases in Ireland.
Identification and clinical characteristics of individuals with ADTKD-MUC1
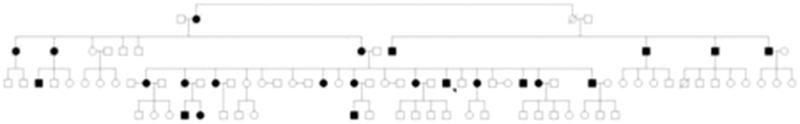
This group included families with a confirmed MUC1 frameshift mutation and families with mutant MUC1fs protein identified on urinary cell smears. This category includes 5/16 families (23%) and is comprised of 23 individuals. In 12/23 (52%) individuals, we identified a pathogenic mutation in the MUC1 gene and seven others were categorized as ADTKD-MUC1 based on clinical characteristics and detection of a pathogenic in MUC1 in a relative. In each of these cases, a frame shift mutation (either a 27dupC or 28dupA in the VNTR region) in the MUC1 gene was identified. A pedigree of the largest family affected by ADTKD-MUC1 is shown in Figure 4.
Figure 4.
Family tree demonstrating a large Irish family with cases of ADTKD over four generations due to MUC1 genetic mutations. Shaded = Affected; Unshaded = Unaffected; Box = Male; Circle = Female; Strikethrough = Deceased.
We identified MUC1fs protein in urinary cell smears from three individuals (two from the same family and in one other individual). These families were not included in the previous report of this technique [10]. We reclassified these individuals and one other affected first degree relative as ADTKD-MUC1. To date we have been unable to identify a pathogenic mutation on genomic sequencing of the MUC1 gene. Age at presentation in these four patients was slightly older (42.5 years).
In the ADTKD-MUC1 group, 8/23 (35%) individuals were male. These 23 individuals presented at a mean of 37 years and with a mean serum creatinine of 136 µmol/L. Twelve of 23 (52%) patients have reached ESRD at a mean age of 42 years.
Identification and clinical characteristics of individuals with ADTKD-UMOD
This category includes 5/16 (31%) families and is comprised of 19 individuals. In 9/19 (47%) individuals we identified a pathogenic mutation in the UMOD gene. The remaining individuals were categorized as ADTKD-UMOD based on clinical characteristics and detected pathogenic gene mutation in a relative. Details of identified mutations are given in Table 3 and have been previously reported [18,26–28]. Of note, three of the families included here were previously reported (Families 8–10 in Table 3 or B2337, P193, and P232 in the report by Connaughton et al. [18]. The others had been previously identified by testing in the Wake Forest School of Medicine. Eighty percent of families had a history of gout (4/5). A pedigree of the largest family affected by ADTKD-UMOD is shown in Figure 5.
Table 3.
Each family included in this cohort showing genetic mutation identified. Details of numbers of affected individuals with confirmed mutation and compatible biopsy are shown. The number of 1st degree relatives affected is shown for the ADTKD-NOS families.
| Family number | ADTKD category | Genetic mutation | Protein | Testing laboratory | Number individuals in family | Number with confirmed mutation | Number with compatible renal biopsy | Number 1st degree relatives (ADTKD-NOS) |
|---|---|---|---|---|---|---|---|---|
| 1 | MUC1 | Insertion in VNTR region | frameshift | BI | 13 | 7 | 5 | |
| 2 | MUC1 | Insertion in VNTR region | frameshift | BI | 4 | 2 | 0 | |
| 3 | MUC1 | Insertion in VNTR region | frameshift | BI | 2 | 1 | 1 | |
| 4 | MUC1* | NA | frameshift | CU | 3 | 0 | 2 | |
| 5 | MUC1* | NA | frameshift | CU | 1 | 0 | 1 | |
| 6 | UMOD | c.821A > G | p.Y274C | BG | 2 | 2 | 1 | |
| 7 | UMOD | c.668G > A | p.C223Y | BG | 5 | 3 | 4 | |
| 8 | UMOD | c.317G > A | p.C106Y | BG | 7 | 2 | 1 | |
| 9 | UMOD | c.1382C > A | p.A461E | BCH | 4 | 1 | 1 | |
| 10 | UMOD | c.280T > C | p.C94R | BCH/RCSI** | 1 | 1 | 0 | |
| 11 | HNF1B | c.1333_1334delGC | p.Ala445fs*105 | BCH | 4 | 4 | 2 | |
| 12 | HNF1B | c.544 + 3_544 + 6 | Del 75% ESS | BCH | 3 | 3 | 0 | |
| 13 | NOS | NA | NA | NA | 2 | 0 | 1 | 2 |
| 14 | NOS | NA | NA | NA | 1 | 0 | 1 | 1 |
| 15 | NOS | NA | NA | NA | 1 | 0 | 1 | 2 |
| 16 | NOS | NA | NA | NA | 1 | 0 | 1 | 2 |
ADTKD: autosomal dominant tubulointerstitial kidney disease; BCH: Boston Children’s Hospital; BI: broad institute; BG: Bowman Grey Institute; CU: Charles University, Prague; del: deletion; ESS: essential splice site; fs: frameshift; NA: non-applicable; RCSI: Royal College of Surgeons Ireland (gene panel analysis); VUS: variant of uncertain significance. *Urinary MUC1fs protein detected, no detected genetic mutation to date; **Mutation identified in both centres.
Figure 5.
Family tree demonstrating a large Irish family with cases of ADTKD over four generations due to UMOD mutations. Shaded = Affected; Unshaded = Unaffected; Square within a circle = Status Unknown; Box = Male; Circle = Female; Strikethrough = Deceased.
In the ADTKD-UMOD group, 13/19 (68%) individuals were male. These 19 individuals presented at a mean age of 35.3 years and with a mean serum creatinine of 272 µmol/L. In this group, 12/19 (63%) patients have reached ESRD at a mean age of 46.9 years.
Identification and clinical characteristics of individuals with ADTIKD-HNF1B
This category includes 2/16 (12.5%) families and is comprised of seven individuals. All seven individuals have had genetic testing confirming a pathogenic mutation in the HNF1B gene. These families were previously included in a report by Connaughton et al. [18].
In the ADTKD-HNF1B group, 3/7 (43%) individuals were male. These seven patients presented at a mean age of 39 years with a mean serum creatinine of 171 µmol/L. Five of these patients (71%) progressed to ESRD at a mean age of 47 years. Patients with pathogenic mutation in HNF1-B also had concomitant conditions including diabetes (n = 2) and abnormal LFTs (n = 2). Hypomagnesemia was not ascertained in this cohort. Hyperuricemia or gout occurred in 4/7 individuals (57%). One individual (from family 11) had a renal ultrasound showing renal cysts in the native kidneys.
Of note, both of these families (Family 11 & 12 in Table 3) have been reported previously (B2342 and P306 in the report by Connaughton et al. [18]). One member of family 12 had had prior unilateral nephrectomy due to vesicoureteric reflux and was previously classified as CAKUT (Congenital Anomalies of the Kidney and Urinary Tract). Given that genetic testing led to a reclassification of the diagnosis we have included that family in this report.
Identification and clinical characteristics of individuals with ADTKD-NOS
This category includes 4/16 (37.5%) of families and is comprised of five individuals who meet KDIGO criteria for a definite diagnosis of ADTKD based on family history and renal biopsy findings in whom we have not yet identified a causative mutation. This is despite gene panel testing for 227 genes, including UMOD, MUC1, HNF1B and REN mutations and examination or urine smears for mutant MUC1fs (in 3/4 families). These families were small with only one available for testing in all but one case and so did not lend themselves to linkage studies.
In this group, 4/5 (80%) of individuals were male. The mean age at presentation in this group was 57 years, compared to 37 in those individuals with an identified mutation (p = 0.049). The mean age of the five patients in this group, who progressed to ESRD was 68 years, which was older than a mean of 46 years for individuals with an identified mutation (p = 0.047).
Discussion
ADTKD is a relatively recently described clinical entity and clinical knowledge of the disease is growing [2,5,29]. This is the first published report of ADTKD in Ireland. Pathogenic mutations in the MUC1, UMOD, and HNF1B genes have been identified in a 10 Irish families to date and a further two families have had MUC1fs protein identified in urinary cell smears.
Knowledge regarding the prevalence of ADTKD is limited. A previous Austrian study estimated that ADTKD due to UMOD mutations accounted for 0.073% of cases of ESRD in Austria [30]. A Spanish study recently reported 131 individuals from 56 families affected by ADTKD [31] but did not comment on how this related to national prevalence. In the UK, a survey of nephrology patients attending a tertiary referral centre estimated prevalence of 9 cases per million of ADTKD-UMOD and 17.5 cases per million of all causes ADTKD – based on family history and genotyping for UMOD mutations [32]. This study estimated that ADTKD-UMOD alone accounts for 2% of cases of ESRD.
We sought to estimate the frequency of ADTKD as a cause of ESRD in Ireland. The 24 individuals currently living with ESRD included in this case series would alone account for 0.54% of ESRD in Ireland [15]. Although we sought to identify all cases of ADTKD by identifying individuals and their relatives through the IKGP study and by seeking referrals to the national renal genetics clinic this may still be an underestimate due to incomplete case ascertainment. Variation between estimates of prevalence may reflect regional or ascertainment differences.
A key recommendation of the KDIGO 2015 consensus report was the establishment of registries of patients with ADTKD to facilitate further research [7]. We have created a database of such patients in Ireland. In the future, increased awareness of the condition may lead to further individuals being identified.
This study further emphasizes the complexity in genetic testing for ADTKD. In 2/5 (40%) families with ADTKD-MUC1 diagnosis was possible only due to the detection of MUC1fs protein in urinary cell smears.
The clinical characteristics of this group are similar to other cohorts [2,5,6]. In other reports, age at ESRD is variable, but typically occurs in middle age. The mean ages of ESRD for ADTKD-MUC1, ADTKD-UMOD, ADTKD-HNF1B, and ADTKD-NOS in this Irish cohort were 44, 47, 47, and 68 years, respectively. These differences were not statistically significant. Thirty-seven percent (20/54) of these patients had histological evidence of ADTKD. As expected, gout was a frequent comorbidity in the ADTKD-UMOD group (15/19, 78%).
Age at presentation was not significantly different between groups although ADTKD-NOS patients presented with higher creatinine and ADTKD-MUC1 patients presented with lower creatinine. No patients in the ADTKD-MUC1 group presented with ESRD at diagnosis. These differences may be, in part, due to earlier screening of family members in a particularly large ADTKD-MUC1 family and demonstrates the value of accurate diagnosis of the disease in facilitating family-based screening.
In 4/16 (25%) families, we did not identify a pathogenic mutation in the genes known to cause ADTKD. This is not unique to our cohort – one Italian centre performed genetic testing in 21 families with suspected ADTKD and found a cytosine duplication in the VNTR of the MUC1 gene in only 10% of cases [12]. Possible reasons for failure to identify a causative mutation include the complexity of sequencing and identifying mutations in the MUC1 gene and existence of yet unrecognized causative gene mutations. Identification of MUC1fs protein in urinary smears is an important diagnostic tool in reducing cases of ADTKD without an established genetic diagnosis. In one of the still unsolved families we were unable to obtain fresh urine samples for immunohistochemical analysis for MUC1fs protein.
Awareness of ADTKD as a cause of CKD and ESRD is important for several reasons. Accurate diagnosis allows patients with CKD to be given prognostic information and planning for progression to ESRD [33]. Genetic diagnosis may allow transplantation from living related donors by determining that a potential donor is not themselves affected [34]. Greater awareness of ADTKD among clinicians will lead to better case ascertainment which allow for more accurate estimation of disease prevalence.
Establishing a renal genetics clinic led to diagnosis of ADTKD in twelve individuals from four families (Figure 1). This clinic was recently established and continues to receive referrals for individuals with a suspected genetic cause of ADTKD. Centralizing genetic referrals allows for nationwide access to testing for rare genetic variants. This testing has formerly been largely done outside Ireland, in collaboration with the Broad Institute, Cambridge, Massachusetts, and Wake Forest School of Medicine, Winston-Salem, North Carolina, and the First Faculty of Medicine, Charles University, Prague, Czech Republic.
In a research lab, we have recently tested individuals with suspected ADTKD using a gene panel testing approach. Continuing to resource the renal genetics clinic and strengthening national capacity to test for genetic causes of kidney disease are important measures to ensure Irish patients with kidney disease receive accurate, timely genetic diagnoses.
There are a number of limitations to this study. The IKGP did not include the whole Irish population. While we sought to identify other families with ADTKD through referrals through the renal genetics clinic and liaison with other nephrology units it is possible that other individuals and families with CKD due to ADTKD exist. ADTKD may therefore be considered to account for a minimum of 0.54% of cases of ESRD, actual prevalence may be higher. Genetic testing for ADTKD continues to evolve and it is possible that this will allow identification of other individuals and families in the future. Finally, this study may not be generalizable to other geographic regions where the prevalence of ADTKD, and underlying genetic mutations, are likely to be different.
In summary, we describe an Irish cohort of patients with ADTKD. Characteristics in our cohort are similar to previously reported data. This report highlights the need to consider a diagnosis of ADTKD in individuals with unspecified tubulointerstitial disease or a family history of kidney disease. This work has established an Irish database of patients with ADTKD who could be included in future work to identify novel mutations or in clinical trials of disease modifying agents.
Supplementary Material
Funding Statement
This work was supported by funding from the Punchestown Kidney Research Fund, Beaumont Nephrology Research Fund, a Meath Foundation grant [Grant no.:205229.13987] and SFI grant [Grant number: 11/Y/B2093). SC is currently supported by an academic training grant under the Irish Clinical Academic Training (ICAT) Programme, supported by the Wellcome Trust and the Health Research Board [Grant Number 203930/B/16/Z]. DMC is funded by the Health Research Board, Ireland [HPF-206–674], the International Pediatric Research Foundation Early Investigators’ Exchange Program, and the Amgen Irish Nephrology Society Specialist Registrar Research Bursary. MŽ and SK were supported by grant NV17-29786A from the Ministry of Health of the Czech Republic and by institutional programs of Charles University in Prague [UNCE/MED/007 and PROGRES-Q26/LF1] and they thank The National Centre for Medical Genomics [LM2015091] for help in DNA sequencing and bioinformatic support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Bleyer AJ, Kmoch S. Autosomal dominant tubulointerstitial kidney disease: of names and genes. Kidney Int. 2014;86:459–461. [DOI] [PubMed] [Google Scholar]
- 2.Faguer S, Decramer S, Chassaing N, et al. Diagnosis, management, and prognosis of HNF1B nephropathy in adulthood. Kidney Int. 2011;80:768–776. [DOI] [PubMed] [Google Scholar]
- 3.Medicine M-N. Online Mendelian inheritance in man. An online catalog of human genes and genetic disorders [Website]. John Hopkins University: OMIM; 2018 [updated 2018 Sep 7]. Available from: https://www.omim.org.
- 4.Bolar NA, Golzio C, Živná M, et al. Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am J Hum Genet. 2016;99:174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleyer AJ, Kmoch S, Antignac C, et al. Variable clinical presentation of an MUC1 mutation causing medullary cystic kidney disease type 1. Clin J Am Soc Nephrol. 2014;9:527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollée G, Dahan K, Flamant M, et al. Phenotype and outcome in hereditary tubulointerstitial nephritis secondary to UMOD mutations. Clin J Am Soc Nephrol. 2011;6:2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckardt KU, Alper SL, Antignac C, et al. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management – a KDIGO consensus report. Kidney Int. 2015;88:676–683. [DOI] [PubMed] [Google Scholar]
- 8.Kirby A, Gnirke A, Jaffe DB, et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet. 2013;45:299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto S, Kaimori JY, Yoshimura T, et al. Analysis of an ADTKD family with a novel frameshift mutation in MUC1 reveals characteristic features of mutant MUC1 protein. Nephrol Dial Transplant. 2017;32:2010–2017. [DOI] [PubMed] [Google Scholar]
- 10.Živná M, Kidd K, Přistoupilová A, et al. Noninvasive immunohistochemical diagnosis and novel MUC1 mutations causing autosomal dominant tubulointerstitial kidney disease. J Am Soc Nephrol. 2018;29:2418–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blumenstiel B, DeFelice M, Birsoy O, et al. Development and validation of a mass spectrometry–based assay for the molecular diagnosis of mucin-1 kidney disease. J Mol Diagn. 2016;18:566–571. [DOI] [PubMed] [Google Scholar]
- 12.Musetti C, Babu D, Fusco I, et al. Testing for the cytosine insertion in the VNTR of the MUC1 gene in a cohort of Italian patients with autosomal dominant tubulointerstitial kidney disease. J Nephrol. 2016;29:451–455. [DOI] [PubMed] [Google Scholar]
- 13.Gale DP, Kleta R. MUC1 makes me miserable. J Am Soc Nephrol. 2018;29:2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stack AG, Casserly LF, Cronin CJ, et al. Prevalence and variation of chronic kidney disease in the Irish health system: initial findings from the National Kidney Disease Surveillance Programme. BMC Nephrol. 2014;15. DOI: 10.1186/1471-2369-15-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plant L. editor Epidemiology of ESKD in Ireland. Irish Nephrology Society Annual Meeting. Kildare (Ireland): Carton House; 2018. [Google Scholar]
- 16.Green A, Allos M, Donohoe J, et al. Prevalence of hereditary renal disease. Ir Med J. 1990;83:11–13. [PubMed] [Google Scholar]
- 17.Connaughton DM, Bukhari S, Conlon P, et al. The Irish Kidney Gene Project – prevalence of family history in patients with kidney disease in Ireland. Nephron. 2015;130:293–301. [DOI] [PubMed] [Google Scholar]
- 18.Connaughton DM, Kennedy C, Shril S, et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95:914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleyer AJ Jr, Hart PS, Kmoch S Autosomal dominant tubulointerstitial kidney disease, UMOD-Related Seattle: University of Washington; 2007 [updated 2016; cited 2018 July 11]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1356/.
- 20.Mann N, Braun DA, Amann K, et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol. 2019;30:201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duitama J, Quintero JC, Cruz DF, et al. An integrated framework for discovery and genotyping of genomic variants from high-throughput sequencing experiments. Nucleic Acids Res. 2014;42:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calado J, Gaspar A, Clemente C, et al. A novel heterozygous missense mutation in the UMOD gene responsible for Familial Juvenile Hyperuricemic Nephropathy. BMC Med Genet. 2005;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DH, Kim JK, Oh SE, et al. A case of familial juvenile hyperuricemic nephropathy with novel uromodulin gene mutation, a novel heterozygous missense mutation in Korea. J Korean Med Sci. 2010;25:1680–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaucke F, Boehnlein JM, Steffens S, et al. Uromodulin is expressed in renal primary cilia and UMOD mutations result in decreased ciliary uromodulin expression. Human Mol Genet. 2010;19:1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scolari F, Caridi G, Rampoldi L, et al. Uromodulin storage diseases: clinical aspects and mechanisms. Am J Kidney Dis. 2004;44:987–999. [DOI] [PubMed] [Google Scholar]
- 30.Lhotta K, Piret SE, Kramar R, et al. Epidemiology of uromodulin-associated kidney disease – results from a nation-wide survey. Nephron Extra. 2012;2:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayasreh N, Bullich G, Miquel R, et al. Autosomal dominant tubulointerstitial kidney disease: clinical presentation of patients with ADTKD-UMOD and ADTKD-MUC1. Am J Kidney Dis. 2018;72:411–418. [DOI] [PubMed] [Google Scholar]
- 32.Gast C, Marinaki A, Arenas-Hernandez M, et al. Autosomal dominant tubulointerstitial kidney disease-UMOD is the most frequent non polycystic genetic kidney disease. BMC Nephrol. 2018;19:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie BW, Morgenstern H, Hedgeman E, et al. Nephrology care prior to end-stage renal disease and outcomes among new ESRD patients in the USA. Clin Kidney J. 2015;8:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soloukides AP, Moutzouris DAD, Papagregoriou GN, et al. Renal graft outcome in autosomal dominant medullary cystic kidney disease type 1. J Nephrol. 2013;26:793–798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.